Abstract
Jumonji domain-containing protein 1A (JMJD1A) play a key role in the development and progression of several malignancies. The present study investigated the expression and clinical significance of JMJD1A in gastric cancer. JMJD1A was found to be upregulated in gastric cancer tissues and cell lines. Furthermore, the upregulated expression of JMJD1A was significantly correlated with the results of the invasion depth (P=0.006), lymph node metastatic status (P<0.001), and TNM stage (P<0.001). JMJD1A was also shown to be an independent prognostic predictor of overall survival (HR3.988; 95% CI 1.948-8.167; P<0.001) for patients with gastric cancer. In addition, in vitro experiment revealed that knockdown of JMJD1A expression inhibited the gastric cancer cell proliferation, and further study suggested that JMJD1A knockdown suppressed MAPK pathway via transcriptional downregulation the expression of long noncoding RNA MALAT1. Therefore, we speculated that JMJD1A-MALAT1-MAPK signaling might participate in the JMJD1A-induced cell proliferation of gastric cancer. Collectively, our data demonstrate for the first time that JMJD1A gene has an important regulatory role in gastric carcinogenesis, and could function as a novel prognostic indicator and a potential therapeutic target for gastric cancer.
Keywords: Gastric cancer, JMJD1A, immunohistochemistry, prognosis, proliferation
Introduction
Gastric cancer is one of the most aggressive and lethal human malignant diseases. It still remains among the leading causes of death from cancer in less developed countries in spite of its declining incidence worldwide [1,2]. The development and progression of gastric cancer involves in a large number of genetic and epigenetic alterations of tumor tumor-related and suppressor genes [3]. A better understanding of the underlying molecular mechanism is essential for the development of novel therapeutic strategies.
Jumonji domain-containing protein 1A (JMJD1A), also called KDM3A or JHDM2A, belongs to iron- and 2-oxoglutarate-dependent dioxygenases [4]. It is an important epigenetic mark associated with transcription repression by specifically demethylating mono- and dimethylated H3 ‘Lys-9’ residue (H3K9me1/2) [4]. The functional relevance of JMJD1A has not been fully clarified, but it appears to cause transcriptional activation of various downstream targeted genes and amplifies hypoxia-induced gene expression [5,6]. Recent studies also indicate that the upregulation of JMJD1A expression is associated with cancer development [6], such as renal cell carcinoma [7] and hepatocellular carcinoma [8]. However, the relationship between JMJD1A and the development of gastric cancer remains unclear. In this study, we would like to explore the expression pattern of JMJD1A and its correlation with clinicopathological factors in gastric cancer. Furthermore, the prognostic significance of JMJD1A was assessed. In addition, the potential effect of JMJD1A on gastric cancer cell proliferation was also investigated.
Materials and methods
Patients and pathological data
This study was approved by the ethics committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. Tissue microarrays (TMAs) purchased from ShGnghGi Outdo Biotech Company (China) were well-documented with clinicopathological information, including patient age, sex, tumor size, pathological grade and tumor staging (detailed in Table 2), as well as follow-up data. Ninety gastric cancer patients included in the TMAs underwent surgery operation between August 2008 and March 2009, and postoperative follow-up was finished by September 2014.
Table 2.
Relationship between JMJD1A protein expression and clinicopathologic parameters of gastric cancer (n = 90)
| Variables | Cases | JMJD1A expression | P-value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Total | 90 | 48 | 42 | |
| Gender | ||||
| Male | 53 | 26 | 27 | 0.472a |
| Female | 37 | 21 | 16 | |
| Age (years) | ||||
| ≤60 | 38 | 17 | 21 | 0.224a |
| >60 | 52 | 30 | 22 | |
| Tumor size (cm) | ||||
| ≤5 | 48 | 27 | 21 | 0.413a |
| >5 | 42 | 20 | 22 | |
| Histopathological grading | ||||
| Well/Moderately | 40 | 24 | 16 | 0.186a |
| Poorly | 50 | 23 | 27 | |
| Tumor invasion (T) | ||||
| T1 + T2 | 14 | 12 | 2 | 0.006a,* |
| T3 + T4 | 76 | 35 | 41 | |
| Nodal status (N) | ||||
| N0 | 23 | 21 | 2 | <0.001a,* |
| N1 | 12 | 10 | 2 | |
| N2 | 18 | 8 | 10 | |
| N3 | 37 | 8 | 29 | |
| Metastasis status (M) | ||||
| M0 | 85 | 46 | 39 | 0.189b |
| M1 | 5 | 1 | 4 | |
| TNM stages | ||||
| I + II | 36 | 34 | 5 | <0.001a,* |
| III + IV | 54 | 13 | 38 | |
Chi-square test;
Fisher’s exact test;
indicates P<0.05.
Immunohistochemistry
Citrate buffer (0.01 M, pH 6.0) was used for antigen retrieval of the paraffin-embedded sections. The slides were then incubated with a rabbit polyclonal antibody against JMJD1A (dilution 1:100; Sigma-Aldrich, USA) at 4°C overnight and then with horseradish peroxidase (HRP) (Gene Tech GTVision III Detection Kit, Shanghai, China) at room temperature for 40 min. Following washing with PBS for 3 times, the signal was detected with 3, 3’-diaminobenzidine (DAB) solution. For negative controls, adjacent sections were carried out as described above, with exception that they were incubated overnight at 4°C in blocking solution without primary antibody.
Score of immunohistochemistry
The staining was scored according to the staining intensity and percentage as previously described [9]. Staining intensity was scored as 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strongly positive). The percentages of cells were scored into five categories: 0 (0%), 1 (1-25%), 2 (26%-50%), 3 (51-75%), and 4 (76-100%). The final staining scores were calculated by staining intensity × percentages of stained cells, ranging from 0 to 12. To evaluate the association between JMJD1A expression and clinicopathological parameters, patients were grouped into two categories: low expression (0-4) and high expression (5-12). Immunostaining was independently scored by two pathologist blinded to the clinicopathological characteristics.
Cell culture
Human gastric cancer cells including BGC-823, MKN-45, MGC80-3, SGC7901, and NCI-N87 and gastric epithelial cell line GES-1, were obtained from Shanghai Institute for Life Science, Chinese Academy of Sciences, and cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS, both from Gibco, Carslbad, CA, USA) at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Transfection of small interfering RNA (siRNA)
Gastric cancer cell lines MKN-45 was transfected with the siRNA in Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s protocols. SiRNA (AGAAGAAUUCAAGAGAUUCCGGAGG) were designed against human JMJD1A and obtained commercially (GeneChem, Shanghai, China). The ability of the siRNA to inhibit JMJD1A expression was assessed at 48 hours after transfection.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the cultured cells with Trizol reagent (Invitrogen), and first-strand cDNA was synthesized from 500 ng of total RNA using random primers and the M-MLV Reverse Transcriptase (Takara). The following primers were used to detect the expression level of JMJD1A, MALAT1 and GAPDH (internal control): JMJD1A (sense): 5’-GGCGGACTTTAGACGTTCCA-3’; JMJD1A (antisense): 5’-AGATGAGCCTTCCACTTGGC-3’; MALAT1 (sense): 5’-GAATTGCGTCATTTAAAGCCTAGTT-3’; MALAT1 (antisense): 5’-GTTTCATCCTACCACTCCCAATTAAT-3’; GAPDH (sense): 5’-AGAAGGCTGGGGCTCATTTG-3’; GAPDH (antisense): 5’-AGGGGCCATCCACAGTCTTC-3’. RNA expression was measured by qRT-PCR using the SYBR-Green method (Takara) according to the manufacturer’s instructions. The results were normalized to the expression of GAPDH.
Western blotting
Western blotting was performed as previously described [10]. Briefly, membranes were incubated with primary antibodies. Following overnight incubation, membranes were washed 3 times in Tris buffered saline-Tween 20 (TBST) for 10 min each. The membranes were then incubated with the secondary antibody in 5% bovine serum albumin in TBST for 1 h. Then targeted proteins were detected by chemiluminescence (Thermo Scientific, Hudson, NH, USA). GAPDH levels were used as loading controls. Antibodies used for Western blot analyses were as follows: JMJD1A (Sigma-Aldrich, USA), ERK, p-ERK, P38, p-P38, JNK, p-JNK, MEK, p-MEK (Cell Signaling Technology, Beverly, MA, USA); GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Cell proliferation assay
The effect of JMJD1A knockdown on gastric cancer cell proliferation in vitro was determined by measuring the absorbance at 450 nm. Briefly, untreated cells, cells treated with siRNA or non-target siRNA in 96-well plates (1×103 cells/well) in triplicate was added (10 μl) by Cell counting kit-8 (CCK-8, Dojindo Molecular Technologies, Kumamoto, Japan) at 24, 48, and 72 h. The absorbance at 450 nm was measured 1 h later.
Statistical analysis
The data are expressed as the means ± standard deviations (SD) of three independent experiments. The differences between two groups were compared with the Student’s t-test, Pearson’s χ2 test, or Fisher’s exact test, as appropriate. The patients’ survival curve was analyzed using the Kaplan-Meier method with the log-rank test. A Cox proportional hazards model was used to calculate univariate and multivariate hazard ratios for the variables. All analyses were carried out using the SPSS 13.0 software (SPSS Inc., Chicago, IL). A P-value of less than 0.05 was considered statistically significant.
Results
JMJD1A expression is upregulated in gastric cancer
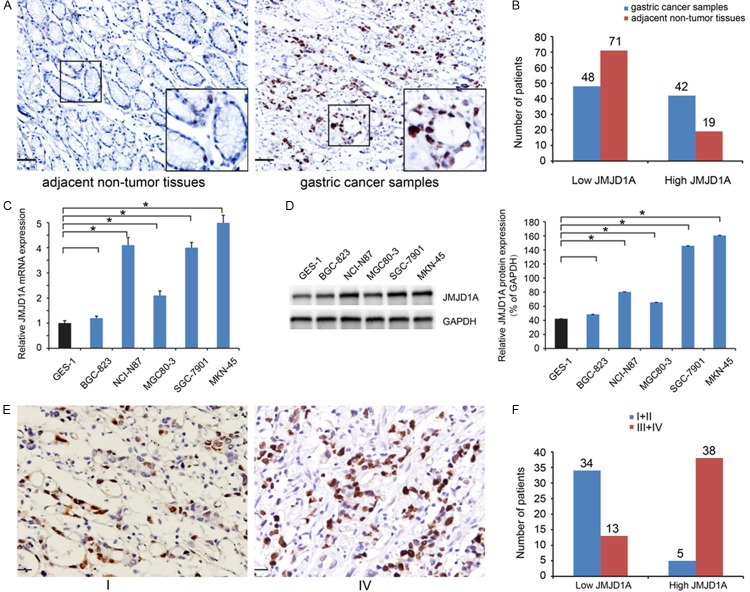
We evaluated the expression of JMJD1A in a cohort of 90 patients diagnosed with gastric cancer and paired adjacent normal tissues by immunohistochemistry. Representative expression patterns in both cancer and noncancerous samples were shown in Figure 1A. JMJD1A expression was found to be localized in the nucleus. And higher JMJD1A expression was observed in gastric cancer samples compared to the matched adjacent non-tumor tissues. In brief, for tumor samples, 42 cases (46.7%) displayed high expression, while only 21.1% in adjacent tissue samples (Figure 1B; Table 1). Furthermore, the five gastric cancer cell lines exhibited higher expression levels of JMJD1A than the GES-1 cells, and the MKN-45 cells exhibited relatively high levels of expression in the five cancer cell lines (Figure 1C, 1D).
Figure 1.
JMJD1A expression is upregulated in gastric cancer. (A) Representative photomicrographs showed JMJD1A expression, which was found to be localized in the nucleus (Scale bar, 50 μm). (B) Higher JMJD1A expression in gastric cancer samples in comparisons with matched adjacent non-tumor tissues. (C, D) Compared to gastric epithelial cell line GES-1, the five gastric cancer cell lines exhibited higher mRNA (C) and protein (D) expression levels of JMJD1A, and the MKN-45 cells exhibited relatively high levels in five cancer cell lines. (E, F) The expression of JMJD1A was increased in tissue with advanced stage as compared to early stage of gastric cancer (Scale bar, 20 μm).
Table 1.
JMJD1A protein is higher in gastric cancer tissues than in paired adjacent normal tissues (n=90)
| Cases | JMJD1A expression | P-value | ||
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Gastric cancer | 90 | 48 | 42 | <0.001* |
| Adjacent normal tissue | 90 | 71 | 19 | |
Indicates P<0.05.
Association between JMJD1A expression and the clinicopathological features of gastric cancer patients
Then, we investigated association of JMJD1A expression and clinicopathological features. As summarized in Table 2, JMJD1A expression showed a positive results with the invasion depth (P=0.006), lymph node metastatic status (P<0.001), and TNM stage (P<0.001, Figure 1E, 1F). These data indicate that JMJD1A may be involved in gastric cancer progression.
High JMJD1A expression is associated with a poor clinical outcome in human gastric cancer
Kaplan-Meier analysis with a log rank test for overall survival (OS) was performed to assess the possible association between tumor expression of and patient survival. Patients with tumors expressing high levels of JMJD1A had a poorer OS than that of patients with JMJD1A-low tumors (log-rank test, P<0.001, Figure 2).
Figure 2.
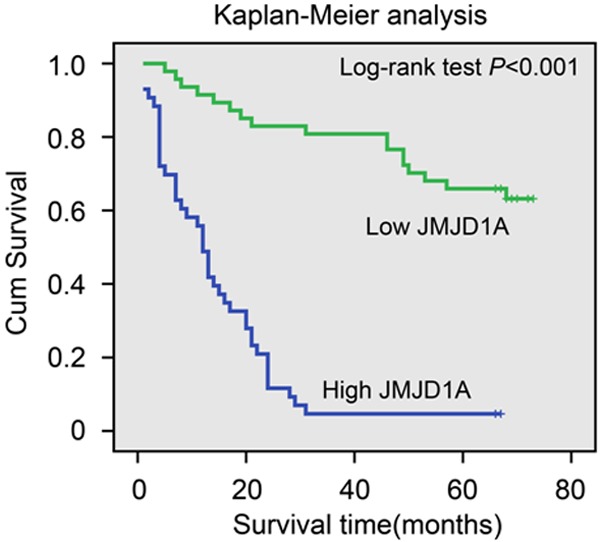
Kaplan-Meier plots with log rank test of overall survival (OS). OS is significantly lower in patients with tumors expressing high levels of JMJD1A than that in patients with tumors expressing low levels of JMJD1A.
Using a univariate analysis in the Cox proportional hazards model, a decreased OS was associated with the following characteristics: tumor depth, nodal status, TNM stage, and the JMJD1A expression (Table 3). Multivariate analysis revealed that the expression of JMJD1A is an independent prognostic factor for OS of gastric cancer patients (HR3.988; 95% CI 1.948-8.167; P<0.001, Table 3).
Table 3.
Univariate and multivariate analysis of overall survival
| Clinicopathological parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| JMJD1A (High/Low) | 8.446 | 4.480-15.923 | <0.001* | 3.988 | 1.948-8.167 | <0.001* |
| Gender (Female/Male) | 1.452 | 0.866-2.435 | 0.157 | |||
| Age (years) (>60/≤60) | 0.622 | 0.371-1.045 | 0.073 | |||
| Tumor size (cm) (>3/≤3) | 1.497 | 0.893-2.510 | 0.126 | |||
| Tumor differentiation (Poorly/Well+Moderately) | 1.666 | 0.972-2.854 | 0.063 | |||
| Tumor invasion (T3+T4/T1+T2) | 1.943 | 1.310-2.881 | 0.001* | |||
| Nodal status (N3/N2/N1/N0) | 2.772 | 2.038-3.770 | <0.001* | 2.507 | 1.623-3.874 | <0.001* |
| Metastasis status (M1/M0) | 1.942 | 0.700-5.392 | 0.203 | |||
| TNM stages (III+IV/I+II) | 6.92 | 3.581-13.373 | <0.001* | |||
HR hazard ratio, 95% CI 95% confidence interval;
indicates P<0.05.
Knockdown of JMJD1A expression inhibits cell proliferation of gastric cancer by suppressing MALAT1-MAPK signaling
To further investigate the potential effects of JMJD1A on gastric cancer cell proliferation, we used JMJD1A-siRNA to down-regulate its endogenous expression in transfected MKN-45 cell lines. The efficacy of JMJD1A knockdown was confirmed by qRT-PCR and western blot analyses (Figure 3A, 3B). As shown in Figure 3C, JMJD1A knockdown was associated with significantly decreased cell proliferation compared with that of control-siRNA. To determine the possible mechanism by which JMJD1A regulated cell proliferation, we performed western blot analysis to investigate the effects of JMJD1A knockdown on the mitogen-activated protein kinase (MAPK) pathway, which is often aberrantly activated in human cancers and contributes to increased cell proliferation ability [11-13]. The results showed that JMJD1A downregulation significantly reduced the levels of phosphorylated P38, ERK, MEK, and JNK, while no detectable changes were observed in the total levels of P38, ERK, MEK, and JNK (Figure 3D). In addition, previous study showed that JMJD1A could up-regulate long noncoding RNA MALAT1 gene transcription [14]. And MALAT1 could promote the tumor cell proliferation by activating MAPK pathway [15]. We also found that the expression of MALAT1 was decreased in response to JMJD1A knockdown (Figure 3E). Taken together, we speculated that JMJD1A-MALAT1-MAPK signaling might participate in the JMJD1A-induced cell proliferation of gastric cancer.
Figure 3.
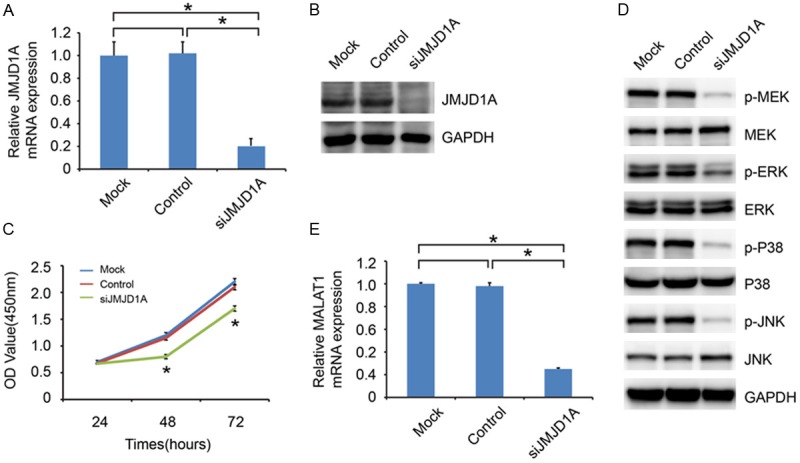
JMJD1A knockdown inhibits gastric cancer cell proliferation. qRT-PCR (A) and western blot analysis (B) were performed to detect the knockdown effect on JMJD1A expression in MKN-45 cell. (C) Effects of JMJD1A knockdown on proliferation was evaluated by Cell Counting Kit-8 assays. (D) Western blot analysis was performed to detect the expression of key molecular factors of MAPK pathways, including ERK, p-ERK, P38, p-P38, MEK, p-MEK, JNK and p-JNK. (E) The mRNA expression of MALAT1 was decreased in response to JMJD1A knockdown.
Discussion
Gastric cancer is a heterogeneous disease that demands continued attention and research with regard to prevention, early detection and novel therapeutic options. A large number of data have demonstrated that histone modifications such as methylation and acetylation play important regulatory roles in processes such as gene transcription and DNA damage repair, and misregulation of histone modification, actively contributes to human cancer [16].
JMJD1A is an H3K9me1/2 histone demethylase [4]. As H3K9 methylation in promoter regions is inhibitory to transcription [17,18], JMJD1A demethylase activity tends to enhance gene expression [5,6,19]. Recent studies show that JMJD1A have important regulatory roles in the development and progression of some types of tumors, such as colorectal cancer [20], neuroendocrine prostate tumors [21], and hepatocellular carcinoma [8]. However, studies on JMJD1A expression during tumorigenesis and progression of gastric cancer remain to be elucidated.
The present study revealed for the first time that JMJD1A expression were upregulated in primary gastric cancer tissues and immortalized cancer cell lines. Further statistical analysis found that high JMJD1A expression in gastric cancer tissues was significantly associated with other malignant tumor characteristics, such as invasion depth, lymph node metastatic and TNM stage.
Moreover, Kaplan-Meier survival analysis demonstrated that high JMJD1A expression was significantly associated with poor prognosis of gastric cancer patients. Finally, univariate and multivariate Cox model analyses suggested that JMJD1A was an independent prognostic factor, indicating that JMJD1A was a new predictor to the prognosis of patients with gastric cancer.
To further understand the role of JMJD1A in tumorigenesis, RNAi was used to knockdown the expression of JMJD1A in MKN-45 cells. Then CCK8 assays were implemented, and the results revealed that knockdown of JMJD1A inhibited cell proliferation activity. To elucidate the possible mechanism by which JMJD1A regulates gastric cancer cell proliferation, western blot analysis of the key molecular factors of MAPK pathways were performed. MAPK pathways are most important signal transduction pathways by activating the related signaling cascade [22-25]. We observed that JMJD1A knockdown significantly reduced the expression phosphorylated MEK, ERK, P38, and JNK. However, no detectable changes in total MEK, ERK, P38, or JNK protein expression were observed. The direct link between JMJD1A and the MAPK pathway remain to be elucidated. Tee AE et al [14] found that JMJD1A could up-regulate MALAT1 gene transcription by demethylating histone H3K9 at the MALAT1 gene promoter. Wu XS et al [15] revealed that MALAT1 promoted the cell proliferation of gallbladder cancer cells by activating MAPK pathway. In our study, we also found that inhibition of JMJD1A expression could suppress MALAT1 expression in gastric cancer. Therefore, we can hypothesize that JMJD1A activates MAPK pathway via transcriptional regulation the expression of MALAT1, which need to be further verified.
In conclusion, our data supported that JMJD1A is an important factor in gastric cancer cell growth and progression and its upregulated expression is common in gastric cancer tissue. Further studies are required to confirm our hypothesis that JMJD1A is a potential target for gastric cancer therapy.
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (grant nos. 81101846, 81171887, 91229117 and 31101016), by Program of Shanghai Subject Chief Scientist (grant no. 12XD1404200), by Shanghai International Science and Technology Cooperation Project (grant no. 12410709000), by Shanghai Science and Technology Committee (grant no. 11DZ1922002), and by National Key Clinical Discipline-Oncology.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes A, Roda D, Tarazona N, Rosello S, Perez-Fidalgo JA. Current questions for the treatment of advanced gastric cancer. Cancer Treat Rev. 2013;39:60–67. doi: 10.1016/j.ctrv.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 6.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo X, Shi M, Sun L, Wang Y, Gui Y, Cai Z, Duan X. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma. 2011;58:153–157. doi: 10.4149/neo_2011_02_153. [DOI] [PubMed] [Google Scholar]
- 8.Yamada D, Kobayashi S, Yamamoto H, Tomimaru Y, Noda T, Uemura M, Wada H, Marubashi S, Eguchi H, Tanemura M, Doki Y, Mori M, Nagano H. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann Surg Oncol. 2012;19(Suppl 3):S355–S364. doi: 10.1245/s10434-011-1797-x. [DOI] [PubMed] [Google Scholar]
- 9.Jin Z, Jiang W, Jiao F, Guo Z, Hu H, Wang L, Wang L. Decreased expression of histone deacetylase 10 predicts poor prognosis of gastric cancer patients. Int J Clin Exp Pathol. 2014;7:5872–5879. [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao F, Hu H, Han T, Yuan C, Wang L, Jin Z, Guo Z, Wang L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci. 2015;16:6677–6693. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong SK, Yoon S, Moelling C, Arthan D, Park JI. Noncatalytic function of ERK1/2 can promote Raf/MEK/ERK-mediated growth arrest signaling. J Biol Chem. 2009;284:33006–33018. doi: 10.1074/jbc.M109.012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Zhang L, Guo H, Wysham WZ, Roque DR, Willson AK, Sheng X, Zhou C, Bae-Jump VL. Glucose promotes cell proliferation, glucose uptake and invasion in endometrial cancer cells via AMPK/mTOR/S6 and MAPK signaling. Gynecol Oncol. 2015;138:668–675. doi: 10.1016/j.ygyno.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Ren L, Liu C, Xia T, Zha X, Wang S. Phenformin Induces Cell Cycle Change, Apoptosis, and Mesenchymal-Epithelial Transition and Regulates the AMPK/mTOR/p70s6k and MAPK/ERK Pathways in Breast Cancer Cells. PLoS One. 2015;10:e0131207. doi: 10.1371/journal.pone.0131207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, Pasquier E, Haber M, Norris MD, Suzuki T, Marshall GM, Liu T. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5:1793–1804. doi: 10.18632/oncotarget.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, Cao Y, Bao RF, Mu JS, Tan ZJ, Tao F, Liu YB. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Parrish JK, Sechler M, Winn RA, Jedlicka P. The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene. 2015;34:257–262. doi: 10.1038/onc.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, Mori M. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: In vivo identification from hypoxic tumor cells. Clin Cancer Res. 2010;16:4636–4646. doi: 10.1158/1078-0432.CCR-10-0407. [DOI] [PubMed] [Google Scholar]
- 21.Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R, Krajewski S, Mercola D, Carpenter PM, Bowtell D, Ronai ZA. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, Li W, Atwood SX, Whitson RJ, Chang AL, Li J, Oro AE, Chan JA, Kelleher JF, Segal RA. RAS/MAPK activation drives resistance to Smo inhibition, metastasis and tumor evolution in Shh pathway-dependent tumors. Cancer Res. 2015;75:3623–35. doi: 10.1158/0008-5472.CAN-14-2999-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo G, Zhou Y, Yi W, Yi H. Lactotransferrin expression is downregulated and affects the mitogen-activated protein kinase pathway in gastric cancer. Oncol Lett. 2015;9:2409–2413. doi: 10.3892/ol.2015.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, Mccormick F, Chu S. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc Natl Acad Sci U S A. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishio Y, Gojoubori T, Kaneko Y, Shimizu N, Asano M. Cancer cell-derived IL-8 induces monocytic THP1 cells to secrete IL-8 via the mitogen-activated protein kinase pathway. Tumour Biol. 2015 doi: 10.1007/s13277-015-3641-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]