Abstract
The family Picornaviridae is a large and diverse group of viruses that infect humans and animals. Picornaviruses are among the most common infections of humans and cause a wide spectrum of acute human disease. This study began as an investigation of acute flaccid paralysis (AFP) in a small area of eastern Bolivia, where surveillance had identified a persistently high AFP rate in children. Stools were collected and diagnostic studies ruled out poliovirus. We tested stool specimens from 51 AFP cases and 34 healthy household or community contacts collected during 2002–2003 using real-time and semi-nested RT-PCR assays for enterovirus, parechovirus, cardiovirus, kobuvirus, salivirus, and cosavirus. Anecdotal reports suggested a temporal association with neurologic disease in domestic pigs, so six porcine stools were also collected and tested with the same set of assays, with the addition of an assay for porcine teschovirus. A total of 126 picornaviruses were detected in 73 of 85 human individuals, consisting of 53 different picornavirus types encompassing five genera (all except Kobuvirus). All six porcine stools contained porcine and/or human picornaviruses. No single virus, or combination of viruses, specifically correlated with AFP; however, the study revealed a surprising complexity of enteric picornaviruses in a single community.
Introduction
Picornaviruses are among the most common viruses infecting humans. Enteroviruses (EV), including EV species A to D, and rhinovirus species A to C, are the most thoroughly characterized human picornaviruses and cause a wide spectrum of acute clinical disease, including undifferentiated febrile illness, respiratory illness, aseptic meningitis, and acute flaccid paralysis (AFP). Human parechoviruses (HPeV) have a similar disease spectrum as the EVs, but routine clinical testing is less common despite increasing evidence of a significant disease burden in infants (Harvala et al., 2010). Zoonotic infection with encephalomyocarditis virus (EMCV) has been reported (Oberste et al., 2009) but testing for this virus is rarely performed in human clinical virology laboratories. No routine testing is available for newer human picornaviruses, which include Saffold viruses (SAFV) (Cardiovirus), cosaviruses (CoSV) (also published as “dekavirus”; Cosavirus), and “salivirus” (SALV) (also published as “klassevirus”; Salivirus) (Adams et al.; Knowles et al., 2012).
AFP is a manifestation of many different disease processes with different mechanisms, and multiple causes, both infectious and non-infectious, including paralytic poliomyelitis, certain arboviruses, Guillain-Barré syndrome (GBS), transverse myelitis, certain toxins, and trauma. Surveillance for AFP is recommended by the World Health Organization (WHO) as the standard approach for poliovirus surveillance for the purposes of Global Polio Eradication (Anonymous, 2006). A wide variety of non-polio enteroviruses and other picornaviruses have been identified in stool specimens of AFP cases (Bingjun et al., 2008; Kapoor et al., 2001; Kapoor et al., 2008; Oberste et al., 2006; Saeed et al., 2007; Santos et al., 2002; Shoja et al., 2007). Detection of high-prevalence agents from a non-sterile site is not sufficient to conclusively establish an etiologic link of picornavirus infection with disease; however, given the known ability of many picornaviruses to cause central nervous system (CNS) disease, it is likely that these viruses contribute to at least a fraction of AFP cases. From 1996 to 2006, AFP surveillance activities in Bolivia identified persistently high AFP rates in Hernando Siles province, Chuquisaca Department, a relatively poor, largely agricultural area in southern Bolivia. These unusually high AFP rates—40 to 140 per 100,000 children under age 15, compared to 1.2 to 2.3 per 100,000 for Bolivia as a whole—have raised considerable local public concern. In all cases, wild poliovirus has been ruled out by laboratory testing through the routine AFP surveillance program (Anonymous, 2006); environmental contamination of water and effects of agricultural chemicals were also ruled out. There were anecdotal reports of concurrent outbreaks of paralytic illness in local pigs, though no direct evidence linking human and porcine diseases was available.
The plausibility of a picornavirus etiology and the availability of archived fecal specimens presented an opportunity to assess the role of non-polio picornaviruses in pediatric AFP and the diversity of enteric picornaviruses in children in rural Bolivia.
Results
A total of 49 AFP cases (32 from Chuquisaca, and 17 from Santa Cruz) and 32 contacts (17 from Chuquisaca and 15 from Santa Cruz) were included in the clinical analyses. Of the 49 AFP cases, 26 were reported in 2002 and 23 in 2003. Detailed clinical and epidemiologic information was available for 45 cases. The median age of cases was 5 years (range, 1–21 years) with >90% occurring among children aged ≤8 years; 19 (42%) were male. There was significant geographic difference in seasonality, as all cases from Chuquisaca had onset during February-May, while cases from Santa Cruz were distributed throughout the year, with February to May accounting for 58.8% (10 cases) (p < 0.001). Most (33 or 75.0%) of the 44 cases for whom available clinical details were sufficient for classification presented with GBS-like paralysis, one case (2.3%) had enterovirus/polio-like paralysis and 10 cases (22.7%) had a mixed clinical picture (Table 1). Clinical presentations of AFP cases in both departments were comparable.
Table 1.
Clinical characteristics of AFP cases from Chuquisaca and Santa Cruz departments, Bolivia, 2002–2003.
| Characteristics | N (%) |
|---|---|
| Duration of progression, mean (range) | 6 (1–15) days |
| Ascending paralysis | 33/43 (76.7%) |
| Descending paralysis | 2/43 (4.7%) |
| Simultaneous paralysis in all 4 limbs | 8/43 (18.6%) |
| Symmetric paralysis | 36/43 (83.7%) |
| Paresthesias | 5/24 (20.8%) |
| Sensory deficits | 19/30 (63.3%) |
| Muscle pain | 36/40 (90.0%) |
| Febrile/”infectious” prodrome | 24/43 (55.8%) |
| Fever at paralysis onset | 20/43 (46.7%) |
| Residual paralysis/weakness (>70 days after onset) |
28/43 (65.1%) |
| Death | 2/43 (4.7%) |
All of the human stools collected were analyzed for picornaviruses (N = 85 total; 51 AFP and 34 contacts). At least one picornavirus was detected in specimens of 73 (85.9%) study subjects, including 37 apparent coinfections with >1 picornavirus, with evidence of up to six different viruses (five different picornaviral genera) in a single patient (Table 2). There were no significant differences between virus detection rates by case/contact status or between departments, suggesting that picornavirus infection was not associated with AFP in these cases. Nevertheless, the study presented an opportunity to examine the diversity of enteric picornaviruses in this population.
Table 2.
Picornavirus detection rates in stool specimens of AFP cases and their contacts, Chuquisaca and Santa Cruz departments, Bolivia, 2002–2003.
| Categories | Total | Chuquisaca | Santa Cruz | |||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Total subjects | 85 | 100.0 | 51 | 100.0 | 34 | 100.0 |
| Any picornavirus (+) | 73 | 85.9 | 47 | 92.2 | 26 | 76.5 |
| Enterovirus (+) | 44 | 51.8 | 30 | 58.8 | 14 | 41.2 |
| Parechovirus (+) | 16 | 18.8 | 10 | 19.6 | 6 | 17.6 |
| Cardiovirus (+) | ||||||
| SAFV | 16 | 18.8 | 9 | 17.6 | 7 | 20.6 |
| EMCV | 5 | 5.9 | 3 | 5.9 | 2 | 5.9 |
| Cosavirus (+) | 44 | 51.8 | 30 | 58.8 | 14 | 41.2 |
| Kobuvirus (+) | 0 | 0 | 0 | 0 | 0 | 0 |
| Salivirus (+) | 1 | 1.2 | 1 | 2.0 | 0 | 0 |
| >1 picornavirus (+) | 37 | 43.5 | 21 | 41.2 | 16 | 47.0 |
| AFP cases | 51 | 100.0 | 32 | 100.0 | 19 | 100.0 |
| Any picornavirus (+) | 43 | 84.3 | 28 | 87.5 | 15 | 79.0 |
| Enterovirus (+) | 22 | 43.1 | 15 | 46.9 | 7 | 36.8 |
| Parechovirus (+) | 8 | 15.7 | 7 | 21.9 | 1 | 5.3 |
| Cardiovirus (+) | ||||||
| SAFV | 11 | 21.6 | 8 | 25.0 | 3 | 15.8 |
| EMCV | 4 | 7.8 | 3 | 9.4 | 1 | 5.3 |
| Cosavirus (+) | 27 | 52.9 | 17 | 53.1 | 10 | 52.6 |
| Kobuvirus (+) | 0 | 0 | 0 | 0 | 0 | 0 |
| Salivirus (+) | 1 | 2.0 | 1 | 3.1 | 0 | 0 |
| >1 picornavirus (+) | 20 | 39.2 | 14 | 43.8 | 6 | 31.6 |
|
Contacts of AFP cases |
34 | 100.0 | 19 | 100.0 | 15 | 100.0 |
| Any picornavirus (+) | 30 | 88.2 | 19 | 100.0 | 11 | 73.3 |
| Enterovirus (+) | 22 | 64.7 | 15 | 78.9 | 7 | 46.7 |
| Parechovirus (+) | 8 | 23.5 | 3 | 15.8 | 5 | 33.3 |
| Cardiovirus (+) | ||||||
| SAFV | 5 | 14.7 | 1 | 5.3 | 4 | 26.7 |
| EMCV | 1 | 2.9 | 0 | 0 | 1 | 6.7 |
| Cosavirus (+) | 17 | 50.0 | 13 | 68.4 | 4 | 26.7 |
| Kobuvirus (+) | 0 | 0 | 0 | 0 | 0 | 0 |
| Salivirus (+) | 0 | 0 | 0 | 0 | 0 | 0 |
| >1 picornavirus (+) | 17 | 50.0 | 10 | 52.6 | 7 | 46.7 |
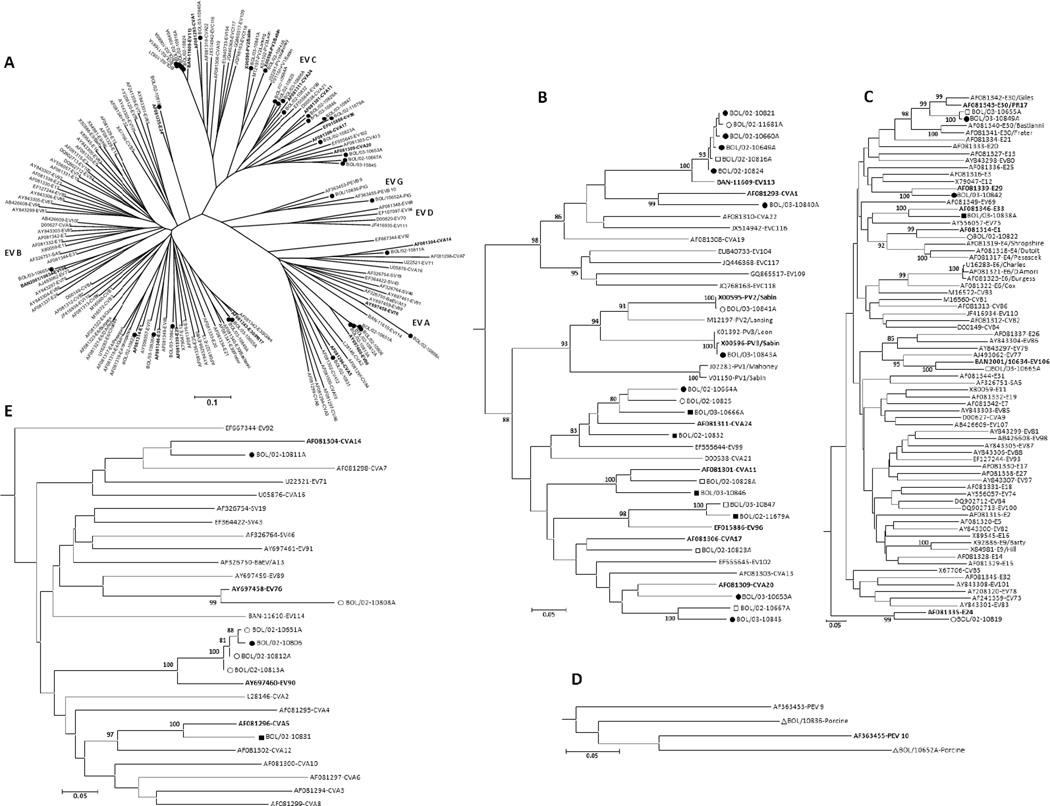
Picornaviruses detected in human stools included members of the Enterovirus, Parechovirus, Cardiovirus, Salivirus, and Cosavirus genera (Tables 2 and 3). EVs were detected in 44 specimens, representing 19 different types from three different EV species (Table 3; Figure 1). Members of EV-A, including coxsackievirus (CV) A5, CVA14, EV76, and EV90, were identified in nine instances (Figure 1E). Nine specimens yielded EV-B, including echovirus (E) 1, E24, E29, E30, E33, and EV106 (Figure 1C). Poliovirus vaccine strains, commonly found in pediatric fecal samples in countries that use live-attenuated, oral polio vaccine, were detected in two samples, one PV2 and one PV3. Other EV-C viruses were also detected, including CVA1, CVA11, CVA17, CVA20, CVA24, EV96, and EV113 (22 detections) (Figure 1B). In two cases, VP1 sequences appeared to be mixtures of EVs and the individual sequences could not be resolved. Two additional specimens were 5’NTR real-time positive with high Ct values near the limit of detection, and negative with the confirmatory/typing EV VP1 assay.
Table 3.
Picornaviruses detected and sequenced (VP1 gene region) from human stool specimensa.
| Genus/Species | Type | No. Sequenced | Chuquisaca | santa Cruz |
AFP | Contact |
|---|---|---|---|---|---|---|
| Enterovirus/EV-A | CVA5 | 1 | 0 | 1 | 1 | 0 |
| Enterovirus/EV-A | CVA14 | 1 | 1 | 0 | 1 | 0 |
| Enterovirus/EV-A | EV-A76 | 1 | 1 | 0 | 0 | 1 |
| Enterovirus/EV-A | EV-A90 | 6 | 6 | 0 | 1 | 5 |
| Enterovirus/EV-B | E1 | 1 | 1 | 0 | 0 | 1 |
| Enterovirus/EV-B | E24 | 3 | 3 | 0 | 1 | 2 |
| Enterovirus/EV-B | E29 | 1 | 1 | 0 | 1 | 0 |
| Enterovirus/EV-B | E30 | 2 | 1 | 1 | 1 | 1 |
| Enterovirus/EV-B | E33 | 1 | 0 | 1 | 1 | 0 |
| Enterovirus/EV-B | EV-B106 | 1 | 0 | 1 | 0 | 1 |
| Enterovirus/EV-C | PV2 b | 1 | 1 | 0 | 0 | 1 |
| Enterovirus/EV-C | PV3 b | 1 | 1 | 0 | 1 | 0 |
| Enterovirus/EV-C | CVA1 | 1 | 1 | 0 | 1 | 0 |
| Enterovirus/EV-C | CVA11 | 3 | 1 | 2 | 2 | 1 |
| Enterovirus/EV-C | CVA17 | 1 | 0 | 1 | 0 | 1 |
| Enterovirus/EV-C | CVA20 | 3 | 2 | 1 | 2 | 1 |
| Enterovirus/EV-C | CVA24 | 4 | 2 | 2 | 3 | 1 |
| Enterovirus/EV-C | EV-C96 | 2 | 0 | 2 | 1 | 1 |
| Enterovirus/EV-C | EV-C113 | 6 | 5 | 1 | 4 | 2 |
| Parechovirus/HPeV | HPeV1 | 2 | 2 | 0 | 1 | 1 |
| Parechovirus/HPeV | HPeV2 | 2 | 1 | 1 | 1 | 1 |
| Parechovirus/HPeV | HPeV3 | 1 | 0 | 1 | 0 | 1 |
| Parechovirus/HPeV | HPeV4 | 4 | 3 | 1 | 2 | 2 |
| Parechovirus/HPeV | HPeV7 | 3 | 1 | 2 | 2 | 1 |
| Parechovirus/HPeV | HPeV9 | 3 | 3 | 0 | 2 | 1 |
| Parechovirus/HPeV | HPeV12 | 1 | 0 | 1 | 0 | 1 |
| Cardiovirus/TMEV | SAFV 1 | 5 | 3 | 2 | 3 | 2 |
| Cardiovirus/TMEV | SAFV 2 | 2 | 2 | 0 | 1 | 1 |
| Cardiovirus/TMEV | SAFV 4 | 2 | 1 | 1 | 2 | 0 |
| Cardiovirus/TMEV | SAFV 9 | 2 | 1 | 1 | 1 | 1 |
| Salivirus | SALV A1 | 1 | 1 | 0 | 1 | 0 |
| Cosavirus | CoSV A3 | 3 | 3 | 0 | 2 | 1 |
| Cosavirus | CoSV A7 | 1 | 1 | 0 | 1 | 0 |
| Cosavirus | CoSV A8 | 3 | 3 | 0 | 3 | 0 |
| Cosavirus | CoSV A17 | 2 | 2 | 0 | 2 | 0 |
| Cosavirus | CoSV A24 | 2 | 0 | 2 | 1 | 1 |
| Cosavirus | CoSV D4 | 2 | 2 | 0 | 0 | 2 |
| Cosavirus | New (15 genotypes) | 22 | 14 | 8 | 14 | 8 |
Only viruses detected and successfully sequenced (VP1 gene region) from human specimens are listed here.
All polioviruses were identified as Sabin-like by sequencing.
Figure 1.
Phylogenetic relationships among human enteroviruses detected in Hernando Siles province. The VP1 nucleic acid tree was constructed by the neighbor-joining method implemented in MEGA, version 4, using the Kimura 2-parameter substitution model (Jones et al., 1992) and 1000 bootstrap pseudoreplicates. E, echovirus; EV, enterovirus; CV, coxsackievirus. A. The radial tree shows the distribution of EV species (EV-A, -B, -C, -D, and porcine EV-G) typed from Bolivia (filled circles). B-E. Subtrees of EV-C, EV-B, EV-G, and EV-A respectively. Chuquisaca: solid circle = AFP case, hollow circle = contact, hollow triangle = porcine; Santa Cruz: solid square = AFP case, hollow square = contact. Prototype viruses detected in Bolivia are in bold. Only full length, double-stranded partial VP1 sequences were included for phylogenetic analyses.
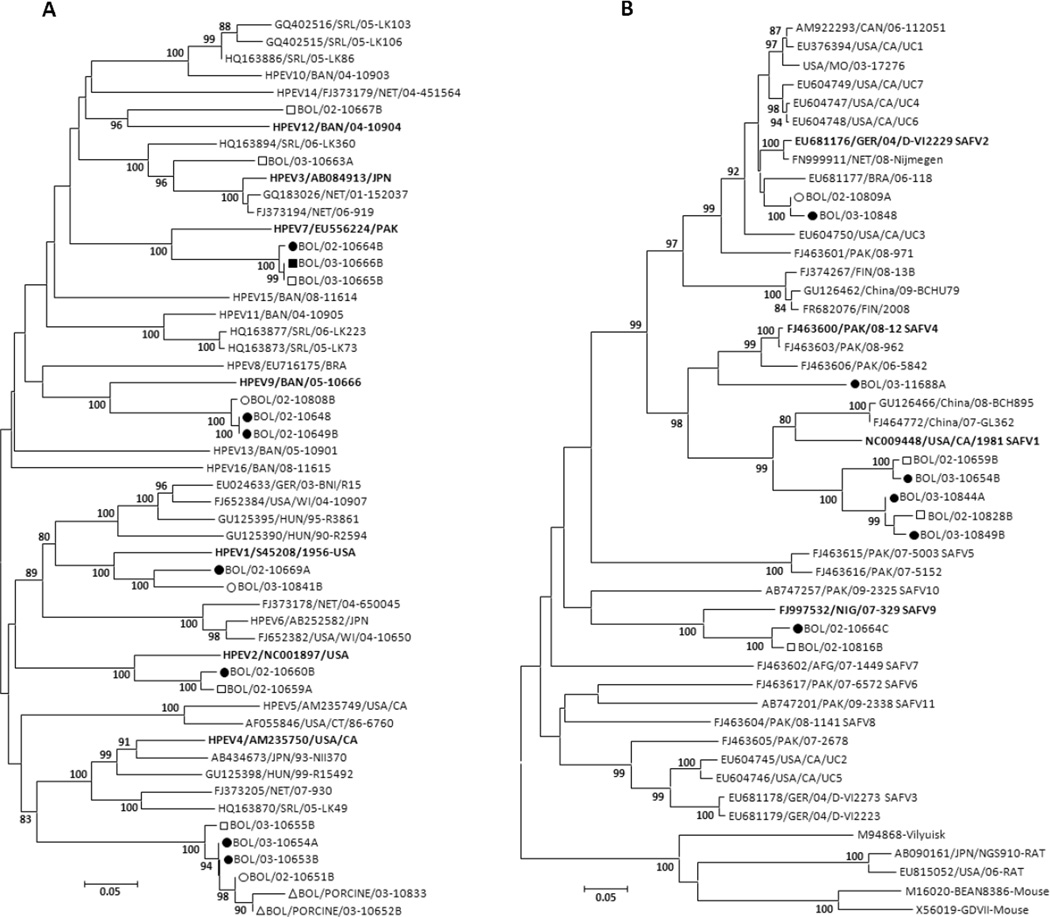
Sixteen human specimens yielded seven types of human parechoviruses (HPeV), including HPeV1, HPeV2, HPeV3, HPeV4, HPeV7, HPeV9, and HPeV12 (Tables 2 and 3; Figure 2A). HPeV4 was the most common HPeV (four human specimens), followed by HPeV7 and HPeV9. The Bolivia HPeV1 VP1 sequences were most closely related to the HPeV1 prototype strain Harris/USA-1956 (84% nucleotide identity [%NT ID] and 95% amino-acid identity [% AA ID]), unlike most recent HPeV1s, which have split into a separate HPeV1 clade (Figure 2A). The two HPeV2 strains from Bolivia are most closely related to the HPeV2 prototype strain Williamson/USA-1956 (86% NT ID and 96% AA ID). Notably, the Bolivia HPeV4 VP1 carboxyl-termini lacked the RGD motif found in all other HPeV4s in GenBank (data not shown), and had instead a variant consensus carboxyl-terminal end (17 amino-acids). We were unable to grow these variant HPeV4s in cell culture, unlike the prototype HPeV4 that grows readily in Vero and other cell types. The Bolivia HPeV4s shared six identical amino acids at the putative VP1-2A cleavage site (NPFEDE / S) and significant amino acid similarity in the remainder of the carboxyl-tail (data not shown) with HPeV1 and HPeV5 variants from the Netherlands and with HPeV15 from Bangladesh (Oberste et al., 2013). The single HPeV3 found in Bolivia also lacked the RGD motif. We did not attempt to grow this virus in culture. HPeV1s and HPeV2s found in Bolivia all contained the RGD motif.
Figure 2.
A. Phylogenetic relationships among human parechoviruses detected in Hernando Siles province. B. Phylogenetic relationships among human cardioviruses detected in Hernando Siles province. The VP1 nucleic acid trees were constructed by the neighbor-joining method implemented in MEGA, version 4, using the Kimura 2-parameter substitution model (Jones et al., 1992) and 1000 bootstrap pseudoreplicates. Chuquisaca: solid circle = AFP case, hollow circle = contact, hollow triangle = porcine; Santa Cruz: solid square = AFP case, hollow square = contact. Prototype viruses detected in Bolivia are in bold. Only full length, double-stranded partial VP1 sequences were included for phylogenetic analyses.
Cardiovirus was detected in 21 human specimens (Table 2), including 16 detections of members of the species Theilovirus, and five detections of members of the Encephalomyocarditis virus (EMCV) species. Partial VP1 sequencing was successful for 11 of 16 theilovirus-positive specimens and all were identified as SAFV (Table 3; Figure 2B). SAFV1 was the most common type identified, with five positive stools, followed by SAFV2, SAFV9, and SAFV4 (Figure 2B). Phylogenetically, SAFV1s split into two Bolivia clades, sharing 87.9 to 89.1% NT ID between them (99.1 to 100 % AA ID). Both clades had from 77.3 to 79.2 % NT ID (94.4 to 95.8 % AA ID) with the 1981 SAFV1 prototype strain from the United States (NC009448). The two SAFV2 strains clustered with SAFV2 from Brazil. One SAFV sequence appeared to be a mixture and four other specimens were SAFV 5’NTR real-time positive, but negative in the SAFV VP1 assay. All EMCV-positive specimens had very high threshold cycle (Ct) values (from 40 to 45) near the limit of detection (Ct of 45), indicating very low genome copy number, and could not be independently confirmed with the EMCV VP1 sequencing/typing assay.
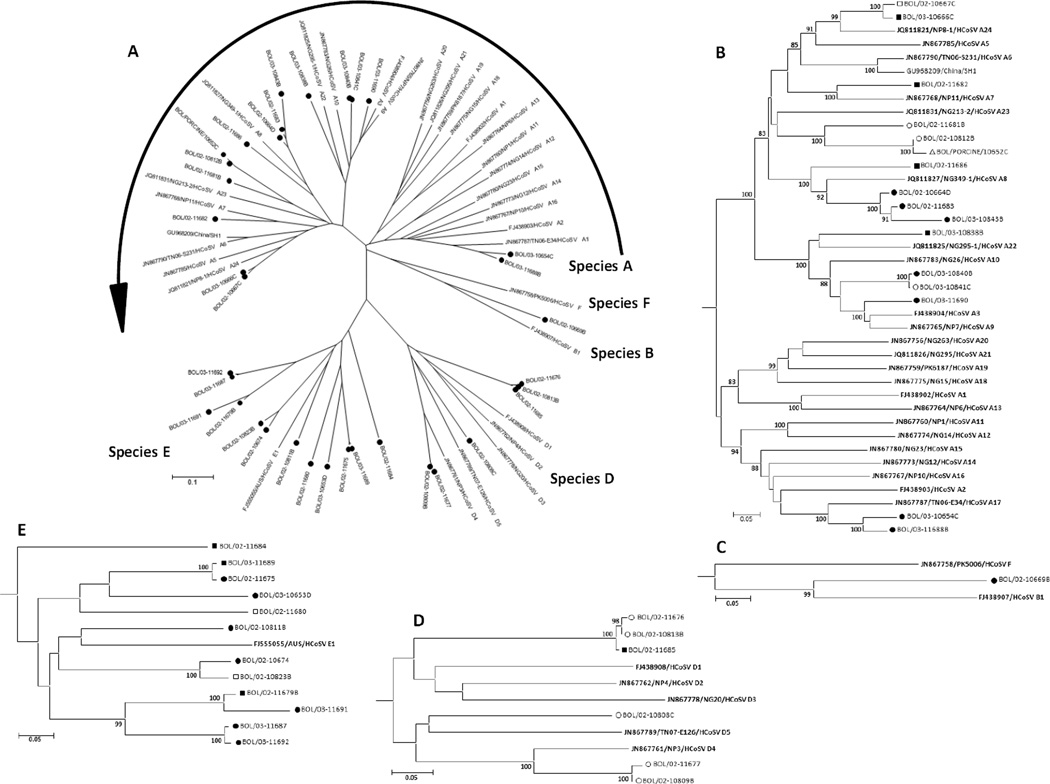
Human cosaviruses (CoSV) were detected in 44 human stool specimens (Ct values from 30 to 44.7), 35 of which yielded clean partial VP1 sequences (Tables 2 and 3, Figure 3A–E). Five CoSV sequences appeared to be mixtures, which could not be resolved and four specimens with high CoSV real-time Ct values were not amplified by the CoSV VP1 sequencing assay. Three CoSVs were typed as CoSV A3 (77 to 85.2% NT ID and 93.2 to 100% AA ID), three as CoSV A8 (76–94.5% NT ID and 88–97.6% AA ID), two as CoSV A17 (78.8–90% NT ID and 93.4–98.2% AA ID), one as CoSV A7 (77.9% NT ID and 95% AA ID) and two as CoSV D4 (80–99.5% NT ID and 88.5–90.7% AA ID). CoSV species A was the most common CoSV detected (15 viruses; eleven types), followed by CoSV E (13 viruses; nine types), CoSV D (six viruses; three types), and CoSV B (one virus). Analysis of the partial VP1 sequences of the Bolivia CoSVs suggests that 22 of them can be classified as 15 new types (nine CoSV E, three CoSV A, two CoSV D and one CoSV B). All of their partial VP1 sequences have <75% NT ID and <88% AA ID to all previously characterized CoSV types (Holtz et al., 2008; Kapoor et al., 2008).
Figure 3.
Phylogenetic relationships among human cosaviruses (CoSV) detected in Hernando Siles province. The VP1 nucleic acid tree was constructed by the neighbor-joining method implemented in MEGA, version 4, using the Kimura 2-parameter substitution model (Jones et al., 1992) and 1000 bootstrap pseudoreplicates. A. The radial tree shows the distribution of CoSV species (CoSV-A, -B, -D, and -G) typed from Bolivia (filled circles). B-E. Subtrees of CoSV-A, -B, -D, and -E respectively. Chuquisaca: solid circle = AFP case, hollow circle = contact, hollow triangle = porcine; Santa Cruz: solid square = AFP case, hollow square = contact. All proposed prototype CoSVs are shown in bold. Only full length, double-stranded partial VP1 sequences were included for phylogenetic analyses.
A single specimen was positive by 5’NTR real-time RT-PCR targeting human kobuvirus and salivirus but not canine, murine, porcine or bovine kobuviruses (in silico analyses only) (Tables 2 and S1), with the partial VP1 sequence having 79.8% NT ID (88% AA ID) to the prototype SALV A1 (GQ507022) and 92.4% NT ID (96% AA ID) to a candidate SALV A2 (JQ898343) from Thailand (2009), sequenced directly from sewage (Ng et al., 2012). Based on the partial VP1 sequence analyses, all three of these viruses are SALV A1. None of the human specimens tested positive for teschovirus.
Picornaviruses were detected in all six pig specimens (Table 2). Partial VP1 sequences from two of these were porcine enteroviruses (PEV) (Figure 1D), one of which may represent a new PEV type, having only 73% NT ID (76% AA ID) to the closest PEV in GenBank (GQ502355), while the other PEV shared 78% NT ID (86% AA ID) with PEV-10 (GQ502355). Parechovirus was detected in three pig specimens; two were identified as HPeV4 (Figure 2A), but the third (Ct = 38.6) produced no VP1 amplicon, so the type could not be determined. One cardiovirus of TMEV species was detected (Ct = 38), but VP1 sequence could not be obtained. CoSV with a partial VP1 sequence that was a close match to a Bolivia CoSV sequence from a human specimen (98.9% NT ID and 97.6% AA ID) was detected in one pig stool (Figure 3B). Teschovirus was detected and identified in three of six pigs with no further characterization. No human kobuviruses or saliviruses were detected in the pigs. Four picornaviruses were detected in a single pig, including teschovirus, PEV (Ct = 25), CoSV (Ct = 42.5), and HPeV4 (Ct = 35.4).
The four human and one porcine HPeV4 with complete VP1 sequences were 97 to 100% NT ID (97.4 to 100% AA ID) to one another and 73.0 to 73.9% NT ID (84.1 to 85.8% AA ID) to the HPeV4 prototype (AM235750). With <75% NT ID and <88% AA ID to the HPeV4 prototype, the Bolivia HPeV4s could possibly be classified as a new type (Nix et al., 2010); however, most of the difference between the prototype and the Bolivia HPeV4s was due to variation in 17 amino acids (51 nucleotides) at the carboxyl-terminal end. When these 17 amino acids were omitted from the analysis, the Bolivia HPeV4s were 89.3 to 91.1% AA ID (75.9 to 76.8% NT ID) to the HPeV4 prototype strain.
Discussion
Our study demonstrates that picornavirus diversity in human and animal feces is clearly much more complex than previously known. Nevertheless, our findings suggest that picornaviruses are not a major contributor to the very high AFP rates observed in the Hernando Siles province of Bolivia. The basis for this conclusion includes the predominance of GBS-like presentation usually not consistent with picornavirus etiology, tremendous diversity of identified picornaviruses without any predominant agents, substantial overlap of results between cases and contacts, or between clinical subtypes, and lack of significant differences between the departments with high and low incidence. The involvement of picornaviruses in individual cases cannot be ruled in or out based on the available data.
Molecular methods detect a wider range of picornaviruses than cell culture alone (Begier et al., 2008) and several of the picornaviruses found in this study have not been cultured to date (CoSV, SALV, and new HPeV types), thereby confirming the superior performance of molecular approaches for surveillance, diagnostics, and genotyping of picornaviruses. Several other studies, using both classical and molecular methods, have detected EV, HPeV, SAFV, and CoSV in fecal samples from AFP cases (Bingjun et al., 2008; Junttila et al., 2007; Kapoor et al., 2001; Oberste et al., 2006; Saeed et al., 2007). Most previous studies, including our own, used specific PCR assays to detect members one or two genera (usually only Enterovirus), due to specificity, low cost, and potential for high throughput, though some investigators are starting to use metagenomics methods to expand the range of viruses that can be detected (Victoria et al., 2009), with limitations of higher per-sample cost and lower overall throughput. Our approach was a compromise between specific PCR and metagenomics, allowing the possibility of detecting seven different genera within the family Picornaviridae. In areas where conditions are favorable for fecal-oral transmission, one should expect to find a wide variety of enteric picornaviruses. As metagenomic approaches become more cost effective, a wider range of potential pathogens can be detected in large studies, facilitating better assessment of disease association.
A diverse range of EVs was identified in this study (Figure 1), including 19 different types, among them a newly identified type, EV-C113, which along with EV-A90 were the most common viruses identified in the study, and another recently discovered type, EV-B106. Of the four EV species EV-C was the most common. No EV-D viruses were identified.
Our study also demonstrated substantial diversity of HPeVs in Bolivia (Figure 2A). One of the seven different types identified had a distinct genetic characteristic, specifically, the absence of the classical RGD integrin-binding motif, shown to be the receptor ligand for HPeV1 and possibly the receptor ligand for HPeV2, 4, 5, and 6 as well. This is distinct from all other previously described HPeV4, but similar to HPeV3 and HPeVs 7 through 16, which lack the RGD motif in all VP1 sequences published to date (Benschop et al., 2008). The absence of the RGD motif in these viruses suggests that these viruses may interact with integrins by a different mechanism or use a different receptor altogether. The carboxyl-terminal differences between the HPeV4 prototype and the Bolivia viruses may be the result of intertypic recombination and another HPeV. Natural intertypic recombination at the 3’ end of the P1 region has been reported for polioviruses (Blomqvist et al., 2003; Blomqvist et al., 2010).
Very few human clinical virology laboratories are capable of testing for cardiovirus. With the exception of the recent reports of SAFV (TMEV species) in human feces and respiratory specimens (Abed & Boivin, 2008; Drexler et al., 2011; Drexler et al., 2008; Jones et al., 2007; Nielsen et al.; Zoll et al., 2009), almost all cardiovirus testing is for animal specimens in the context of EMCV epizootics (Maurice et al., 2005). Recent studies have reported EMCV infection in two human cases from Peru (Oberste et al., 2009) and 21% EMCV seroprevalence among persons sampled in the same area of Peru (Czechowicz et al., 2011). Both cardiovirus species were identified in Bolivia human samples. All EMCV-positive specimens in our study had very high Ct values approaching the assay’s limit of detection (Ct of 45 or approximately 10 EMCV genomic copies), suggesting a very low viral load and no type data was acquired from these specimens. The low levels of EMCV detected may be indicative of environmental contamination with rodent materials, resulting in “pass-through” virus and not indicative of virus replication in the human intestinal tract. SAFV1, the most common cardiovirus genotype identified (Figure 2B), was the overall third most common virus found among study subjects. SAFV2, detected in North and South America, Europe, and Asia, has been the most commonly identified genotype to date (Abed & Boivin, 2008; Blinkova et al., 2009; Chiu et al., 2008; Drexler et al., 2008; Itagaki et al., 2010; Zoll et al., 2009). SAFV2 has been associated with respiratory disease (Abed & Boivin, 2008; Chiu et al., 2008), and with invasive disease, including two fatalities (Drexler et al., 2011; Nielsen et al.). All SAFV types have been associated with gastroenteritis, often in coinfections with other known viral pathogens. Additional studies are needed to determine the prevalence of SAFV and zoonotic EMCV infection in humans, define the associations with human disease, and to estimate the human disease burden due to cardioviruses.
CoSV, found in a large proportion of study subjects, were equal to EV in prevalence. Quantitative real-time RT-PCR showed high CoSV viral load in stool, ranging from approximately 4 × 104 to 4 × 109 per gram stool (data not shown), indicating probable CoSV replication in the intestinal tract. CoSV diversity, based on partial VP1 sequences, was the broadest of all picornavirus genera identified, encompassing 21 types in CoSV-A, -B, -D, and -E (Table 3; Figure 3). Picornavirus typing using partial VP1 sequences is generally reliable; however, because the ICTV Picornavirus Study Group requires at least complete VP1 sequence for consideration of new types, the new CoSV types from Bolivia must be considered tentative until complete VP1 sequences have been acquired. These studies are in progress.
A single salivirus, SALV A1 was identified in the Bolivia samples. SALV A1 has not been isolated in culture, and represents the only definitive type identified so far worldwide. SALV seroprevalence in the United States, investigated using an IgG ELISA, was 8.6% in a pediatric cohort and 21.8% in an adult cohort (Greninger et al., 2009), suggesting infection is relatively common.
The inclusion of pig specimens was related to anecdotal reports of frequent paralytic illness in pigs in communities with high human AFP rates. Three pig illnesses which are associated with encephalitis and can present with paralysis have been repeatedly documented in the area: pseudorabies (Aujeszky’s disease), caused by porcine herpesvirus 1; classical swine fever, caused by a flavivirus; and Teschen-Talfan disease, caused by teschoviruses (Anonymous, 2011; 2012a; b; World Organization for Animal Health). None of these viruses infects humans but, because people in these communities often have very high exposure to pigs due to their economic importance, potential zoonotic transmission of other porcine viruses (e.g. EMCV) must be considered. Our study did not identify porcine viruses in any of the human specimens. However, we detected carriage of human picornaviruses, including HPeV, CoSV, and SAFV, in pigs. The HPeV4 and CoSV found in pig specimens were clearly the same respective viruses found in the human specimens (Figure 2A–B, and 3B) but the possibility of human picornavirus replication in the pigs can only be systematically addressed through animal studies. Though the study is limited by the amount and quality of clinical and epidemiologic data available for diseased pigs and the small number of specimens, the data suggest zoonotic porcine picornaviruses are an unlikely cause of the disease in humans
The limitations of this study include lack of true controls as only contacts of AFP cases were tested, which are likely to share exposures, and consequently viral flora, with cases. In addition, stool is not the optimal specimen for picornavirus disease association studies due to the high proportion of asymptomatic infections and because disease generally affects other organ systems. The zoonotic transmission aspect of the study was limited by the lack of clinical and epidemiologic data for the pigs and the small number of specimens.
The cause of very high AFP rates in Hernando Siles province of Chuquisaca Department in Bolivia remains unclear, but most likely is not due to picornavirus infections. A potential non-infectious cause worth additional exploration could be the toxins from Karwinskia plants common in Central and South America. The K. humboldtiana fruits contain toxins known to cause GBS-like illness, and their consumption has been associated with relatively high rates of AFP in some Central American countries (Martinez et al., 1998; Ocampo-Roosens et al., 2007). A representative of Karwinskia genus, K. oblongifolia grows in this area of Bolivia (Serrano & Terán, 2000) and its fruit is commonly consumed by children.
Materials and Methods
Study subjects
AFP cases and their contacts from Hernando Siles province reported to the national AFP surveillance system during 2002–2003 were included in the study. Hernando Siles is a poor, difficult to access, agricultural region with mountainous terrain, a subtropical climate, and a population of approximately 36,000 (~0.4% of the national population, 34.6% aged <15 years; 2001 census, http://apps.ine.gob.bo/censo/entrance.jsp). Most people live in isolated communities established in river canyons and in the towns of Huacareta and Monteagudo, the provincial capital. Available specimens from AFP cases and their contacts from Santa Cruz Department in eastern Bolivia, which does not have a high AFP rate, were tested for comparison.
Clinical and epidemiologic data for the AFP cases were obtained from national AFP surveillance records during a 2007 site visit. Details for the contacts of cases were not available. The available clinical data were limited because of the lack of specialist neurology examinations for all but two cases and the absence of imaging or nerve conduction studies to confirm the diagnosis and further characterize the subtype of AFP. Additionally, no cerebrospinal fluid was taken for laboratory examination or virus diagnostic studies. Clinical subtypes of AFP were classified as GBS-like, enterovirus/poliovirus-like, and “mixed.” Cases with ascending, symmetric, more distal than proximal paralysis which progressed over several days and often had sensory involvement were considered as having GBS-like-paralysis. Cases with descending, asymmetric, more proximal than distal paralysis were considered as having EV/polio-like paralysis. Cases with features of both categories were classified as having a “mixed” subtype. Chi-square and Fisher’s test were used for statistical comparisons, as appropriate.
Specimen collection
Stool specimens included in the study were collected from AFP cases and their household or community contacts by the Ministry of Health, aided by PAHO-Bolivia, following World Health Organization guidelines for AFP case investigation (Anonymous, 2006). Additionally, six fecal specimens were collected from domestic pigs in the affected communities in Hernando Siles.
Picornavirus molecular detection and identification
RNA was extracted from stool suspensions, as previously described (Nix et al., 2006), and tested for enterovirus, parechovirus, cardiovirus, kobuvirus, salivirus, and cosavirus by genus- or genus/species-specific Taqman® real-time RT-PCR assays targeting the 5’ non-translated region (5’NTR) (Kilpatrick et al., 2009; Nix et al., 2008; Oberste et al., 2009) (Table S1). Teschovirus testing was performed by using a 5’-NTR nested RT-PCR (Zell et al., 2000), with the addition of PCR amplicon sequencing to confirm the identity of the correct-sized PCR product.
In vitro transcribed single-stranded RNA controls were produced from PCR products as described previously (Nix et al., 2008). RNA control transcripts were used to generate standard curves for estimating genome copy numbers in the stool specimens. RT-PCR primers and the viral RNAs used to make DNA templates for the in vitro RNA transcription reactions are shown in Table S2.
Real-time PCR-positive specimens were confirmed by nested or semi-nested RT-PCR targeting a portion of the genome encoding the VP1 capsid protein (S3–S6), followed by amplicon sequencing for EV (Nix et al., 2006), HPeV (Nix et al., 2010), cardiovirus (Oberste et al., 2009), kobuvirus, salivirus, and cosavirus. Virus type was determined by comparison of the VP1 amplicon nucleotide sequences with VP1 sequences of the reference strains for each genus by script-driven sequential pairwise comparison using the program Gap (Wisconsin Sequence Analysis Package, version 11.0, Accelrys, Inc., San Diego, CA). Deduced VP1 amino acid sequences were compared by a similar method. VP1 nucleic acid sequences were aligned using the Pileup program (Wisconsin Package) and phylogenetic relationships were inferred by the neighbor-joining method implemented in MEGA, version 4.0 (Kumar et al., 2001), using the Kimura 2-parameter method (Jones et al., 1992; Kumar et al., 2001). Support for specific tree topologies was estimated by bootstrap analysis with 1000 pseudo-replicate data sets. Nucleotide and amino acid identity matrices were calculated using MegAlign (DNAStar, version 8, Madison, WI).
The nucleotide sequences described here have been deposited in the GenBank sequence database, accession no. JX219484-JX219590. Four nucleotide sequences used to identify virus types were rejected by GenBank due to their short lengths (<200 base pairs) (Table S9).
Supplementary Material
Acknowledgements
We thank the staff of the Bolivia EPI program for their work on identification and investigation of the AFP cases and support of the study specimen collection; Dr. Ramon D. Ibanez Calderon of LIDIVET, Santa Cruz, for his assistance in exploring potential etiologies of pig diseases in Hernando Siles; Dr. Noemi Waksman de Torres for discussions of Karwinskia toxicology and assistance with exploring its role in AFP in Bolivia; ZC was supported by a fellowship from the Hungarian-American Enterprise Scholarship Fund, Budapest, Hungary.
Footnotes
“The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of CDC.”
References
- Abed Y, Boivin G. New Saffold cardioviruses in 3 children, Canada. Emerg Infect Dis. 2008;14:834–836. doi: 10.3201/eid1405.071675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch Virol. 2013 doi: 10.1007/s00705-013-1688-5. In Press. [DOI] [PubMed] [Google Scholar]
- Anonymous . Poliomyelitis Eradication Field Guide. Washington, D.C: Pan American Health Organization; 2006. [Accessed: June 11, 2012]. http://www.paho.org/english/ad/fch/im/fieldguide_polio.pdf. [Google Scholar]
- Anonymous . Porcine Enteroviral Encephalomyelitis (Teschen disease, Talfan disease, Porcine polioencephalomyelitis) In: Kahn CM, editor. The Merck Veterinary Manual. Whitehouse Station, NJ Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc; 2011. [Google Scholar]
- Anonymous . Bolivia/Aujeszky's disease. Multiannual animal disease status. Paris: World Organization for Animal Health; 2012a. [Accessed: January 16, 2013]. http://web.oie.int/hs2/sit_pays_mald_pl.asp?c_pays=25&c_mald=21. [Google Scholar]
- Anonymous . Bolivia/Classical swine fever. Multiannual animal disease status. Paris: World Organization for Animal Health; 2012b. [Accessed: January 16, 2013]. http://web.oie.int/hs2/sit_pays_mald_pl.asp?c_pays=25&c_mald=14. [Google Scholar]
- Begier EM, Oberste MS, Landry ML, Brennan T, Mlynarski D, Mshar PA, Frenette K, Rabatsky-Ehr T, Purviance K, Nepaul A, Nix WA, Pallansch MA, Ferguson D, Cartter ML, Hadler JL. An outbreak of concurrent echovirus 30 and coxsackievirus A1 infections associated with sea swimming among a group of travelers to Mexico. Clin Infect Dis. 2008;47:616–623. doi: 10.1086/590562. [DOI] [PubMed] [Google Scholar]
- Benschop K, Thomas X, Serpenti C, Molenkamp R, Wolthers KC. High prevalence of human parechovirus genotypes in the Amsterdam region and the identification of specific HPeV variants by direct genotyping of stool samples. J Clin Microbiol. 2008;46:3965–3970. doi: 10.1128/JCM.01379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingjun T, Yoshida H, Yan W, Lin L, Tsuji T, Shimizu H, Miyamura T. Molecular typing and epidemiology of non-polio enteroviruses isolated from Yunnan Province, the People's Republic of China. J Med Virol. 2008;80:670–679. doi: 10.1002/jmv.21122. [DOI] [PubMed] [Google Scholar]
- Blinkova O, Kapoor A, Victoria J, Jones M, Wolfe N, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M, Zaidi S, Delwart EL. Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J Virol. 2009;83:4631–4641. doi: 10.1128/JVI.02085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist S, Bruu A-L, Stenvik M, Hovi T. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J Gen Virol. 2003;84:573–580. doi: 10.1099/vir.0.18708-0. [DOI] [PubMed] [Google Scholar]
- Blomqvist S, Savolainen-Kopra C, Paananen A, El Bassioni L, El Maamoon Nasr EM, Firstova L, Zamiatina N, Kutateladze T, Roivainen M. Recurrent isolation of poliovirus 3 strains with chimeric capsid protein Vp1 suggests a recombination hot-spot site in Vp1. Virus Res. 2010;151:246–251. doi: 10.1016/j.virusres.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Chiu CY, Greninger AL, Kanada K, Kwok T, Fisher KF, Runckel C, Louie JK, Glaser CA, Yagi S, Schnurr DP, Haggerty TD, Parsonnet J, Ganem D, DeRisi JL. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proceedings of the National Academy of Sciences (USA) 2008;105:14124–14129. doi: 10.1073/pnas.0805968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowicz J, Huaman JL, Forshey BM, Morrison AC, Castillo R, Huaman A, Caceda R, Eza D, Rocha C, Blair PJ, Olson JG, Kochel TJ. Prevalence and risk factors for encephalomyocarditis virus infection in Peru. Vector borne and zoonotic diseases. 2011;11:367–374. doi: 10.1089/vbz.2010.0029. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Baumgarte S, Eschbach-Bludau M, Simon A, Kemen C, Bode U, Eis-Hübinger A-M, Madea B, Drosten C. Human cardioviruses, meningitis, and sudden infant death syndrome in children. Emerg Infect Dis. 2011;17:2313–2315. doi: 10.3201/eid1712.111037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Kleber de Souza Luna L, Stöcker A, Silva Almeida P, Medrado Ribeiro TC, Petersen N, Herzog P, Pedroso C, Hupperz HI, da Costa Ribeiro H, Jr, Baumgarte S, Drosten C. Circulation of 3 lineages of a novel human Saffold cardiovirus in humans. Emerg Infect Dis. 2008;14:1398–1405. doi: 10.3201/eid1409.080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, Derisi JL. The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol J. 2009;6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvala H, Wolthers KC, Simmonds P. Parechoviruses in children: understanding a new infection. Curr Opin Infect Dis. 2010;23:224–230. doi: 10.1097/qco.0b013e32833890ca. [DOI] [PubMed] [Google Scholar]
- Holtz LR, Finkbeiner SR, Kirkwood CD, Wang D. Identification of a novel picornavirus related to cosaviruses in a child with acute diarrhea. Virol J. 2008;5:159. doi: 10.1186/1743-422X-5-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itagaki T, Abiko C, Ikeda T, Aoki Y, Seto J, Mizuta K, Ahiko T, Tsukagoshi H, Nagano M, Noda M, Mizutani T, Kimura H. Sequence and phylogenetic analyses of Saffold cardiovirus from children with exudative tonsillitis in Yamagata, Japan. Scand J Infect Dis. 2010;42:950–952. doi: 10.3109/00365548.2010.496791. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jones MS, Lukashov VV, Ganac RD, Schnurr DP. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J Clin Microbiol. 2007;45:2144–2150. doi: 10.1128/JCM.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila N, Leveque N, Kabue JP, Cartet G, Mushiya F, Muyembe-Tamfum JJ, Trompette A, Lina B, Magnius LO, Chomel JJ, Norder H. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J Med Virol. 2007;79:393–400. doi: 10.1002/jmv.20825. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Ayyagari A, Dhole TN. Non-polio enteroviruses in acute flaccid paralysis. Indian Journal of Pediatrics. 2001;68:927–929. doi: 10.1007/BF02722583. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M, Wang C, Shafer RW, Zaidi SZ, Delwart E. A highly prevalent and genetically diversified Picornavirus genus in South Asian children. Proceedings of the National Academy of Science. 2008;105:20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick DR, Yang C-F, Ching K, Vincent A, Iber J, Campagnoli R, Mandelbaum M, De L, Yang S-J, Nix A, Kew OM. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol. 2009;47:1939–1941. doi: 10.1128/JCM.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Skern T, Stanway G, Yamashita T, Zell R. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. pp. 855–880. [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Martinez HR, Bermudez MV, Rangel-Guerra RA, de Leon Flores L. Clinical diagnosis in Karwinskia humboldtiana polyneuropathy. J Neurol Sci. 1998;154:49–54. doi: 10.1016/s0022-510x(97)00212-8. [DOI] [PubMed] [Google Scholar]
- Maurice H, Nielen M, Brocchi E, Nowotny N, Kassimi LB, Billinis C, Loukaides P, O'Hara RS, Koenen F. The occurrence of encephalomyocarditis virus (EMCV) in European pigs from 1990 to 2001. Epidemiol Infect. 2005;133:547–557. doi: 10.1017/s0950268804003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AC, Bottiger B, Banner J, Hoffmann T, Nielsen LP. Serious invasive Saffold virus infections in children. Emerg Infect Dis. 2009;18:7–12. doi: 10.3201/eid1801.110725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix WA, Maher K, Niklasson B, Lindberg M, Johansson S, Pallansch MA, Oberste MS. Detection of all known parechoviruses by real time-PCR. J Clin Microbiol. 2008;46:2519–2524. doi: 10.1128/JCM.00277-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix WA, Maher K, Pallansch MA, Oberste MS. Parechovirus typing in clinical specimens by nested or semi-nested VP1 PCR coupled with sequencing. J Clin Virol. 2010;48:202–207. doi: 10.1016/j.jcv.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Feeroz MM, Maher K, Nix WA, Engel GA, Chowdhury AH, Pallansch MA, Jones-Engel L. Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007–2008. J Virol. 2013:87. doi: 10.1128/JVI.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Gotuzzo E, Blair PJ, Nix WA, Ksiazek TG, Comer JA, Goldsmith CA, Olson JG, Kochel T. Human encephalomyocarditis virus disease in Perú. Emerg Infect Dis. 2009;15:640–646. doi: 10.3201/eid1504.081428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Maher K, Williams AJ, Dybdahl-Sissoko N, Brown BA, Gookin MS, Penaranda S, Mishrik N, Uddin M, Pallansch MA. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J Gen Virol. 2006;87:119–128. doi: 10.1099/vir.0.81179-0. [DOI] [PubMed] [Google Scholar]
- Ocampo-Roosens LV, Ontiveros-Nevares PG, Fernandez-Lucio O. Intoxication with buckthorn (Karwinskia humboldtiana): report of three siblings. Pediatr Dev Pathol. 2007;10:66–68. doi: 10.2350/06-03-0067.1. [DOI] [PubMed] [Google Scholar]
- Saeed M, Zaidi SZ, Naeem A, Masroor M, Sharif S, Shaukat S, Angez M, Khan A. Epidemiology and clinical findings associated with enteroviral acute flaccid paralysis in Pakistan. BMC Infect Dis. 2007;7:6. doi: 10.1186/1471-2334-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AP, Costa EV, Oliveira SS, Souza MC, Da Silva EE. RT-PCR based analysis of cell culture negative stools samples from poliomyelitis suspected cases. J Clin Virol. 2002;23:149–152. doi: 10.1016/s1386-6532(01)00211-6. [DOI] [PubMed] [Google Scholar]
- Serrano M, Terán J. Identificación de Especies Vegetales en Chuquisaca --Teoría, Práctica y Resultados. Sucre, Bolivia: PLAFOR, Intercooperación, Fundación Ceibo; 2000. [Google Scholar]
- Shoja ZO, Tabatabai H, Sarijloo M, Shahmahmoodi S, Azad TM, Nategh R. Detection of enteroviruses by reverse-transcriptase polymerase chain reaction in cell culture negative stool specimens of patients with acute flaccid paralysis. J Virol Methods. 2007;142:95–97. doi: 10.1016/j.jviromet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organization for Animal Health. World Animal Health Information Database. http://web.oie.int/wahis/public.php.
- Zell R, Krumbholz A, Henke A, Birch-Hirschfeld E, Stelzner A, Doherty M, Hoey E, Dauber M, Prager D, Wurm R. Detection of porcine enteroviruses by nRT-PCR: differentiation of CPE groups I-III with specific primer sets. J Virol Meth. 2000;88:205–218. doi: 10.1016/s0166-0934(00)00189-0. [DOI] [PubMed] [Google Scholar]
- Zoll J, Hulshof SE, Lanke K, Lunel FV, Melchers WJG, Schoondermark-van de Ven E, Roivainen M, Galama JMD, van Kuppeveld FJM. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLOS Pathogens. 2009;5:e1000416. doi: 10.1371/journal.ppat.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.