Abstract
Long non-coding RNA (lncRNA) has an important role in carcinoma progression and prognosis. However, little is known about the pathological role of lncRNA HOTTIP (HOXA transcript at the distal tip) in colorectal cancer (CRC) patients. This study attempted to investigate the association of lncRNA HOTTIP expression with progression and prognosis in CRC patients. LncRNA HOTTIP expression was measured in 156 CRC tissues and 21 adjacent non-malignant tissues using qRT-PCR. In present study, our results indicated that lncRNA HOTTIP was highly expressed in CRC compared with adjacent non-malignant tissues (P<0.001), and positively correlated with T stage (T1-2 vs. T3-4, P = 0.001), clinical stage (I-II stages vs. III-IV stages, P = 0.003), and distant metastasis (absent vs. present, P = 0.014) in CRC patients. Furthermore, we also observed that increased lncRNA HOTTIP expression was an unfavorable prognostic factor in CRC patients (P = 0.001), regardless of T stage, distant metastasis and clinical stage. Finally, overexpression of lncRNA HOTTIP was supposed to be an independent poor prognostic factor for CRC patients through multivariate analysis (P = 0.017). In conclusion, lncRNA HOTTIP overexpression maybe serves as an unfavorable prognosis predictor for CRC patients. However, a further larger sample size investigation is needed to support our results.
Keywords: lncRNA, HOTTIP, colorectal cancer, prognosis
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer deaths in the world [1]. The prognosis for early stage CRC (Dukes’ Stages A and B) is superior, with a 5-year survival estimate of 97%. Unfortunately, about 60% of colorectal cancer patients are first diagnosed at an advanced stage (Dukes’ Stages C and D) with an expected 5-year survival rate of 5% [2-4]. Colorectal carcinogenesis is a complicated, multistep and multifactorial progress which is caused by the interaction of many factors such as dietary, lifestyle and genetic susceptibility [5,6]. Therefore, cancer screening and early detection have major importance in the survival of CRC patients. Identification of novel and improved markers is of great clinical value for the diagnosis and treatment of CRC.
Recently, many studies highlighted a number of long non-coding RNAs (lncRNAs) play essential roles in carcinogenesis, and suggested that these genes might be used as biomarkers in carcinoma [7-9]. LncRNAs comprise the mainstream of transcripts that are larger than 200 nucleotides (nt) and not translated into proteins [10], and also show emerging roles in the regulation of critical cellular functions, including transcriptional, posttranscriptional, and epigenetic mechanisms of gene regulation [7,11-13]. The HOTTIP lncRNA, located at the 5’ end of the HOXA cluster, was recently functionally characterized [14]. Recently, it was reported by Quagliata et al that HOTTIP is a negative prognostic factor in hepatocellular carcinoma (HCC) patients, and increased HOTTIP expression was associated with enhanced HCC metastasis [15]. To date, however, little is known about the significance of HOTTIP expression and CRC prognosis.
In the present study, we investigated expression level of lncRNA HOTTIP in human CRC tissue, and then explored the association between HOTTIP expression and clinicopathological characteristics. Our results suggest that HOTTIP may represent a novel indicator of poor prognosis and may be a potential target for the diagnosis and gene therapy of CRCs.
Materials and methods
Sample collection
156 freshly frozen colorectal cancer samples and 21 adjacent non-malignant samples were obtained from Department of general surgery, Affiliated Cancer Hospital of Zhengzhou University, between January 2010 and May 2012. All samples had been collected before any kind of therapeutic measures, and fresh samples were immediately preserved in liquid nitrogen. None of the patients received treatment prior to radical surgical treatment. The median duration of follow-up time was 46 months (range, 33-65 months). Written informed consent was obtained from all participants. The study protocol was approved by the Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University in accordance with the Declaration of Helsinki. The histopathological diagnosis of all samples was, respectively, diagnosed by two pathologists. The clinical staging was based on the 7th edition of the AJCC Cancer Staging Manual.
Quantitative real-time PCR
Expressions of lncRNA HOTTIP in CRC and non-malignant tissues were detected. Total RNA was extracted from cells using Trizol reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s protocol. The quantitative real-time PCR (qRT-PCR) was carried out using a Roche Light-Cycler (Roche, Basel, Switzerland) and SYBR Green reaction mix (Qiagen, Germany) to detect the level of lncRNA HOTTIP, with β-actin as a normalizing control. The PCR primers for lncRNA HOTTIP or β-actin were as follows: lncRNA HOTTIP forward: 5’-GTGGGGCCCAGACCCGC-3’; lncRNA HOTTIP reverse: 5’-AATGATAGGGACACATCGGGGAACT-3’; β-actin forward: 5’-GAAATCGTGCGTGACATTAA-3’; β-actin reverse: 5’-AAGGAAGGCTGGAAGAGTG-3’. The relative expression of lncRNA HOTTIP was calculated and normalized using the delta-delta CT (2-ΔΔCt) method relative to β-actin. Independent experiments were done in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS Statistics 13.0 (IBM Chicago, IL, USA). The unpaired t test was applied to test the differential expression of lncRNA HOTTIP in cancer tissues compared to adjacent non-malignant tissues. The chi-square test was applied to the examination of relationship between lncRNA HOTTIP expression levels and clinicopathologic characteristics. Overall survival was defined as the interval from the date of diagnosis to pancreatic cancer-related death. Survival curves were plotted using the Kaplan-Meier method and the log-rank test. The significance of survival variables was analyzed using the Cox multivariate proportional hazards model. P value of less than 0.05 was considered statistically significant.
Results
LncRNA HOTTIP is highly expressed in CRC
In order to assess the role of lncRNA HOTTIP in CRC, we performed qRT-PCR to examine the status of lncRNA HOTTIP expression in 42 clinical fresh samples of CRC tissues and non-malignant tissues. Compared with adjacent non-malignant tissues, colorectal cancer tissues showed increased expression levels of lncRNA HOTTIP (P<0.001, Figure 1).
Figure 1.
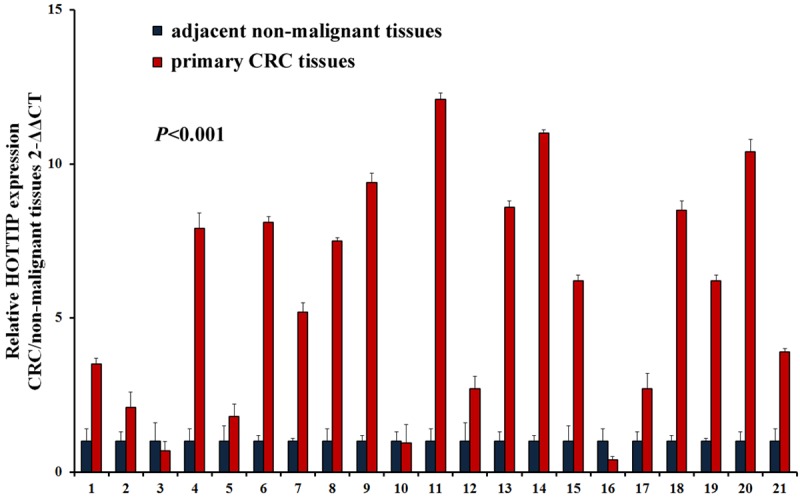
Expression of lncRNA HOTTIP is increased in CRC tissues compared with non-malignant tissues through qRT-PCR.
Relationship between lncRNA HOTTIP expression and clinicopathological characteristics in CRC patients
In the 156 CRC cases, there were 107 males and 49 females with age ranging from 25 to 81 years. We further investigated the association between lncRNA HOTTIP expression and clinicopathological characteristics of CRC patients. Based on a previous study [16], CRC tissue samples were classified into low expression group (n = 79) and high expression group (n = 77), according to the median expression level of all CRC samples (median ΔCT value 7.52). The association between lncRNA HOTTIP expression levels and clinicopathological characteristics in patients with CRC was showed in Table 1. Overall, no statistically significant association was observed between lncRNA HOTTIP expression levels and patient’s age, gender, smoking status, drinking status and differentiated (P = 0.764, 0.776, 0.415, 0.894 and 0.418, respectively). Although high lncRNA HOTTIP expression was more common in advanced nodal stage patients compared with low lncRNA HOTTIP expression cases (47/77 vs. 36/79), this result was not statistically significant (P = 0.053). However, lncRNA HOTTIP was positively associated with clinical stage (P = 0.003), T stage (P = 0.001) and distant metastasis status (P = 0.014) in CRC patients.
Table 1.
Associations between lncRNA HOTTIP expression and clinicopathological characteristics in CRC
| Characteristics | n | High expression | Low expression | P |
|---|---|---|---|---|
|
| ||||
| 77 | 79 | |||
| Age (year) | ||||
| <60 | 89 | 43 | 46 | 0.764 |
| ≥60 | 67 | 34 | 33 | |
| Gender | ||||
| Female | 53 | 27 | 26 | 0.776 |
| Male | 103 | 50 | 53 | |
| Smoker | ||||
| Yes | 72 | 33 | 39 | 0.415 |
| No | 84 | 44 | 40 | |
| Drinker | ||||
| Yes | 92 | 45 | 47 | 0.894 |
| No | 64 | 32 | 32 | |
| T stage | ||||
| T1-2 | 69 | 24 | 45 | 0.001 |
| T3-4 | 87 | 53 | 34 | |
| Nodal stage | ||||
| Negative | 73 | 30 | 43 | 0.053 |
| Positive | 83 | 47 | 36 | |
| Distant metastasis | ||||
| Absent | 115 | 50 | 65 | 0.014 |
| Present | 41 | 27 | 14 | |
| Clinical stage | ||||
| I-II | 97 | 39 | 58 | 0.003 |
| III-IV | 59 | 38 | 21 | |
| Differentiation | ||||
| Well | 34 | 16 | 18 | 0.418 |
| Moderate | 71 | 32 | 39 | |
| Poor | 51 | 29 | 22 | |
LncRNA HOTTIP expression is associated with overall survival in CRC patients
In order to assess the prognostic value of lncRNA HOTTIP expression for CRC, we investigated the association between lncRNA HOTTIP expression levels and overall survival (OS) through Kaplan-Meier analysis and log-rank test. In 156 CRC cases, we observed that lncRNA HOTTIP expression was significantly associated with CRC patients’ OS (P<0.001, Figure 2). Moreover, we also observed that lncRNA HOTTIP overexpression was an unfavorable prognostic factor in CRC patients (P = 0.001, Table 2), regardless of T stage, distant metastasis and clinical stage. Finally, multivariate analysis showed that increased lncRNA HOTTIP expression was an independent poor prognostic factor for CRC patients (P = 0.017, Table 2).
Figure 2.
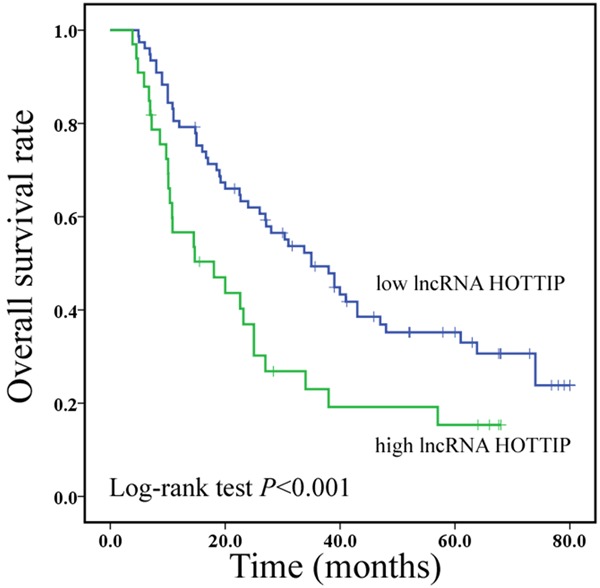
Increased lncRNA HOTTIP expression predicts a poor prognosis in CRC patients.
Table 2.
Univariate and multivariate Cox regression of prognostic factors for overall survival in pancreatic cancer
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (year) | ||||||
| <60 vs. ≥60 | 0.879 | 0.561-1.373 | 0.652 | |||
| Gender | ||||||
| Female vs. male | 1.325 | 0.568-1.676 | 0.440 | |||
| Differentiation | ||||||
| Well vs. moderate vs. poor | 1.223 | 0.673-1.542 | 0.659 | |||
| T stage | ||||||
| T1-2 vs. T3-4 | 3.536 | 1.458-5.519 | 0.001 | 2.648 | 1.423-3.384 | 0.045 |
| Nodal stage | ||||||
| Negative vs. positive | 2.573 | 1.578-4.127 | 0.005 | 1.320 | 0.563-2.893 | 0.522 |
| Distant metastasis | ||||||
| Absent vs. present | 3.153 | 1.357-7.475 | 0.003 | 1.672 | 0.458-3.349 | 0.220 |
| Clinical stage | ||||||
| I-II vs. III-IV | 3.451 | 1.354-5.620 | 0.005 | 1.463 | 0.562-2.375 | 0.136 |
| LncRNA HOTTIP | ||||||
| Low vs. high | 2.896 | 1.635-4.378 | 0.001 | 2.151 | 1.306-3.415 | 0.017 |
HR: hazard ratio; 95% CI: 95% confidence interval.
Discussion
To date, it has been estimated that approximately 15,000 lncRNAs are present in the human genome [17]. Recent studies have also demonstrated that lncRNAs play important roles in carcinogenesis and cancer metastasis and abnormal expression of lncRNAs has been identified in CRC [8,18]. In the present study, we examined the expression of lncRNA HOTTIP and its clinicopathological/prognostic significance in 156 specimens of primary CRCs and 21 samples of adjacent non-malignant samples.
Recently, application of lncRNAs as cancer diagnostic or prognostic biomarkers has been reported in several studies [19]. Zheng and colleagues investigated lncRNA MALAT-1 expression in 146 CRC patients and 23 paired normal colonic mucosa samples. Their results showed that expression of lncRNA MALAT-1 was up-regulated in CRC tissues, and a higher expression level of MALAT-1 might serve as a negative prognostic marker in CRC patients [20]. In another study, Svoboda and colleagues also observed that CRC patients had higher lncRNA HOTAIR expression in circulation than healthy controls. HOTAIR expression levels positively correlated between circulation and tumor, indicating circulation HOTAIR levels may serve as a potential prognostic marker in CRC [21]. However, there are still a number of common cancer-related lncRNAs, such as ANRIL [22], HOTTIP [14], HULC [23] and MEG3 [24], which functions associated with CRC have not been reported.
HOTTIP is located at the 5’ tip of the HOXA locus and coordinates the activation of multiple 5’ HOXA genes [14]. In one previous study, Li and colleagues observed that HOTTIP was one of the most significantly upregulated lncRNAs in pancreatic ductal adenocarcinoma (PDAC) tissues compared with pancreatic tissues by microarray analyses. Furthermore, this study also showed that HOTTIP silencing resulted in cell proliferation arrest by altering cell-cycle progression, and impaired cell invasion by inhibiting epithelial-mesenchymal transition in pancreatic cancer [25]. A previous study also showed that patients with higher lncRNAs HOTTIP/HOXA13 expression had poorer prognosis in liver cancer [15]. A similar trend was also seen in this study. We observed that lncRNA HOTTIP was highly expressed in 21 CRC tissues compared with adjacent non-malignant tissues. Furthermore, we analyzed the association between lncRNA HOTTIP expression and clinicopathological characteristics in 156 CRC patients. We found that lncRNA HOTTIP was positively associated with clinical stage, tumor size and distant metastasis in CRC patients.
In conclusion, our findings indicated that HOTTIP expression level has the potential to be an independent unfavorable prognostic indicator for CRC patients. However, the functional consequences of altered HOTTIP expression, the different feature of HOTTIP expression between CRC and other malignancies, and the underlying mechanisms of the heterogeneous expression levels need to be extensively investigated in the future.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Booth RA. Minimally invasive biomarkers for detection and staging of colorectal cancer. Cancer Lett. 2007;249:87–96. doi: 10.1016/j.canlet.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Moreaux J, Catala M. Carcinoma of the colon: long-term survival and prognosis after surgical treatment in a series of 798 patients. World J Surg. 1987;11:804–809. doi: 10.1007/BF01656605. [DOI] [PubMed] [Google Scholar]
- 4.Burt RW. Strategies for colon cancer screening with considerations of cost and access to care. J Natl Compr Canc Netw. 2010;8:2–5. doi: 10.6004/jnccn.2010.0002. [DOI] [PubMed] [Google Scholar]
- 5.Arends MJ. Pathways of colorectal carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:97–102. doi: 10.1097/PAI.0b013e31827ea79e. [DOI] [PubMed] [Google Scholar]
- 6.Tannapfel A, Neid M, Aust D, Baretton G. The origins of colorectal carcinoma: specific nomenclature for different pathways and precursor lesions. Dtsch Arztebl Int. 2010;107:760–766. doi: 10.3238/arztebl.2010.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 11.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hammerle M, Tornillo L, Heim MH, Diederichs S, Cillo C, Terracciano LM. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 17.Walsh AL, Tuzova AV, Bolton EM, Lynch TH, Perry AS. Long noncoding RNAs and prostate carcinogenesis: the missing ‘linc’? Trends Mol Med. 2014;20:428–436. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic Roles of Noncoding RNAs in Lung Cancer Biology and Their Clinical Implications. Genet Res Int. 2012;2012:737416. doi: 10.1155/2012/737416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 21.Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z, Protivankova M, Vycital O, Liska V, Schwarzova L, Vodickova L, Vodicka P. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 22.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 (INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


