Abstract
MicroRNAs (miRNAs) act as key regulators of multiple cancers. MicroRNA-506 (miR-506) functions as a tumor suppressor in various types of cancers. However, its role in esophageal cancer remains unclear. In our study, we found that miR-506 was significantly down-regulated in esophageal cancer tissues and cell lines. In vitro assay, our results showed that ectopic over-expression of miR-506 inhibited esophageal cancer cells proliferation, meanwhile, cells proliferation was promoted by miR-506 inhibition. In exploring mechanisms underlying the inhibitive role, we found that miR-506 significantly decreased the expression and transcription activity of cAMP responsive element binding protein 1 (CREB1). CREB1, tumor oncogene, exhibited significantly promote effect on esophageal cancer cell proliferation. Taken together, our data identify a new role of miR-506 in esophageal cancer involving CREB1 suppression.
Keywords: CAMP responsive element binding protein 1, miR-506, esophageal cancer, proliferation
Introduction
Esophageal cancer is one of the most common upper gastrointestinal tract cancers worldwide and esophageal squamous cell carcinoma (ESCC) is the most predominant type of esophageal cancer in eastern Asia and southern Africa [1,2]. Traditionally, surgery is considered the best treatment in early stage. In advanced esophageal cancer, a multidisciplinary approach that includes surgery, radiotherapy, and chemotherapy, alone or in combination, has been developed [3]. Despite the development of improved therapeutic techniques, the overall five-year survival rate of ESCC is only 20-30% [4]. Thus, understanding the mechanism of ESCC development is essential for improving disease diagnosis, treatment and prevention.
MicroRNAs (miRNAs) are a class of highly-conserved, non-coding 18-25 nucleotide RNAs that function as negative regulators of gene expression at the post-transcription level, binding to the 3’-untranslated region (3’-UTR) of mRNAs transcripts and targeting them for degradation [5,6]. Dependenting on their target genes, miRNAs can function as both oncogenes and tumor suppressor genes by affecting cell proliferation, migration, apoptosis, differentiation, and metabolism [7-9]. For example, Su et al showed that miR-152 was down-regulated in non-small cell lung cancer (NSCLC), furthermore, they indicated that miR-152 significantly reduced cell proliferation, colony formation, migration and invasion of NSCLC cells via targeting ADAM17 [10]. Wang et al revealed that miR-22 was decreased in gastric cancer, and over-expression of miR-22 suppressed cell proliferation and colony formation by inhibiting CD151 [11]. In contrast, Chen et al showed miR-103/107 promoted metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4 [12]. Sun et al showed that miRNA-221 accelerated the proliferation of laryngeal cancer cell by suppressing Apaf-1 [13]. However, the role of miR-506 in ESCC is unclear.
To understand the role of miR-506 in esophageal cancers, we introduced miR-506 over-expression and underexpression models in the present study. Using these models, we investigated the effect of miR-506 on esophageal cancer cell proliferation. cAMP responsive element binding protein 1 (CREB1), a tumor oncogene has complementary binding site of miR-506 at 3’-UTR, which indicated CREB1 is putatively potential target of miR-506. Thus, the effect of miR-506 on CREB1 was explored.
Materials and methods
Clinical samples
Matched ESCC tissues and adjacent non-tumor tissues were obtained from 28 patients who underwent esophagectomy for primary ESCC in the Department of Thoracic Surgery, The First Affiliated Hospital of Xinxiang Medical University. None of the patients had received preoperative chemotherapy or radiotherapy. All tissue samples were frozen in liquid nitrogen immediately after resection and stored at -80°C. The histological diagnosis of these tissue samples was confirmed by a pathologist. This study was approved by the Committee for Ethical Review of Research at First Affiliated Hospital of Xinxiang Medical University, and informed consent was obtained from all the patients involved.
Cell culture and transfection
The human ESCC cell lines, Eca109, Kyse30, TE-1, and normal esophageal cell line NEEC were obtained from the American Type Culture Collection (USA) and grown in RPMI-1640 medium, with 10% fetal bovine serum (FBS, Gibco), 1% antibiotics (100 μ/ml penicillin and 100 mg/ml streptomycin sulfates) and 1% glutamate (Gibco), and then incubated at 37°C in a humidified chamber containing 5% CO2.
The miR-506 mimic, miR-506 inhibitor (anti-miR-506) and CREB1-targeted small interfering RNA (si-CREB1) were synthesized by RiboBio Corporation (China), and transfected at a final concentration of 50-100 nM with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency was detected by qRT-PCR after 48 h of incubation.
RNA extraction and quantitative real-time PCR
RNA was isolated from ESCC tissues and cells using TRIzol (Invitrogen) and cDNA was synthesized using a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems of Thermo Fisher Scientific) according to the manufacturer’s instructions. The expression level of miR-506 in the esophageal cancer tissues or cells was confirmed by a TaqMan microRNA assay (Applied Biosystems). Expression of CREB1 mRNA in esophageal cancer cells were quantified using SYBR Green real-time PCR master mix (Applied Biosystems). The relative expression levels of miR-506 and CREB1 mRNA were calculated by the 2-ΔΔCt method and normalized to U6 snRNA and GAPDH mRNA levels, respectively. All PCR reactions were performed on a StepOne Plus RT-PCR instrument (Applied Biosystems).
Cell proliferation assay
Cell proliferation was analyzed with MTT assay. Briefly, 2000 cells of each group were plated in 96-well microplates in 150 μl of medium. After different time of culture (24 h, 48 h, 72 h, 96 h), 20 μl of MTT substrate (5 mg/ml) was added to each well, and the plates were incubated for an additional 4 h. The medium was then removed, and the cells were solubilized in 150 μl of dimethyl sulfoxide. The culture plates were then shaken for 15 min and the optical absorbance values were read at 490 nm.
Flow cytometry analysis
Flow cytometry analysis was performed according to previous report [13]. Cells were harvested by trypsinization, washed in ice-cold PBS, and fixed in 80% ice-cold ethanol. Before staining, the cells were pelleted in a cooled centrifuge and re-suspended in cold PBS. Cells were then incubated in 20 μg/ml propidium iodide for 20 min at room temperature, and analyzed on a flow cytometry (FACSCalibur, BD Biosciences).
Luciferase assay
The CREB1 3’-UTR was amplified from human genomic DNA and subcloned into the pGL3 basic luciferase reporter plasmid using MluI and XhoI sites. CREB1 3’-UTR luciferase reporter plasmids was co-transfected with miR-506 mimics using lipofectamine 2000 reagent. Transfected cells were serum shocked (20%) for 2 h and maintained in 2% media for 24 h before harvest. Dual-Luciferase Reporter Assay was performed according to the manufacturer’s instructions. Data were presented as relative luciferase activity.
Western blot
Total protein from cells was extracted in T-PER Tissue Protein Extraction Reagent (Pierce) with protease inhibitors phenylmethanesulfonyl fluoride. Protein were subjected to SDS-PAGE and then electrophoretically transferred to a PVDF membrane (Millipore). After blocking with 5% nonfat dry milk, the membranes were incubated with primary antibody, and then the membranes incubated with a horseradish peroxidase-conjugated secondary antibody. The immunoreactive proteins were detected by ECL Detection Systems (Thermo Scientific).
Statistic analysis
All data are expressed as the mean ± SD from at least three separate experiments. All data were analyzed by the SPSS 19.0 statistical software. The differences between two groups were analyzed using a Student’s t-test or one-way ANOVA. Results were considered statistically significant with P<0.05.
Results
miR-506 is down-regulated in human esophageal cancer tissues and cell lines
To determine the expression level of miR-506, qRT-PCR was performed in 28 paired ESCC tissues and adjacent non-tumor tissues (ANT). As shown in Figure 1A, the expression of miR-506 was significantly down-regulated in ESCC tissues compared with that in ANTs (P<0.05). We also analyzed the expression of miR-506 in ESCC cell lines (Eca109, Kyse30 and TE-1) and normal esophageal cell line NEEC. As shown in Figure 1B, expression level of miR-506 was also significantly lower in Eca109, Kyse30 and TE-1 cells than NEEC cells (P<0.05). These results suggested that miR-506 was associated with the development and progression of esophageal cancer.
Figure 1.
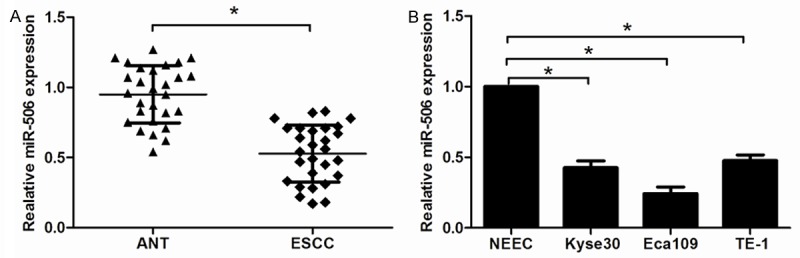
miR-506 expression was decreased in ESCC tissues and cell lines. A: The expression of miR-506 in ESCC tissues and adjacent non-tumor tissues (ANT) was detected by qRT-PCR. B: The expression of miR-506 in three ESCC cell lines, Eca109, Kyse30, and TE-1, and normal esophageal cell line (NEEC) was measured by qRT-PCR. Data are mean ± SD of three independent experiments. *P<0.05.
miR-506 inhibits esophageal cancer cell proliferation
To explore the function of miR-506 in esophageal cancer, esophageal cancer cell line Eca109 were transfected with miR-506 mimics or miR-506 inhibitor (anti-miR-506). The transfection efficiency was confirmed by qRT-PCR (P<0.05, Figure 2A, 2B). MTT assay showed that the rate of cell proliferation in the miR-506 mimics transfected group was significantly decreased compared with miR-NC groups (P<0.05, Figure 2C). In contrast to miR-506 over-expression, the proliferation rate of Eca109 cells transfected with anti-miR-506 was significantly increased in comparison to cells transfected with anti-miR-NC (P<0.05, Figure 2D). In addition, we analyzed the cell cycle by flow cytometry, our results showed that ectopic over-expression of miR-506 markedly induce G1 phase arrest of esophageal cancer cells (P<0.05, Figure 2E). Consistent with this result, silencing of miR-506 resulted in a significant decrease in the cellular population in G0/G1 phase but an increase in S phase (P<0.05, Figure 2F). All these results suggested that miR-506 could inhibit the proliferation of Eca109 cells.
Figure 2.
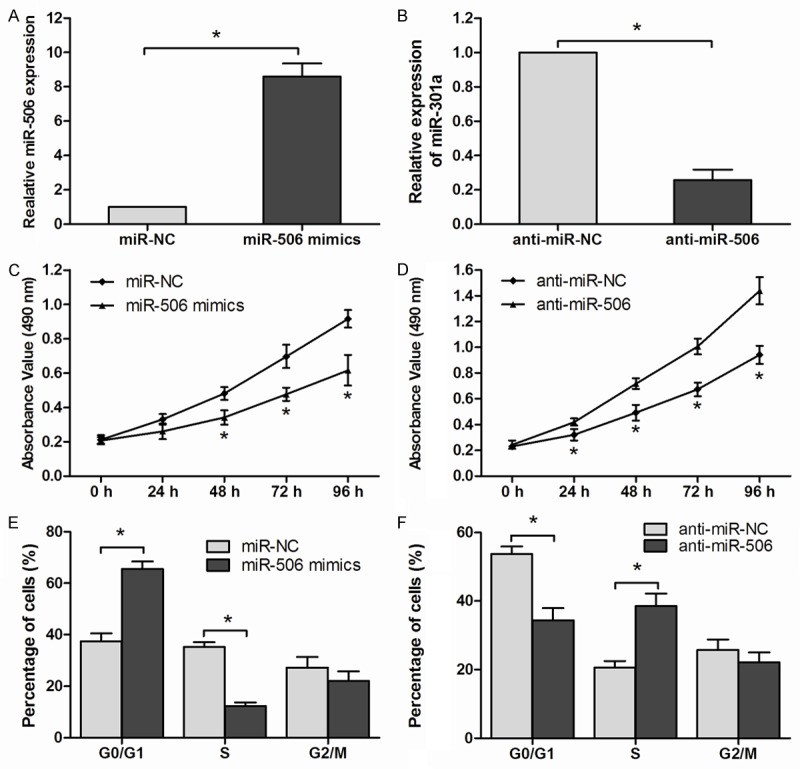
miR-506 suppressed ESCC cell proliferation. A, B: Relative expression of miR-506 determined by qRT-PCR in Eca109 cells after transfection with miR-506 mimics or miR-506 inhibitors (anti-miR-506). C, D: Cell proliferation of Eca109 cells transfected with miR-506 mimics or anti-miR-506 was detected by MTT assay. E, F: Cell cycle distribution of transfected Eca109 cells assessed by flow cytometry. Data are mean ± SD of three independent experiments. *P<0.05.
CREB1 is a target of miR-506
To investigate the mechanisms by which miR-506 influences cellular proliferation, we employed TargetScan to search for putative protein-coding gene targets of miR-506, The gene for CREB1 was identified as a potential target (Figure 3A), To determine whether CREB1 was directly regulated by miR-506, luciferase reporter assay was performed in Eca109 cells. As showed in Figure 3B, the relative luciferase activity of wide type plasmids containing putative binding site was significantly decreased when transfected with miR-506 mimics in Eca109 cells (P<0.05). In addition, western blot showed miR-506 over-expression decreased CREB1 protein level, but miR-506 inhibition increased CREB1 protein level (P<0.05, Figure 3C). These results indicated that CREB1 is a potential target of miR-506, and that miR-506 negatively regulated CREB1 gene expression in ESCC.
Figure 3.
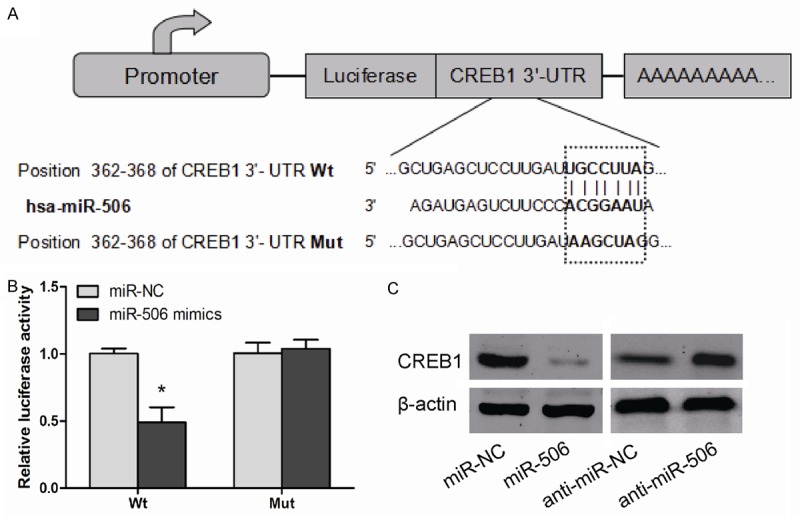
CREB1 was a target of miR-506 in ESCC cells. A: The potential miR-506 binding sites of CRE1 3’-UTR and the mutant (Mut). B: Eca109 cells were co-transfected with miR-506 mimics or miR-NC with Wt or Mut CREB1 3’-UTR. Luciferase reporter assay was performed. C: Protein level of CREB1 was detected by western blot in Eca109 cells transfected with miR-506 mimics or anti-miR-506. β-actin was used as an internal control. Data are mean ± SD of three independent experiments. *P<0.05.
CREB1 knockdown inhibits esophageal cancer cell proliferation
To explore the function of CREB1 in esophageal cancer, Eca109 cells were transfected with si-CREB1. qRT-PCR was used to determinate the efficiency of si-CREB1 (Figure 4A). The MTT assay showed that knockdown of CREB1 inhibited esophageal cancer cell proliferation (P<0.05, Figure 4B). Furthermore, flow cytometry results also suggested that silence of CREB1 significantly induce G1 phase arrest of esophageal cancer cells (P<0.05, Figure 4C). CREB1 silencing induces a very similar phenotype to miR-506 over-expression in esophageal cancer cells. These data demonstrated that miR-506 down-regulated CREB1, thus inhibiting esophageal cancer cell proliferation.
Figure 4.
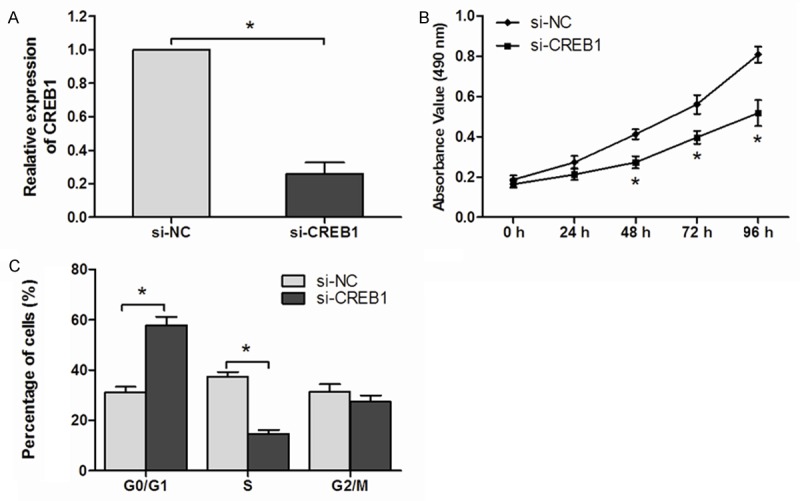
CREB1 knockdown inhibited ESCC cell proliferation. A: Expression of CREB1 was detected by qRT-PCR in Eca109 cells transfected with si-CREB1 or si-NC. B: Cell proliferation of Eca109 cells transfected with si-CREB1 or si-NC was detected by MTT assay. C: Cell cycle distribution of transfected Eca109 cells assessed by flow cytometry. Data are mean ± SD of three independent experiments. *P<0.05.
Discussion
MicroRNAs play important roles in multiple aspects of tumor biology. Yang et al reported that miR-506 was down-regulated in renal cancer, and inhibited cell growth and metastasis via targeting FLOT1 [14]. Deng et al showed that miRNA-506 suppressed gastric cancer proliferation and invasion by directly targeting Yap1 [15]. Furthermore, Liu et al suggested that miR-506 inhibited proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer [16]. Here, we found that miR-506 was markedly down-regulated in ESCC tissues and esophageal cancer cell lines, indicating a potential tumor suppressive function of miR-506. Our results showed that miR-506 suppressed cell proliferation of esophageal cancer cells in vitro, and provided that these effects were partially mediated by inhibiting CREB1 expression. Our data revealed that CREB1 knockdown performed a similar effect of miR-506 over-expression on esophageal cancer cells. These data indicated that CREB1 act as a direct target gene of miR-506, important in regulating esophageal cancer progression.
The CREB1 gene is localized at human chromosome 2q32.3-q34, a region known to be a hot spot for translocations or deletions in various diseases including cancers [17]. Wang et al reported that CREB1 was high expressed in gastric cancer, and correlated with tumor stage, lymph node metastasis and distant metastasis. Furthermore, they demonstrated that CREB1 could serve as a potential prognostic maker in gastric cancer [18]. Li et al reported that loss of CREB1 suppressed the growth of urinary bladder urothelial carcinoma cells [19]. Moreover, Peng et al found that miR-200b suppressed the growth and motility of human malignant glioma by targeting CREB1 [20]. In addition, Chen et al reported that miRNA-181b suppressed tumor proliferation and invasion by targeting CREB1 in gastric adenocarcinomas [21]. In the present study, we found that inhibition of CREB1 significantly inhibited the proliferation of esophageal cancer cells. Our study expanded the function of CREB1 in esophageal cancer.
In conclusion, our study found that miR-506 was decreased in ESCC tissues and cell lines, and enforced over-expression of miR-506 substantially suppressed proliferation of esophageal cancer cells. CREB1 was identified as a target of miR-506, and CREB1 knockdown performed similar effects of miR-506 over-expression, indicating that miR-506 might act as a novel therapeutic strategy for the treatment of esophageal cancer.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Wieder HA, Brucher BL, Zimmermann F, Becker K, Lordick F, Beer A, Schwaiger M, Fink U, Siewert JR, Stein HJ, Weber WA. Time course of tumor metabolic activity during chemoradiotherapy of esophageal squamous cell carcinoma and response to treatment. J. Clin. Oncol. 2004;22:900–908. doi: 10.1200/JCO.2004.07.122. [DOI] [PubMed] [Google Scholar]
- 3.Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis. 2001;22:1737–1746. doi: 10.1093/carcin/22.11.1737. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 8.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 9.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Y, Wang Y, Zhou H, Lei L, Xu L. MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett. 2014;588:1983–1988. doi: 10.1016/j.febslet.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Yu H, Lu X, Zhang P, Wang M, Hu Y. MiR-22 suppresses the proliferation and invasion of gastric cancer cells by inhibiting CD151. Biochem Biophys Res Commun. 2014;445:175–179. doi: 10.1016/j.bbrc.2014.01.160. [DOI] [PubMed] [Google Scholar]
- 12.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, Shyy JY, Liang JT, Chen RH. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 13.Sun X, Liu B, Zhao XD, Wang LY, Ji WY. MicroRNA-221 accelerates the proliferation of laryngeal cancer cell line Hep-2 by suppressing Apaf-1. Oncol Rep. 2015;33:1221–1226. doi: 10.3892/or.2015.3714. [DOI] [PubMed] [Google Scholar]
- 14.Yang FQ, Zhang HM, Chen SJ, Yan Y, Zheng JH. MiR-506 Is Down-Regulated in Clear Cell Renal Cell Carcinoma and Inhibits Cell Growth and Metastasis via Targeting FLOT1. PLoS One. 2015;10:e0120258. doi: 10.1371/journal.pone.0120258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng J, Lei W, Xiang X, Zhang L, Yu F, Chen J, Feng M, Xiong J. MicroRNA-506 inhibits gastric cancer proliferation and invasion by directly targeting Yap1. Tumour Biol. 2015 doi: 10.1007/s13277-015-3364-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D, Parker Kerrigan BC, Shmulevich I, Chen K, Sood AK, Xue F, Zhang W. MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol. 2014;233:308–318. doi: 10.1002/path.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang YW, Chen X, Gao JW, Zhang H, Ma RR, Gao ZH, Gao P. High expression of cAMP-responsive element-binding protein 1 (CREB1) is associated with metastasis, tumor stage and poor outcome in gastric cancer. Oncotarget. 2015;6:10646–57. doi: 10.18632/oncotarget.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wu W, Wu W, Liao Y, Chen L, Huang C, Li C, Li W, Huang H, Chen Y. The cAMP responsive element binding protein 1 transactivates epithelial membrane protein 2, a potential tumor suppressor in the urinary bladder urothelial carcinoma. Oncotarget. 2015;6:9220–9239. doi: 10.18632/oncotarget.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng B, Hu S, Jun Q, Luo D, Zhang X, Zhao H, Li D. MicroRNA-200b targets CREB1 and suppresses cell growth in human malignant glioma. Mol Cell Biochem. 2013;379:51–58. doi: 10.1007/s11010-013-1626-6. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Yang Q, Kong WQ, Liu T, Liu M, Li X, Tang H. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012;64:628–635. doi: 10.1002/iub.1030. [DOI] [PubMed] [Google Scholar]


