Abstract
Macrophages recognize microbes through Pattern Recognition Receptors (PRRs), and then release pro-inflammatory and anti-inflammatory cytokines. Recent studies have highlighted that collaboration between different PRRs. However, these studies have neglected the crosstalk between various PRRs on macrophages. In the present study, we investigated the interplay of nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) (NOD1, NOD2) and TLRs (TLR1, 2, 3, 4, 5, 6, 7, 8) in terms of macrophage activation, the expression and production of cytokines. The macrophages were stimulated with a single PRR ligand or a combination of TLR and NOD ligands. After 8 h of incubation, the mRNA expression of interleukin-1β (IL-1β), IL-4, IL-6, IL-10, IL-12p35, IL-12p40, IL-13, and interferon-γ (IFN-γ) was evaluated. The production of these cytokines was also measured. NOD2 synergized with TLR3 agonists on enhancement of IL-10 release. However, the combination of NOD1 with TLR3 ligands showed little effect on IL-10 production. Moreover, NOD2 inhibited the percentages of CD11b + F4/80 + cells activated by TLR3 agonist.
Keywords: Macrophage, pattern recognition receptor, NLRs, toll-like receptor, interleukin
Introduction
The innate immune response is the first line of defense against microbial infections and can specifically recognize invading microorganisms. The targets of innate immune recognition are the conserved molecular patterns of microorganisms (pathogen-associate molecular patterns, PAMPs), so the receptors in the innate immune system are called pattern recognition receptors (PRRs). PRRs such as like Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain NOD-like receptors (NLRs) are important sensors of microbial products, and they have been found not only to play key roles in the innate immune system but also to serve as an important bridge linking innate immune system and the adaptive immune system. TLRs are one of the most important types of PRRs and are capable of sensing organisms ranging from bacteria to fungi, protozoa, and viruses. NLRs, as a group of signal transduction cytosolic signal transducers with a NOD, broadly participate in inflammatory responses. Whereas TLRs sense PAMPs in the extracellular space and in endosomes, NLRs function exclusively as intracytoplasmic pathogen sensors [1,2]. In recent years, several studies have described different levels of crosstalk between NLR- and TLR-dependent signaling pathways in innate immune cells. NLRs and TLRs can act in complementary, additive, or even synergistic ways [3-6]. It has been shown that in dendritic cells and monocytes, NOD1 and NOD2 agonists can synergize with TLR agonists to induce pro-inflammatory mediators and cell maturation. Furthermore, NLR-induced immune responses may represent an alternative host defense mechanism when pro-inflammatory responses are impaired by TLR-induced to polarization.
As an essential component of innate immunity, macrophages play a central role in inflammation and host defense [1]. Cells of the monocyte-macrophage lineage are characterized by considerable diversity and plasticity, and certain studies have indicated that the initial interaction of macrophages with specific cytokines determines their functional phenotype. Fully polarized macrophages, or M1-polarized and M2-polarized (or alternatively activated) macrophages, are the extremes of a continuum of functional states. These cells interact with other local molecules to regulate immunoreactions against microbial pathogens and are involved in tissue remodeling. M1-polarized macrophages mediate the initiation of inflammatory responses and tissue damage [7,8] and, M2-polarized macrophages play a key role in tissue repair.
As the first line of host defense against infections, macrophages are activated through their surface PRRs by a variety of pathogenic bacteria. Murine macrophages are equipped with all types of TLRs except TLR9, and also express the NLRs NOD1 and NOD2. Recent studies have revealed the crucial role of TLRs and NLRs in activating macrophages during the inflammatory response and the initiation of adaptive immunity. In the context of an infection, host cells exposed to complex pathogens are simultaneously challenged by several microbial products, and bacteria likely activate the immune response through several PRMs simultaneously. Thus, the overall immune response will be the sum of all of the TLRs and PRRs that a pathogen is able to stimulate, which means that the crosstalk between TLRs and NLRs should be more important than either TLR or NLR stimulation alone in the initiation and developing of inflammation. However, in macrophages, few studies have investigated the interactions between NLRs and TLRs, and the effects of different combinations of PRRs ligands on distinct inflammatory cytokine expression and macrophage polarization are still unknown.
An interesting hypothesis is that, various combinations of NLR and TLR ligands would lead to different modulation of immune reactions, with different cytokines’ profiles and different pathways of macrophage activation, compared with when these ligands are used alone. Moreover, the dynamic process and phenotype of polarized macrophages induced by TLRs might be altered during NLR stimulation. Because NOD1 and NOD2 are two important members of the NLR family, to confirm this hypothesis, we investigated the effects of NLR-TLR crosstalk triggered by NOD1/NOD2 ligands and different types of TLR agonists on the pro-inflammatory cytokine profile in macrophages and the polarization of macrophages. Therefore, we sought to investigate the possible mechanisms of bacterial infection-related inflammation via the PRRs in macrophages and to provide a new method for the modulation of excessive inflammatory responses.
Materials and methods
Reagents
Ligands were purchased from In vivo Gen (San Diego, CA, USA) and were used to stimulate the murine macrophage cell line RAW264.7. These ligands were iE-DAP, MDP, Pam3CSK4, FSL-1, Poly (I:C), LPS, recFLA-ST and R848. The concentrations of these ultrapure ligands were based on published studies showing what concentrations are effective in eliciting cytokine production and also nontoxic to cell growth [9-12]. Table 1 provided details about the ligands’ corresponding PRRs and working concentrations.
Table 1.
Ultrapure PRR ligands used in stimulation experiments with RAW264.7 cells
| PRR | Ligand | Working concentration |
|---|---|---|
| NOD1 | iE-DAP | 10 μg/ml |
| NOD2 | MDP | 10 μg/ml |
| TLR1/2 | Pam3CSK4 | 10 μg/ml |
| TLR2/6 | FSL-1 | 10 μg/ml |
| TLR3 | Poly (I:C) | 10 μg/ml |
| TLR4 | LPS | 1 ng/ml |
| TLR5 | recFLA-ST | 10 μg/ml |
| TLR7/8 | R848 | 10 μg/ml |
RAW264.7 cell stimulation
The murine RAW264.7 macrophage cell line was cultured in complete medium consisting of Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum (FBS) and 1% penicillin, 1% streptomycin at 37°C in the presence of 5% CO2. Then 1 × 106 cells/ml was stimulated with a single ultrapure PRR ligand or a combination of two ligands.
mRNA detection
RNA isolation and quantitative real-time PCR (qRT-PCR) were then performed after 8 h of stimulation. Total RNA was extracted from the RAW264.7 cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and reverse transcribed using Rever Tra Aceq PCR RT Master Mix (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. qRT-PCR was performed in an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster, CA, USA) using KODSYBR qPCR Mix (Toyobo, Osaka, Japan). The primers are listed in Table 2. The specificity of the qRT-PCR reactions was observed via melting curves for the PCR products. We used GAPDH as the reference gene. Relative mRNA expression levels were analyzed using the formula: 2-∆∆Ct, where-∆∆Ct equals the difference between the ΔCt values of stimulated and unstimulated cells [13].
Table 2.
Primers sequences
| Target gene | Primer sequences forward/reverse | Size of amplified products (bp) |
|---|---|---|
| IL-1β | GCAACTGTTCCTGAACTCAACT/ | 89 |
| ATCTTTTGGGGTCCGTCAACT | ||
| IL-4 | ACAGGAGAAGGGACGCCAT/ | 289 |
| GAAGCCCTACAGACGAGCTCA | ||
| IL-6 | GAGGATACCACTCCCAACAGACC/ | 141 |
| AAGTGCATCATCGTTGTTCATACA | ||
| IL-10 | GGTTGCCAAGCCTTATCGGA/ | 191 |
| ACCTGCTCCACTGCCTTGCT | ||
| IL-12p35 | GCCTCACCCTCGGCATCCAGCA/ | 82 |
| GGGTGGCCAAAAAGAGGAGGTAGCG | ||
| IL-12p40 | GGAAGCACGGCAGCAGAATA/ | 180 |
| AACTTGAGGGAGAAGTAGGAATGG | ||
| IL-13 | GGTGGTCTCGCCGCCCCAGG/ | 160 |
| CACAGAACCCGCCAGCGGCCA | ||
| IFN-γ | TGACGTCACTGGAGTTGTACGG/ | 92 |
| GGTTCATGTCATGGATGGTGC | ||
| GAPDH | GTGAAGGTCGGTGTGAACGG/ | 227 |
| TCCTGGAAGATGGTGATGGG |
Cytokine detection
After 24 h of stimulation, the cell supernatants were collected to measure cytokine secretion using ELISA kits for IL-4, IL-10, IL-13 (all from R & D systems, Minneapolis, MN, USA), IL-6, IL-12p70, and IFN-γ (all from eBioscience, San Diego, CA, USA). The measurements were carried out according to the manufacturers’ protocols.
Flow cytometry
RAW264.7 cells were collected after they were stimulated for 24 h. The cells were washed in phosphate-buffered saline (PBS) and then simultaneously stained with phycoerythrin (PE)-conjugated rat anti-mouse CD11b antibodies and PerCP-Cyanine5.5-conjugated rat anti-mouse F4/80 antibodies at 4°C for 1 h. (eBioscience, San Diego, CA, USA). After three more washes with PBS, the samples were measured on a FACSC aliburand analyzed using Cell Quest software (Becton Dickinson, Fullerton, CA, USA).
Calculations and statistic analysis
The criteria according to which interactions between the types of PRRs were considered synergistic or inhibitory were defined before the start of the experiments. A “synergistic effect” was defined as a cytokine response (protein secretion) to a combination of ligands that was at least 1.5-foldhigher than the sum of the cytokine responses induced by each of the single ligands. An “inhibitory effect” was defined as a cytokine response to a combination of ligands that was less than or equal to 0.75-foldthe sum of the cytokine responses to each of the single ligands and lower than the cytokine response to an individual ligand or to both of the ligands. All other patterns were considered as “no effect/additive effect” [6]. Cytokine production and the mRNA expression of target genes were analyzed using Students’ t-test. P-values of ≤ 0.05 were considered statistically significant.
Results
Stimulation of RAW264.7 cells with single PRR ligand had distinct effects on production of different cytokines
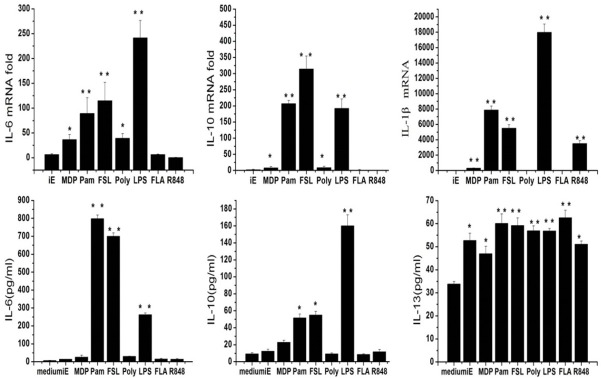
The cytokine responses after each type of PRR ligands were observed based on mRNA expression and cytokine production. Detailed results are shown in Figure 1. The most remarkable elevation was that of IL-6 and IL-10. For IL-6, the amount secreted after the cells were activated by Pam3CSK4, FSL-1 or LPS significantly increased to 800, 750 and 300 pg/ml, respectively (P < 0.01), compared with the nearly absent secretion in the control group (Figure 1B). The mRNA changes induced by these ligands exhibited the same tendency. It was also found that Pam3CSK4, FSL-1 or LPS each enhanced the production of IL-10 by nearly 2-, 2- and 7-folds, respectively (Figure 1B). The P-values (P < 0.01) showed that these ligands all obviously enhanced normal production. The mRNA level exhibited a similar change (Figure 1A). Additionally, nearly all types of PRR ligands enhanced IL-13 (Figure 1B) production. By contrast, IL-1β secretion was observed to change only little after single TLR ligand stimulation (data not shown). However, this elevation was not observable at the mRNA level (Figure 1A). Apart from these changes, the secreted amounts of IL-4, IL-12p70 and IFN-γ seemed unchanged after stimulation with individual PRR ligands (data not shown).
Figure 1.
The qRT-PCR and ELISA results of IL-6, IL-10, IL-13 and IL-1β when RAW264.7 cells were stimulated with iE-DAP, MDP, Pam3CSK4, FSL-1, Poly (I:C), LPS, Rec FLA-ST and R848 alone. Data showed the mean and standard deviation values of results, with statistically significant difference, *and **, P < 0.05, and P < 0.01, respectively, versus normal secretion level (medium only).
Interestingly, iE-DAP and MDP each only increased the IL-13 (Figure 1B) production, but did not enhance other cytokines’ secretion.
NOD1 and NOD2 ligands each interacted with TLR ligands with different effects on cytokine secretion
The combination of iE-DAP or MDP with each TLR ligand was also investigated. Using the definitions of “synergy” and “inhibition”, the interactions between TLR ligands and NOD ligands were studied. Compared with stimulation with single TLR ligand, co-stimulation with NOD ligand resulted in diverse responses with respect of different cytokines. Regarding the secretion of IL-6, iE-DAP had a synergistic effect on both recFLA-ST and R848-activated IL-6 production (Figure 2A), which increased from dozens of pg/ml to 200 and 450 pg/ml, respectively. An inhibitory effect was found between iE-DAP and both Pam3CSK4 and LPS (Figure 2A), with production decreasing by nearly 200 pg/ml in both cases. By contrast, MDP synergized with FSL-1, LPS and R848 (Figure 2A), with production increases of 1000, 1500 and 2500 pg/ml, respectively. Inhibitory interactions between MDP and other TLR ligands were not found in our study.
Figure 2.
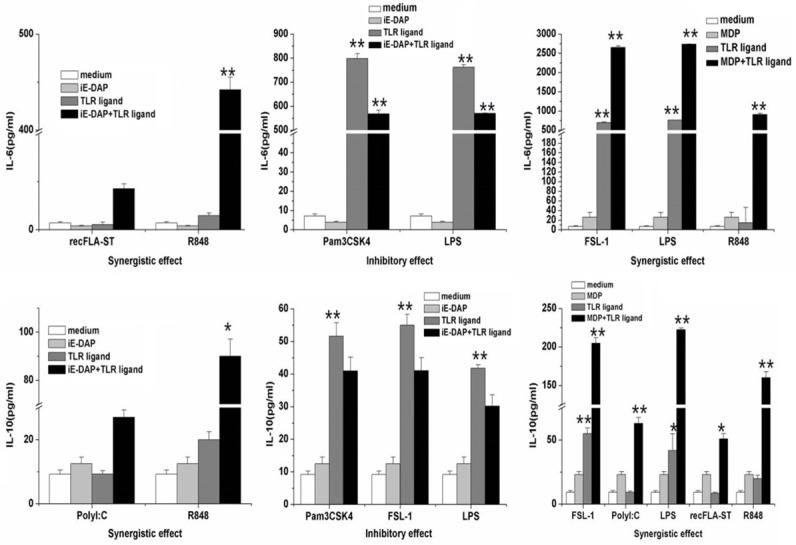
iE-DAP or MDP had synergistic or inhibitory interactions with each type of TLR ligand on IL-6 and IL-10 production. The supernatant was collected after RAW264.7 cells were stimulated 24 h with combination of iE-DAP or MDP with single TLR ligand. The amount of these cytokines was measured via ELISA. Data showed the mean and standard deviation values of ELISA results with statistically significant difference, * and **, P < 0.05, and P < 0.01, respectively, versus normal secretion level (medium only).
For IL-10, R848 and Poly (I:C) each had synergistic interactions with iE-DAP (Figure 2B), which IL-10 production elevated 4-folds in both cases. However, iE-DAP exerted inhibitory actions on the production induced by Pam3CSK4, FSL-1 and LPS (Figure 2B), with a decrease of approximately 10 pg/ml. Compared with iE-DAP, MDP synergized with FSL-1, Poly (I:C), LPS, recFLA-ST and R848 in increasing IL-10 secretion (Figure 2B). The increases for these TLR ligands were 150, 100, 175, 40 and 100 pg/ml, respectively.
In Table 3, the details of synergy and inhibition between TLR ligands and NOD1 or NOD2 ligand are shown. iE-DAP synergized with recFLA-ST and R848 in increasing IL-6 production and also synergized with R848 in increasing IL-10 secretion. By contrast, iE-DAP had and inhibitory on IL-6 production, when combined with either Pam3CSK4 or LPS. Moreover, iE-DAP had an inhibitory effect on IL-10 secretion when combined with Pam3CSK4, FSL-1 or LPS. The co-application of iE-DAP with FSL-1 or Poly (I:C) had no obvious effect on IL-6 levels. This was also found for IL-10 after co-stimulation with iE-DAP and Poly (I:C) or recFLA-ST.
Table 3.
Synergistic and Inhibitory effects between TLR and NOD ligands
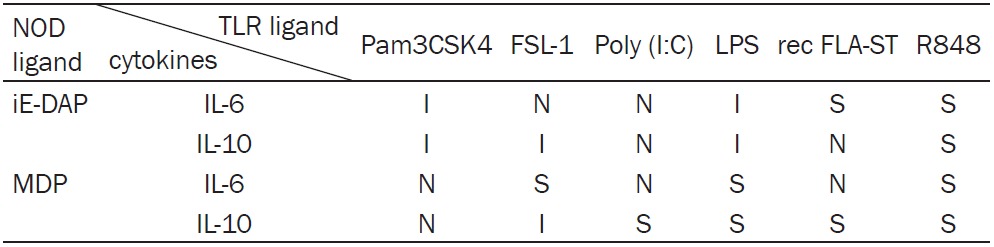
|
MDP synergized with FSL-1, LPS and R848 in enhancing the production of IL-6. The synergistic effect was observed on IL-10 levels, when MDP was used in combination with Poly (I:C), LPS, recFLA-ST or R848. An inhibitory effect between MDP and FSL-1 was only found for secretion of IL-10. The combination of MDP with Pam3CSK4, Poly (I:C) or recFLA-ST had no significant effect on IL-6 secretion. Meanwhile, MDP combined with Pam3CSK4 showed neither synergy nor inhibition.
Combination of NOD1 or NOD2 ligand with TLR ligand had different effects on percentage of CD11b + F4/80 + RAW264.7 cells compared with single TLR ligand
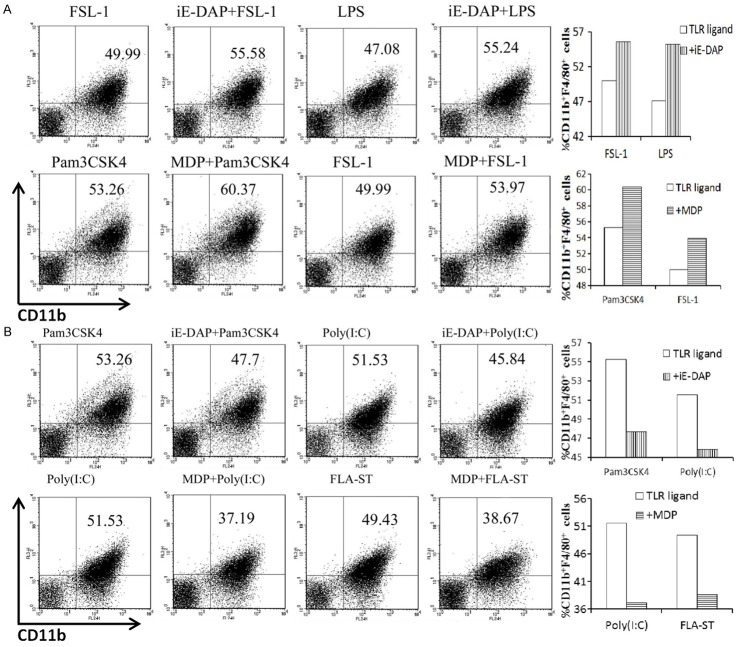
In addition to cytokine release, the polarization of stimulated RAW264.7 cells was analyzed by investigating cell-surface expression of CD11b and F4/80. iE-DAP acted cooperatively with FSL-1 and LPS in increasing the percentage of CD11b + F4/80 + cells (Figure 3A). However, iE-DAP combined with Pam3CSK4 or Poly (I:C) inhibited the increase in double-positive cells (Figure 3B). For MDP, synergy with Pam3CSK4 and FSL-1 was observed in terms of orienting cells toward the CD11b + F4/80 + subtype (Figure 3A). By contrast, MDP inhibited the expansion of double-positive cells caused by Poly (I:C) and recFLA-ST (Figure 3B). It was also found when combined with R848, neither iE-DAP nor did MDP have an obvious effect on the increased CD11b + F4/80 + ratio (data not shown).
Figure 3.
FACS plots showed percentages of CD11b + F4/80 + cells in upper right quadrant. A. iE-DAP promoted the double-positive cells activated by FSL-1 and LPS, while MDP enhanced the ratio of CD11b + F4/80 + cells induced by Pam3CSK4 and FSL-1. B. iE-DAP decreased the percentage of double-positive cells which were elicited by Pam3CSK4and Poly (I:C). By contrast, MDP inhibited the percentages of CD11b + F4/80 + cells induced by Poly (I:C) and recFLA-ST.
Discussions
Macrophages are the first defense the threat of against microbes, and they can recognize pathogens via PRRs. After receiving signals through various PRRs, macrophages may be activated differentially. The present study compared the cellular responses and the type of macrophage stimulated following stimulation NOD1 or, NOD2 ligand and several TLR agonists, first individually and then in combination. Each TLR and NOD agonist was found to induce a distinct type of response. Remarkably, co-stimulation with a NOD1/NOD2 agonist was able to affect the induction of certain cytokines and the macrophage polarization induced by TLR agonists.
When the patterns of cytokines induced by each TLR agonist were separately analyzed, the secretion of cytokines was inconsistent due to the activation of different PRRs. Consistent with the expression levels of all TLRs examined, the levels of IL-6 and IL-10 induced by Pam3CSK4, FSL-1 or LPS were much higher than those induced by the other TLR agonists. These findings suggest that macrophages are sensitive to TLR2, TLR4, TLR6 agonists, and that the recognition of microbial structures through these TLRs is crucial for the initiation of inflammation and the activation of the adaptive immune response. Recent evidence has reinforced the important roles of TLRs in modulating the nature of the inflammatory response induced by macrophages. It has been shown that RAW264.7 cells constitutively express TLR2, and that the expression of TLR2 can be stimulated further when macrophages encounter microorganisms [14]. For example, macrophages may respond to Candida. albicans’ PAMPs through TLR2 receptors, leading to expression of IL-6 to induce inflammation [15], and of IL-10 to induce immune regulation [16].
Mycobacterial molecules, e.g., LprA, LprG, PhoS1, LAM, LM and PIMs, act as ligands for TLR2, leading to the production of IL-10 and, IL-4 by macrophages, allowing Mycobacterium tuberculosis to evade the host innate and adaptive immune responses [17]. Other mycobacterial antigens, e.g., the 38 kD a glycolipo protein and PIM6, are sensed by TLR4/TLR2 and generate Th1-polarized cytokine responses in Mycobacterium tuberculosis-infected mouse lungs [18,19]. Mycoplasmal lipoproteins trigger TLR6-mediated sequential responses: NF-κB activation and pro-inflammatory cytokine production [20].
Other studies have demonstrated that TLR1, TLR2, TLR4 and TLR6 recognize triacyllipo peptides, PG, LPS and LTA from microbes, involved in infection with mycobacteria, Group B Streptococcus and Gram-negative bacteria [21,22]. As TLRs were discovered before NLRs, the role of TLRs expressed on macrophages in the bacteria-activated immune response has already been described. However, studies focused on NLRs were only started recently, and moreover, the effects of NLRs on macrophages have rarely been covered in these studies. Similar to TLRs, NLRs sense characteristic microbial products and possibly “danger signals” but, in the present study, an NLR agonist alone had few effects on cytokine mRNA levels in and secretion by macrophages. In our study, when macrophages were stimulated by iE-DAP or MDP alone, it was observed that only MDP stimulated macrophages showed a slightly enhanced level of IL-10 transcripts, whereas iE-DAP- and MDP-mediated IL-6 and IL-10 levels did not show obvious enhancement compared with the levels secreted by control macrophages. Certain studies have reported that NOD1 and NOD2 only affected systemic infection when mice were previously or simultaneously stimulated by TLR ligands [23]. Fritz et al. [3] and Kim et al. [6] also found that NOD1 and NOD2 ligands were not able to induce an increase in the production of IL-6 and IL-10, which is consistent with the results of our study.
Although the secretion of IL-6 and IL-10 by macrophages is predominantly driven by TLR agonist, the marginal effect of additional treatment with iE-DAP or MDP could not be ignored. Interestingly, the secretion of IL-6 and IL-10 was dramatically changed in macrophages co-stimulated with a TLR agonist and iE-DAP or MDP. Compared with macrophages stimulated with a TLR agonist alone, it is well known that IL-6 is pro-inflammatory cytokine that is involved in microbicidal tissue damage, whereas IL-10 is considered to be an anti-inflammatory cytokine that suppresses the development of inflammation [24].
Based on the secretion of IL-6 and IL-10 and the polarization of macrophages, our study suggested a possible interaction between NOD1 or NOD2 and TLRs. The details are shown in the Figure 2 and Table 2. First, NOD1 ligand inhibited activation by TLR1/2 ligands, decreased IL-6 and IL-10 secretion, and reduced the percentage of CD11b + F4/80 + macrophages. These results suggested that NOD1 and TLR1/2 may be antagonistic. Compared with stimulation with NOD1 ligand, co-stimulation with NOD2 and TLR1/2 ligands did not change cytokine production, but, it did promote double-positive cells among macrophages. All of the mentioned results implied that NOD1 can weaken the inflammation elicited by activation of TLR1/2 on macrophages, whereas NOD2 may be beneficial for tissue repair and inflammation control.
Regarding TLR2/6 ligands, the results demonstrated that NOD1 or NOD2 ligand reduced the IL-10 secretion elicited by TLR2/6 activation. For IL-6, NOD1 ligand caused no change, whereas NOD2 ligand improved the production of IL-6 induced by TLR2/6 ligands. Co-application of NOD1 or NOD2 with TLR2/6 ligands enhanced the percentage of CD11b + F4/80 + macrophages. These data may suggest that both NOD1 and NOD2 can promote or decrease the inflammation caused by activation of TLR2/6 on macrophages. Kim et al. [6] found that NOD2 ligand synergistically enhanced the production of IL-6 induced by TLR2/6 ligands on dendritic cells. Tang et al. [25] also reported that the co-application of NOD2 and TLR2/6 ligands elicited a higher level of IL-6 among periodontal ligament fibroblasts than when used alone.
In terms of TLR3 ligand, combination with NOD1 ligand induced no change in the amount of IL-6 or IL-10. NOD2 ligand showed no effect on TLR3-induced IL-6 secretion, whereas it promoted IL-10 secretion. Regarding the percentage of CD11b + F4/80 + macrophages, both NOD1 and NOD2 ligands decreased the percentage of TLR3-activated double-positive cells. These data may demonstrate that NOD1 and NOD2 weaken the macrophage activation induced by TLR3, and alleviate inflammation.
As far as TLR4 is concerned, our study considered LPS as its agonist. NOD1 ligand decreased the IL-6 and IL-10 production induced by LPS, and reduced the percentage of CD11b + F4/80 + macrophages. This change is advantageous to control inflammation and promote tissue repair. In contrast, NOD2 ligand enhanced the amount of IL-6 and IL-10 elicited by LPS. Interestingly, this ligand exerted no obvious effect on the percentage of CD11b + F4/80 + macrophages. The above results implied that NOD2 may facilitate LPS-induced inflammation. Moreover, the interaction between NOD1 and TLR4 is possibly contrary to that between NOD2 and TLR4 in terms of the effects on cytokine secretion and the polarization of macrophages. Additionally, certain studies have found that NOD1 and NOD2 play different roles in activating immune responses; more specifically, they reported that NOD2 had more remarkable synergy with TLRs in promoting inflammation [26,27]. The possible mechanisms may include the structural differences between NOD1 and NOD2. NOD1 detects only PGN fragments containing DAP at the third position, which relies only on the peptidic moiety [28]. Furthermore, NOD2 expression has been found at low levels in unstimulated cells, but NOD2 mRNA is increased in inflammatory tissue and LPS-induced cells [29-31].
Conclusions
This study demonstrated that NOD1 and NOD2 had interplay with various TLRs, and played different roles in the initiation and development of inflammation and in the tissue repair. Thus, these findings suggested that interactions between multiple PRRs are involved in the elaborate regulation of inflammation induced by macrophages. The combination of NOD2 and with TLR4 ligands alleviated inflammation, which may be related to the development of systemic inflammation. This result implied that we can avoid systemic responses and aberrant inflammation by aiming to stimulate at least two types of PRRs simultaneously. Our findings also revealed that co-stimulation of NOD1 with TLR3 ligands depressed inflammation, allowing the control of inflammation and promotion of tissue repair. Above all, the possible mechanisms of these findings are still unknown. These results will help us to find a new way to control inflammatory responses and to promote tissue repair once we explore the mechanism of the antagonism between NLRs and TLRs.
Acknowledgements
The authors acknowledge the assistance of Prof. Yu Hong Li and Miss Xi Xi Cao for help with real-time PCR analysis. This work was supported by grants from the National Natural Science Foundation of China (No. 30672325).
Disclosure of conflict of interest
None.
References
- 1.Bourhis LL, Werts C. Role of Nods in bacterial infection. Microbes Infect. 2007;9:629–636. doi: 10.1016/j.micinf.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz JH, Girardin SE, Fitting C, Werts C, Mengin-Lecreulx D, Caroff M, Cavaillon JM, Philpott DJ, Adib-Conquy M. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005;35:2459–2470. doi: 10.1002/eji.200526286. [DOI] [PubMed] [Google Scholar]
- 4.Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic Effect of Nod1 and Nod2 Agonists with Toll-Like Receptor Agonists on Human Dendritic Cells To Generate Interleukin-12 and T Helper Type 1 Cells. Infect Immun. 2005;73:7967–7976. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negativeregulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 6.Timmermans K, Plantinga TS, Kox M, Vaneker M, Scheffer GJ, Adema GJ, Joosten LA, Netea MG. Blueprints of Signaling Interactions between Pattern Recognition Receptors: Implications for the Design of Vaccine Adjuvants. Clin Vaccine Immunol. 2013;20:427–432. doi: 10.1128/CVI.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 9.Hirschfeld M, Weis JJ, Toshchakov V, Salkowski CA, Cody MJ, Ward DC, Qureshi N, Michalek SM, Vogel SN. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect Immun. 2001;69:1477–82. doi: 10.1128/IAI.69.3.1477-1482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz H, Posselt G, Wurm P, Ulbing M, Duschl A, Horejs-Hoeck J. TLR8 and NOD signaling synergistically induce the production of IL-1β and IL-23 inmonocyte-derived DCs and enhance the expression of the feedback inhibitor SOCS2. Immunobiology. 2013;218:533–42. doi: 10.1016/j.imbio.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, Tamai R, Akashi S, Takeuchi O, Akira S, Sugawara S, Takada H. Synergistic effect of muramyldipeptide with lipopolysaccharide or lipoteichoic acid to induce inflammatory cytokines in human monocytic cells in culture. Infect Immun. 2001;69:2045–2053. doi: 10.1128/IAI.69.4.2045-2053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uehara A, Yang S, Fujimoto Y, Fukase K, Kusumoto S, Shibata K, Sugawara S, Takada H. Muramyldipeptide and diaminopimelic acid-containing desmuramyl peptides in combination with chemically synthesized Toll-like receptor agonists synergistically induced production of interleukin-8 in a NOD2- and NOD1-dependentmanner, respectively, in human monocytic cells in culture. Cell Microbiol. 2005;7:53–61. doi: 10.1111/j.1462-5822.2004.00433.x. [DOI] [PubMed] [Google Scholar]
- 13.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An Overview of Real-Time Quantitative PCR: Applications to Quantify Cytokine Gene Expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 14.Al-Salleeh F, Petro TM. TLR3 and TLR7 are involved in expression of IL-23 subunits while TLR3 but not TLR7 is involved in expression of IFN-b by Theiler’s virus-infected RAW264.7 cells. Microbes Infect. 2007;9:1384–92. doi: 10.1016/j.micinf.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Korn T, Mitsdoerffer M, Croxford AL, Awasthi A, Dardalhon VA, Galileos G, Vollmar P, Stritesky GL, Kaplan MH, Waisman A, Kuchroo VK, Oukka M. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2008;105:18460–5. doi: 10.1073/pnas.0809850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Mosser Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80:298–307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heo DR, Shin SJ, Kim WS, Noh KT, Park JW, Son KH, Park WS, Lee MG, Kim D, Shin YK, Jung ID, Park YM. Mycobacterium tuberculosis lpdC, Rv0462, induces dendritic cell maturation and Th1 polarization. Biochem Biophys Res Commun. 2011;206:642–7. doi: 10.1016/j.bbrc.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Byun EH, Kim WS, Kim JS, Jung ID, Park YM, Kim HJ, Cho SN, Shin SJ. Mycobacterium tuberculosis Rv0577, a novel TLR2 agonist, induces maturation of dendritic cells and drives Th1 immune response. FASEB J. 2012;26:2695–711. doi: 10.1096/fj.11-199588. [DOI] [PubMed] [Google Scholar]
- 19.Byun EH, Kim WS, Shin AR, Kim JS, Whang J, Won CJ, Choi Y, Kim SY, Koh WJ, Kim HJ, Shin SJ. Rv0315, a novel immune stimulatory antigen of Mycobacterium tuberculosis, activates dendritic cells and drives Th1 immune responses. J Mol Med. 2012;90:285–98. doi: 10.1007/s00109-011-0819-2. [DOI] [PubMed] [Google Scholar]
- 20.Into T, Kiura K, Yasuda M, Kataoka H, Inoue N, Hasebe A, Takeda K, Akira S, Shibata K. Stimulation of human Toll-like receptor (TLR)2 and TLR6 with membrane lipoproteins of Mycoplasma fermentans induces apoptotic cell death after NF-kappa B activation. Cell Microbiol. 2004;6:187–99. doi: 10.1046/j.1462-5822.2003.00356.x. [DOI] [PubMed] [Google Scholar]
- 21.Akira S, Uematsu S, Takeuchi O. Pathogen Recognition and Innate Immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 22.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. Nod-like proteins in inflammation and disease. J Pathol. 2008;214:136–48. doi: 10.1002/path.2271. [DOI] [PubMed] [Google Scholar]
- 24.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 25.Tang L, Zhou XD, Wang Q, Zhang L, Wang Y, Li XY, Huang DM. Expression of TRAF6 and pro-inflammatory cytokines through activation of TLR2, TLR4, NOD1, and NOD2 in human periodontal ligament fibroblasts. Arch Oral Biol. 2011;56:1064–1072. doi: 10.1016/j.archoralbio.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 26.Girardin SE, Travassos LH, Hervé M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. Peptidogly can molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nuñez G, Cho JH. A frame shift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 28.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from Gram-negative bacteria peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez O, Pipaon C, Inohara N, Fontalba A, Ogura Y, Prosper F, Nunez G, Fernandez-Luna JL. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor kappa B activation. J Biol Chem. 2002;277:41701–41705. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 30.Berrebi D, Maudinas R, Hugot JP, Chamaillard M, Chareyre F, De Lagausie P, Yang C, Desreumaux P, Giovannini M, Cézard JP, Zouali H, Emilie D, Peuchmaur M. Card15 gene over expression in mononuclear and epithelial cells of the inflamed Crohn’s disease colon. Gut. 2003;52:840–846. doi: 10.1136/gut.52.6.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin Mc. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]