Abstract
Background: Vincristine (VCR) is a chemical that is widely used in tumor therapy. While long-term use can make tumor cells resistant to VCR, the underlying mechanisms of this resistance are still unclear. Objective: This study aimed at investigating the role of microRNA (miRNA) in colon cancer drug resistance. Methods: HCT-8 colon carcinoma cells were cultured and treated with different VCR concentrations to establish an HCT-8/VCR resistant cell line. Whole-genome screens, HiSeq 2500 sequencing, and bioinformatics methods were used to detect and analyze differences in miRNA expression between the drug-resistant HCT-8/VCR cells and non-resistant HCT-8 cells. Differential expression profiles of miRNAs were constructed based on sequencing result. Results: The HCT-8/VCR resistant colon carcinoma cell line was established. With regard to the difference in drug resistance between HCT-8/VCR and HCT-8 cells, 24 miRNAs showed statistically significant differences in their expression (fold change > 4), of which 17 were up-regulated. Seven miRNAs were down-regulated. Conclusion: As abnormal expression of miRNAs was associated with VCR resistance of colon carcinoma cells, differences in miRNA expression may play a key role in VCR resistance of colon cancer cells.
Keywords: Colon cancer, drug resistance, microRNA, HCT-8
Introduction
Colon cancer is one of the most common malignant tumors of the digestive tract. With increasing incidences every year, colon cancer belongs to the most widespread types of cancer overall, together with esophageal cancer, gastric cancer, and lung cancer [1,2]. Radical resection is the most effective method of colon cancer treatment. Postoperative chemotherapy plays an important role in prolonging patient survival and improving quality of life. The chemotherapeutic drug vincristine (VCR) is widely used in the clinical treatment of colon cancer. However, long-term therapy can make cancer cells resistant to VCR [2], causing treatment failure. As 90% of deaths in cancer patients are related to drug resistance, further studies on the mechanism of occurrence and development of drug resistance are required to improve the treatment outcomes in cases of malignant tumors. Mechanisms of tumor drug resistance are related to various aspects of molecular and cell biology such as changes in drug targets, repair of damaged cells, activation or inhibition of the death signaling pathway of the cell, including genetic modifications such as gene mutations, deletions, amplifications, epigenetic changes caused by abnormal DNA methylation, and microRNA (miRNA) for post transcriptional regulation [3-6].
MicroRNA is a new class of regulatory, small, non-coding RNA molecules of about 20-25 nucleotides, associated with many biological mechanisms such as development, differentiation, and apoptosis. As abnormal expression of miRNAs is involved in tumorigenesis [7,8], miRNAs are also related to the sensitivity of tumor cells to radiotherapy and chemotherapy [9,10]. Mutations in and abnormal expression or modification of miRNAs affect their normal function, leading to changes in the expression of target genes, conceivably affecting drug sensitivity of tumor cells.
So far, studies on the involvement of miRNA in colon cancer drug resistance have only focused on 5-fluoruracil (5-Fu) and other drugs, while no data on miRNA regulation of vincristine drug resistance have been reported. Therefore, this study set out to establish a VCR-resistant colon cancer cell line and use HiSeq 2500 sequencing technology to screen miRNAs associated with drug resistance. Bioinformatics analysis was used to detect target genes. The study contributes to finding new ways to reverse drug resistance to vincristine.
Materials and methods
Cells and culture
Colon carcinoma HCT-8 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM containing 10% fetal calf serum, 100 μg/mL penicillin, and 100 μg/mL streptomycin at 37°C with 5% CO2. Cells were subcultured using digestion with 0.02% EDTA, 0.1% trypsin every 2-3 d.
Establishment of VCR resistant colon carcinoma cell
The drug resistant colon carcinoma cell line HCT-8/VCR was established by gradually increasing concentrations of VCR. Sensitive HCT-8 cells were cultured in VCR solution of an initial concentration of 5 ng/mL, which was then gradually increased to 10, 100, 1000, and 2000 ng/mL. When cells had acquired resistance at a certain concentration, growing cells were cloned by limiting dilution before the culture and screening in the next concentration was started. Finally, HCT-8 cells were continually cultured at 2000 ng/mL VCR for more than 20 generations, and VCR treatment was stopped 1 week before the experiment.
MTT assay
Cells were used in logarithmic growth phase, diluted to 1 × 105 cells/mL in serum-free culture medium, and seeded into 96-well plates at 1 × 104 cells/well. Background controls were culture solutions without cells and/or without VCR, while experimental groups were treated with 50 ng/mL, 100 ng/mL, 200 ng/mL, 400 ng/mL, 800 ng/mL, and 2000 ng/mL vincristine in a volume of 100 µL/well.
Cells were incubated at 37°C for 48 h, transferred to the culture plate with 20 µL/well MTT (5 mg/mL) solution, and incubated again at 37°C for 48 h. Supernatants were discarded, and 150 µL/well DMSO was added. Cells were agitated, and absorbance (OD) of each well was read at 570 nm using an ELISA reader (Beijing Tianshi Tianli Medical Device Technology Development Center, Beijing, China).
Mean values of cell survival rates of each group were calculated according to the following formula: (experimental group OD-background group OD)/blank control group OD-background group OD) × 100%. The drug concentration of 50% cell survival was calculated as the concentration of 50% inhibition (IC50).
RNA extraction and cDNA library construction
Total cellular RNA was extracted using a Trizol kit (TakaRa Company, Dalian, China), and the purity and quantity of RNA was determined with a UV spectrophotometer. According to Hafner et al. [11], fragment lengths in the range of 18-30 nt were recovered from total cellular RNA. Sequencing adaptors were added at the 5’ and 3’ ends of the fragments using T4 RNA ligase. Using adaptors as templates, cDNA was synthesized with random primers, 5’ and 3’ adaptor primers were added for PCR amplification, and the cDNA library was completely sequenced. MiRNAs of HCT-8 and HCT-8/V were sequenced using the HiSeq 2500 high-throughput sequencing platform (Illumina, San Diego, CA, USA). HiSeq sequencing results were analyzed after removal of noise, adaptors, and low-quality reads, and high-quality target sequences were used for analysis including statistical length distribution of fragments and classification of target sequences. Finally, reliable fragment sequences were selected for subsequent analysis.
Species, quantity, and length distribution of small RNA (sRNA) were counted. In general, the length of fragments was 18-30 nt with a peak of length distribution at 21-22 nt for siRNA and 24-30 nt for piRNA.
Sequence alignment
Sequences were aligned to those in reference genome sequence databases such as Rfam, Repbase sequence, and miRBase by using bowtie software (Bow Tie Pro, Aberdeen, UK). Sequence alignment was set to allow for only one mismatch, and alignments and annotations of all small RNAs to all kinds of RNAs were summarized. Small RNAs were annotated according to the priority order of miRNA > rRNA > tRNA > snRNA > snoRNA > repeat.
Statistical analysis
MiRNA expression levels were calculated as transcripts per million (TPM) = (reads of each miRNA alignment)/(reads of samples total alignment) × 106. TPM uses pairwise sequence per million as miRNA expression index with reads of total matching pairs as expression of normalized reads. Differential expression of miRNAs was assessed using DESeq software. Sample expression was calculated using TPM and log2. Differences in miRNA expression were screened from P ≤ 0.05 to greater than or equal to 2-fold difference.
Results
Establishment of drug resistant cell line
Increasing drug concentrations gradually, a VCR resistant cell line was established and named HCT-8/VCR, growing well in the presence of 2 × 103 ng/mL VCR. Inhibition rates of HCT-8 and HCT-8/VCR cells are shown in Tables 1 and 2, leading to 50% inhibitory concentrations (IC50) of 140.265 and 2350.469, respectively.
Table 1.
The inhibition rate of HCT-8 cells with different concentrations of VCR
| VCR concentration (ng/ml) | Inhibition rate (%) |
|---|---|
| 50 | 8.5 ± 1.3 |
| 10 | 16.8 ± 3.0 |
| 100 | 29.5 ± 3.8 |
| 200 | 52.8 ± 6.7 |
| 1000 | 79.3 ± 8.9 |
| 2000 | 93.4 ± 9.6 |
Table 2.
The inhibition rate of HCT-8/VCR cells with different concentrations of VCR
| VCR concentration (ng/ml) | Inhibition rate (%) |
|---|---|
| 100 | 2.5 ± 0.31 |
| 500 | 3.8 ± 0.56 |
| 1000 | 8.5 ± 2.81 |
| 2000 | 26.8 ± 4.73 |
| 3000 | 69.3 ± 8.92 |
| 4000 | 96.2 ± 11.6 |
Cell growth
Cells were used in logarithmic growth phase, diluted to 1 × 105 cells/mL in serum-free culture medium, seeded into 96-well plates at 1 × 104 cells/well, cultured, and counted continuously for 6 days. A growth curve was sketched using culture time as abscissa and number of cells as ordinate, showing that HCT-8/VCR cells grow slowly compared to sensitive cell line HCT-8 (Figure 1).
Figure 1.
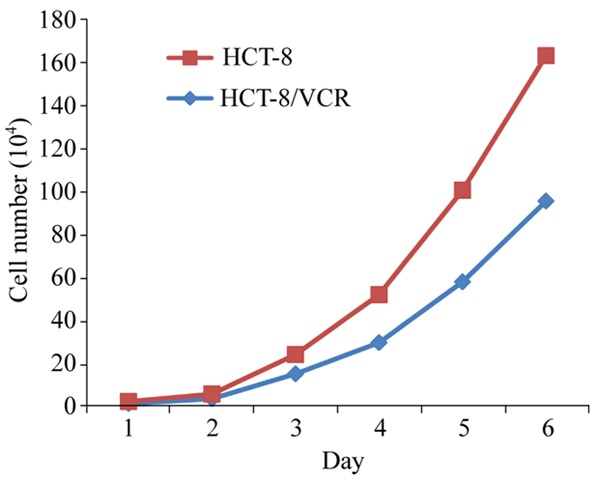
Growth curve of resistant and susceptible cells.
Sequencing results and statistics
RNA samples extracted from HCT-8 and HCT-8/VCR cells were sequenced in triplicate. Statistical data showed that the total numbers of reads and the ratio of high quality read were high, reflecting a better quality and quantity of sequencing (Table 3).
Table 3.
The number of total reads and high quality reads of samples
| Sample | Total reads | High quality reads |
|---|---|---|
| HCT8 | 16009298 | 15455682 |
| HCT8V | 21122967 | 19911807 |
Read lengths distributions
Both cell samples had the highest abundance at 22 nt and 23 nt fragments. In HCT-8 cells, 22 nt and 23 nt fragments accounted for 39.87% and 15.24%, respectively, while 21 nt and 24 nt fragments were represented at 12.11% and 9%, respectively. In HCT-8/VCR cells, 22 nt and 23 nt fragments accounted for 29.77% and 11.93%, respectively, whereas 21 nt and 24 nt fragments were represented at 10.43% and 7.78%, respectively.
sRNA location in the genome
After sequencing and data cleaning, all clear reads were compared to the human genome, revealing that HCT-8 and HCT-8/VCR match 67.56% and 68.04% on average, respectively (Table 4).
Table 4.
miRNA location on the genome
| Sample | High quality reads | Total number of matching | Ratio (%) |
|---|---|---|---|
| HCT-8 | 15455682 | 456123 | 67.56 |
| HCT-8/VCR | 19911807 | 1043295 | 68.04 |
sRNA annotation
sRNA library annotation shows that rRNA sequences of HCT-8 cells account for 41.4%, while rRNA sequences of HCT-8/VCR represent 47.0% of total sequences.
Library sequencing produced results well, returning 14527 unique miRNAs of HCT-8 cells, accounting for 11.4% of the total number of sequences, and 18543 unique miRNAs of HCT-8/VCR cells, accounting for 7% of the total number of sequences.
miRNA differential expression analysis
Each sample sequence was compared to the sequence in the new prediction miRNA library, and miRNA expression levels were calculated as TPM. As shown in Table 5, 1651 miRNAs were sequenced, revealing 48 miRNAs differing in their expression. Comparing HCT-8/VCR to non-resistant colon carcinoma cell line HCT-8, the difference in the expression of 24 miRNAs was statistically significant (P < 0.05), including 17 miRNAs with upregulated expression (P < 0.05), of which hsa-miR-675-3p was the most upregulated (up to 7.497681 times), followed by hsa-miR-100-3p, hsa-miR-125b-1-3p, hsa-miR-582-5p, hsa-miR-3141, chr15-10055, hsa-miR-582-5p, hsa-miR-582-3p, and hsa-miR-3687. Seven miRNAs were downregulated (P < 0.05) of which hsa-miR-146a-5p was the most downregulated (7.75199 times), followed by hsa-miR-1247-3p, hsa-miR-181a-3p, and hsa-miR-1247-5p.
Table 5.
24 differential expressed miRNAs in HCT-8/VCR versus HCT-8
| miRNA | Fold change | P value | Q value |
|---|---|---|---|
| hsa-miR-3141 | 6.113252 | 0.002181 | 0.257258286 |
| hsa-miR-100-5p | 5.091838 | 0.000115 | 0.027008707 |
| hsa-miR-675-5p | 5.447769 | 0.000562 | 0.077341102 |
| hsa-miR-125b-1-3p | 6.513067 | 7.76E-05 | 0.027008707 |
| hsa-miR-181a-3p | 0.06097 | 0.00353 | 0.329145753 |
| chr16_11501 | 4.346158 | 0.005961 | 0.468641386 |
| hsa-miR-1247-3p | 0.04064 | 0.000317 | 0.058212705 |
| hsa-miR-125b-5p | 4.777906 | 0.001258 | 0.159787932 |
| hsa-miR-145-5p | 5.079829 | 0.009196 | 0.607296551 |
| hsa-miR-582-5p | 6.297921 | 0.000201 | 0.041544052 |
| hsa-miR-675-3p | 7.497681 | 0.000441 | 0.066165275 |
| hsa-miR-92b-3p | 0.02415 | 0.019997 | 0.971040534 |
| hsa-miR-1247-5p | 0.06976 | 0.003153 | 0.325335878 |
| hsa-miR-126-5p | 0.2049 | 0.01128 | 0.642556044 |
| hsa-miR-218-5p | 4.93781 | 0.019194 | 0.971040534 |
| hsa-miR-101-5p | 0.09523 | 0.011124 | 0.642556044 |
| hsa-miR-100-3p | 0.04389 | 2.43E-05 | 0.013362807 |
| hsa-miR-215-5p | 3.814881 | 0.006797 | 0.490879344 |
| hsa-miR-146a-5p | 0.04368 | 2.22E-09 | 3.67E-06 |
| hsa-miR-582-3p | 5.862238 | 0.000102 | 0.027008707 |
| hsa-miR-3687 | 5.780269 | 0.012261 | 0.67477536 |
| hsa-miR-6087 | 4.954298 | 0.004517 | 0.392492364 |
| chr19_16126 | 4.524614 | 0.0103 | 0.642556044 |
| chr15_10055 | 6.067251 | 0.000363 | 0.059994689 |
Discussion
Colon cancer is a common malignant digestive tract tumor whose incidence is increasing every year due to improvement of living standards and changing food structures. Onset age is trending towards younger, and overall growth rates are higher in women than in men. Colorectal cancer is one of the most common types of cancer together with esophageal cancer, gastric cancer, and lung cancer. As surgical treatment alone cannot cure this cancer, chemotherapy plays an important role in postoperative treatment and became one of the main methods for colon cancer therapy. However, clinical practice experienced drug resistance of tumor cells to cause ineffective chemotherapy for some patients. Postoperative recurrence and metastasis still reflect a lack of effective means of control, in addition to adverse side effects for the patient. Therefore, improving the therapeutic effects of cancer chemotherapy by overcoming drug resistance of tumor cells is a medical problem that needs solving.
A large amount of research on drug resistant tumors has shown that resistance to chemotherapy is related to many factors. Changes in the molecular biology of tumor cells are often associated with aberrant gene expression. Therefore, extensive research on the mechanisms of tumor resistance to chemotherapy has been focused on the gene and protein levels, returning initial results.
As the mechanisms underlying the drug resistance of tumor cells are highly complicated, studying them requires the establishment of suitable resistant cell models [12]. An in vitro induction method has been used to obtain drug resistant tumor cells since in vitro dosages are small, the study source is convenient, operation is easy, and experimental results are quick to obtain. Therefore, drug resistance of tumor cells and its related mechanisms cannot be studied without in vitro experiments, mainly using sustained drug effect methods. In this study, we established the VCR drug resistant cell line HCT-8/VCR by using a gradually increasing drug concentration method. The cell line is resistant to a VCR concentration of 2 × 103 ng/mL. Its drug resistance is 19.7 times higher than that of the parental HCT-8 cell line. When cell morphology was examined using light microscopy, obvious differences between the drug resistant cell line HCT-8/VCR and the sensitive cell line HCT-8 were recorded: Under low concentrations of VCR, HCT-8 cells grew slowly and the morphology of adherent cells appeared irregular. The adherent cells appeared triangular, polygonal, and had small protrusions, and the cell surface became burr shaped. When subcultured in fresh medium not containing VCR, cell growth accelerated considerably, and the morphology of cells gradually changed to normal. HCT-8 cells have a clear boundary and are singly dispersed. Induced HCT-8/VCR resistant cells are bigger and dark in color. There are obvious changes like intracellular appearance, projections, and burrs. Growth curves indicated a slower growth rate of HCT-8/VCR cells than that of the parental HCT-8 cells.
One of the important characteristics of malignant tumors is that the regulation of the cell cycle is out of control, causing cells to grow disorderly, lacking differentiation [13]. Flow cytometry data showed that the ratio of S-phase cells was reduced and that of G1-stage cells significantly (P < 0.05) increased in HCT-8/VCR resistant cells, which may be the reason why drug-resistant cells grow slowly.
In summary, the vincristine resistant colon carcinoma cell line HCT-8/VCR was successfully established using the sustained drug effects method. Resistant cells displayed moderate resistance, and VCR showed some influence on the colon cancer cell cycle. The establishment of this model lays the foundation for further studies on the mechanism of drug resistance in colon cancer cells.
Small RNAs include miRNA, piRNA, and siRNA. In this study, we found that small RNA could regulate gene expression and affect individual development, metabolism, incidence of disease, and other physiological processes. MicroRNA (miRNA) comprises small single-stranded RNA of about 21-25 nt fragment length, widely occurring in eukaryotic organisms. Mature miRNAs have a hairpin structure and are made from a single-stranded RNA precursor (pre-miRNA) by Dicer enzyme processing. As target gene expression can be regulated by binding to (imperfectly) complementary target genes, miRNA has extensive applications in cell development, multiple biomarker fields, and tumor therapy.
Numerous studies show that miRNAs can regulate the expression of oncogenes or tumor suppressor genes and thereby play roles of oncogenes or tumor suppressor genes. Using gene chip technology to detect the expression of miRNA in tumor tissue samples, it was found that the expression of a small portion of miRNAs was increased, while most of the miRNAs in the tumor samples appeared to be downregulated [14]. Studies on lung cancer samples showed that the expression of Let-7 was downregulated, showing positive correlation with prognosis. While the expression of miR-125b, miR-21, miR-155, and miR-145 was significantly downregulated in breast cancer [15] that of miR-17, miR-18, and miR-19 was upregulated in lung cancer samples [16]. The expression level of precursor miR-155 was found to be upregulated in Burkitt lymphoma [17].
In recent years, more and more evidence has shown that miRNA is not only closely related to tumorigenesis but also plays an important role in the drug resistance of tumor cells. MiR-122 showed specifically low expression in hepatocellular carcinoma (HCC) cells. Through genetic engineering, miR-122 could be transferred into HCC cells for overexpression and downregulate the expression of GST, MRP, MDR-1, and cyclin Bl. CKDl can block cancer cells in the G2/M phase and increase their sensitivity to doxorubicin and vincristine [18]. Adriamycin (ADM) concentrations required for chronic myelogenous leukemia cells of the adriamycin-resistant line K562/A02 are 180 times those required for the parental cell line K562, which is concomitant with the significant upregulation of miR-451, miR-155, and miR-221 expression and downregulation of let-7f and miR-424 expression in the resistant line. The expression rate of the cell membrane protein PgP is only 0.2% in K562 cells, while in the K562/A02 cell line it is 86% [19]. These differences suggest that miRNAs may play a role in the drug resistance of leukemia cells by regulating the expression of PgP, but the mechanism is still not clear. Downregulation of miR-27a in esophageal squamous cell carcinoma induced the downregulation of PgP expression, thus improving drug sensitivity. Pan et al. found that the protein BCRP/A1G2 in human breast cancer cells was associated with resistance to multiple drugs [20]. MiR-328 acts on 3’-UTRs of ABCG2 and enhances the chemosensitivity of tumor cells by inhibiting ABGG2 expression.
Using HiSeq 2500 sequencing and bioinformatics methods, we detected miRNA expression levels in colon cancer drug resistant cells in this study. First, we extracted total RNA of high quality using the Trizol method, clearly detecting bands by electrophoresis, indicating complete total RNA extraction. Bands of 28S (about 4.5 kb), 18S (about 2.1 kb), and 5S (about 0.1-0.3 kb) of small molecule RNA were clearly detected on the gel, and no RNA degradation was noticed. After removal of adaptors and quality inspection of fragmental base composition and lengths, sequencing results showed high numbers of total reads and a high proportion of high quality reads, ensuring the quality of sequencing. Among of the samples, 22 nt and 23 nt fragments shared the highest abundance, with 22 nt fragments accounting for 39.87% and 23 nt accounting for 15.24%. For the remaining length fragments, 21 nt and 24 nt fragments showed proportions of 12.11% and 9%, respectively. All clear reads were compared to the human genome, with HCT-8 covering an average proportion of matches of 67.56%, while the average of HCT-8/VCR amounted to 68.04%. sRNA library annotation showed that the rRNA of HCT-8 cells accounted for 41.4% of the total RNA, while the rRNA of HCT-8/VCR accounted for 47% of the total RNA. The number of unique miRNAs of HCT-8 cells was 14527, accounting for 11.4% of the total number of sequences, while the number of unique HCT-8/VCR miRNAs was 18543, accounting for 7% of the total number of sequences. This shows a significant difference in miRNA expression between VCR-resistant and -sensitive colon cancer cells.
Conclusions
In this study, we established the VCR-resistant colon cancer cell line HCT-8/VCR using a concentration gradient of VCR. HCT-8/VCR cells grew healthy when treated with a concentration of 2 × 103 ng/mL VCR. HiSeq 2500 sequencing technology was used for the screening of differentially expressed miRNA. A total of 1651 miRNAs were sequenced, resulting in the identification of 48 miRNAs with differential expression. Compared with HCT-8 colon cancer cells with no drug resistance, 24 miRNAs of the VCR resistant cell line HCT-8/VCR showed statistically significant differences in their expression (P < 0.05), including 17 miRNAs with upregulated expression (P < 0.05).
Acknowledgements
This study was supported by the Project from Henan Collaborative Innovation Center of Molecular Diagnosis and Laboratory Medicine (No. XTCX-2015-ZD1).
Disclosure of conflict of interest
None.
References
- 1.Li CL, Liu JW, Sa XY, Deng WL, Xu JH, Fan ZZ. Preliminary study on establishment of a VCR-resistant colon carcinoma cell line and its underlying mechanism of drug-resistance. Tumor. 2009;29:31–34. [Google Scholar]
- 2.Guo YS. The screening for oxaliplatin resistant microRNAs and research on their mechanisms in colon cancer cell. Doctor Dissertation of Tianjin Medical University; 2013. [Google Scholar]
- 3.Kimura M, Endo H, Inoue T, Nishino K, Uchida J, Kumagai T, Kukita Y, Kato K, Imamura F, Inoue M. Analysis of ERBB ligand-induced resistance mechanism to crizotinib by primary culture of lung adenocarcinoma with EML4-ALK fusion gene. J Thorac Oncol. 2015;10:527–530. doi: 10.1097/JTO.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Ngo JA, Wetzel MD, Marchetti D. Heparanase mediates a novel mechanism in lapatinib-resistant brain metastatic breast cancer. Neoplasia. 2015;17:101–113. doi: 10.1016/j.neo.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borges S, Döppler HR, Storz P. A combination treatment with DNA methyltransferase inhibitors and suramin decreases invasiveness of breast cancer cells. Breast Cancer Res Treat. 2014;144:79–91. doi: 10.1007/s10549-014-2857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma J, Fang B, Zeng F, Ma C, Pang H, Cheng L, Shi Y, Wang H, Yin B, Xia J, Wang Z. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740–1749. doi: 10.18632/oncotarget.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sianou A, Galyfos G, Moragianni D, Andromidas P, Kaparos G, Baka S, Kouskouni E. The role of microRNAs in the pathogenesis of endometrial cancer: a systematic review. Arch Gynecol Obstet. 2015;292:271–282. doi: 10.1007/s00404-015-3660-y. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. doi: 10.5812/hepatmon.18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao J, Peng F, Yu C, Wang M, Li X, Li Z, Jiang J, Sun C. microRNA-137 modulates pancreatic cancer cells tumor growth, invasion and sensitivity to chemotherapy. Int J Clin Exp Pathol. 2014;7:7442–7450. [PMC free article] [PubMed] [Google Scholar]
- 10.Matuszcak C, Haier J, Hummel R, Lindner K. MicroRNAs: promising chemoresistance biomarkers in gastric cancer with diagnostic and therapeutic potential. World J Gastroenterol. 2014;20:13658–13666. doi: 10.3748/wjg.v20.i38.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burden C, Rose D, Daniels IR. Bowel Cancer: the outcome is improving. J R Soc Promot Health. 2005;25:255–258. doi: 10.1177/146642400512500606. [DOI] [PubMed] [Google Scholar]
- 13.Dictor M, Ehinger M, Mertens F, Akervall J, Wennerberg J. Abnormal cell cycle regulation in malignancy. Am J Clin Pathol. 1999;12:S40–S52. [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 17.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 18.Cao YX, Dai CW, Zhang GS. Screening for drug resistance related microRNAs in K562 and K562/A02 cell lines. Zhonghua Xue Ye Xue Za Zhi. 2010;31:361–365. [PubMed] [Google Scholar]
- 19.Zhang H, Li M, Han Y, Hong L, Gong T, Sun L, Zheng X. Down-regulation of miR-27a might reverse multidrug resistance of esophageal squamous cell carcinoma. Dig Dis Sci. 2010;55:2545–2551. doi: 10.1007/s10620-009-1051-6. [DOI] [PubMed] [Google Scholar]
- 20.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]


