Abstract
Background: MicroRNAs (miRNA) have been documented playing a critical role in cancer progression. Although miR-338-3p has been implicated in several cancers, its role in gastric cancer is still unknown. The aim of our study was to investigate the role of miR-338-3p in gastric cancer progression. Methods: Expression levels of miR-338-3p in gastric cancer cell lines and tissues were determined by quantitative real-time PCR (qRT-PCR). The effect of miR-338-3p on proliferation was evaluated by MTT assay, cell migration and invasion were evaluated by transwell migration and invasion assays. Furthermore, luciferase reporter assay was conducted to confirm the target gene of miR-338-3p, and the results were validated in gastric cancer cells. Results: In the present study, we found that miR-338-3p was down-regulated in both gastric cancer cell lines and tissues. Enforced expression of miR-338-3p inhibited proliferation, migration and invasion of gastric cancer cells in vitro. Moreover, we identified A disintegrin and metalloproteinase 17 (ADAM17) gene as potential target of miR-338-3p. Importantly, ADAM17 rescued the miR-338-3p mediated inhibition of cell proliferation, migration and invasion. Conclusions: Our study suggested that miR-338-3p is significantly decreased in gastric cancer, and inhibits cell proliferation, migration and invasion partially via the downregulation of ADAM17. Thus, miR-338-3p may represent a potential therapeutic target for gastric cancer intervention.
Keywords: Gastric cancer, miR-338-3p, ADAM17
Introduction
Gastric cancer is the fourth most common cancer and the second leading cause of cancer-related death in the world. Almost two thirds of cases occur in developing countries, of which 42% occur in China [1,2]. Despite the significant achievements in the treatment, the prognosis for gastric cancer remains poor with an overall 5-year survival rate below 30% [3]. Thus, it is of great significance to further elucidate the molecular mechanisms of gastric cancer and look for new therapeutic targets for this disease.
MicroRNAs (miRNA), a class of small non protein coding RNAs, have been identified as a new kind of gene expression regulators through binding to the 3’ untranslated regions (UTR) of target mRNA [4]. miRNAs have been demonstrated to play critical roles in the biologic processes such as proliferation, apoptosis, differentiation and metastasis [5-7]. Accumulating evidence showed that many miRNAs have significant regulatory functions in gastric cancer. For example, Wang et al showed that miR-199a-3p may function as a novel tumor promoter in gastric cancer and its oncogenic activity may involve the direct targeting and inhibition of ZHX1 [8]. Li et al found that miR-409-3p was significantly down-regulated in gastric cancer and overexpression of miR-409-3p in gastric cancer cells dramatically suppressed cell proliferation and induced cell apoptosis by direct targeting PHF10 [9]. Duan et al suggested that miR-24 was significantly decreased in gastric cancer. Furthermore, they revealed that ectopic expression of miR-24 inhibited gastric cancer cell proliferation, migration and invasion via targeting RegIV [10]. Although plenty of miRNAs in relation to gastric cancer have been studied, the underlying mechanism of miRNAs in carcinogenesis of gastric cancer remains to be investigated.
Previous study showed that miR-338-3p was transcribed from the intron 8 of apoptosis-associated tyrosine kinase (AATK) gene, located on chromosome 17q25, playing a critical role in promoting cell death, neuronal differentiation and neurite extension [11]. Recent studies indicated that miR-338-3p could act as a tumor suppressor in types of cancers. For example, Sun et al indicated that miR-338-3p was decreased expression in non-small cell lung cancer (NSCLC). Ectopic miR-338-3p expression significantly suppressed cell proliferation and colony formation of NSCLC cells by inhibiting RAB14 expression [12]. Chen et al found that miR-338-3p was down-regulated in neuroblastoma and inhibited neuroblastoma proliferation, migration and invasion through targeting PREX2a [13]. Wang et al demonstrated that miR-338-3p was down-regulated in hepatocellular carcinoma and overexpression of miR-338-3p could inhibit cell proliferation by targeting FOXP4 [14]. However, the specific roles and underlying mechanisms of miR-338-3p in gastric cancer is still unknown.
In this study, we demonstrated that miR-338-3p was decreased in gastric cancer, and ectopic overexpression of miR-338-3p significantly inhibited the proliferation, migration and invasion of gastric cancer cells. Furthermore, we identified ADAM17 as a direct target of miR-338-3p, and ADAM17 overexpression partially attenuated the tumor suppressive effect of miR-338-3p in gastric cancer cells.
Materials and methods
Clinical samples
Human gastric cancer tissues and their corresponding non-tumor tissues were collected at the time of surgical resection from 36 patients from 2013 to 2014 at the Department of General Surgery, Huaihe Hospital of Henan University. Human tissues were immediately frozen in liquid nitrogen and stored at -80°C refrigerator. Signed informed consent was obtained from all patients and the study was approved by the Clinical Research Ethics Committee of Huaihe Hospital of Henan University.
Cell culture
Human gastric cancer cell lines (AGS, HGC-27, MKN-45 and BGC-823), and immortalized normal gastric mucosal epithelial cell line (GES-1) were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences. All cells were cultured in RPMI-1640 medium (Gibco) and supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 10 U/ml penicillin-streptomycin (Sigma). All cells were maintained in a humidified incubator at 37°C with 5% CO2.
Transient transfection
miR-338-3p mimics, negative control (miR-NC) were purchased from GenePharma (Shanghai, China). The pEGFP-ADAM17 plasmid was generated using the following primers: forward, 5’-GGGGTACCGAATCTTCCCAGTAGGCG-3’ and reverse, 5’-TTGCGGCCGCAAAGGAGAAGGGCCAAAC-3’. The transfection was performed using Lipofectamine 2000 (Invitrogen) according to the instructions provided by the manufacturer. Transfection efficiency was monitored by qRT-PCR.
Cell proliferation assay
Cell viability was measured using the MTT assay. The viability of BGC-823 cells transfected with miR-338-3p mimics or negative control was detected every 24 h after transfection. 100 μL MTT (10 mg/mL) was added to each well of the 96-well plates and incubated for 4 h, and then the reaction was terminated by DMSO. The optical density (OD) at 490 nm was detected using a Microplate Reader (Bio-Tek Instruments).
Cell migration and invasion assays
For the transwell migration assay, 5 × 104 BGC-823 cells were placed in the upper chamber of each insert (Corning). For the invasion assay, 1 × 105 cells were placed on the upper chamber of each insert coated with 40 μl matrigel (Clontech), which was diluted to 2 μg/μl with RPMI-1640 medium. Medium supplemented with 20% FBS (600 μl) was added to the lower chambers. After several hours of incubation, the upper surface of the membrane was wiped with a cotton tip and cells attached to the lower surface were stained for 20 min with crystal violet. Cells in 5 random fields of view at 100 × magnification were counted and expressed as the average number of cells per field of view.
Luciferase reporter assay
The 3’UTR of ADAM17 containing the predicted miR-338-3p binding site was constructed by RiboBio (China). The mutant ADAM17 3’UTR was created by mutating multiple nucleotides complementary to the miR-338-3p seed region. HEK293T cells were cultured in 96-well plates with 50-70% confluence 24 h before transfection. A mixture of 100 ng pmiR-RB-Report™ h-ADAM17 wild type (Wt) or mutant (Mut) reporter plasmid vector together with 50 nM miR-338-3p mimics or negative control were co-transfected. The luciferase activity was measured 48 h post-transfection using Dual-Glo Luciferase Assay System (Promage) with Renilla luciferase activity as the reporter gene and firefly luciferase as the reference gene.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from tissue samples and cell lines using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. The concentration and quality of total RNA were assessed via spectrophotometric and electrophoretic assay. First-strand cDNA was synthesized using the SuperScript III first-strand synthesis system (Invitrogen). Real-time PCR was carried out in the ABI 7900 system (Applied Biosystems) using the SYBR Premix Ex Taq™ (TaKaRa). Relative expression levels of miR-338-3p and ADAM17 were calculated by the 2-ΔΔCT method after normalization with reference to expression of U6 and GAPDH.
Western blot
Total protein from cells were lyzed by RIPA buffer. Equal amount of proteins were separated by 12% SDS-PAGE, and transferred to a nitrocellulose membrane (Bio-Rad). After being blocked with 5% non-fat milk, the membrane was incubated with anti-ADAM17 antibody or anti-GAPDH (Abcam). The membrane was then washed extensively and incubated with a goat anti-mouse secondary antibody (Pierce). The proteins were detected with ECL reagents (Pierce).
Statistical analysis
All statistical analyses were performed using SPSS 18.0 software (IBM). Data are expressed as the mean ± SD from at least three separate experiments. Differences between groups were analyzed using Student’s t test or one-way ANOVA analysis. A P-value less than 0.05 was considered statistically significant.
Results
Down-regulation of miR-338-3p in human gastric cancer cell lines and tissues
To determine whether miR-338-3p was involved in the tumorigenesis of gastric cancer, miR-338-3p expression was examined by qRT-PCR in gastric cancer cell lines and tissues. As shown in Figure 1A, miR-338-3p was significantly decreased in all gastric cancer cells compared with GES-1 cells. The expression of miR-338-3p was further evaluated by qRT-PCR in tumor samples and their matched non-tumor samples from 36 gastric cancer patients. The average expression level of miR-338-3p was significantly decreased in gastric cancer tissues compared with adjacent non-tumor tissues (Figure 1B). These results strongly suggested that miR-338-3p might play a key role in gastric cancer progression.
Figure 1.
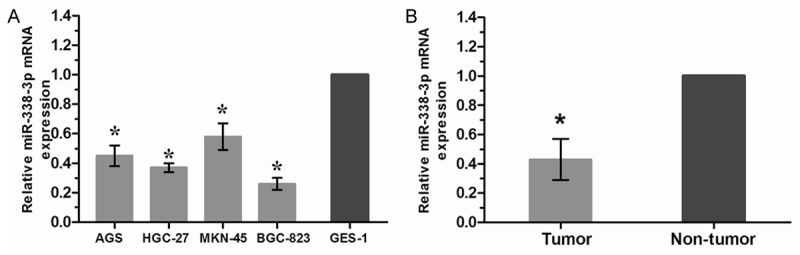
miR-338-3p is down-regulated in gastric cancer cell lines and tissues. A. qRT-PCR was performed to examine miR-338-3p expression in gastric cancer cell lines (AGS, HGC-27, MKN-45 and BGC-823) and normal gastric mucosal epithelial cell line (GES-1). B. qRT-PCR analysis of miR-338-3p expression in 36 paired human gastric cancer tissues and adjacent non-tumor tissues. *P<0.05.
miR-338-3p inhibits gastric cancer cell proliferation, migration and invasion in vitro
To investigate the function of miR-338-3p in the gastric cancer cells, BGC-823 were transfected with miR-338-3p mimics. The efficiency of miR-338-3p mimics was determined by qRT-PCR (Figure 2A). MTT assay showed overexpression of miR-338-3p significantly suppressed cell proliferation of BGC-823 cells compared with the negative control (Figure 2B). Furthermore, we investigated the effect of miR-338-3p on migration and invasion of gastric cancer cells. Transwell migration assay revealed that increased expression of miR-338-3p dramatically inhibited tumor cell migration in BGC-823 cells compared with the negative control (Figure 2C). Similarly, transwell invasion assay demonstrated that miR-338-3p markedly decreased the invasive capacity of BGC-823 cells (Figure 2D).
Figure 2.
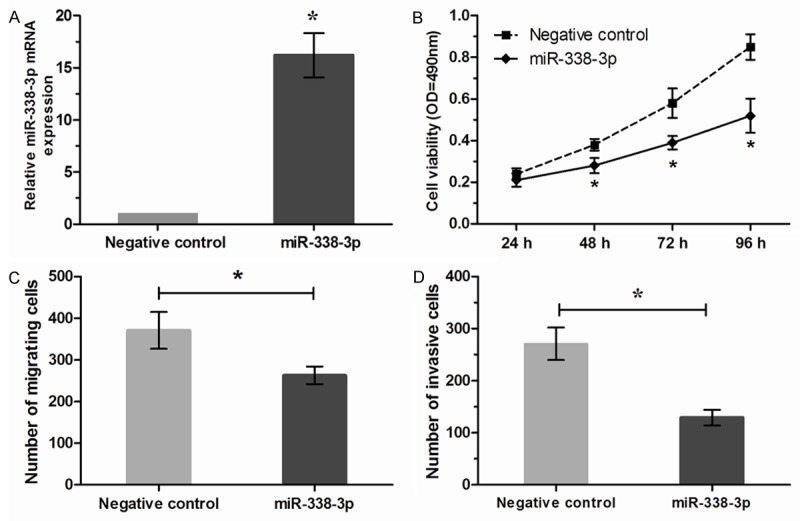
miR-338-3p inhibits gastric cancer cell proliferation, migration and invasion in vitro. A. Expression of miR-338-3p in BGC-823 cells transfected with miR-338-3p mimics or negative control was detected by qRT-PCR. B. Cell proliferation of BGC-823 cells transfected with miR-338-3p mimics or negative control was detected by MTT assay. C. Transwell migration assay of BGC-823 cells transfected with miR-338-3p mimics or negative control. D. Transwell invasion assay of BGC-823 cells transfected with miR-338-3p mimics or negative control. *P<0.05.
ADAM17 is a direct target of miR-338-3p
To investigate the molecular mechanisms of miR-338-3p in gastric cancer cells, we screened the potential target gene of miR-338-3p using TargetScan 6.2. We found that ADAM17 contained the potential binding sites of miR-338-3p (Figure 3A). Luciferase activity assay showed that miR-338-3p significantly suppressed the Wt but not the Mut 3’UTR luciferase activity in HEK293 cells (Figure 3B). Furthermore, overexpression of miR-338-3p significantly inhibited ADAM17 protein levels in BGC-823 cells (Figure 3C). These results indicated that ADAM17 was a direct target of miR-338-3p in gastric cancer cells.
Figure 3.
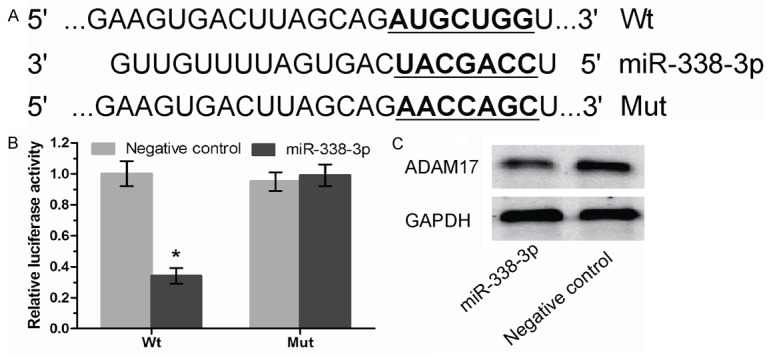
ADAM17 is identified as a target of miR-338-3p in gastric cancer. A. The wild type and mutant ADAM17 3’UTR sequences are shown with the miR-338-3p sequence. B. Luciferase assay in HEK293T cells with wild-type or mutant ADAM17 3’UTR vectors and miR-338-3p mimics or negative control. C. ADAM17 expression in BGC-823 cells transfected with miR-338-3p mimics or negative control was detected by Western blot. *P<0.05.
Overexpression of ADAM17 abolished the tumor suppressive effects of miR-338-3p
We further investigated whether overexpression of ADAM17 could reverse the suppressive effect of miR-338-3p. MTT assay, transwell migration assay and transwell invasion assay showed that increased expression of ADAM17 could significantly reverse the suppressive effect of miR-338-3p on BGC-823 cells (Figure 4A-C). The effect of pEGFP-ADAM17 was confirmed by qRT-PCR (Figure 4D).
Figure 4.
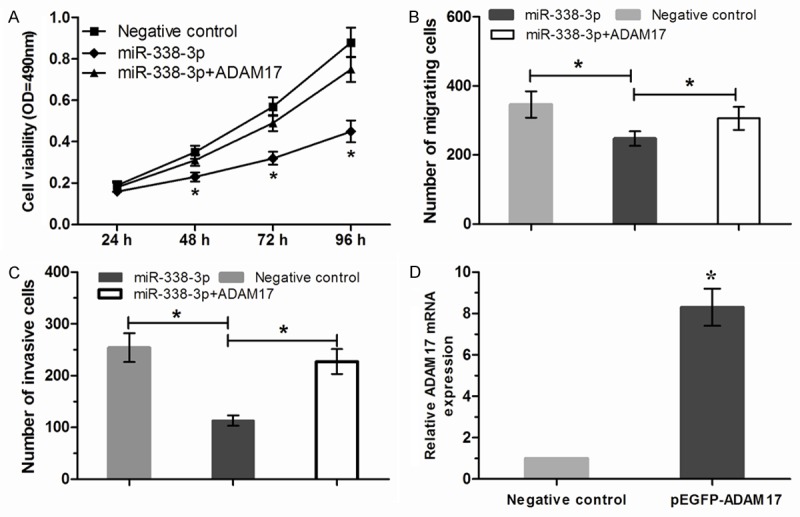
ADAM17 overexpression attenuated the suppressive effect of miR-338-3p. A. BGC-823 cells were co-transfected with miR-338-3p and/or ADAM17. MTT assay was used to assay proliferation. B. Transwell migration assay of BGC-823 cells co-transfected with miR-338-3p and/or ADAM17. C. Transwell invasion assay of BGC-823 cells co-transfected with miR-338-3p and/or ADAM17. D. Expression of ADAM17 was detected by qRT-PCR in BGC-823 cells transfected with pEGFP-ADAM17 or negative control. *P<0.05.
Discussion
Growing evidence showed that miRNAs hold great promise for novel therapeutic approaches for treating human cancers [15]. Deregulation of miR-338-3p has been reported for many different cancers. Even though recent evidence indicated the inhibitory effect of miR-338-3p on human cancers, such as NSCLC, neuroblastoma and hepatocellular carcinoma, there is little knowledge about miR-338-3p and its targets in gastric cancer.
In this study, we revealed that miR-338-3p expression was decreased in gastric cell lines and tissues. Further studies revealed that miR-338-3p overexpression was able to suppress cell proliferation, migration and invasion in BGC-823 cells. Therefore, our study demonstrated that miR-338-3p could act as a tumor-suppressor in the progression of gastric cancer.
A disintegrin and metalloproteinase 17 (ADAM17) is a membrane anchored metalloproteinase that controls the release of tumor necrosis factor α (TNF-α) and ligands of the epidermal growth factor receptor (EGFR) from cells [16,17]. Accumulating evidence suggested that ADAM17 was increased in a various cancers including gastric cancer, glioma, NSCLC and renal cancer and was involved in tumor growth and invasion. Thus, ADAM17 has been considered as a potential therapeutic target in several tumors. For example, Shou et al showed that the expressions of ADAM17 was upregulated in gastric cancer tissues and associated with invasion, TNM stage and overall survival of gastric cancer patients [18]. Zheng et al showed that high level expression of ADAM17 was found in high grade glioma samples and knockdown of ADAM17 inhibited glioma cell proliferation and invasion [19]. Su et al showed that ADAM17 was identified as a target of miR-152 in NSCLC cells, and miR-152-induced suppression of proliferation and motility of NSCLC cells was partially mediated by silencing of ADAM17 expression. Furthermore, they showed that ADAM17 was inversely correlated with miR-152 in NSCLC tissues [20]. Doberstein et al indicated that ADAM17 was upregulation in renal cancer and miR-145 inhibits renal cancer cell proliferation and migration by targeting ADAM17 [21]. However, the molecular determinants of ADAM17 expression remain largely unexplored. In the present study, we identified that ADAM17 was a direct target of miR-338-3p in gastric cancer cells, and supplement of ADAM17 remarkably attenuated the tumor suppressive effects of miR-338-3p on gastric cancer cells.
In conclusion, the present study indicated that miR-338-3p was down-regulated in gastric cancer, and enforced overexpression of miR-338-3p significantly inhibited proliferation, migration and invasion of gastric cancer cells. Furthermore, ADAM17 was identified as a target of miR-338-3p and overexpression of ADAM17 partially attenuated the suppressive effect of miR-338-3p, indicating that miR-338-3p might be a novel therapeutic strategy for the treatment of gastric cancer.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009;472:467–477. doi: 10.1007/978-1-60327-492-0_23. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 7.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Ma X, Cai Q, Wang X, Yu B, Liu B, Zhu Z, Li C. MiR-199a-3p promotes gastric cancer progression by targeting ZHX1. FEBS Lett. 2014;588:4504–4512. doi: 10.1016/j.febslet.2014.09.047. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Nie H, Wang M, Su L, Li J, Yu B, Wei M, Ju J, Yu Y, Yan M, Gu Q, Zhu Z, Liu B. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett. 2012;320:189–197. doi: 10.1016/j.canlet.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Duan Y, Hu L, Liu B, Yu B, Li J, Yan M, Yu Y, Li C, Su L, Zhu Z, Xiang M, Yang Q. Tumor suppressor miR-24 restrains gastric cancer progression by downregulating RegIV. Mol Cancer. 2014;13:127. doi: 10.1186/1476-4598-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kos A, Olde Loohuis NF, Wieczorek ML, Glennon JC, Martens GJ, Kolk SM, Aschrafi A. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS One. 2012;7:e31022. doi: 10.1371/journal.pone.0031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Feng X, Gao S, Xiao Z. microRNA-338-3p functions as a tumor suppressor in human non-small cell lung carcinoma and targets Ras-related protein 14. Mol Med Rep. 2015;11:1400–1406. doi: 10.3892/mmr.2014.2880. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Pan M, Han L, Lu H, Hao X, Dong Q. miR-338-3p suppresses neuroblastoma proliferation, invasion and migration through targeting PREX2a. FEBS Lett. 2013;587:3729–3737. doi: 10.1016/j.febslet.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Sun Y, He Y, Ji C, Hu B. MicroRNA-338-3p inhibits cell proliferation in hepatocellular carcinoma by target forkhead box P4 (FOXP4) Int J Clin Exp Pathol. 2015;8:337–344. [PMC free article] [PubMed] [Google Scholar]
- 15.Mirnezami AH, Pickard K, Zhang L, Primrose JN, Packham G. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur J Surg Oncol. 2009;35:339–347. doi: 10.1016/j.ejso.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Black RA. Tumor necrosis factor-alpha converting enzyme. Int J Biochem Cell Biol. 2002;34:1–5. doi: 10.1016/s1357-2725(01)00097-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee DC, Sunnarborg SW, Hinkle CL, Myers TJ, Stevenson MY, Russell WE, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, Chang A, Jackson LF. TACE/ADAM17 processing of EGFR ligands indicates a role as a physiological convertase. Ann N Y Acad Sci. 2003;995:22–38. doi: 10.1111/j.1749-6632.2003.tb03207.x. [DOI] [PubMed] [Google Scholar]
- 18.Shou ZX, Jin X, Zhao ZS. Upregulated expression of ADAM17 is a prognostic marker for patients with gastric cancer. Ann Surg. 2012;256:1014–1022. doi: 10.1097/SLA.0b013e3182592f56. [DOI] [PubMed] [Google Scholar]
- 19.Zheng X, Jiang F, Katakowski M, Lu Y, Chopp M. ADAM17 promotes glioma cell malignant phenotype. Mol Carcinog. 2012;51:150–164. doi: 10.1002/mc.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y, Wang Y, Zhou H, Lei L, Xu L. MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett. 2014;588:1983–1988. doi: 10.1016/j.febslet.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 21.Doberstein K, Steinmeyer N, Hartmetz AK, Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R, Pfeilschifter J, Gutwein P. MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients. Neoplasia. 2013;15:218–230. doi: 10.1593/neo.121222. [DOI] [PMC free article] [PubMed] [Google Scholar]


