Abstract
Long non-coding RNAs (lncRNAs) have been shown to play key roles in cancer development and progression. In this study, we focused on lncRNA HOTTIP and investigated its expression pattern, clinical significance, and biological function in osteosarcoma (OS). In the present study, lncRNA HOTTIP expression in OS tissues was examined and its correlation with clinicopathological features and patient prognosis was analyzed. In vitro assays were performed to understand the biological roles of lncRNA HOTTIP in OS progression. In the study, we found that HOTTIP expression was up-regulated in OS tissues, and correlated with advanced clinical stage and distant metastasis. OS patients with high HOTTIP expression level had poorer overall survival than those with low HOTTIP expression. Multivariable Cox proportional hazards regression analysis suggested that increased HOTTIP expression was an independent prognostic factor of overall survival in OS patients. Moreover, the results of in vitro assays showed that the suppression of HOTTIP in OS cells significantly reduced cell proliferation, migration and invasion ability. Our study demonstrated that lncRNA HOTTIP play critical roles in OS progression and could represent a novel prognostic marker and potential therapeutic target in OS patients.
Keywords: Osteosarcoma, long non-coding RNAs, HOTTIP, prognosis
Background
Osteosarcoma (OS) is the most frequent malignant bone tumor in children and adolescents, comprising 2.4% of all malignancies in pediatric patients [1]. The 5-year survival rate of OS patients has significantly improved over the past decades to approximately 60-70% since the introduction of combinatorial chemotherapy [2]. However, a significant proportion of OS patients still respond poorly to chemotherapy, and they have a risk of local relapse or distant metastasis even after curative resection of the primary tumor [3]. Therefore, there is an urgent need to identify biomarkers which could predict metastasis and prognosis for OS patients to improve the individual treatment.
Long non-coding RNAs (lncRNAs) are evolutionarily conserved non coding RNAs that are longer than 200 nucleotides in length with no protein-coding capacity [4]. Emerging evidence suggested that lncRNAs have important roles in diverse biological processes, such as cell growth and apoptosis, as well as in cancer progression and metastasis [5,6]. For example, Gupta et al. showed that lncRNA HOTAIR expression was increased in primary breast tumors, and HOTAIR expression level in breast tumors was a powerful predictor of eventual metastasis and death [7]. Ren et al. indicated that lncRNA MALAT1 was up-regulated in prostate cancer and correlated with Gleason score, prostate specific antigen, tumor stage and castration resistant prostate cancer. Moreover Decreased expression of MALAT1 could inhibit prostate cancer cell proliferation, migration and invasion [8]. Sun et al. revealed that lncRNA GAS5 expression was markedly down-regulated in gastric cancer and associated with tumor size and pathologic stage, enforced expression of GAS5 was demonstrated to inhibit cell proliferation and induce cell apoptosis [9]. Ma et al. demonstrated that lncRNA LET was down-regulated in gallbladder cancer and patients with low expression of lncRNA LET have significantly poorer prognosis, ectopic expression of LncRNA LET could suppress gallbladder tumor growth [10]. However, to our knowledge the clinical significance and biological function of lncRNA HOTTIP in OS remains unclear.
In the present study, we investigated the expression level of HOTTIP in OS tissues and adjacent non-tumor tissues, as well as analyzed the correlation between HOTTIP expression levels, clinicopathological features, and OS patients’ prognosis. Furthermore, we investigated the biological function of HOTTIP on OS cell proliferation, migration and invasion during OS progression in a series of in vitro assays. Our results suggested that lncRNA HOTTIP could represent a novel indicator of poor prognosis and might be a potential therapeutic target in OS patients.
Materials and methods
Patients and specimens
A total of 68 osteosarcomas tissues and matched adjacent non-tumor tissues were obtained from patients who underwent surgery in the Department of Trauma and Orthopedics, Shanghai First People’s Hospital, Shanghai Jiao Tong University from 2005 to 2009. None of the patients received radiotherapy or chemotherapy before surgery. After surgical resection, OS tissues and matched adjacent non-tumor tissues were collected and immediately stored in liquid nitrogen until use. All OS tissues and paired adjacent non-tumor tissues were confirmed by a senior pathologist. This study was approved by the Research Ethics Committee of Shanghai Jiao Tong University, Shanghai, China. Written informed consent was obtained from all of the patients. All specimens were handled and made anonymous according to the ethical and legal standards.
Cell culture and cell transfection
The human osteosarcoma cell lines (MG-63 and HOS) were purchased from the American Type Cell Culture Collection (ATCC). All cells were maintained in DMEM, which was supplemented with 10% fetal bovine serum (FBS, Invitrogen), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) in an incubator at 37°C with 5% CO2.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from tissues and cell lines using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The expression levels of HOTTIP was determined by quantitative real-time PCR (qRT-PCR) using the SYBR® Green (TaKaRa) dye detection method on ABI Step One PCR instrument, with GAPDH as an internal control. The primers are as follows: HOTTIP, forward primer: 5’-CCTAAAGCCACGCTTCTTTG-3’; reverse primer: 5’-TGCAGGCTGGAGATCCTACT-3’; GAPDH, forward primer 5’-GTCAACGGATTTGGTCTGTATT-3’ and reverse primer: 5’-AGTCTTCTGGGTGGCAGTGAT-3’. Comparative Ct method was used for quantification of the transcripts. The fold-change for target genes normalized by internal control was determined by the formula 2-ΔΔCt.
MTT assay
MTT assay was used to detect cell proliferation. Briefly, cells were seeded at a density of 2 × 103 cells per well in 100 μl culture medium in 96-well culture plates. The proliferation of cell was assessed after transfection for 24, 48, 72 and 96 h. One-tenth volume of 5 mg/ml MTT was added to each well, and the plate was further incubated at 37°C for another 2 h, next, the medium was removed and the formazan crystals formed were dissolved in 100 μl dimethyl suophoxide with shaking for another 10 min. Absorbance was then measured by a multiwell spectrophotometer (BioTek) at 490 nm.
Scratch assay
Scratch assay was used to detect cell migration. Briefly, cells were seeded into 12-well plates. When cell confluence reached approximately 80%, wounds were created in the confluent cells using a 200 μl pipette tip. The cells were then rinsed with medium to remove free-floating cells and debris. Medium was added, and the culture plates were incubated at 37°C. Different stages of wound healing were observed along the scrape line, and representative scrape lines were imaged.
Transwell invasion assay
Transwell invasion assay was used to determine cell invasion, Matrigel-coated invasion chambers (8 μm, Costar) were used according to the manufacturer’s protocol. Briefly, cells were resuspended (1 × 105 cells/well) in 200 μl serum free medium and transferred to the upper chamber of the Matrigel-coated inserts, culture medium containing 10% FBS was placed in the bottom chamber. The cells were incubated for 48 h at 37°C. After incubation, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed and stained using 0.1% crystal violet. Numbers of invaded cells were counted in five randomly selected fields under a microscope (Nikon).
Statistical analysis
Statistical analyses were performed using SPSS version 18.0. All data are presented as the mean ± SD and differences between groups were analyzed using Student’s t test or chi-square test analysis. Overall survival curves were plotted following the Kaplan-Meier method, and the log-rank test was used to compare the survival in the different groups. A Cox proportional hazards regression analysis was used for univariate and multivariate analyses of prognostic values. P<0.05 was considered statistically significant.
Results
HOTTIP is up-regulated in OS and correlated with clinicopathological features
We analyzed the expression levels of HOTTIP in 68 pairs of OS tissues and adjacent non-tumor tissues. As revealed by qRT-PCR, HOTTIP expression was significantly increased in OS tissues compared with adjacent non-tumor tissues (P<0.05, Figure 1A). Next, the clinicopathological significance of HOTTIP expression in OS was analyzed. We divided the 68 OS patients into a high expression group (n=34) and a low expression group (n=34) according to the median value of HOTTIP expression levels in OS tissues. As shown in Table 1, high HOTTIP expression levels were correlated with advanced clinical stage and distant metastasis (P<0.05, Table 1), but not with other clinicopathological features such as gender, age, tumor size and anatomic location (P>0.05, Table 1).
Figure 1.
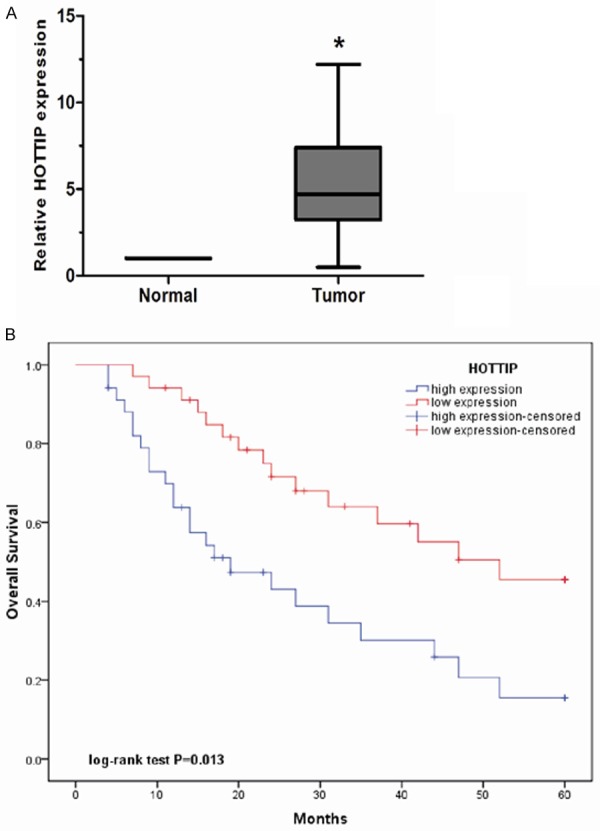
Expression of HOTTIP was up-regulated in OS. A. The relative expression of HOTTIP was measured by qRT-PCR in 68 OS tissues and paired adjacent non-tumor tissues. The expression of HOTTIP was calculated with the 2-ΔΔCt method using GAPDH levels for normalization. B. Kaplan-Meier plots showed overall survival according to the level of HOTTIP in OS patients who underwent surgery. The data represent the mean ± SD of three independent experiments. *P<0.05.
Table 1.
Association of HOTTIP expression with clinicopathological features of OS patients
| Clinicopathological features | Group | Total | HOTTIP expression | P value | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Gender | Male | 37 | 20 | 17 | 0.465 |
| Female | 31 | 14 | 17 | ||
| Age (years) | <25 | 41 | 23 | 18 | 0.215 |
| ≥25 | 27 | 11 | 16 | ||
| Tumor size (cm) | <8 cm | 46 | 26 | 20 | 0.120 |
| ≥8 cm | 22 | 8 | 14 | ||
| Anatomic location | Tibia/femur | 51 | 28 | 23 | 0.161 |
| Elsewhere | 17 | 6 | 11 | ||
| Clinical stage | IIA | 30 | 21 | 9 | 0.003 |
| IIB/III | 38 | 13 | 25 | ||
| Distant metastasis | Absence | 54 | 31 | 23 | 0.016 |
| Presence | 14 | 3 | 11 | ||
Prognostic values of HOTTIP expression in OS
To evaluate the prognostic value of HOTTIP in OS, survival curves were calculated by Kaplan-Meier method and compared by the log-rank test. Our data showed that OS patients with high HOTTIP expression had a significantly shorter overall survival than those with low HOTTIP expression (P<0.05, Figure 1B). Univariate and multivariate analyses were performed to identify the impact of HOTTIP expression and other clinicopathological features on prognosis of OS patients. Univariate analysis showed that HOTTIP expression, clinical stage and distant metastasis were significantly correlated with overall survival of OS patients (P<0.05, Table 2). Moreover, HOTTIP expression, clinical stage and distant metastasis were independent predictors for overall survival of OS patients in multivariate analysis (P<0.05, Table 2). These data indicated that HOTTIP expression represent a powerful independent prognostic factor for overall survival of OS patients and increased expression of HOTTIP might play key roles in the progression of OS.
Table 2.
Univariate and multivariate analysis of overall survival in OS patients
| Clinicopathological features | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Hazard ratio | 95% CI | P | Hazard ratio | 95% CI | P | |
| Gender | 0.936 | 0.641-2.513 | 0.482 | |||
| Male vs. Female | ||||||
| Age (years) | 1.214 | 0.753-2.906 | 0.376 | |||
| ≥25 vs. <25 | ||||||
| Tumor size | 2.427 | 0.815-5.186 | 0.043 | |||
| ≥8 cm vs. <8 cm | ||||||
| Anatomic location | 0.864 | 0.572-2.356 | 0.294 | |||
| Elsewhere vs. Tibia/femur | ||||||
| Clinical stage | 3.784 | 2.417-7.768 | 0.002 | 3.271 | 2.293-6.579 | 0.005 |
| IIB/III vs. IIA | ||||||
| Distant metastasis | 4.726 | 1.943-9.675 | 0.004 | 4.375 | 1.801-8.739 | 0.009 |
| Presence vs. Absence | ||||||
| HOTTIP | 3.018 | 1.487-7.241 | <0.001 | 2.887 | 1.367-7.061 | 0.007 |
| High vs. Low | ||||||
HOTTIP silencing inhibits OS cell proliferation, migration and invasion in vitro
To investigate the functional role of HOTTIP in OS, we constructed specific HOTTIP siRNAs (siR-HOTTIP) to silence HOTTIP expression in MG-63 and HOS cells. The transfection efficiency of the MG-63 and HOS cells was determined by qRT-PCR (P<0.05, Figure 2A). Cell proliferation assay showed that down-regulated expression of HOTTIP significantly de-creased the proliferation ability of MG-63 and HOS cells compared to their negative controls (P<0.05, Figure 2B). Furthermore, the effects of HOTTIP on OS cell migration and invasion were tested. The scratch assay showed that the decreased expression of HOTTIP significantly inhibited cell migration in MG-63 and HOS cells (P<0.05, Figure 2C). As well as scratch assay, transwell invasion assay showed that the knockdown of HOTTIP decreased the invasive ability of MG-63 and HOS cells (P<0.05, Figure 2D). Taken together, these results demonstrated that HOTTIP expression was involved in the progression and development of OS.
Figure 2.
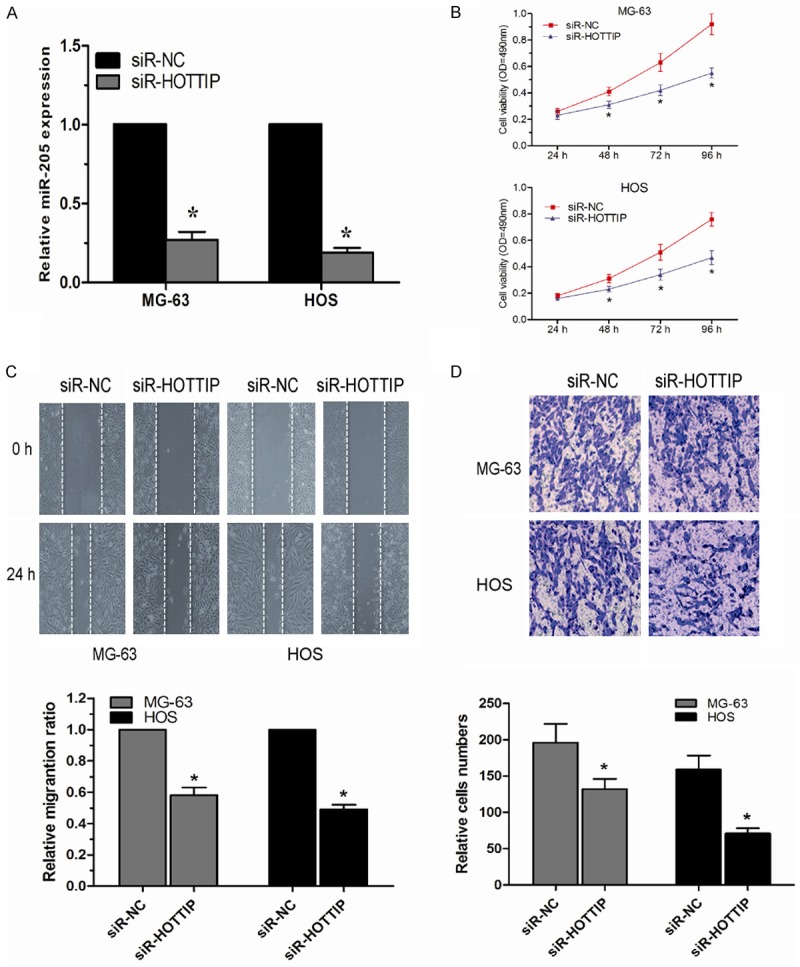
The knockdown of HOTTIP inhibited OS cell proliferation, migration and invasion. A. qRT-PCR revealed that HOTTIP expression was efficiently knocked down by treatment with siR-HOTTIP in MG-63 and HOS cells. B. Suppression of HOTTIP in MG-63 and HOS cells significantly reduced their proliferative capacities, as determined by MTT assay. C. Knockdown of HOTTIP reduced migration capacities of MG-63 and HOS cells, as determined by scratch assay. D. Decreased expression of HOTTIP inhibited invasion abilities of MG-63 and HOS cells, as determined by transwell invasion assay. The data represent the mean ± SD of three independent experiments. *P<0.05.
Discussion
The prognosis of OS is still poor; therefore, understanding the mechanisms underlying OS pathogenesis may help yield novel biomarkers for early detection and treatment [11]. Recent studies suggested that dysregulated expression of lncRNAs in OS progression and may independently predict patient outcome [12,13]. In the present study, our data indicated that lncRNA HOTTIP was associated with OS progression and OS patient’s outcome.
HOTTIP is an antisense non-coding transcript located at the distal end of HOXA gene cluster [14]. HOTTIP has been shown to be associated with the WDR5/MLL complex to drive the H3K4 methylation and transcriptional activation of the distal HOXA genes during embryonic development [15]. Recent studies demonstrated that HOTTIP was involved in the progression of cancers. For example, Quagliata et al. showed that HOTTIP was significantly up-regulated in hepatocellular carcinoma (HCC); high expression of HOTTIP was associated with metastasis and poor survival of HCC patients [16]. Tsang et al. revealed that HOTTIP was a novel oncogenic lncRNA, which negatively regulated by miR-125b in HCC [17]. Li et al. found that HOTTIP was up-regulated in pancreatic ductal adenocarcinoma (PDAC) and promoted cell proliferation, invasion and chemoresistance by modulating HOXA13 expression, indicating that HOTTIP/HOXA13 axis is a potential therapeutic target and molecular biomarker for PDAC [18]. However, to our knowledge, no studies have investigated the clinical significance or biological role of HOTTIP in OS progression.
In this study, we found that HOTTIP expression levels were higher in OS tissues than in adjacent non-tumor tissues, and elevated HOTTIP expression was associated with advanced clinical stage and distant metastasis, indicating that HOTTIP overexpression may promote an aggressive phenotype of OS. Furthermore, we analyzed the correlation of HOTTIP expression with prognosis of OS patients; we found that OS patients with high HOTTIP expression had a shorter overall survival than those with low HOTTIP expression. More importantly, multivariate analysis showed that high HOTTIP expression was an unfavorable prognostic factor for OS patients. These results highlight the clinical significance of HOTTIP in OS patients and imply a potentially critical role for HOTTIP in predicting the progression of OS patients. To further understand the mechanism of HOTTIP in OS progression, in vitro experiments were conducted. We found that knock-down HOTTIP expression could inhibit proliferation, migration and invasion capability of OS cells compared with the negative control group, suggesting that increased HOTTIP expression could promote the metastasis capability of OS cells.
In summary, we demonstrated that lncRNA HOTTIP was up-regulated in human OS tissues and could be considered an independent prognostic factor in OS patients. Knock-down expression of HOTTIP could inhibit OS cell proliferation, migration and invasion in vitro. These results indicated HOTTIP could be a promising biomarker and a therapeutic target in the treatment of OS.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:959248. doi: 10.1155/2011/959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190:2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, Quan ZW. Long noncoding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2015;54:1397–406. doi: 10.1002/mc.22215. [DOI] [PubMed] [Google Scholar]
- 11.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 12.Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, Wang H, Zhang R, Liu Y. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol Cells. 2015;38:432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–2315. doi: 10.7314/apjcp.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 14.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T. Long noncoding RNA HOTTIP HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. Long noncoding RNA HOTTIP is frequently upregulated in--hepatocellular carcinoma and is targeted by tumour suppressive miR125b. Liver Int. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


