Abstract
Despite advances made in the diagnosis and treatment of human colorectal cancer (CRC), the long-term survival for CRC remains poor. Long non-coding RNA anti-differentiation ncRNA (lncRNA DANCR) was identified to be involved in carcinogenesis of hepatocellular carcinoma. While its expression in CRC and potential role in tumor progression is still unknown. In the present study, we investigated the expression level of lncRNA DANCR as well as its association with CRC progression and prognosis. The expression of lncRNA DANCR was detected by quantitative real-time PCR (qRT-PCR) in 104 CRC specimens. The prognostic value of lncRNA DANCR was further analysis. Our results showed that lncRNA DANCR expression was increased in CRC tissues compared with that in adjacent normal tissues (P<0.05). In addition, tumors with high lncRNA DANCR expression was correlated with TNM stage, histologic grade, and lymph node metastasis (P<0.05). Kaplan-Meier analysis showed that patients with high lncRNA DANCR expression had a shorter overall survival (OS) and disease-free survival (DFS) compared with the low lncRNA DANCR expression group (P<0.05). Moreover, in a multivariate Cox model, our results showed that lncRNA DANCR expression was an independent poor prognostic factor for both OS and DFS in CRC. Our data indicated that lncRNA DANCR expression might be a novel potential biomarker for CRC prognosis.
Keywords: Colorectal cancer, lncRNA DANCR, quantitative real-time PCR, prognosis
Introduction
Colorectal cancer is one of the leading causes of cancer mortality and the third most common malignant neoplasm all over the world [1]. Due to the chemotherapy and radiation therapy, the incidence and mortality of CRC are decreased in recent years. But the 5-year survival rate of CRC patients remains unsatisfactory due to metastasis leading to poor outcomes [2,3]. Therefore, it is important to identify novel biomarkers that can accurately identify the biological characteristics of tumors and predict the prognosis in patients with CRC.
The human genome sequencing project has found that 70% of the genome is transcribed, but only up to 2% of the human genome serves as blueprints for proteins [4]. Long non-coding RNAs (lncRNAs) are defined as endogenous cellular RNAs more than 200 nucleotides in length that lack an open reading frame of significant length [5]. In recent years, lncRNAs have been shown to be involved in carcinogenesis and cancer progression [6,7]. Long non-coding RNA, anti-differentiation ncRNA (lncRNA DANCR or ANCR), was found to be required for the dedifferentiation of epidermal cells [8]. Zhu et al. found that downregulated lncRNA ANCR expression could promote osteoblast differentiation by targeting EZH2 and regulating Runx2 expression [9]. Jia et al. indicated that down-regulated lncRNA ANCR could promote osteogenic differentiation of periodontal ligament stem cells [10]. Recently, Yuan et al. found that high expression of lncRNA DANCR was involved in the progression of hepatocellular carcinoma [11]. However, the role of lncRNA DANCR in CRC is still unknown.
In the present study, we investigated the expression level of lncRNA DANCR in CRC tissues and adjacent non-tumor tissues. Then, the association of lncRNA DANCR with clinicopathological characteristics and outcome of the CRC patients was investigated.
Materials and methods
Tissue specimens
A total of 104 fresh colorectal cancer tissues and paired adjacent non-tumor tissues were obtained from patients who had undergone surgical resection of colorectal cancer between 2007 and 2008 at the Qilu Hospital of Shandong University, China. The colorectal cancer diagnosis was confirmed by an experienced pathologist. All of the tissue samples were washed with sterile phosphate-buffered saline before being snap frozen in liquid nitrogen and stored at -80°C until total RNA was extracted. No patients had been treated with radiotherapy or chemotherapy before surgery. This study was approved by the Ethics Committee of Shandong University and informed consent was obtained from each patient involved in the study.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from CRC tissues and cell lines using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The expression level of DANCR in CRC tissues and cell lines was measured by qRT-PCR using the SYBR-Green method (Takara) according to the manufacturer’s protocol and normalized using GAPDH. The primers were as follows: DANCR sense: 5’-GCGCCACTATGTAGCGGGTT-3’; DANCR antisense: 5’-TCAATGGCTTGTGCCTGTAGTT-3’; GAPDH sense: 5’-AGAAGGCTGG GGCTCATTTG-3’; GAPDH antisense: 5’-AGGGGCCATCCACAGTCTTC-3’. All experiments were performed using the 2-ΔΔCt method. Each experiment was performed in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS version 13.0 software. The measurement data were analyzed by one-way ANOVA. Randomized block design ANOVA was used to analyze the statistical difference among different tissue types. Associations between DANCR expression and clinicopathological characteristics were analyzed by Chi-square test. Survival curves were plotted using the Kaplan-Meier method, and differences between survival curves were tested using the log-rank test. Cox’s proportional hazards model was used to identify the factors that have significant influence on survival. All data are presented as the mean ± SD from at least three independent experiments. P values lower than 0.05 was considered statistically significant.
Results
lncRNA DANCR is significantly up-regulated in CRC tissues
We explored the expression levels of lncRNA DANCR in 104 pairs of CRC tissues and adjacent non-tumor tissues. As revealed by qRT-PCR analysis, lncRNA DANCR expression was remarkably higher in CRC tissues compared with adjacent non-tumor tissues (P<0.05, Figure 1).
Figure 1.
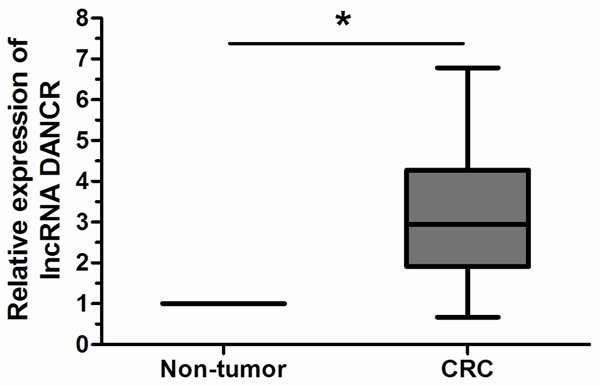
LncRNA DANCR expression levels assessed by qRT-PCR in CRC tissue and adjacent non-tumor tissues. LncRNA DANCR expression levels were normalized to GAPDH. Data are means ± SD, *P<0.05.
Correlations of lncRNA DANCR expression with clinicopathologic features of CRC patients
To further investigate the association of lncRNA DANCR with clinicopathological features of CRC patients. The median value of lncRNA DANCR in all CRC tissues was 2.94 and used as a cutoff value, and all patients were divided into two groups: high DANCR expression group (≥2.94; n=52) and low DANCR expression group (<2.94; n=52). The relationship of lncRNA DANCR with various clinical features of CRC was analyzed and is summarized in Table 1. The results showed that expression of lncRNA DANCR was significantly associated with TNM stage, histologic grade and lymph node metastasis (P<0.05). However, there was no significant correlation of DANCR expression with other clinical features such as gender, age, tumor size, and local invasion (P>0.05). These results indicated that lncRNA DANCR expression may play an oncogenic role in colorectal cancer progression.
Table 1.
Correlation between lncRNA DANCR expression and clinicopathological characteristics of colorectal cancer
| Parameters | Group | Total | lncRNA DANCR expression | P value | |
|---|---|---|---|---|---|
|
| |||||
| High | Low | ||||
| Gender | Male | 64 | 33 | 31 | 0.687 |
| Female | 40 | 19 | 21 | ||
| Age (years) | <60 | 48 | 20 | 28 | 0.116 |
| ≥60 | 56 | 32 | 24 | ||
| Tumor size (cm) | <5 cm | 59 | 28 | 31 | 0.553 |
| ≥5 cm | 45 | 24 | 21 | ||
| Histological grade | Well and moderately | 61 | 23 | 38 | 0.003 |
| Poorly | 43 | 29 | 14 | ||
| Local invasion | T1-T2 | 43 | 17 | 26 | 0.073 |
| T3-T4 | 61 | 35 | 26 | ||
| Lymph nodes metastasis | Negative | 75 | 31 | 44 | 0.004 |
| Positive | 29 | 21 | 8 | ||
| TNM stage | I-II | 37 | 12 | 25 | 0.008 |
| III-IV | 67 | 40 | 27 | ||
Prognostic values of lncRNA DANCR expression in CRC
To further investigate the correlation of lncRNA DANCR expression with overall survival and disease-free survival of CRC patients, Kaplan-Meier analyses were performed. We found that overall survival time of high lncRNA DANCR expression group was significantly shorter than that of low lncRNA DANCR expression group (P<0.05, Figure 2A). In addition, our results showed disease-free survival of high lncRNA DANCR expression group was also significantly shorter than that of low lncRNA DANCR expression group (P<0.05, Figure 2B). Furthermore, in multivariate Cox model, our results revealed that lncRNA DANCR expression was an independent prognostic indicator for overall survival (HR=2.131, 95% CI, 1.157-7.058; P=0.009) and disease-free survival (HR=2.397, 95% CI, 1.385-7.279; P=0.006) in patients with colorectal cancer (Table 2).
Figure 2.
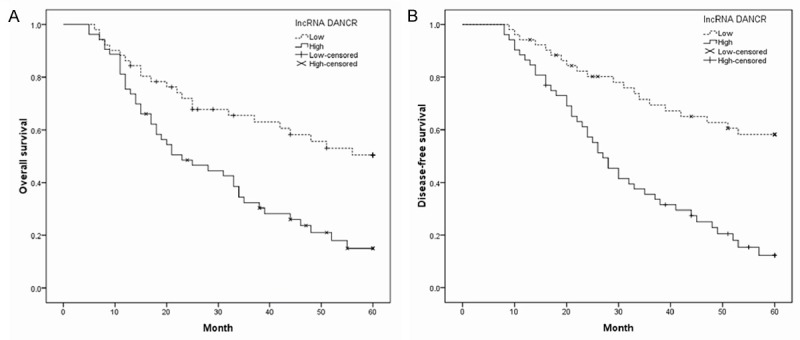
The correlation between lncRNA DANCR expression and the overall survival or disease-free survival of colorectal cancer patients. Kaplan-Meier analysis of overall survival (A) or disease-free survival (B) was analyzed according to lncRNA DANCR expression levels.
Table 2.
Univariate and multivariate Cox regression analyses of overall survival and disease-free survival in CRC patients
| Overall survival | Disease-free survival | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Hazard ratio | 95% CI | P | Hazard ratio | 95% CI | P | |
| Univariate analyses | ||||||
| Gender | 1.274 | 0.739-3.107 | 0.254 | 1.196 | 0.712-2.959 | 0.198 |
| Age (years) | 1.764 | 0.525-4.928 | 0.197 | 1.558 | 0.486-4.527 | 0.136 |
| Tumor size (cm) | 1.582 | 0.605-4.035 | 0.312 | 1.375 | 0.574-3.856 | 0.322 |
| Histological grade | 2.916 | 1.662-6.952 | 0.013 | 2.743 | 1.792-7.174 | 0.009 |
| Local invasion | 2.675 | 0.804-7.112 | 0.081 | 2.541 | 0.856-6.784 | 0.073 |
| Lymph nodes metastasis | 3.172 | 1.417-8.629 | 0.023 | 3.429 | 1.767-9.264 | 0.016 |
| TNM stage | 2.822 | 1.014-6.275 | 0.019 | 2.725 | 1.145-7.152 | 0.014 |
| DANCR expression | 2.491 | 1.335-7.264 | 0.008 | 2.614 | 1.572-7.715 | 0.004 |
| Multivariate analyses | ||||||
| Histological grade | 2.604 | 1.537-6.408 | 0.017 | 2.434 | 1.625-6.791 | 0.011 |
| Lymph nodes metastasis | 3.052 | 1.375-8.309 | 0.028 | 3.215 | 1.631-8.657 | 0.019 |
| TNM stage | 2.502 | 0.974-6.014 | 0.022 | 2.417 | 1.027-6.827 | 0.017 |
| DANCR expression | 2.131 | 1.157-7.058 | 0.009 | 2.397 | 1.385-7.279 | 0.006 |
Discussion
CRC is a highly heterogeneous disease. Mainstream tumorigenic processes involved in CRC are characterized by phenotypic multistep progression cascades [12]. The reliable identification of CRC progression-specific targets has huge implications for its prevention and treatment [13,14]. However, identification of the molecular mechanisms underlying tumorigenesis still remains a challenge.
LncRNAs are a class of non-coding RNA transcripts longer than 200 nucleotides and are implicated in a number of important events, such as epigenetic regulation, transcriptional regulation, and post-transcriptional regulation [15,16]. Emerging evidence showed that lncRNAs play an important role in tumor progression and may serve as prognostic marker [17]. For example, Zhang et al. showed that upregulation of lncRNA MALAT1 correlated with tumor progression and poor prognosis in clear cell renal cell carcinoma [18]. Wang et al. found that overexpression of lncRNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma [19]. Cao et al. showed that lncRNA GAS5 was downregulated in cervical cancer and association with cervical progression, moreover, they showed that decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer patients [20]. Shi et al. suggested that downregulated lncRNA BANCR promotes the proliferation of colorectal cancer cells via downregualtion of p21 expression [21]. However, the roles of lncRNA DANCR in the carcinogenesis of CRC are still unknown.
In the present study, we investigated the expression and clinical significance of lncRNA DANCR in human colorectal cancer. Our results showed that lncRNA DANCR expression in CRC tissues was significantly high than that in matched adjacent non-tumor tissues. Then the relationships of lncRNA DANCR with various clinical features of CRC were analyzed, we found that lncRNA DANCR expression was proven to be associated with TNM stage, histologic grade, and lymph node metastasis, suggesting that lncRNA DANCR might be involved in the carcinogenesis of CRC. Furthermore, Kaplan-Meier analysis with the log-rank test indicated that patients with a high level of lncRNA DANCR expression had significantly shorter overall survival and disease-free survival compared to those with a low level of lncRNA DANCR expression. In a multivariate Cox model, our results suggested that lncRNA DANCR expression level was independent prognostic factors for overall survival and disease-free survival of CRC patients, indicating that high lncRNA DANCR level was a promising non-invasive biomarker for prognosis of CRC patients.
In conclusion, our data suggested that lncRNA DANCR upregulation was associated with aggressive progression and poor prognosis in colorectal cancer. LncRNA DANCR was identified for the first time as an independent marker for predicting the clinical outcome of colorectal cancer patients. Further studies are needed to elucidate the mechanisms of action of lncRNA DANCR in colorectal cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, NaisThadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 3.Meyerhardt JA, Giovannucci EL, Holmes MD, Chan AT, Chan JA, Colditz GA, Fuchs CS. Physical activity and survival after colorectal cancer diagnosis. J. Clin. Oncol. 2006;24:3527–3534. doi: 10.1200/JCO.2006.06.0855. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, Rinn JL, Chang HY, Siprashvili Z, Khavari PA. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Jia Q, Jiang W, Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. 2015;60:234–241. doi: 10.1016/j.archoralbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma via de-repression of CTNNB1. Hepatology. 2015 doi: 10.1002/hep.27893. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LCG, Lannon WA, Grotzinger C, Del Rio M. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe T, Itabashi M, Shimada Y, Tanaka S, Ito Y, Ajioka Y, Hamaguchi T, Hyodo I, Igarashi M, Ishida H, Ishiguro M, Kanemitsu Y, Kokudo N, Muro K, Ochiai A, Oguchi M, Ohkura Y, Saito Y, Sakai Y, Ueno H, Yoshino T, Fujimori T, Koinuma N, Morita T, Nishimura G, Sakata Y, Takahashi K, Takiuchi H, Tsuruta O, Yamaguchi T, Yoshida M, Yamaguchi N, Kotake K, Sugihara K. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2010 for the treatment of colorectal cancer. Int J Clin Oncol. 2012;17:1–29. doi: 10.1007/s10147-011-0315-2. [DOI] [PubMed] [Google Scholar]
- 14.Uccello M, Malaguarnera G, Basile F, D’Agata V, Malaguarnera M, Bertino G, Vacante M, Drago F, Biondi A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012;12(Suppl 1):S35. doi: 10.1186/1471-2482-12-S1-S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 16.Clark M. Transcriptional complexity and post-transcriptional regulation of long noncoding RNAs. 2012. [Google Scholar]
- 17.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, Mrozek K, Bucci D, Carroll AJ, Baer MR, Wetzler M, Carter TH, Powell BL, Kolitz JE, Moore JO, Eisfeld AK, Blachly JS, Blum W, Caligiuri MA, Stone RM, Marcucci G, Croce CM, Byrd JC, Bloomfield CD. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111:18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, Wang H, Zhang R, Liu Y. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol Cells. 2015;38:432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776–6783. [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W, Yan L, Wang K, Feng J. Downregulated Long Noncoding RNA BANCR Promotes the Proliferation of Colorectal Cancer Cells via Downregualtion of p21 Expression. PLoS One. 2015;10:e0122679. doi: 10.1371/journal.pone.0122679. [DOI] [PMC free article] [PubMed] [Google Scholar]


