Abstract
microRNAs (miRs) play critical roles in the progression of glioma. Previous in vitro studies have described the anti-tumor role of miR-149 in cancer cells including glioma. In this study, we aimed to investigate whether miR-149 is associated with the prognosis of glioma patients. A total of 163 glioma patients who underwent tumor resection were included in our follow-up study. We found that the miR-149 expression was significantly lower in tumor tissues compared with that in normal tissues (P<0.05). Kaplan-Meier and analysis showed that the miR-149 expression status was significantly associated with the survival duration (logrank test, P<0.001), and multivariate Cox regression revealed that patients with low miR-149 expression were exposed to a 1.825 fold higher death risk (HR=1.825, 95% CI=1.031-3.229, P=0.039) compared with those with high miR-149 expression. Further study showed that Akt/mTOR signaling was hyperactive in low miR-149 expressing tissues. Our study thus demonstrates that miR-149 expression in glioma tissues is critically associated with the prognosis of patients, suggesting its potential clinical significance.
Keywords: Glioma, miR-149, prognosis, Akt, mTOR
Introduction
Glioma, as the one of the most common malignancies in central nervous system, is characterized by high invasiveness, early recurrence and poor prognosis [1]. Despite the recently achieved advances in cancer diagnosis and treatment, the dismal survivalrate of high grade glioma still represents one of the major challenges in clinical practice. The unmet demand for the early prediction of prognosis, which might guide the therapeutic strategy for glioma, has underlined the importance of developing new diagnostic and prognostic approaches.
In recent years, microRNAs, a class of non-coding short single RNA strands, have emerged as key regulators of carcinogenesis and tumor progression [2-5]. Multiple microRNAs have exhibited their potency in governing the critical biological processes of tumor cells such as proliferation, apoptosis and differentiation by post-transcriptionally repressing their target genes [6]. The biology of glioma is also controlled by a microRNA network without exception [5,7-9], a large spectrum of microRNAs in glioma has been extensively investigated; for example, miR-10 and miR-16 have been shown to control glioma cell migration and invasion through regulating the epithelial-mesenchymal transition process [10,11], and several miRs have been shown to be associated with the prognosis of glioma [12-14]. Among the miRs that play pivotal roles in the pathophysiology of glioma, miR-149 is believed to act as a versatile tumor-suppressionmiRand is downregulatedin several cancer types [15-17]. Recent studies have addressed its anti-proliferation role by targeting FOXM1 [15]; and importantly, it has been reported to inhibit the invasiveness of glioma cell through blockade of AKT1 signaling [18]. Although these studies have implied its potential values in the diagnosis and treatment of glioma, the clinical evidence of this microRNA is still lacking and remain to be identified; more importantly, elucidating its distinct expression profilein glioma patients may broaden our knowledge on the potentialrole of microRNAs in the emerging field of biomarker screen.
In the present study, we launched a follow-up study on 113 patients to examine whether mir-149 is associated with the clinical outcome of glioma patients. Our pilot data revealed its prognostic function in glioma patients.
Patients, materials and methods
Study subjects
We enrolled 163 glioma patients who underwent tumor resection from January 2005 to December 2012 in Tianjin Medical University General Hospital. Among these patients, 88 patients (mean age: 56.6±8.7 years) were male and 75 (mean age: 55.2±8.4 years) were female. Fresh tumor specimens were stored in -80°C. All the patients’ diagnoses were confirmed based on the histopathological examination. The histological grade was determined according to the criteria formulated by the World Health Organization in 2007. The patients were followed-up every 3 months, and the follow-up was ended on December 31st 2014. 26 normal brain tissue samples were collected from the surgical waste from other surgeries and used as controls. We obtained a writing consent from each patient, and this study was approved by the Ethics Committee of Tianjin Medical University General Hospital.
RNA isolation and miR-149 quantification
The RNA was isolated using Trizol Reagent (Invitrogen) according to the instructions provided by the manufacturer. To assess the levels of miR-149 in each group, we utilized a TaqMan based quantitative PCR method. The TaqMan PCR assay kits were purchased from Applied Biosystems. The expression of U6 was used as an internal control to adjust miR-149 expression. And the relative expression of miR-149 was determined by the 2-ΔΔCt method.
Western blot
Tissues were homogenized with RIPA lysis buffer and were subsequently subjected to electrophoresis, followed by transferring onto a NC membrane. Primary antibodies were all purchased from Cell Signaling Technology Inc. (Beverly, MA, USA). The bands were visualized by an ECL detection kit (P0018, Beyotime, Shanghai China). The band density was determined by ImageJ software (version 1.44, NIH).
Statistical analysis
The data in this study were analyzed by SPSS 19.0 software package, Wilcoxon-Mann-Whitney test was used to compare the difference of miR-149 expression between glioma tumor samples and normal brain samples. The patient population was dichotomized according to miR-149 expression level. Patients who had a 0.5-fold or above miR-149 expression than the mean expression of control group were categorized as high miR-149 expression, otherwise categorized as low miR-149 expression. Chi square test was performed to examine the association between miR-149 expression status and clinical-pathological characteristics. Survival analysis was performed by Kaplan-Meier estimator, comparison of the overall survival between two groups was performed by logrank test. Multivariate Cox regression analysis was also used to determine the hazard ratio of each covariate. A two tailed probability less than 0.05 was considered statistically significant.
Results
MiR-149 is down-regulated in gliomatissue
We first compared the miR-149 expression profile in normal brain tissues and glioma tissues. As shown in Figure 1, miR-149 level was found to be significantly higher in normal brain tissues than that in glioma tissues (P<0.05).
Figure 1.
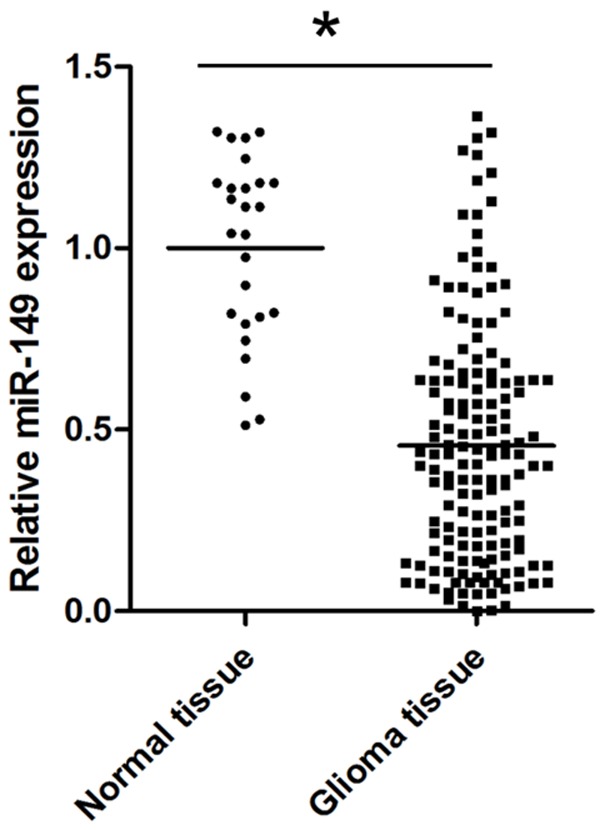
The differential expression of miR-149 in normal brain tissues and glioma tissues *P<0.05 n=26 for nomal tissue and n=163 for glioma tissue.
Association between miR-149 expression and clinical characteristics
To examine the possible association between miR-149 expression and clinical characteristcs, we first divided the patients into high-expression group and low-expression group based one the criteria described above. 103 patients were identified as low expression, and 60 patients were identified as high expression. We studied the association between miR-149 expression and general clinical characteristics in the study cohort. As presented in Table 1, miR-149 expression was not associated with age, sex, tumor location, tumor size, resection range and adjuvant therapy. By contrast, association between miR-149 expression and WHO grade, Karnofsky score, recurrence time or survival duration was observed.
Table 1.
Association between miR-149 expression and clinical characteristics
| Clinical characteristics | N | miR-149 expression | |
|---|---|---|---|
| Low | High | ||
| Sex | |||
| Male | 88 | 57 | 31 |
| Female | 75 | 46 | 29 |
| Age (years) | |||
| ≤60 | 106 | 68 | 38 |
| >60 | 57 | 35 | 22 |
| Tumor location | |||
| Parenchyma | 104 | 68 | 36 |
| Ventricle | 59 | 39 | 20 |
| Tumor size | |||
| ≤3 cm | 84 | 52 | 32 |
| >3 cm | 79 | 51 | 28 |
| WHO grade* | |||
| I+II | 80 | 36 | 44 |
| III+IV | 83 | 67 | 16 |
| Karnofsky score* | |||
| ≤80 | 94 | 88 | 6 |
| >80 | 69 | 15 | 54 |
| Resection range | |||
| Total resection | 93 | 55 | 38 |
| Local resection | 70 | 48 | 22 |
| Adjuvant therapy | |||
| Chemotherapy only | 100 | 63 | 37 |
| Radiotherapy and chemotherapy | 63 | 40 | 23 |
| Recurrence time* | |||
| ≤3 months | 97 | 68 | 29 |
| >3 months | 66 | 33 | 33 |
| Survival duration* | |||
| <15 months | 81 | 71 | 10 |
| ≥15 months | 82 | 32 | 50 |
P<0.05, performed by χ2 test.
MiR-149 correlates with the survival of glioma patients
To investigate whether miR-149 expression is correlated with the survival of glioma patients, we performed Kaplan-Meier survival analysis. All the cases were followed-up after surgery, and 7 patients lost follow-up due to the changes of their telephone number. As presented in Figure 2, patients with high expression of miR-149 exhibited greater overall survival than those with low miR-149 expression. Moreover, multivariate Cox regression analysis showed that the adjusted hazard ratio of miR-149 low expression was 1.825 (Table 2). Taken together, these data suggested that low expression of miR-149 is associated with a more aggressive type of glioma.
Figure 2.
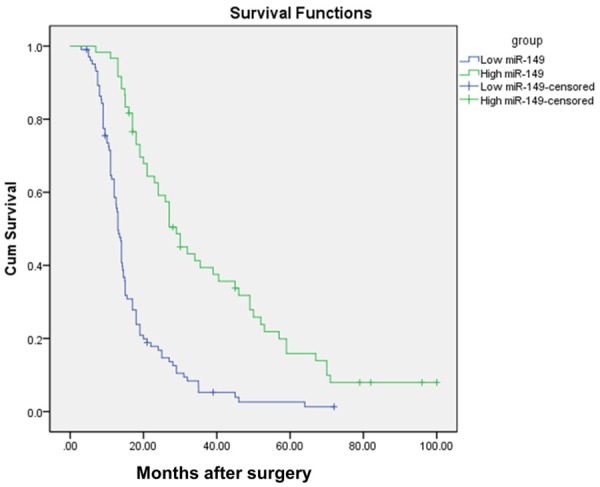
Survival of gliomapatients with different miR-149 expression statuses.
Table 2.
Multivariate Cox regression analysis of risk factors associated with overall survival of glioma patients
| Clinical characteristics | Adjusted HR | 95% CI | P |
|---|---|---|---|
| Age (>60) | 1.369 | 0.958-1.956 | 0.085 |
| Sex (Male) | 1.096 | 0.783-1.535 | 0.593 |
| Tumor size (>3 cm) | 0.779 | 0.541-1.121 | 0.179 |
| WHO grade (III+IV) | 1.475 | 1.010-2.154 | 0.044 |
| Karnofsky score (<80) | 1.899 | 1.093-3.267 | 0.023 |
| Resection range (≤3 cm) | 1.050 | 0.743-1.484 | 0.780 |
| Adjuvant therapy (Chemotherapy only) | 1.255 | 0.845-1.863 | 0.261 |
| miR-149 expression (Low) | 1.825 | 1.031-3.229 | 0.039 |
MiR-149 is associated with Akt/mTOR signaling in glioma patients
Previous in vitro study has demonstrated that miR-149 targets Akt. To test the potential role of miR-149 in regulating Akt/mTOR signaling in the clinical setting, we assayed the protein level of the Akt/mTOR axis. As shown in Figure 3A, both the phosphorylated and total AKT level were lower in high miR-149 expressing tissues, consequently, mTOR, the downstream effector of AKT, was less phosphorylated. Correlation analysis confirmed the association between miR-149 expression and mTOR signaling, which suggested that the upregulatedAkt/mTOR might mediate the unfavorable effect of low miR-149 expression on prognosis of glioma patients (Figure 3B).
Figure 3.
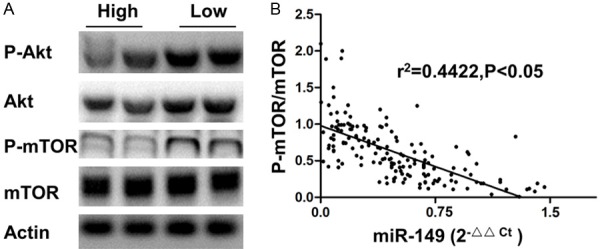
A. The representative western blot image of Akt/mTOR axis in low and high miR-149 expressing glioma tissues. B. Correlation between mTOR phosphorylation and miR-149 expression.
Discussion
In the current follow-up study, we evaluated the prognostic value of miR-149 in glioma patients. Our data showed that the differential expression of miR-149 in glioma tissues and normal tissues, Kaplan-Meier survival analysis and Cox regression analysis together showed that patients with low expression of miR-149 have a worse clinical outcome, which confirmed our hypothesis that miR-149 might play a favorable role in the survival of glioma patients. Therefore, our study demonstrates that miR-149 expression might be an independent indicator of prognosis of patients with glioma.
MicroRNAs have been recognized as indispensable regulators of normal cell function. Aberrant microRNA expression has been found in several diseases including glioma [19]. Several microRNAs have been implicated in the multifacetregulatory network of the pathogenesis of this cancer type. For example, a recent study shows that miR-218 functions as a tumor suppressor by affecting a series of critical biological processes of glioma including cell invasion, migration, proliferation and the maintenance of cancer cell stemness [20]. Particularly, growing numbers of studies have revealed their potential clinical values, circulating miR-128 has been identified as a diagnostic marker [21], and miR-218 has also been reported to show prognostic significance [22]. Although these studies revealed the pivotal role of microRNA in glioma biology, whether other microRNAs are also implicated in this issue is still to be explored. Moreover, given the broad effects of microRNAs are exhibiting, it is reasonable to speculate that a large number of previously uncharacterized microRNAs may show macro effects in glioma patients.
Accumulating evidences have unraveled the role of miR-149 in cancer development, it is believed that this microRNA probably acts as a nodal point to control the apoptotic program. To date, a number of target genes have been validatedsuch as PUMAand FOXM1 [15,23]. Although most of the studies on miR-149 was conducted in other cancers withfew studies in the setting of central nervous system, a recent in vitro study suggesting blockade of AKT1 by miR-149 in glioma cell has implicated its possible involvement in clinical samples [18]. Intriguingly, She X et al. described enhanced chemo sensitivity against temozolomide in miR-149 overexpressed glioma cell [24]. Our study, which deciphered the association between low miR-149 expression and poor prognosis, corroborates these in vitro findings. Although the overall miR-149 expression status of the principal cohort is low in our study, it is interesting to find out that a small proportion of these patients still presented high level of miR-149, which is comparable to that in normal tissues. Recent study by Ding et al. highlighted the crucial role of polymorphism within pre-miR-149 in its maturation, in their study, lower levels of miR-149 production was observed in rs71428439 pre-miR-149 expressing HEK293 cells [23]. More importantly, several studies also reported the association between miR-149 polymorphisms and cancer susceptibility or prognosis [25-27]. Therefore, it is conceivable that pre-miR-149 polymorphism at least partly account for its lowered expression level in glioma patients. Whether miR-149 polymorphisms are associated with glioma progression is expected to be an interesting topic awaiting to be uncovered.
AKT/mTOR signaling plays a central role in gliomacell proliferation. Considering that AKT1 has already been identified as the target of miR-149 in glioma cells, our finding could be expected to reinforce these in vitro mechanistic studies. Importantly, the association between miR-149 and AKT/mTOR signaling in the clinical sample may represent the possible mechanism that drives the unfavorable clinical outcome of low miR-149 expression. However, it should be noted that miR-149 was also described to be related with myocardium infarction [28,29], and functional tests revealed its anti-apoptotic action in myocardium [23], which is contrary to the reports in cancers. This discrepancy may be explained by the reprogrammed signaling network in cancer cells. Due to the several mutations occurring in pro-apoptotic genes in cancer cells, the overall effect of miR-149 may be death-prone, making it as a potential tumor suppressor in glioma.
The current prognostic model is largely dependent upon the WHO histological grading and immunohistochemistry analysis. However, histological examinations are often subjective and call for rich clinical diagnostic experience of pathologists, and the false positive of immunohistochemistry also limits its diagnostic power. The application of microRNA in the clinical practice would be expected to meet the needs of clinical practitioner. MicroRNA detection in the paraffin section by the RNA in situ method might represents a more reliable diagnostic approach. Moreover, as several circulating microRNAs are emerging as the biomarkers for cancer diagnosis, investigating whether circulating miR-149 level correlates with its expression in tumor site should represent a more convenient alternative to assess the prognosis of patients. Identifying miR-149 as a prognostic marker enables the quantitative evaluation of the death risk at early diagnosis, which might be helpful to tailoring individualized treatment. To go further, concerning that we have identified low miR-149 expression is associated with poorer prognosis, it might be an effective therapeutic approach to complement miR-149 in low expression patients.
In summary, although our study population is relatively small, we demonstrate the aberrant miR-149 expression in glioma patients for the first time; more importantly, the expression status is significantly associated with the survival duration of patients. In the future, large scale studies and more detailed experimental studies are still needed to confirm the prognostic significance and the mechanism of miR-149 in glioma.
Disclosure of conflict of interest
None.
References
- 1.Ahmed R, Oborski MJ, Hwang M, Lieberman FS, Mountz JM. Malignant gliomas: current perspectives in diagnosis, treatment, and early response assessment using advanced quantitative imaging methods. Cancer Manag Res. 2014;6:149–170. doi: 10.2147/CMAR.S54726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301–308. doi: 10.1016/j.canlet.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 4.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low SY, Ho YK, Too HP, Yap CT, Ng WH. MicroRNA as potential modulators in chemoresistant high-grade gliomas. J Clin Neurosci. 2014;21:395–400. doi: 10.1016/j.jocn.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Katsushima K, Kondo Y. Non-coding RNAs as epigenetic regulator of glioma stem-like cell differentiation. Front Genet. 2014;5:14. doi: 10.3389/fgene.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, Bian EB, Li J, Li J. New advances of microRNAs in glioma stem cells, with special emphasis on aberrant methylation of microRNAs. J Cell Physiol. 2014;229:1141–1147. doi: 10.1002/jcp.24540. [DOI] [PubMed] [Google Scholar]
- 9.Palumbo S, Miracco C, Pirtoli L, Comincini S. Emerging roles of microRNA in modulating cell-death processes in malignant glioma. J Cell Physiol. 2014;229:277–286. doi: 10.1002/jcp.24446. [DOI] [PubMed] [Google Scholar]
- 10.Yan Y, Wang Q, Yan XL, Zhang Y, Li W, Tang F, Li X, Yang P. miR-10a controls glioma migration and invasion through regulating epithelial-mesenchymal transition via EphA8. FEBS Lett. 2015;589:756–765. doi: 10.1016/j.febslet.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Li X, Zhu Y, Yang P. MicroRNA-16 suppresses epithelial-mesenchymal transitionrelated gene expression in human glioma. Mol Med Rep. 2014;10:3310–3314. doi: 10.3892/mmr.2014.2583. [DOI] [PubMed] [Google Scholar]
- 12.Sun G, Yan S, Shi L, Wan Z, Jiang N, Li M, Guo J. Decreased Expression of miR-15b in Human Gliomas Is Associated with Poor Prognosis. Cancer Biother Radiopharm. 2015;30:169–73. doi: 10.1089/cbr.2014.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Y, Wei Y, Wang J, Gong K, Zhang Y, Zuo H. Correlation of microRNA-10b upregulation and poor prognosis in human gliomas. Tumour Biol. 2015;36:6249–6254. doi: 10.1007/s13277-015-3310-9. [DOI] [PubMed] [Google Scholar]
- 14.Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112:1241–1246. doi: 10.1038/bjc.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu K, Liu X, Mao X, Xue L, Wang R, Chen L, Chu X. MicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem. 2015;35:499–515. doi: 10.1159/000369715. [DOI] [PubMed] [Google Scholar]
- 16.He DX, Gu XT, Li YR, Jiang L, Jin J, Ma X. Methylation-regulated miR-149 modulates chemoresistance by targeting GlcNAc N-deacetylase/N-sulfotransferase-1 in human breast cancer. FEBS J. 2014;281:4718–4730. doi: 10.1111/febs.13012. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Rivas LG, Jerez JM, Carmona R, de Luque V, Vicioso L, Claros MG, Viguera E, Pajares B, Sanchez A, Ribelles N, Alba E, Lozano J. A microRNA signature associated with early recurrence in breast cancer. PLoS One. 2014;9:e91884. doi: 10.1371/journal.pone.0091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan SJ, Zhan SK, Pei BG, Sun QF, Bian LG, Sun BM. MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. Int J Immunopathol Pharmacol. 2012;25:871–881. doi: 10.1177/039463201202500405. [DOI] [PubMed] [Google Scholar]
- 19.Speranza MC, Frattini V, Pisati F, Kapetis D, Porrati P, Eoli M, Pellegatta S, Finocchiaro G. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget. 2012;3:723–734. doi: 10.18632/oncotarget.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73:6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 21.Sun J, Liao K, Wu X, Huang J, Zhang S, Lu X. Serum microRNA-128 as a biomarker for diagnosis of glioma. Int J Clin Exp Med. 2015;8:456–463. [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng MW, Wang LL, Hu GY. Expression of microRNA-218 and its Clinicopathological and Prognostic Significance in Human Glioma Cases. Asian Pac J Cancer Prev. 2015;16:1839–1843. doi: 10.7314/apjcp.2015.16.5.1839. [DOI] [PubMed] [Google Scholar]
- 23.Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, Liu F, Li Q, Gao J, Zhou QY, Gu DF, Li PF. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. J Biol Chem. 2013;288:26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Xiang J, Wu M, Li G. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol Rep. 2014;32:957–964. doi: 10.3892/or.2014.3318. [DOI] [PubMed] [Google Scholar]
- 25.Wei WJ, Lu ZW, Li DS, Wang Y, Zhu YX, Wang ZY, Wu Y, Wang YL, Ji QH. Association of the miR-149 Rs2292832 polymorphism with papillary thyroid cancer risk and clinicopathologic characteristics in a Chinese population. Int J Mol Sci. 2014;15:20968–20981. doi: 10.3390/ijms151120968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia L, Ren Y, Fang X, Yin Z, Li X, Wu W, Guan P, Zhou B. Prognostic role of common microRNA polymorphisms in cancers: evidence from a meta-analysis. PLoS One. 2014;9:e106799. doi: 10.1371/journal.pone.0106799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Wei M, Ren Y, Liu H, Wang M, Shi K, Jiang H. miR149 rs71428439 polymorphism and risk of clear cell renal cell carcinoma: a case-control study. Tumour Biol. 2014;35:12127–12130. doi: 10.1007/s13277-014-2517-5. [DOI] [PubMed] [Google Scholar]
- 28.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Yang D, Xie P, Ren G, Sun G, Zeng X, Sun X. MiR-106b and MiR-15b modulate apoptosis and angiogenesis in myocardial infarction. Cell Physiol Biochem. 2012;29:851–862. doi: 10.1159/000258197. [DOI] [PubMed] [Google Scholar]


