Abstract
Objective: The study aimed to investigate the function of uromodulin (UMOD) gene and its effect on inflammatory cytokines in serum of essential hypertension patients. Methods: The online database and software of computer were used for bioinformatics analysis on UMOD gene as well as the structure and function of its encoding proteins. Moreover, radioimmunoassay and enzyme linked immunosorbent assay was adopted to validate the content of urine UMOD protein of essential hypertension patients and their serum inflammatory cytokines. Results: As an alkaline and hydrophilic protein, UMOD has no transmembrane region, but it does have a signal peptide sequence. It is mainly located extracellularly, belonging to a secreted protein, whose secondary structure was based mainly on Random coil which account for 58.44%. According to function prediction, it is found that the UMOD protein has stress response which may be participate in the inflammatory reaction. It has been observed from the experiment which was designed on the basis of the correlation between inflammation reaction and essential hypertension that the content of urine UMOD protein of essential hypertension patients who is in stage I was (28.71±10.53) mg/24 h and when compared with the control group’s content (30.15±14.10 mg/24 h), the difference was not obviously; The content of urine UMOD protein of essential hypertension patients who’s in stage II and III was (18.24±6.12) mg/24 h and (9.43±3.16) mg/24 h, respectively, which were obviously lower than that of the control group (P<0.01). Additionally, the serum inflammatory cytokines, such as TNF-α, IL-6 and IL1-α content of essential hypertension patients were all markedly higher than that of control group (P<0.05). Conclusion: For essential hypertension patients, there’s a close relationship between the expression level of UMOD gene and inflammatory cytokines, which were manifested as the negative correlation between the level of the gene’s expression and inflammatory cytokines. That has certain reference value to realize the targeted treatment for essential hypertension through regulated blood pressure conversely in the view of expression level of inflammatory cytokines.
Keywords: UMOD, essential hypertension, inflammatory cytokines, gene ontology, protein function analysis
Introduction
Hypertension, sustaining high blood pressure disease, is often accompanied with systemic disease of functional change in heart, blood vessel, brain and kidney, etc. It was divided into essential hypertension and secondary hypertension. As a polygenic hereditary disease caused by the joint action of genetic and environmental factors, Essential hypertension (EH) is an independent risk factor which could bring about myocardial infarction, cardiac failure, cerebrovascular disease and chronic renal failure, and accounting for about 90% to 95% of the total number of patients with hypertension [2]. With the rapid development of molecular biology techniques and bioinformatics, discussion on EH susceptibility genes has become the hot topic of its pathogenesis research in both current and future studies. Currently, dozens of related genes of EH have been identified using candidate gene approach.
As an identified candidate gene that closely related to EH, uromodulin (UMOD) gene located on the short arm of chromosome 16p11-13 promoter region, consists of 11 exons (39777 bp). The protein, which was expressed by it also known as Tamm-Horsfall (THP), one of the mucin in the urine, was separated from the urine via salting out method by Igor Tamm and Frank Horsfall in 1950 [3]. UMOD is primarily synthesized and secreted into the urine by thick ascending limb (TAL) of kidney tubules medullary loop and epithelial cells in the initial segment of the distal convoluted tubule. The daily secretion of the healthy is about 20~70 mg. It is the most abundant protein in normal urine. However, at present, the physiological function of UMOD protein is not very clear, and there are researches indicated that the UMOD gene knockout mice has decreased urine concentration ability while the incidence of urinary tract infections as well as kidney stones increased [4,5], which conducted to the conjecture that the gene may be related to the lithangiuria formation, immune response, renal tubular epithelial cell protection. Up to now, more than 60 disease-causing mutations have been identified, and almost all of them are resulted by missense mutations. Mutation sites are mostly located No. 4, 5 and 8 exons. In addition, the vast majority of mutations occur in exon 4. All of them are involved the highly conservative base sequence in the evolution, and the tertiary structure of UMOD was destroyed by that and result in its functional loss.
In 2010, as a genome-wide association analysis (GWAS) showed, there is a significant correlation between blood pressure and the single nucleotide polymorphism of rs13333226 locus in UMOD gene promoter region, and people who carry G allelic fragments suffered lower incidence of hypertension and abnormal renal function [6-9]. Trudu et al. [10] found that after UMOD gene mutations, the expression of uromodulin increased, leading to salt-sensitive hypertension and renal damage. The research of Trudu et al. about the role of NKCC2 between urine regulation and hypertension established link of hereditary susceptibility between hypertension and chronic nephrosis. Studies of Graham LA et al. [11] have shown that the UMOD gene is the candidate gene of essential hypertension, which can be regarded as a novel drug target and provides a new way in the prevention and treatment of essential hypertension. However, as a new finding in studying essential hypertension, the specific functions or possible signal pathway of UMOD gene in the diagnosis and treatment of primary hypertension is still not clear. In order to get some functional information from the gene and protein sequential structure, this paper performed bioinformatics analysis of UMOD gene and the structure as well as the function of its proteins. The influence of UMOD gene on serum Inflammatory cytokines was investigated by the use of experiment, thus offering reference for the practical research to regulated blood pressure conversely along with the level change of inflammatory cytokines.
Materials and methods
Bioinformatics analysis of UMOD gene and encoding product
UMOD gene sequences came from Genbank (Accession: M17778.1). The ORF Finder program of NCBI was used to analyze UMOD gene open reading frame. The ProtParam, Compute pI/Mw and ProtScale software in ExPASy server was utilized to predict amino acid composition, molecular mass, isoelectric point, hydrophobicity/hydrophilicity, instability coefficient and fat coefficient etc. of UMOD protein. TMHMM software was put to use to predict transmembrane region of UMOD protein. Then, SignalP 4.1 Server tools were adopted to predict amino acid sequence signal peptide of the human UMOD encoded products. Afterwards, TargetP software was employed to predict localization of UMOD protein in cells. Hopfield neural network was used to predict the secondary structure of UMOD protein, and Protfun software and Gene Ontology database was used separately to predict the function of UMOD protein. Its interactional proteins were analyzed using STRING online database of protein interactions, and then the signaling pathways of the UMOD interactional proteins were analyzed in KEGG database.
Regulation of UMOD gene on essential hypertension patients
This study was carried out in accordance with the “Ethical Guidelines and Regulations” and the international standards for editors and authors press release.
EH case group: fifty-two EH patients who received treatment in hospital during September, 2011 to September, 2014 were enrolled as case group, among which were 28 males and 24 females, their age ranged within 54 to 80 years old and share an average age of 64.6 years old, an average blood pressure of 170±14/105±10 mm Hg as well. There are 13 essential hypertension patients who were in stage I, and 18 patients in stage II and 21 patients in stage III. All of them meet the WHO hypertension diagnosis and grading standard, and none of them suffered from disease like immunogenic, diabetes and hepatic renal dysfunction. Other 24 healthy volunteer were treat as control group, including 13 males and 11 females, their age were within 53 to 79 years old and the average age was 63.2 years old.
Content determination of UMOD protein
Sampling patients urine in 24 hours and the content of UMOD protein in it was tested through radioimmunoassay for both EH case group and control group. The kit was provided by Chinese Isotopes Company North Immunoreagent Institution, and the experimental procedures were carried out in strict accordance with the kit instructions.
Contents determination of serum inflammatory cytokines TNF-α, IL-6 and IL1-α
2 ml fasting venous blood from control group and case group were collected, respectively, and the blood was centrifuged for 10 minutes in speed of 3000 r/min, and then the supernatant was collected and preserved in -20°C for preparation. TNF-α, IL1-α and IL-6 levels in serum were detected by using enzyme-linked immunosorbent assay (ELLSA). The experimental procedures were carried out in strict accordance with the kit instructions, and the kit was purchased from Roche, Germany.
Statistical analysis
Notable difference analysis of experimental data were run via statistics software SPSS17.0, and the measurement data were written as (x̅ ± s), then the t test was used to determine. There’s statistical significance for the differences when P < 0.05.
Results
GO functional analysis of UMOD gene
Taking the function of gene and protein in cells as the measurement basis, Gene Ontology (GO) has provided three kinds of ontological categories: biological process, molecular function and cellular components. UMOD plays a vital role in biological process, molecular function and cellular components. As it shown, what was contained in Table 1 is the GO analysis results of UMOD protein in biological process, and there is where it can be observed that the UMOD protein has such functions like maintaining systemic chemical equilibrium and cellular defense, which has further explanation about the crucial role of UMOD protein played in inflammatory reaction.
Table 1.
The GO analysis results of UMOD gene
| Acc | Taxon | Accession | Name |
|---|---|---|---|
| UMOD | Homo sapiens | GO:0048878 | chemical homeostasis |
| UMOD | Homo sapiens | GO:0007588 | excretion |
| UMOD | Homo sapiens | GO:0010033 | response to organic substance |
| UMOD | Homo sapiens | GO:0072218 | metanephric ascending thin limb development |
| UMOD | Homo sapiens | GO: 072233 | metanephric thick ascending limb development |
| UMOD | Homo sapiens | GO:2000021 | regulation of ion homeostasis |
| UMOD | Homo sapiens | GO:0072221 | metanephric distal convoluted tubule development |
| UMOD | Homo sapiens | GO:0006968 | cellular defense response |
| UMOD | Homo sapiens | GO:0008285 | negative regulation of cell proliferation |
| UMOD | Homo sapiens | GO:1990266 | neutrophil migration |
| UMOD | Homo sapiens | GO:0007159 | leukocyte cell-cell adhesion |
| UMOD | Homo sapiens | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules |
Physicochemical properties and structure analysis of UMOD of protein
Such bio-information software as ProtParam, Compute pI/Mw, ProtScale and TMHMM were used to analyze the physicochemical properties and structure of UMOD protein. The results indicated the atomic composition of UMOD protein was C3011H4654N832O952S63; relative molecular weight: about 69760.86 Da; theoretical isoelectric point: 5.0; the unstable coefficient was 40.53; the fat coefficient was 70.69; total average hydrophobicity was -0.111. No typical spiral transmembrane region in UMOD protein, and the possibility to function outside the cell was nearly 100%. Thus, it was speculated as an alkaline and unstable hydrophilia protein which contains a signal peptide sequence, located extracellularly, belonging to a secreted protein. In its secondary structure, Alpha helix accounted for 23.44%, extended strand accounted for 18.12%, Random coil accounted for 58.44%.
Prediction of UMOD protein function
The prediction results of UMOD protein which obtained through software Protfun were as followed. Functional category displayed UMOD protein had functions of purines and pyrimidines (Prob = 0.541, Odds = 2.227). Enzyme/nonenzyme displayed that UMOD protein was lyase (Prob = 0.057, Odds = 1.211). Gene Ontology category displayed that UMOD protein had stress response (Prob = 0.188, Odds = 2.140).
Analysis of UMOD protein cross-linking function
The interacting proteins of UMOD were analyzed using STRING protein interaction online database, and the results were show in Figure 1. It was indicated that what interacting with UMOD protein were mainly ten kinds of protein, they are IGK@, MMP8, ENSG00000226979, PKHD1, HNF1B, IL4R, IFNGR1, SH3GL1, MCL1 and LTA. Molecular signaling pathway analysis showed that these above mentioned proteins participated in inflammatory bowel disease, NF-κB signaling pathway, TNF signaling pathway, cytokines-cytokine receptor interaction, PI3K-Akt signaling pathway, etc. The results of present researches show that both inflammatory bowel disease and TNF signaling pathway were relative to the level of TNF-α [12]; the activated NF-κB canonical signaling gave rise to the increased level of IL-6 [13] and the restrained of it could result in the decreased level of IL-6 [14], in addition, there’s a close link between the signaling pathway of cytokines-cytokine receptor interaction and the level of TNF-α [15]; the level of TNF-α and IL-6 deduced when P13k-Akt signaling pathway was activated [16]; when it combined with the prediction results of UMOD protein function, the important role UMOD protein played in the inflammatory response should be speculated.
Figure 1.
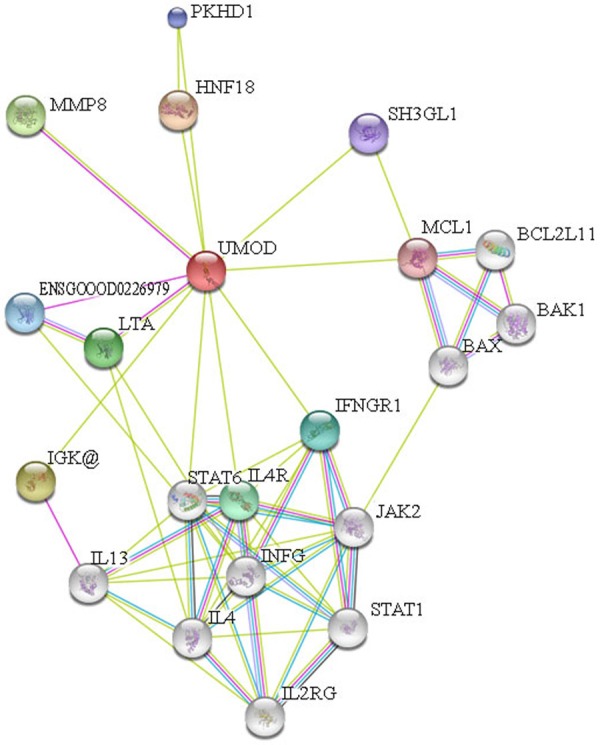
Analysis figure of UMOD protein cross-linking function.
Detection of urine UMOD protein in essential hypertension patients
The detection results of urine UMOD protein in essential hypertension patients were shown in the Table 2. It can be observed from it that the content of urine UMOD protein of essential hypertension patients who is in stage I was (28.71±10.53) mg/24 h and when compared with the control group’s content (30.15±14.10 mg/24 h), the difference was not significant; The content of urine UMOD protein of essential hypertension patients who are in stage II and III was (18.24±6.12) mg/24 h and (9.43±3.16) mg/24 h, respectively, which were obviously lower than that of the control group (P<0.01). Besides, the content of urine UMOD protein of patients in stage II and III has dramatic decrease when comparing to that of control group and patients in stage I (P<0.01).
Table 2.
The detection results of urine UMOD protein in Essential hypertension patients
| Group | Cases | The content of UMOD protein mg/24 h | |
|---|---|---|---|
| Control group | 24 | 30.15±14.10 | |
| EH group | Stage I | 13 | 28.71±10.53 |
| Stage II | 18 | 18.24±6.12**,ΔΔ | |
| Stage III | 21 | 9.43±3.16**,ΔΔ |
Note: when comparing to control group the
means P<0.01;
comparing to stage I the
means P<0.01.
Content differences of serum inflammatory cytokines in essential hypertension patients
The level of serum inflammatory cytokines TNF-α, IL-6 and IL1-α from both EH case group and control group were shown in the Figure 2. It can be noticed that the content of serum inflammatory cytokines TNF-α, IL-6 and IL1-α of EH patients was obviously higher than that of the control group (P < 0.05). Combining the functional analysis for UMOD gene and its protein which mentioned in the last chapter, the conclusion can be drawn as the UMOD gene brings enormous influence to EH patients’ multiple inflammatory cytokines during its expression.
Figure 2.
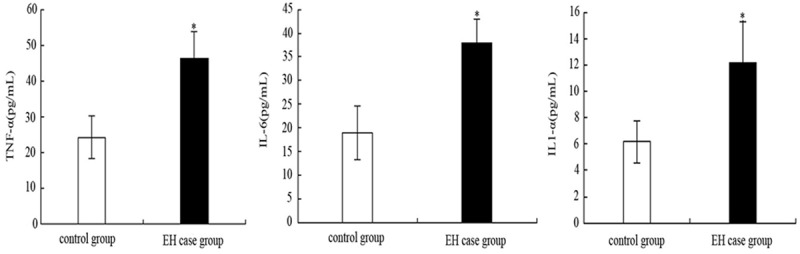
The level of TNF-α, IL-6 and IL1-α in serum from both EH case group and control group.
Discussion
Essential hypertension is one of the common and frequently-occurring diseases which has severely damaged people’s health, it is the disease of blood under supplied or relatively under supplied in important organs like heart, brain and kidney, which could blame it on systemic arteriolar constriction and the increased blood viscosit caused by many reasons as psychic factors. The blood pressure got rise to satisfy the supply requirement of organs through the blood regulation mechanism. The pathology of EH was not clear by now, and the excavation of EH susceptibility gene developed rapidly. As a new-found candidate gene with a high relevance to EH [17], UMOD gene has provide a new research direction for the prevention and treatment of EH.
Essential hypertension is a systemic metabolic disorder disease. In a sense, it is also a low-grade systemic inflammatory reaction, taking the form of the abnormal inflammatory cytokines and the activation of inflammatory signaling pathways [18]. Inflammation factors play a vital role in the occurrence, development and outcome of hypertension. What’s worse, they are closely related to the occurrence and development of many cardiovascular diseases [19], while hypertension will promote the inflammation response through its bloodstream biomechanical stimulation [20]. In this paper, bioinformatics methods were used for prediction and analysis of UMOD protein structure and hence its function. We conclude that UMOD protein was a secreted protein and the proportion of Random coil in its secondary structure was the highest, reaching 58.44%. Additionally, after the functional prediction and analysis of UMOD protein, it turns out that it may be involved NF-κB signaling pathway, TNF signaling pathway, cytokine-cytokine receptor interactions and other signaling pathways. As a series of present researches about inflammatory cytokines has proved, a number of molecules in each stage of inflammatory response, including: TNF-α, IL-1β, IL-2, IL-6, IL-8, IL-12, etc. were regulated by these signaling pathways. The activation of signaling pathway can strengthen transcription of inflammation factors and enhance inflammatory signals [21]. In this study, after the note analysis UMOD function through GO database, it has been found that UMOD protein takes part in cellular defense, which means UMOD gene play an important role in the inflammatory response. Several studies indicated that it is obviously that some cytokines and inflammatory cytokines have close relevance to the occurrence as well as the development of hypertension. Inflammatory cytokines, a biologically active peptide which were synthesized and excreted after the injury or stimulation of immunocyte and non immune cells, had the central function in inflammatory reaction [22]. At present, the report about regulating the level of inflammatory cytokines to control blood pressure is quite rarely. In this study, the EH patients’ level of inflammatory cytokines is obviously higher than that of control group, which is in accordance with the domestic research finding that the level of inflammatory cytokines in serum of EH patients increased with the rising of blood pressure. It is such inflammatory cytokines as TNF-α, IL1, IL-6 that functioned among variety of them [23]. Serum inflammatory cytokines TNF-α and IL-1 can promote the proliferation of smooth muscle cells from a variety of ways, thereby enabling calcium ions within the smooth muscle cells rise quickly, causing the constriction of blood vessels and then the elevation of blood pressure [24]. Serum IL-6 can change the rheological properties of leukocytes, make them adhere to vascular endothelium more easily, increase the vascular resistance and increase the number of platelets. In addition, it may also interact with angiotensin II in the blood vessels and elevate the blood pressure [25,26]. A study of one hundred and ninety six healthy people conducted by Bautista et al. [27] found that IL-1 and TNF-α levels was positively correlated to systolic blood pressure and diastolic blood pressure, suggesting that IL-1 and TNF-α may be the independent risk factors of hypertension and inflammatory reaction was indirectly involved in the pathogenesis of essential hypertension.
The results of this study show that the content of urine UMOD protein of essential hypertension patients who is in stage I was (28.71±10.53) mg/24 h and when compared with the control group’s content (30.15±14.10 mg/24 h), the difference was not significant; The content of urine UMOD protein of essential hypertension patients who’s in stage II and III was (18.24±6.12) mg/24 h and (9.43±3.16) mg/24 h, respectively, which were obviously lower than that of the control group (P<0.01). Besides, the content of TNF-α, IL-6 and IL1-α in serum of EH patients has dramatic increasing when comparing to that of control group (P<0.05).
To sum up, this study predicated UMOD gene and its encoding proteins’ structure and function using bioinformatics methods, meanwhile, the negative correlation between the expression level of UMOD gene and the level of inflammatory cytokines were verified via the valid experimental data. All of that presaged that through simple antibacterial and anti-inflammatory medicine we can regulate EH conversely from the view of level of inflammatory cytokines, which has reference value for the prevention and treatment of EH.
Acknowledgements
The authors thank the Medical Engineering Technology & Data Mining Institute of Zhengzhou University for their invaluable technical assistance. Moreover, the authors would like to thank Zhengzhou Central Hospital for providing us the experimental base. This work was supported by grant: 20120900, 20130459 and 132102310332.
Disclosure of conflict of interest
None.
References
- 1.Chobanian AV. Mixed messages on blood pressure goals. Hypertension. 2011;57:1039–1040. doi: 10.1161/HYPERTENSIONAHA.111.170514. [DOI] [PubMed] [Google Scholar]
- 2.Wang GY, Wang YH, Xu Q, Tong WJ, Qiu CC, Fang MW, Wang J, Gu ML, Zhang YH. Genetic gene polymorphisms and interactions of hypertension in Mongolian people. Chin J Public Health. 2006;22:1332–1333. [Google Scholar]
- 3.Tamm I, Horsfall FL Jr. Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc Soc Exp Biol Med. 1950;74:106–108. [PubMed] [Google Scholar]
- 4.Bachmann S, Mufig K, Bates J, Welker P, Geist B, Gross V, Luft FC, Alenina N, Bader M, Thiele BJ, Prasadan K, Raffi HS, Kumar S. Renal effects of Tamm-Horsfall protein (uromodulin) deficiency in mice. Am J Physiol Renal Physio. 2005;288:F559–F567. doi: 10.1152/ajprenal.00143.2004. [DOI] [PubMed] [Google Scholar]
- 5.Bates JM, Rattl HM, Prasadan K, Mascarenhas R, Laszik Z, Maeda N, Hultgren SJ, Kumar S. Tamm-Horsfall protein knockout mice are more prone to urinary tract infection: rapid communication. Kidney Int. 2004;65:791–797. doi: 10.1111/j.1523-1755.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- 6.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Paré G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Gene. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köttgen A, Hwang SJ, Larson MG, Van Eyk JE, Fu Q, Benjamin EJ, Dehghan A, Glazer NL, Kao WH, Harris TB, Gudnason V, Shlipak MG, Yang Q, Coresh J, Levy D, Fox CS. Uromodulin levels associate with a common UMOD variant and risk for incident CKD. J Am Soc Nephrol. 2010;21:337–344. doi: 10.1681/ASN.2009070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjögren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JM, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Völzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJ, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P Global BPgen Consortium. Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF. Global BP gen Consortium. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team. Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham LA, Padmanabhan S, Fraser NJ, Kumar S, Bates JM, Raffi HS, Welsh P, Beattie W, Hao S, Leh S, Hultstrom M, Ferreri NR, Dominiczak AF, Graham D, McBride MW. Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension. 2014;63:551–558. doi: 10.1161/HYPERTENSIONAHA.113.01423. [DOI] [PubMed] [Google Scholar]
- 12.Liu TT. The Effect of Pioglitazone on the level of serum TNF-α and hs-CRP in mice inflammatory bowel disease caused by DSS. Shanxi Medical University. 2014 [Google Scholar]
- 13.Gao YN, Guan XL, Liu YZ, editors. The Increasing Expression of IL-6 Caused by Avian Leukosis Virus Subgroup J through the NF-kB signaling pathway; Proceeding of 9th Annuel Meeting of Chinese Society for Immunology; 2014. [Google Scholar]
- 14.Piao MZ. A Study of the Quyin granule Regulates Human Beta Defen sin-2 on Human Keratinocyte in Vitro. Bei Jing University of Chinese Medicine. 2014 [Google Scholar]
- 15.Bi JH. The Study on the Effect of High Calorie Intake on the Directional Differentiation of Mesenchymal Stem Cells in Mice Marrow. Peking Union Medical College. 2014 [Google Scholar]
- 16.Xu XN, Niu ZR, Wang SB, Chen YC, Gao L, Fang LH, Du GH. Effect and mechanism of total flavonoids of bugloss on rats with myocardial ischemia and reperfusion injury. Yao Xue Xue Bao. 2014;49:875–81. [PubMed] [Google Scholar]
- 17.Green L. JNC 7 express: New thinking in hypertension treatment. Am Fam Physician. 2003;68:228–230. [PubMed] [Google Scholar]
- 18.Sesso HD, Buring JE, Rifai N, Blake GJ, Gaziano JM, Ridker PM. C-reactive protein and the risk of developing hypertension. J Am Med Ass. 2003;290:2945–2951. doi: 10.1001/jama.290.22.2945. [DOI] [PubMed] [Google Scholar]
- 19.Badr K, Wainwright CL. Inflammation in the cardiovascular system: Here, there and everwhere. Cart Opin Pharmacol. 2004;4:107–109. doi: 10.1016/j.coph.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Blake GL, Rifai N, Bueing JE, Ridker PM. Blood pressure, C-reactive protein, and risk of future cardiovascular events. Circulation. 2003;108:2993–2999. doi: 10.1161/01.CIR.0000104566.10178.AF. [DOI] [PubMed] [Google Scholar]
- 21.Blackwell TS, Christman JW. The role of nuclear factor-kappa B in cytokine gene regulation. Am J Respir Cell Mol Biol. 1997;17:3–9. doi: 10.1165/ajrcmb.17.1.f132. [DOI] [PubMed] [Google Scholar]
- 22.Guo XP. Analysis on the effect of Irbesartan in inflammatory factors of patients with hypertension. Chin Med Guides. 2013;9:61–62. [Google Scholar]
- 23.Chen HT, Hou WH, Miao YG, et al. The Clinical Significance of serum inflammatory cytokines TNF-α, IL-6, and hs-CRP in essential hypertension patients. J Shanxi Med. 2015 [Google Scholar]
- 24.Wang AL, Yu YX, Xu Y. Studies of the association between angiotensinogen gene regulation and cytokines in essential hypertension. Yi Chuan Xue Bao. 2003;30:978–982. [PubMed] [Google Scholar]
- 25.Yang MX, Tian YX. Effect of Zhitiaokang (ZTK) capsule on the IL-6 in the blood serum of hyperlipemia rats. Chin J Basic Med Tra Chin Med. 2003;9:12–13. [Google Scholar]
- 26.Charnontin B. The best of hypertension 2005. Arch Mal Coeur Vaiss. 2006;99:35–41. [PubMed] [Google Scholar]
- 27.Bautista LE, Vera LM, Arenas IA, Gamarra G. Independent association between inflammatory markers, (C-reactive protein, inter leukin-6, and TNF-alpha) and essential hypertension. J Hum Hypertens. 2005;19:149–154. doi: 10.1038/sj.jhh.1001785. [DOI] [PubMed] [Google Scholar]


