Abstract
Previous studies have investigated the association of mutS homolog 3 (MSH3) rs26279 G > A polymorphism with the risk of different types of cancers including colorectal cancer, breast cancer, prostate cancer, bladder cancer, thyroid cancer, ovarian cancer and oesophageal cancer. However, its association with cancer remains conflicting. We performed a comprehensive meta-analysis to derive a more precise estimation of the relationship between MSH3 rs26279 G > A polymorphism and cancer susceptibility. Systematically searching the PubMed and EMBASE databases yielded 11 publications with 12 studies of 3282 cases and 6476 controls. The strength of the association was determined by crude odds ratios (OR) and 95% confidence intervals (CI). Overall, pooled risk estimates demonstrated that MSH3 rs26279 G > A was significantly associated with an increased overall cancer risk under all the genetic models (GG vs. AA: OR = 1.27, 95% CI = 1.09-1.48, P = 0.002; AG vs. AA: OR = 1.10, 95% CI = 1.00-1.21, P = 0.045; GG vs. AG + AA: OR = 1.23, 95% CI = 1.06-1.42, P = 0.005; AG + GG vs. AA: OR = 1.13, 95% CI = 1.04-1.24, P = 0.006; G vs. A: OR = 1.13, 95% CI = 1.05-1.20, P = 0.001). The association was more evident for colorectal cancer and breast cancer. Moreover, the significant association was also observed in the following subgroups: Europeans, Asians, population-based studies, hospital-based studies, and studies comprising relatively large sample size (≥ 200). Our meta-analysis results demonstrated that MSH3 rs26279 G > A polymorphism is associated with an increased risk of overall cancer, especially for the colorectal cancer and breast cancer.
Keywords: MSH3, polymorphism, cancer, meta-analysis
Introduction
Cancer is a complex multistep disease. Genetic aberrations may contribute to the development of cancer through interaction with environment factors [1]. Genetic aberrations can occur during the process of DNA replication and homologous recombination or as a result of DNA damage, owing to exposure to various exogenous and endogenous factors such as UV, ionizing radiation, and products of lipid peroxidation. In order to protect against DNA aberrations and maintain the integrity of the genome, human body possesses four major DNA repair, including nucleotide excision repair (NER), base excision repair (BER), double-strand break repair (DSBR) and mismatch repair (MMR) pathways [2].
As one of fundamentally biological pathway, the MMR system plays an important role in cell cycle regulation and apoptosis, answerable for a variety of DNA damage [3,4]. It is primarily responsible for recognizing and correcting base-base mismatches and insertion/deletion loops generated during DNA replication and homologous recombination [5,6].
Additionally, a fraction of mismatches can arise from DNA damage. If not properly repaired, these mismatches may enhance spontaneous mutation rates and lead to the occurrence of microsatellite repeat instability in cells, thereby contributing to carcinogenesis. Therefore, MMR plays a critical role in maintaining the integrity of the genome. In humans, there are seven mismatch repair genes including MSH2 (mutS homolog 2), MSH3, MSH6, MLH1 (mutL homolog 1), PMS1 (postmeiotic segregation increased 1), PMS2 and MLH3. The MSH2, MSH3 and MSH6 genes are MutS homologs, while the rest of genes are MutL homologes [7]. Unlike homo-oligomers in prokaryotes, MMR proteins form heterodimers in human. The function of MutS is to recognize and bind to mismatched DNA fragments, and thereafter initiate MMR. There are two kinds of MutS homodimers, MSH2-MSH6 (MutSα) and MSH2-MSH3 (MutSβ) in human. The MutSα searchs for single base mismatches and short insertion/deletion loops, while the MutSβ are able to detect large insertion/deletion loops [6,8].
The MSH3 gene is located at chromosome 5q11-13 and encodes a protein of 1137 amino acid residues [9]. Some potentially functional single-nucleotide polymorphisms (SNPs) of MSH3 may have influence the DNA repair capacity and thereby predispose individuals to a variety of cancers. Thus far, at least 180 SNPs have been reported in MSH3 gene (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=4437). Among all of these SNPs, rs26279 G > A polymorphism are frequently investigated and have been thought to be involved in carcinogenesis in recent years. Some studies suggested that this variant was associated with the risk of cancer including colorectal cancer [10,11], breast cancer [12-14], prostate cancer [15,16], bladder cancer [17], thyroid cancer [18], ovarian cancer [19] and oesophageal cancer [20]. However, the association of interest remains inconclusive. Hence, to provide a more precise estimation of the association between MSH3 rs26279 G > A polymorphism and the risk of cancer, we conducted a meta-analysis with all eligible case-control studies.
Materials and methods
Identification of eligible relevant studies
A systematic literature search was conducted throughout the Pubmed and EMBASE database to find studies assessing the association between MSH3 rs26279 G > A polymorphism and the risk of cancer prior to May 30, 2015. The following keywords were used to identify relevant studies “MSH3 or hMSH3”, “variant or polymorphism” and “cancer or carcinoma or neoplasm or tumor”. Additional related studies have been also identified by hand screening. The search was limited to human studies written in Chinese and English.
Inclusion and exclusion criteria
Inclusion criteria were listed in the following paragraph: (a) a case-control design study evaluating the association between MSH3 rs26279 G > A polymorphism and cancer risk; (b) sufficient raw data for estimating odds ratios (ORs) and their 95% confidence intervals; and (c) the genotype distribution in the control group of the studies should be in accordance with the Hardy-Weinberg equilibrium (HWE). Exclusion criteria included: (a) case-only study; (b) reviews and duplicate data.
Data extraction
After selection of eligible studies according to the inclusion criteria and exclusion criteria, two of the researchers independently collected information from all eligible publications. The following data was extracted from each study: the last name of the first author, publication year, country, ethnicity, cancer type, source of control (hospital-based or population-based), genotyping methods, genotype counts of cases and controls for MSH3 rs26279 G > A polymorphism. The stratification analysis was conducted by cancer type (If one type of cancer was only investigated in a study, the cancer type would be merged into the “other type”), ethnicity (Asians, Europeans and Africans), control source (hospital-based and population-based) and sample size (number of cases < 200 and ≥ 200).
Statistical analysis
Crude ORs with the corresponding 95% CIs were employed to estimate the strength of association between the MSH3 rs26279 G > A polymorphism and cancer risk. The following genetic models were chosen: homozygous model (GG vs. AA), heterozygous model (AG vs. AA), recessive model (GG vs. AG + AA), dominant model (AG + GG vs. AA) and allele comparison (G vs. A). Goodness-of-fit chi-square test was used to check whether there was deviation from Hardy-Weinberg equilibrium in control subjects of each included study. The heterogeneity of studies was tested by Chi squared-based Q-test, with a P value of more than 0.10 denoting a lack of between-study heterogeneity. In the absence of heterogeneity, we used the fixed-effects model (the Mantel-Haenszel method) [21]. Otherwise, we would apply the random-effects model (the DerSimonian and Laird method) to the analysis [22]. The potential publication bias was examined by Egger’s linear regression test [23]. All the analyses were tested with the STATA software (version 11.0; Stata Corporation, College Station, TX). All P values were two-sided, and we referred to statistically significant as P < 0.05 without further special notification.
Results
Study characteristics
A total of 68 articles were initially retrieved from the Pubmed and EMBASE electronic databases. Among these, 46 studies were excluded after preliminary screening of titles and abstracts. The remaining 22 publications underwent full-text reviews. Of these, 3 studies were excluded because they did not contain MSH3 rs26279 G > A polymorphism [24-26], five non-case-control studies [27-31] and 3 studies without detailed information were also eliminated [32-34]. In the study By Jafary et al. genotype distribution of MSH3 rs26279 G > A polymorphism was not in agreement with HWE. However, this study was included in our meta-analysis, because another SNP, rs1805355 in this study was in HWE. Finally, 11 publications with 12 studies, comprising 3282 cases and 6476 controls were included to examine the association of MSH3 rs26279 G > A polymorphism with cancer risk [10-20].
As summarized in Table 1, all publications were case-control studies. Overall, there were 2 studies conducted on colorectal cancer, 4 on breast cancer, 2 on prostate cancer and 4 on others. Regarding ethnicity, 5, 5, and 2 studies were performed among Europeans, Asians, and Africans, respectively. Five studies were population-based, while 7 studies were hospital-based. Moreover, these studies were also stratified by sample size: 6 studies with sample size < 200 and 6 studies with sample size ≥ 200.
Table 1.
Characteristics of studies included in the meta-analysis
| Surname | Year | Country | Ethnicity | Cancer type | Control source | Genotype method | NO. of cases | NO. of control | MAF | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| All | AA | AG | GG | All | AA | AG | GG | |||||||||
| Berndt | 2007 | USA | European | Colorectal | PB | TaqMan | 229 | 101 | 101 | 27 | 2081 | 1063 | 851 | 167 | 0.28 | 0.855 |
| Conde | 2009 | Portugal | European | Breast | HB | TaqMan | 286 | 121 | 129 | 36 | 544 | 246 | 240 | 58 | 0.33 | 0.962 |
| Hirata | 2008 | Japan | Asian | Prostate | HB | SSCP/PCR-RFLP | 110 | 60 | 43 | 7 | 110 | 72 | 31 | 7 | 0.20 | 0.160 |
| Jafary | 2012 | Iran | Asian | Prostate | PB | SSCP | 18 | 12 | 5 | 1 | 60 | 34 | 18 | 8 | 0.28 | 0.043 |
| Kawakami | 2004 | Japan | Asian | Bladder | PB | SSCP | 99 | 53 | 38 | 8 | 101 | 66 | 28 | 7 | 0.21 | 0.112 |
| Orimo | 2000 | Japan | Asian | Colorectal | PB | SSCP | 19 | 10 | 8 | 1 | 89 | 55 | 28 | 6 | 0.22 | 0.360 |
| Shi | 2008 | China | Asian | Thyroid | HB | PCR-RFLP | 204 | 120 | 68 | 16 | 204 | 130 | 60 | 14 | 0.22 | 0.062 |
| Smith | 2008 | USA | European | Breast | HB | MassARRAY | 321 | 154 | 123 | 44 | 409 | 198 | 175 | 36 | 0.30 | 0.762 |
| African | Breast | HB | MassARRAY | 52 | 21 | 23 | 8 | 72 | 33 | 29 | 10 | 0.34 | 0.383 | |||
| Song | 2006 | UK | European | Ovarian | PB | TaqMan | 1310 | 663 | 525 | 122 | 2003 | 1064 | 771 | 168 | 0.28 | 0.093 |
| Terrazzino | 2012 | Italy | European | Breast | HB | TaqMan | 89 | 41 | 36 | 12 | 196 | 93 | 86 | 17 | 0.31 | 0.646 |
| Vogelsang | 2012 | South Africa | African | Oesophageal | HB | TaqMan | 545 | 204 | 256 | 85 | 607 | 230 | 295 | 82 | 0.38 | 0.410 |
SSCP, single-strand conformational polymorphism; PCR-RFLP, polymerase chain reaction restriction fragment length polymorphisms; HB, hospital based; PB, population based; MAF, Minor Allele Frequency; HWE: Hardy-Weinberg equilibrium.
Meta-analysis results
The overall findings of the meta-analysis of the associations between MSH3 rs26279 G > A polymorphism and cancer risk were shown in Table 2. Pooled risk estimates indicated that MSH3 rs26279 G > A polymorphism was associated with an increased risk of cancer under all the genetic models (GG vs. AA: OR = 1.27, 95% CI = 1.09-1.48, P = 0.002; AG vs. AA: OR = 1.10, 95% CI = 1.00-1.21, P = 0.045; GG vs. AG + AA: OR = 1.23, 95% CI = 1.06-1.42, P = 0.005; AG + GG vs. AA: OR = 1.13, 95% CI = 1.04-1.24, P = 0.006; G vs. A: OR = 1.13, 95% CI = 1.05-1.20, P = 0.001, Figure 1). Next, when the analysis was stratified by cancer type, the same study also revealed a significant increase in the risk of colorectal cancer (GG vs. AA: OR = 1.65, 95% CI = 1.06-2.58, P = 0.027, Figure 2; AG + GG vs. AA: OR = 1.33, 95% CI = 1.02-1.74, P = 0.034; G vs. A: OR = 1.28, 95% CI = 1.05-1.56, P = 0.014) and breast cancer (GG vs. AA: OR = 1.42, 95% CI = 1.05-1.91, P = 0.023, Figure 2; GG vs. AG + AA: OR = 1.40, 95% CI = 1.06-1.86, P = 0.020), but not prostate cancer. Stratified analysis by ethnicity further elucidated that the pooled odds ratio for the association with cancer risk was statistically significant among Europeans (GG vs. AA: OR = 1.32, 95% CI = 1.10-1.58, P = 0.003; GG vs. AG + AA: OR = 1.28, 95% CI = 1.07-1.52, P = 0.006; AG + GG vs. AA: OR = 1.12, 95% CI = 1.01-1.25, P = 0.028, Figure 3; G vs. A: OR = 1.13, 95% CI = 1.04-1.22, P = 0.003) and Asians (AG vs. AA: OR = 1.40, 95% CI = 1.06-1.85, P = 0.018, AG + GG vs. AA: OR = 1.35, 95% CI = 1.04-1.75, P = 0.024, Figure 3), while no significant association was observed among African populations. Moreover, there was a significant association with increased cancer risk in population-based studies and hospital-based studies. Lastly, association was also observed in subgroup with studies with no less than 200 cases.
Table 2.
Meta-analysis of the association between rs26279 polymorphism and cancer risk
| Variables | N (case/control) | Homozygous | Heterozygous | Recessive | Dominant | Allele | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| GG vs. AA | AG vs. AA | GG vs. (AG + AA) | (AG +GG) vs. AA | G vs. A | ||||||||||||
|
| ||||||||||||||||
| OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | ||
| All | 12 (3282/6476) | 1.27 (1.09-1.48) | 0.943 | 0.0 | 1.10 (1.00-1.21) | 0.675 | 0.0 | 1.23 (1.06-1.42) | 0.925 | 0.0 | 1.13 (1.04-1.24) | 0.787 | 0.0 | 1.13 (1.05-1.20) | 0.863 | 0.0 |
| Cancer type | ||||||||||||||||
| Colorectal | 2 (248/2170) | 1.65 (1.06-2.58) | 0.593 | 0.0 | 1.27 (0.96-1.68) | 0.676 | 0.0 | 1.48 (0.97-2.26) | 0.542 | 0.0 | 1.33 (1.02-1.74) | 0.856 | 0.0 | 1.28 (1.05-1.56) | 0.921 | 0.0 |
| Breast | 4 (748/1221) | 1.42 (1.05-1.91) | 0.910 | 0.0 | 1.00 (0.83-1.22) | 0.783 | 0.0 | 1.40 (1.06-1.86) | 0.746 | 0.0 | 1.08 (0.90-1.30) | 0.940 | 0.0 | 1.13 (0.98-1.30) | 0.998 | 0.0 |
| Prostate | 2 (128/170) | 0.88 (0.34-2.27) | 0.324 | 0.0 | 1.44 (0.86-2.40) | 0.266 | 19.0 | 0.79 (0.31-2.01) | 0.430 | 0.0 | 1.32 (0.82-2.14) | 0.160 | 49.2 | 1.15 (0.78-1.71) | 0.123 | 58.0 |
| Other | 4 (2158/2915) | 1.18 (0.97-1.43) | 0.986 | 0.0 | 1.09 (0.97-1.23) | 0.391 | 0.1 | 1.15 (0.95-1.38) | 0.995 | 0.0 | 1.11 (0.99-1.25) | 0.470 | 0.0 | 1.10 (1.01-1.20) | 0.642 | 0.0 |
| Ethnicity | ||||||||||||||||
| European | 5 (2235/5233) | 1.32 (1.10-1.58) | 0.580 | 0.0 | 1.08 (0.97-1.21) | 0.650 | 0.0 | 1.28 (1.07-1.52) | 0.500 | 0.0 | 1.12 (1.01-1.25) | 0.752 | 0.0 | 1.13 (1.04-1.22) | 0.735 | 0.0 |
| Asian | 5 (450/564) | 1.14 (0.69-1.88) | 0.851 | 0.0 | 1.40 (1.06-1.85) | 0.730 | 0.0 | 1.02 (0.62-1.67) | 0.902 | 0.0 | 1.35 (1.04-1.75) | 0.619 | 0.0 | 1.23 (0.99-1.52) | 0.574 | 0.0 |
| African | 2 (597/679) | 1.18 (0.84-1.65) | 0.900 | 0.0 | 1.00 (0.79-1.27) | 0.560 | 0.0 | 1.18 (0.86-1.61) | 0.929 | 0.0 | 1.04 (0.83-1.31) | 0.601 | 0.0 | 1.07 (0.91-1.25) | 0.729 | 0.0 |
| Control source | ||||||||||||||||
| PB | 5 (1675/4334) | 1.24 (1.00-1.54) | 0.480 | 0.0 | 1.14 (1.01-1.30) | 0.549 | 0.0 | 1.18 (0.96-1.45) | 0.596 | 0.0 | 1.16 (1.03-1.31) | 0.409 | 0.0 | 1.13 (1.03-1.24) | 0.314 | 15.7 |
| HB | 7 (1607/2142) | 1.30 (1.05-1.61) | 0.976 | 0.0 | 1.05 (0.91-1.21) | 0.601 | 0.0 | 1.28 (1.04-1.57) | 0.908 | 0.0 | 1.10 (0.96-1.26) | 0.830 | 0.0 | 1.12 (1.01-1.23) | 0.968 | 0.0 |
| Sample size | ||||||||||||||||
| ≥200 | 6 (2895/5848) | 1.27 (1.08-1.50) | 0.704 | 0.0 | 1.08 (0.97-1.19) | 0.672 | 0.0 | 1.24 (1.06-1.44) | 0.683 | 0.0 | 1.12 (1.01-1.23) | 0.753 | 0.0 | 1.12 (1.04-1.20) | 0.763 | 0.0 |
| <200 | 6 (387/628) | 1.25 (0.78-2.00) | 0.880 | 0.0 | 1.31 (0.99-1.74) | 0.613 | 0.0 | 1.15 (0.73-1.81) | 0.850 | 0.0 | 1.30 (1.00-1.69) | 0.651 | 0.0 | 1.21 (0.99-1.49) | 0.702 | 0.0 |
HB, hospital based; PB, population based.
Figure 1.
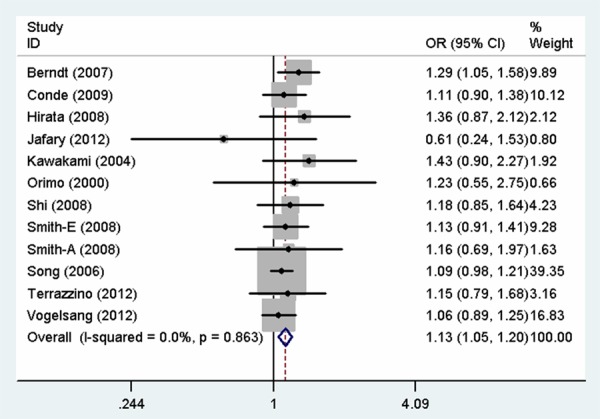
Forest plot for the association between MSH3 rs26279 G > A polymorphism and cancer risk (G vs. A).
Figure 2.
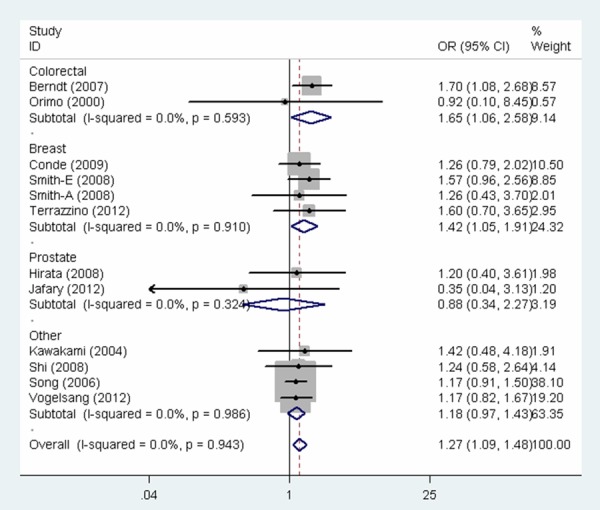
Forest plot for the association between MSH3 rs26279 G > A polymorphism and cancer risk (GG vs. AA).
Figure 3.
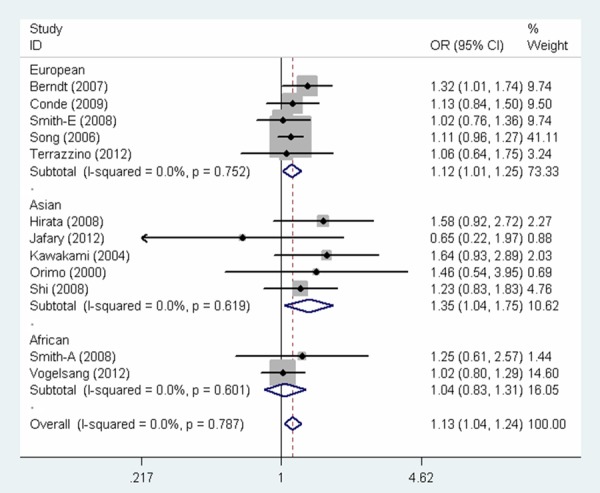
Forest plot for the association between MSH3 rs26279 G > A polymorphism and cancer risk (AG + GG vs. AA).
Heterogeneity and sensitivity analyses
No significant heterogeneity was detected for the meta-analysis of MSH3 rs26279 G > A polymorphism and overall cancer risk (GG vs. AA: P = 0.943; AG vs. AA: P = 0.675; GG vs. AG + AA: P = 0.925; AG + GG vs. AA: P = 0.787; G vs. A: P = 0.863). The sensitivity analysis provided the evidence of the stability of the meta-analysis, since no single study was found to influence the pooled ORs qualitatively.
Publication bias
We used Begg’s funnel plot and Egger’s test to estimate the publication bias of literatures. No evidence of publication bias was observed under all the genetic models (GG vs. AA: P = 0.910; AG vs. AA: P = 0.292; GG vs. AG + AA: P = 0.672; AG + GG vs. AA: P = 0.339 and G vs. A: P = 0.443). These results suggested that no publication bias influenced the evaluation of the association between MSH3 rs26279 G > A polymorphism and cancer susceptibility.
Discussion
Human MSH3 gene is located on chromosome 5q11-13 and encodes MSH3 protein containing 1137 amino acids residus [9]. The MSH3 protein is one of the essential components of mismatch repair (MMR) system that is critical to maintaining genome stability [35]. MMR deficiency has been reported to associate with increased risk of several types of cancer [6]. Loss of MSH3 protein expression is associated with colorectal cancer progression [36]. Until now, there are numerous publications investigating the association of MSH3 polymorphisms and cancer susceptibility. MSH3 rs26279 G > A polymorphism is most wildly studied for its association with cancer risk among those genetic variation. However the results were controversy. For example, Berndt et al. found that MSH3 rs26279 G > A polymorphism significantly increased the risk of developing colorectal cancer [10]. In contrast, Smith et al. did not observe any significant association between MSH3 rs26279 G > A polymorphism and the risk of breast cancer [13].
To the best of our knowledge, this is the first meta-analysis exploring the relationship of MSH3 rs26279 G > A polymorphism with overall cancer susceptibility. In the current study, MSH3 rs26279 G > A polymorphism was found to confer 10% to 27% increase in the risk of overall cancer under different genetic models. These findings further substantiated that MSH3 rs26279 G > A polymorphism may play an important role in carcinogenesis. Germline mutation in the MSH3 gene, leading to inactivation of MMR, can increase the risk for cancer susceptibility [5,37]. Stratified analysis by cancer type found that there was 28% to 65% increase in the risk of colorectal cancer under homozygous, dominant and allele models, including 2 studies of 2418 subjects. Frameshift mutations in MSH3 gene play an important role in the development of colorectal cancer with microsatellite instability [38-40]. Similarly, this variant was also related to a 40% to 42% increase in the risk of breast cancer under homozygous and recessive models (4 studies with 1969 subjects), but no risk effect was found on prostate cancer (2 studies with 298 subjects). Our results suggested that MSH3 rs26279 G > A polymorphism may play a differential role in the carcinogenesis of different types of cancers [16]. Moreover, we did not evaluate the association of this polymorphism and the rest of types of cancers (e.g., prostate cancer), since the relatively small number of studies and subjects could only provide very limited statistical power. Stratified analysis by ethnicity indicated significant association between MSH3 rs26279 G > A polymorphism and around 12% and 35% higher cancer risk among Europeans and Asians respectively, whereas, no association was not found among Africans. Many factors may contribute to the differences between different ethnic groups, such as environment, gene, lifestyle and eating habits [41]. However, these finding should be considered carefully because of small sample size. For instance, only 2 studies conducted among Africans were included in the meta-analysis.
Despite the significant findings, there are still several limitations in the current meta-analysis. Firstly, due to failure to acquire original data, our results were based on single factor estimates without adjustment for other factors, such as individual’s age, smoking and drinking status. Secondly, only 12 studies with 3282 cases and 6476 controls were included to estimate the relationship between MSH3 rs26279 G > A polymorphism and cancer risk. As a result, the study has no sufficient statistical power to evaluate the association with the risk of certain type of cancer, especially for prostate cancer. Finally, because we only searched studies written in Chinese and English language, related publications in other language might be omitted.
Conclusions
This meta-analysis results suggests that the presence of MSH3 rs26279 G > A polymorphism may increase the risk of overall cancer, especially for the colorectal cancer and breast cancer. However, large studies with well matched cancer-free controls, involving different types of cancers, are necessary to verify these finding and provide more information about gene-gene and gene-environment interactions in carcinogenesis.
Disclosure of conflict of interest
None.
References
- 1.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Yu Z, Chen J, Ford BN, Brackley ME, Glickman BW. Human DNA repair systems: an overview. Environ Mol Mutagen. 1999;33:3–20. doi: 10.1002/(sici)1098-2280(1999)33:1<3::aid-em2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 3.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 4.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 6.Xiao X, Melton DW, Gourley C. Mismatch repair deficiency in ovarian cancer--molecular characteristics and clinical implications. Gynecol Oncol. 2014;132:506–512. doi: 10.1016/j.ygyno.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Buermeyer AB, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 8.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 9.Risinger JI, Umar A, Boyd J, Berchuck A, Kunkel TA, Barrett JC. Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair. Nat Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 10.Berndt SI, Platz EA, Fallin MD, Thuita LW, Hoffman SC, Helzlsouer KJ. Mismatch repair polymorphisms and the risk of colorectal cancer. Int J Cancer. 2007;120:1548–1554. doi: 10.1002/ijc.22510. [DOI] [PubMed] [Google Scholar]
- 11.Orimo H, Nakajima E, Yamamoto M, Ikejima M, Emi M, Shimada T. Association between single nucleotide polymorphisms in the hMSH3 gene and sporadic colon cancer with microsatellite instability. J Hum Genet. 2000;45:228–230. doi: 10.1007/s100380070031. [DOI] [PubMed] [Google Scholar]
- 12.Conde J, Silva SN, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Association of common variants in mismatch repair genes and breast cancer susceptibility: a multigene study. BMC Cancer. 2009;9:344. doi: 10.1186/1471-2407-9-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terrazzino S, La Mattina P, Masini L, Caltavuturo T, Gambaro G, Canonico PL, Genazzani AA, Krengli M. Common variants of eNOS and XRCC1 genes may predict acute skin toxicity in breast cancer patients receiving radiotherapy after breast conserving surgery. Radiother Oncol. 2012;103:199–205. doi: 10.1016/j.radonc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Hirata H, Hinoda Y, Kawamoto K, Kikuno N, Suehiro Y, Okayama N, Tanaka Y, Dahiya R. Mismatch repair gene MSH3 polymorphism is associated with the risk of sporadic prostate cancer. J Urol. 2008;179:2020–2024. doi: 10.1016/j.juro.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jafary F, Salehi M, Sedghi M, Nouri N, Sadeghi F, Motamedi S, Talebi M. Association between mismatch repair gene MSH3 codons 1036 and 222 polymorphisms and sporadic prostate cancer in the Iranian population. Asian Pac J Cancer Prev. 2012;13:6055–6057. doi: 10.7314/apjcp.2012.13.12.6055. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami T, Shiina H, Igawa M, Deguchi M, Nakajima K, Ogishima T, Tokizane T, Urakami S, Enokida H, Miura K, Ishii N, Kane CJ, Carroll PR, Dahiya R. Inactivation of the hMSH3 mismatch repair gene in bladder cancer. Biochem Biophys Res Commun. 2004;325:934–942. doi: 10.1016/j.bbrc.2004.10.114. [DOI] [PubMed] [Google Scholar]
- 18.Shi WP, Bian JC, Jiang F, Ni HX, Zhu QX, Tang HW, Shen Q, Wu Y. [Association of genetic polymorphisms and haplotypes in hMLH1 and hMSH3 gene with the risk of papillary thyroid carcinoma] . Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2008;25:390–395. [PubMed] [Google Scholar]
- 19.Song H, Ramus SJ, Quaye L, DiCioccio RA, Tyrer J, Lomas E, Shadforth D, Hogdall E, Hogdall C, McGuire V, Whittemore AS, Easton DF, Ponder BA, Kjaer SK, Pharoah PD, Gayther SA. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006;27:2235–2242. doi: 10.1093/carcin/bgl089. [DOI] [PubMed] [Google Scholar]
- 20.Vogelsang M, Wang Y, Veber N, Mwapagha LM, Parker MI. The cumulative effects of polymorphisms in the DNA mismatch repair genes and tobacco smoking in oesophageal cancer risk. PLoS One. 2012;7:e36962. doi: 10.1371/journal.pone.0036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann A, Hogdall E, Ramus SJ, DiCioccio RA, Hogdall C, Quaye L, McGuire V, Whittemore AS, Shah M, Greenberg D, Easton DF, Ponder BA, Kjaer SK, Gayther SA, Thompson DJ, Pharoah PD, Song H. Mismatch repair gene polymorphisms and survival in invasive ovarian cancer patients. Eur J Cancer. 2008;44:2259–2265. doi: 10.1016/j.ejca.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCullough LE, Santella RM, Cleveland RJ, Millikan RC, Olshan AF, North KE, Bradshaw PT, Eng SM, Terry MB, Shen J, Crew KD, Rossner P Jr, Teitelbaum SL, Neugut AI, Gammon MD. Polymorphisms in DNA repair genes, recreational physical activity and breast cancer risk. Int J Cancer. 2014;134:654–663. doi: 10.1002/ijc.28383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michiels S, Danoy P, Dessen P, Bera A, Boulet T, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis. 2007;28:1731–1739. doi: 10.1093/carcin/bgm111. [DOI] [PubMed] [Google Scholar]
- 27.Dong X, Li Y, Hess KR, Abbruzzese JL, Li D. DNA mismatch repair gene polymorphisms affect survival in pancreatic cancer. Oncologist. 2011;16:61–70. doi: 10.1634/theoncologist.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krajinovic M, Labuda D, Mathonnet G, Labuda M, Moghrabi A, Champagne J, Sinnett D. Polymorphisms in genes encoding drugs and xenobiotic metabolizing enzymes, DNA repair enzymes, and response to treatment of childhood acute lymphoblastic leukemia. Clin Cancer Res. 2002;8:802–810. [PubMed] [Google Scholar]
- 29.Mangoni M, Bisanzi S, Carozzi F, Sani C, Biti G, Livi L, Barletta E, Costantini AS, Gorini G. Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. Int J Radiat Oncol Biol Phys. 2011;81:52–58. doi: 10.1016/j.ijrobp.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 30.Reeves SG, Meldrum C, Groombridge C, Spigelman A, Suchy J, Kurzawski G, Lubinski J, Scott RJ. DNA repair gene polymorphisms and risk of early onset colorectal cancer in Lynch syndrome. Cancer Epidemiol. 2012;36:183–189. doi: 10.1016/j.canep.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Smith TR, Liu-Mares W, Van Emburgh BO, Levine EA, Allen GO, Hill JW, Reis IM, Kresty LA, Pegram MD, Miller MS, Hu JJ. Genetic polymorphisms of multiple DNA repair pathways impact age at diagnosis and TP53 mutations in breast cancer. Carcinogenesis. 2011;32:1354–1360. doi: 10.1093/carcin/bgr117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koessler T, Oestergaard MZ, Song H, Tyrer J, Perkins B, Dunning AM, Easton DF, Pharoah PD. Common variants in mismatch repair genes and risk of colorectal cancer. Gut. 2008;57:1097–1101. doi: 10.1136/gut.2007.137265. [DOI] [PubMed] [Google Scholar]
- 33.Mathonnet G, Krajinovic M, Labuda D, Sinnett D. Role of DNA mismatch repair genetic polymorphisms in the risk of childhood acute lymphoblastic leukaemia. Br J Haematol. 2003;123:45–48. doi: 10.1046/j.1365-2141.2003.04551.x. [DOI] [PubMed] [Google Scholar]
- 34.Mondal P, Datta S, Maiti GP, Baral A, Jha GN, Panda CK, Chowdhury S, Ghosh S, Roy B, Roychoudhury S. Comprehensive SNP scan of DNA repair and DNA damage response genes reveal multiple susceptibility loci conferring risk to tobacco associated leukoplakia and oral cancer. PLoS One. 2013;8:e56952. doi: 10.1371/journal.pone.0056952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 36.Plaschke J, Kruger S, Jeske B, Theissig F, Kreuz FR, Pistorius S, Saeger HD, Iaccarino I, Marra G, Schackert HK. Loss of MSH3 protein expression is frequent in MLH1-deficient colorectal cancer and is associated with disease progression. Cancer Res. 2004;64:864–870. doi: 10.1158/0008-5472.can-03-2807. [DOI] [PubMed] [Google Scholar]
- 37.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 38.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 39.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 40.Ikeda M, Orimo H, Moriyama H, Nakajima E, Matsubara N, Mibu R, Tanaka N, Shimada T, Kimura A, Shimizu K. Close correlation between mutations of E2F4 and hMSH3 genes in colorectal cancers with microsatellite instability. Cancer Res. 1998;58:594–598. [PubMed] [Google Scholar]
- 41.Dick DM. Gene-environment interaction in psychological traits and disorders. Annu Rev Clin Psychol. 2011;7:383–409. doi: 10.1146/annurev-clinpsy-032210-104518. [DOI] [PMC free article] [PubMed] [Google Scholar]


