Abstract
Background: microRNAs (miRNAs) play a significant role in cancer development and progression by regulating the expression of oncogenes or tumor suppressor genes. Previous study using microarrays demonstrated that miR-142-3p was downregulated in patients with Non-small-cell lung carcinoma (NSCLC). However, the functional role of miR-142-3p in NSCLC is still unclear. Material and method: Real-time quantitative PCR was applied to evaluate the expression level of miRNA-142-3p in NSCLC and normal samples. The cell proliferation of NSCLC cells was analyzed by MTT and colony formation assay after miR-142-3p transfection. Luciferase activities assay, cotransfection and Western blot were used to reveal that the predicted target genes of miR-125b were direct and specific. Results: In this study, we demonstrate miR-142-3p was downregulated in NSCLC tissues and cell lines. We demonstrated that the overexpression of miR-142-3p inhibits NSCLC cell proliferation and induced cell apoptosis. Furthermore, we demonstrate HMGB1 was a directly target of miR-142-3p in NSCLC cells, and confirmed the target specificity between miR-142-3p and the HMGB1 3’-untranslated region by luciferase reporter assay. Conclusions: These results suggest that miR-142-3p may be a tumor suppressor through the downregulation of HMGB1 in NSCLC. miR-142-3p may be a tumor suppressor and a potential therapeutic agent for patients with NSCLC.
Keywords: NSCLC (Non-small-cell-carcinoma), microRNA, HMGB1, cell proliferation, apoptosis
Introduction
Non-small cell lung cancer (NSCLC), which including adenocarcinoma and squamous cell carcinoma, is the predominant form of lung cancer, and accounts for the majority of cancer deaths worldwide [1]. Although recent advances in clinical and experimental oncology, the prognosis of lung cancer is still unfavorable, with a 5-year overall survival rate of approximately 11% [2]. Thus, to explore the detailed mechanisms underlying NSCLC development and progression are still need for improving, diagnosis, prevention and treatment of this disease. In these decades, accumulating evidence has shown that microRNAs (miRNAs) may be involved in NSCLC pathogenesis, providing new insights into disease biology.
MicroRNAs (miRNAs) are small, non-coding RNA sequences about 22nt nucleotides long, which bind to complementary sequences generally located within the 3’ untranslated region (UTR) of target transcript RNAs to act as critical post-transcriptional regulators of gene expression, either inhibiting translation or reducing stability and inducing degradation [3]. Approximately half of all miRNAs being located within fragile sites and genomic regions associated with cancer [4,5]. Deregulation of miRNA expression has been associated with carcinogenesis [6] and abnormal expression of miRNAs has been demonstrated in most tumor types [7]. For instance, MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis [8]. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA-328 may decrease their chemoresistance [9]. In addition, MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2 [10]. To date, MiR142-3p has been reported to be down-regulated and serve as a potential tumor suppressor in several distinct cancer types including hepatocellular carcinoma (HCC), acute myeloid leukemia (AML), colon cancer and pancreatic ductal adenocarcinoma (PDAC) [11-13]. Furthermore, Namløs et al. found that miR-142-3p was significantly downregulated in osteosarcoma cell lines compared to normal bone tissues by miRNA microarray analysis [14]. However, the role and molecular mechanism of miR-142-3p in NSCLC remain largely unknown.
In this study, we investigated miR-142-3p expression in NSCLC, and then analyzed the effect of miR-142-3p overexpression on cell proliferation and apoptosis. Furthermore, we identified the downstream target of miR-142-3p and find miR-142-3p was directly targeted HMGB1 in NSCLC.
Material and methods
Patients and specimens
A total of 21 NSCLC and 21 relative Normal specimens were surgically collected after informed consent from patients aged from 55 to 67 years. None of these patients received chemotherapy and radiotherapy before the surgery. Tumor and corresponding non-tumor lung tissue samples were collected and rapidly frozen in liquid nitrogen and stored at -80°C. Ethical approval was obtained from the hospital and fully informed consent from all patients before sample collection. The characteristics of NSCLC patients in this study were shown in Table 1.
Table 1.
Characteristics of NSCLC patients in this study
| Features | Number of cases |
|---|---|
| Tumor size (cm) | |
| <5 | 11 |
| ≥5 | 10 |
| Family history of tumor | |
| Negative | 17 |
| Positive | 4 |
| Age | |
| ≥60 | 15 |
| <60 | 6 |
| Differentiation | |
| Well and moderate | 9 |
| Poor or no differentiation | 12 |
| TNM stage | |
| I+II | 10 |
| III+IV | 11 |
| Lymphatic metastasis | |
| pN (N0) | 13 |
| pN (N1+N2+N3) | 8 |
Cell culture
Two NSCLC cell lines (NCI-H23 and NCI-H838) were cultured in RPMI 1640 (GIBCO-BRL) medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin in humidified air at 37°C with 5% CO2.
Bioinformatics methods
The miRNA targets predicted by computer-aided algorithms were obtained from pictar (http://pictar.mdc-berlin.de/cgi-bin/new_PicTar_vertebrate.cgi), targetscan (http://www.targetscan.org) and miRbase (http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl).
Real-time reverse transcription (qRT) polymerase chain reaction (PCR)
A TaqMan miRNA-assay kit was obtained from Applied Biosystems (Foster City, CA, USA) for the detection of mature miR-142-3p expression. According to the manufacturer’s instructions, the 2-DeltCt method was used in conjunction with the RNU6B gene as a control for normalization. All experiments were performed in triplicate and repeated once. For analysis of HMGB1 mRNA expression, total RNA was extracted from cells and was reverse transcribed using RT-PCR kits (Applied Biosystems) with an oligo d(T). To verify integrity of RAB14 expression, ACTIN gene was used as an internal control. PCR conditions were 30 cycles consisted of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. Each PCR product was separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining.
Western blot assay
Cell protein lysates were separated in 10% sodium dodecyl sulfate polyacrylamide gels, electrophoretically transferred to polyvinylidene difluoride membranes (Roche Diagnostics, Mannheim, Germany), then detected with anti-HMGB1. Protein loading was estimated using mouse anti-ACTIN monoclonal antibody. Lab Works Image Acquisition and Analysis Software (UVP, Upland, CA, USA) were used to quantify band intensities. Antibodies were purchased from Abcam.
Cell proliferation (MTT) assay and colony formation assay
The transfected cells were plated into 96-well plates at a density of 5000 cells/well. At 48 h after transfection, the cells were incubated with MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 4 h at 37°C. Then the cells were agitated with MTT solvent on an orbital shaker for 10 min avoiding light. The absorbance at 570 nm (A570 nm) was measured with a spectrophometer. For the colony formation assay, a total of 500 miR-142-3p transfected NSCLC cells were placed in a fresh six-well plate and maintained in media containing 10% FBS, replacing the medium every 4 days. After 14 days, cells were fixed with methanol and stained with 0.1% crystal violet (sigma). Visible colonies were manually counted. Triplicate wells were measured in each treatment group.
Flow-cytometric analysis of apoptosis
NSCLC cells transiently transfected with miR-142-3p or miR-control, were harvested at 48 h after transfection by trypsinization. After the double staining with FITC-Annexin V and 7-AAD, the cells were analyzed with a flow cytometry (FACScanW; BD Biosciences) equipped with a CellQuest software (BD Biosciences). Cells were discriminated into viable cells, dead cells, early apoptotic cells, and apoptotic cells, and then the relative ratio of early apoptotic cells were compared to control from each experiment. All of the samples were assayed in triplicate.
Results
Decreased expression of miR-142-3p in human NSCLC specimens
To explore the possible role of miR-142-3p in NSCLC development, we first examined the expression of miR-125b in 21 paired NSCLC samples and 4 NSCLC cell lines by SYBR-Green stem-loop qRT-PCR. As shown in Figure 1, the expression level of miR-142-3p in NSCLC samples and cell lines was much lower than that in Normal samples, which provided initial evidence that miR-142-3p may play a role in the development of human NSCLC.
Figure 1.
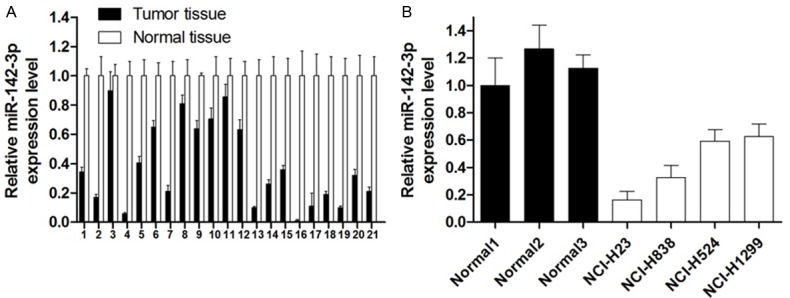
miR-142-3p expression in NSCLC tissues and cell lines. A. Relative expression of miR-142-3p in 21 NSCLC tissues in comparison with corresponding normal lung tissues. B. The miR-142-3p expression was significantly lower in NSCLC cell lines than in those corresponding normal lung tissues.
Effect of miR-142-3p on NSCLC cell proliferation
To assess the biological role of miR-142-3p in NSCLC, we investigated the effect of overexpression of miR-142-3p on cell viability and proliferation. Firstly, qRT-PCT results confirmed miR-142-3p expression level was significant elevated by miR-142-3p transfection. MTT assay revealed that cell growth was significantly impaired in NCI-H23 and NCI-H838 cells transfected with miR-142-3p (Figure 2B, 2C). Similarly, the results of colony-formation assays revealed that clonogenic survival was decreased following overexpression of miR-142-3p in both NSCLC cells (Figure 2D, 2E).
Figure 2.
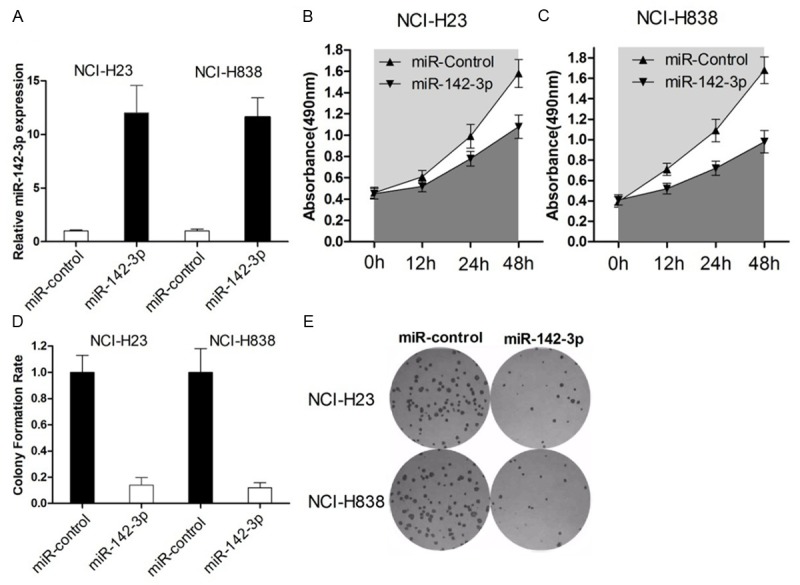
Overexpression of miR-142-3p inhibits in vitro growth of NSCLC cells. A. NCI-H23 and NCI-H838 cells were transfected with miR-142-3p or miR-controls, respectively. qRT-PCR was used to measure the miR-142-3p expression after transfection. B, C. At 48 h after transfection, MTT assay was performed to determine the proliferation of NCI-H23 and NCI-H838 cells. Data represent the mean ± s.d. from three independent experiments. D. Representative results of colony formation of NCI-H23 and NCI-H838 cells transfected with miR-142-3p or miR-control. The results were reproducible in three independent experiments.
Effect of miR-142-3p on NSCLC cell apoptosis
To determine whether apoptosis was a contributing factor to cell growth inhibition, we performed Hochest staining and flow-cytometric analysis of NSCLC cells after transfection with miR-142-3p. As shown in Figure 3A-D, we find that overexpression of miR-142-3p could significantly increased the apoptosis rate in NCI-H23 and NCI-H838 cells. Taken together, these results indicate that ectopic expression of miR-142-3p suppresses cell growth and is associated with induction of apoptosis in NSCLC.
Figure 3.
Ectopic miR-142-3p expression enhances apoptosis of NSCLC cells. At 48 h after transfection with miR-142-3p or miR-control, NCI-H23 and NCI-H838 cells were collected for apoptosis analysis. A-D. Flow cytometry was used to detect the apoptotic cells in each Gate. All experiments were repeated in triplicate with similar results.
HMGB1 was a target for miR-142-3p in NSCLC
We then investigated the mechanisms by which miR-142-3p inhibits NSCLC cell tumorigensis. Online search for miR-142-3p targeting genes by miRBase, TargetScan, miRanda and PicTar revealed that HMGB1, a proto-oncogene could be a potential target of miR-142-3p (Figure 4A). To ascertain direct miRNA-target interaction, we set up a luciferase reporter assay. The 3’-UTR of HMGB1, including miR-142-3p target sites, was cloned into the downstream of pGL3-promotor vector, along with corresponding point mutations within the miRNA seed of these predicted sites. The NCI-H23 and NCI-H838 cells were cotransfected with the reporter vector and miR-142-3p. As shown in Figure 4B, the luciferase activity in NSCLC cells was decreased with WT construct by enhanced miR-142-3p level, which could be restored with mutant constructs. These results suggest that 3’UTR of HMGB1 is a direct target of miR-142-3p.
Figure 4.
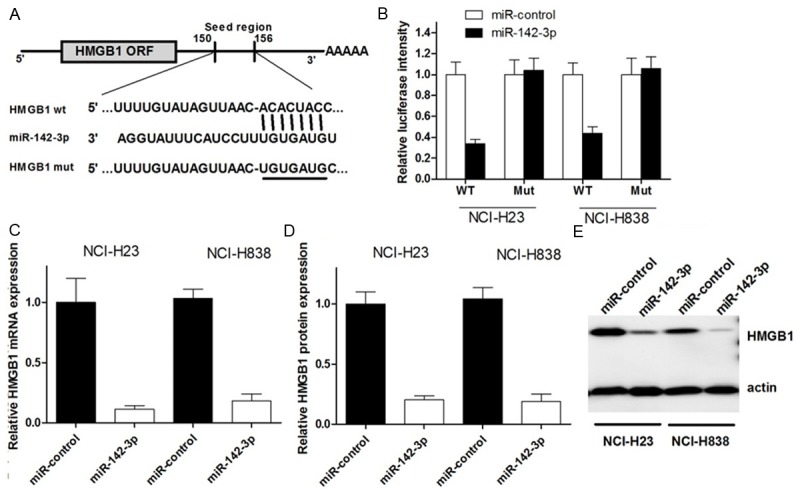
MiR-142-3p directly targets HMGB1 mRNA and inhibits its expression. A. The putative miR-142-3p-binding sites in the 3’-UTR of HMGB1 mRNA was shown. Mutation was generated on the HMGB1 3’-UTR sequence in the complementary site for the seed region of miR-142-3p, as shown. B. The wild type (HMGB1 3’-UTR-WT) or mutant (HMGB1 3’-UTR-mut) reporter plasmids was co-transfected into NCI-H23 and NCI-H838 with miR-142-3p or miR-control. The normalized luciferase activity in the control group was set as relative luciferase activity. C. The expression of HMGB1 mRNA was analyzed by real time PCR assay. ACTIN was used as an internal control. D, E. The expression of HMGB1 protein was analyzed by western blot assay. actin was used as an internal control. All experiments were at least repeated in triplicate with similar results.
We next examined whether miR-142-3p could regulate endogenous HMGB1 expression. The result showed that compared with controls, endogenous HMGB1 mRNA level was significantly decreased in NSCLC cells trasnfected with miR-142-3p (Figure 4C), meanwhile, the HMGB1 protein level was also significantly suppressed with miR-142-3p enhancement (Figure 4D, 4E). Considering these findings together, we can conclude that miR-142-3p represses endogenous HMGB1 expression at both mRNA and protein level, which may partly explain the miR-142-3p induced NSCLC cell tumorigenesis inhibition.
Discussion
Understanding the molecular mechanism of tumor was pivotal for effective therapy. Studies have been report that numerous of miRNAs contributed to cancer tumorigenesis including oncogenic miRNAs and tumor suppressor miRNAs [15]. Therefore, it is crucial to explore the function of deregulated miRNAs in NSCLC. Previous studies have discovered that miR-142-3p is downregulated in kinds of tumors [11-13]. However, the expression pattern of miR-142-3p in NSCLC cancer and cell lines was still poorly explored. In this study, we firstly determined the deregulated expression of miR-142-3p in NSCLC using real time PCR, and confirmed miR-142-3p was significantly down-regulated in NSCLC tissues compared to the corresponding matched normal tissues. We also found the relative expression level of miR-142-3p in NSCLC cell lines was down-regulated. Because of the limited tissue samples in this study, further investigation of a larger patient population is necessary to confirm the clinical significance and prognostic evaluation of miR-142-3p in NSCLC.
HMGB1, which located on the human chromosome 13q12, is belongs to the high mobility group protein super family. It has been reported to participate in various biological processes, including inflammation, DNA and tissue repair, cell mobility [16-20]. Interaction of HMGB1 and its receptor RAGE (receptor for advanced glycation end-products) is involved in the malignant behaviors in several cancers, such as breast cancer, colon cancer, melanoma and lung cancer [21-25]. Studies also demonstrate that HMGB1 is associated with cancer cell migration and invasion. HMGB1 promotes the cell invasion via regulation of MMP-9 in lung cancer [26,27]. HMGB1 can activate caspase-1 and contribute to hepatocellular carcinoma invasion and metastasis [28]. Our results were consistent with the above reports and suggested silencing of HMGB1 inhibited lung cancer migration and invasion. With respect to the mechanism of HMGB1 regulation on cell behaviors, we need to investigate the genes and signaling pathway regulated by HMGB1, like MMP-9, caspase-3 and HMGB1-RAGE axis, which we will focus on in future study.
In summary, we present here the first evidence that HMGB1is potential target for miR-142-3p and that miR-142-3p may modulate NSCLC cell tumorigenesis through targeting HMGB1. This finding not only helps us understand the molecular mechanism of NSCLC carcinogenesis, but also gives us a strong rationale to further investigate miR-142-3p as a potential treatment target for NSCLC.
Acknowledgements
Hunan Provincial Natural Science Foundation of China (14JJ2029).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I EUROCARE-Working Group. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8:784–796. doi: 10.1016/S1470-2045(07)70246-2. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY, Xi QS. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012;18:BR299–308. doi: 10.12659/MSM.883262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li WQ, Li YM, Tao BB, Lu YC, Hu GH, Liu HM, He J, Xu Y, Yu HY. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA-328 may decrease their chemoresistance. Med Sci Monit. 2010;16:HY27–30. [PubMed] [Google Scholar]
- 10.Shang C, Lu YM, Meng LR. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med Sci Monit. 2012;18:BR149–155. doi: 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Cai C, Wang X, Liu M, Li X, Tang H. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 2011;585:1322–1330. doi: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 12.Shen WW, Zeng Z, Zhu WX, Fu GH. MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl) 2013;91:989–1000. doi: 10.1007/s00109-013-1037-x. [DOI] [PubMed] [Google Scholar]
- 13.MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, Subramanian S, Saluja AK. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. 2013;12:1266–1275. doi: 10.1158/1535-7163.MCT-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Namlos HM, Meza-Zepeda LA, Baroy T, Ostensen IH, Kresse SH, Kuijjer ML, Serra M, Burger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang C, Hong Y, Guo Y, Liu YH, Xue YX. MiR-210 up-regulation inhibits proliferation and induces apoptosis in glioma cells by targeting SIN3A. Med Sci Monit. 2014;20:2571–2577. doi: 10.12659/MSM.892994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanuma S, Yagi T, Johnson GS. Endogenous ADP ribosylation of high mobility group proteins 1 and 2 and histone H1 following DNA damage in intact cells. Arch Biochem Biophys. 1985;237:38–42. doi: 10.1016/0003-9861(85)90251-6. [DOI] [PubMed] [Google Scholar]
- 17.Ulloa L, Messmer D. High-mobility group box 1 (HMGB1) protein: friend and foe. Cytokine Growth Factor Rev. 2006;17:189–201. doi: 10.1016/j.cytogfr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari S, Finelli P, Rocchi M, Bianchi ME. The active gene that encodes human high mobility group 1 protein (HMG1) contains introns and maps to chromosome 13. Genomics. 1996;35:367–371. doi: 10.1006/geno.1996.0369. [DOI] [PubMed] [Google Scholar]
- 19.Tanuma S, Johnson GS. ADP-ribosylation of nonhistone high mobility group proteins in intact cells. J Biol Chem. 1983;258:4067–4070. [PubMed] [Google Scholar]
- 20.Rauvala H, Huttunen HJ, Fages C, Kaksonen M, Kinnunen T, Imai S, Raulo E, Kilpelainen I. Heparin-binding proteins HB-GAM (pleiotrophin) and amphoterin in the regulation of cell motility. Matrix Biol. 2000;19:377–387. doi: 10.1016/s0945-053x(00)00084-6. [DOI] [PubMed] [Google Scholar]
- 21.Flohr AM, Rogalla P, Meiboom M, Borrmann L, Krohn M, Thode-Halle B, Bullerdiek J. Variation of HMGB1 expression in breast cancer. Anticancer Res. 2001;21:3881–3885. [PubMed] [Google Scholar]
- 22.Bartling B, Hofmann HS, Weigle B, Silber RE, Simm A. Down-regulation of the receptor for advanced glycation end-products (RAGE) supports non-small cell lung carcinoma. Carcinogenesis. 2005;26:293–301. doi: 10.1093/carcin/bgh333. [DOI] [PubMed] [Google Scholar]
- 23.Brezniceanu ML, Volp K, Bosser S, Solbach C, Lichter P, Joos S, Zornig M. HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J. 2003;17:1295–1297. doi: 10.1096/fj.02-0621fje. [DOI] [PubMed] [Google Scholar]
- 24.Volp K, Brezniceanu ML, Bosser S, Brabletz T, Kirchner T, Gottel D, Joos S, Zornig M. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55:234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poser I, Bosserhoff AK. Transcription factors involved in development and progression of malignant melanoma. Histol Histopathol. 2004;19:173–188. doi: 10.14670/HH-19.173. [DOI] [PubMed] [Google Scholar]
- 26.Liu PL, Tsai JR, Hwang JJ, Chou SH, Cheng YJ, Lin FY, Chen YL, Hung CY, Chen WC, Chen YH, Chong IW. High-mobility group box 1-mediated matrix metalloproteinase-9 expression in non-small cell lung cancer contributes to tumor cell invasiveness. Am J Respir Cell Mol Biol. 2010;43:530–538. doi: 10.1165/rcmb.2009-0269OC. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Fei G, Liu Z, Li Q, Xu Z, Ren T. HMGB1 was a pivotal synergistic effecor for CpG oligonucleotide to enhance the progression of human lung cancer cells. Cancer Biol Ther. 2012;13:727–736. doi: 10.4161/cbt.20555. [DOI] [PubMed] [Google Scholar]
- 28.Yan W, Chang Y, Liang X, Cardinal JS, Huang H, Thorne SH, Monga SP, Geller DA, Lotze MT, Tsung A. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55:1863–1875. doi: 10.1002/hep.25572. [DOI] [PMC free article] [PubMed] [Google Scholar]



