Abstract
Small-cell lung cancer (SCLC) is the most aggressive subtype of lung cancer in its clinical behavior, with a 5-year overall survival as low as 5%. Despite years of research in the field, molecular determinants of SCLC behavior are still poorly understood, and this deficiency has translated into an absence of specific diagnostics and targeted therapeutics. We hypothesized that tumor DNA copy number alterations would allow the identification of molecular pathways involved in SCLC progression. Array comparative genomic hybridization was performed on DNA extracted from 46 formalin-fixed paraffin-embedded SCLC tissue specimens. Genomic profiling of tumor and sex-matched control DNA allowed the identification of 70 regions of copy number gain and 55 regions of copy number loss. Using molecular pathway analysis, we found a strong enrichment in these regions of copy number alterations for 11 genes associated with the focal adhesion pathway. We verified these findings at the genomic, gene expression and protein level. Focal Adhesion Kinase (FAK), one of the central genes represented in this pathway, was commonly expressed in SCLC tumors and constitutively phosphorylated in SCLC cell lines. Those were poorly adherent to most substrates but not to laminin-322. Inhibition of FAK phosphorylation at Tyr397 by a small-molecule inhibitor, PF-573,228, induced a dose-dependent decrease of adhesion and an increase of spreading in SCLC cell lines on laminin-322. Cells that tended to spread also showed a decrease in focal adhesions, as demonstrated by a decreased vinculin expression. These results support the concept that pathway analysis of genes in regions of copy number alterations may uncover molecular mechanisms of disease progression and demonstrate a new role of FAK and associated adhesion pathways in SCLC. Further investigations of FAK at the functional level may lead to a better understanding of SCLC progression and may have therapeutic implications.
Keywords: array CGH, copy number alterations, pathway analysis, FAK, adhesion
Introduction
Small-cell lung cancer (SCLC) and non-small-cell lung cancer account for approximately 15 and 85% of annual lung cancer cases, respectively (Jemal et al., 2009). SCLC is the most aggressive subtype of lung cancer in its clinical behavior, with a five-year overall survival as low as 5% (Jemal et al., 2009). SCLC is characterized by rapid doubling time and early development of metastasis (Jackman and Johnson, 2005). To date, no targeted therapy is available in SCLC. This is in sharp contrast to progress made in non-small-cell lung cancer, in which identification of key gene amplifications, fusion oncogenes and somatic oncogene mutations has led to the development of successful targeted therapies (Greulich et al., 2005).
Chromosomal imbalances were first studied in SCLCs with low-throughput analytical platforms (Sozzi et al., 1987; Miura et al., 1992; Levin et al., 1994; Balsara and Testa, 2002). More recently, few genomic studies analyzed SCLCs with greater resolution arrays (Ullmann et al., 1998; Peng et al., 2005; Zhao et al., 2005; Coe et al., 2006). In one of them, analysis of 19 SCLC tumors and 5 SCLC lines with 115 000 single-nucleotide polymorphism arrays led to the identification of new regions of interest (Zhao et al., 2005). Later, submegabase resolution tiling-set arrays compared DNA from 14 SCLCs with those from 27 non-small-cell lung cancer lines and showed differential disruption of cell cycle pathways (Coe et al., 2006). Despite their important contribution to the field, these studies have not identified specific molecular targets for SCLC diagnostics or therapeutics.
We hypothesized that tumor DNA copy number alterations would allow the identification of molecular pathways of SCLC progression. We therefore undertook a genome-wide approach using gene copy number assessment in SCLC tumors, identified significant regions of copy number alterations and determined that these regions were enriched for the focal adhesion pathway and, among the genes represented, PTK2 (Focal Adhesion Kinase, FAK). Because of its role in cell adhesion, spreading, migration, invasion, survival and anchorage-independent growth (Hanks et al., 2003; Parsons, 2003; Mitra et al., 2005; Siesser and Hanks, 2006), all relevant to SCLC behavior, we investigated the role of FAK in SCLC cell lines with respect to adhesion, spreading and migration.
Results
Identification of recurrent gene copy number alterations in SCLC by array CGH
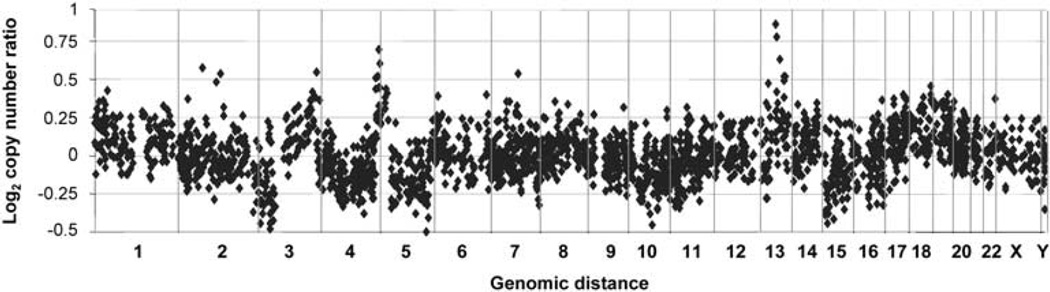
Forty-six pathologically confirmed formalin-fixed paraffin-embedded SCLCs were used in genomic profiling experiments. Patient characteristics are described in Supplementary Table 1. The tumors displayed numerous chromosomal regions of copy number gain and loss (Figure 1). Seventy regions of copy number gain and 55 regions of copy number loss were identified as significant (log2 copy number ratio > 0.3 or < −0.3) and recurrent (prevalence > 50%). A total of 329 and 99 genes were described in these regions, respectively (Supplementary Tables 2A and 2B).
Figure 1.
Gene copy number alterations in 46 primary small-cell lung cancers (SCLCs). Each dot represents the average log2 copy number ratio in 46 SCLC tumors at a given genomic locus across the genome tested by array comparative genomic hybridization (CGH). The value of 0 represents an equal fluorescence intensity ratio between SCLC and normal reference genomic DNA. Log2 ratios > 0.3 or < −0.3 are defined as copy number gain or loss. Vertical lines represent chromosomal boundaries.
To validate our findings, we compared these alterations with those of eight previously published studies in SCLC: Ried et al. (1994) (13 tumors, comparative genomic hybridization (CGH)), Levin et al. (1994) (10 tumors, CGH), Lindblad-Toh et al. (2000) (17 tumors, 1500 single-nucleotide polymorphism arrays), Peng et al. (2005) (10 tumors, 800 BAC clone arrays); Zhao et al. (2005) (19 tumors, 5 cell lines, 115 000 single-nucleotide polymorphism arrays); Coe et al. (2006) (14 cell lines, 97 299 elements submegabase resolution tiling-set arrays); Kim et al. (2006) (24 cell lines, ~22 000 genes complementary DNA microarrays); and Olejniczak et al. (2007) (23 cell lines, ~114 000 single-nucleotide polymorphism arrays). Overall, we did not identify new regions of copy number alterations, but our results confirmed the regions of copy number gain and loss that were published in literature in the largest number of primary SCLCs (n = 46). Comparison with Coe et al.’s data was performed in greater depth, as we obtained their detailed dataset. We found that 184 of 212 (87%) of their probes matching the 70 BAC clones with copy number gain and 136 of 159 (86%) of their probes matching the 55 BAC clones with copy number loss identified in our arrays presented a very similar copy number trend.
Enrichment for genes associated with the focal adhesion and the neuroactive ligand–receptor interaction pathways in SCLC
To identify molecular pathways enriched in this genomic data set, we compared the 428 genes with significant and frequent copy number gain or loss in SCLCs with the reference gene set that corresponds to all 7913 genes represented in this array. We found a strong enrichment in these regions for 11 genes associated with the focal adhesion pathway (P = 0.0018) (Table 1) and for eight genes associated with the neuroactive ligand–receptor interaction pathway (P = 0.0481) (Table 1). In this report, we focus on the focal adhesion pathway. Among genes represented in this pathway, FAK is a non-receptor tyrosine kinase that is overexpressed in many cancers. Because of its role in cell adhesion, spreading, migration, invasion, survival, proliferation and anchorage-independent growth (Hanks et al., 2003; Parsons, 2003; Mitra et al., 2005; Siesser and Hanks, 2006), we seek to determine its potential contribution to SCLC progression.
Table 1.
Enrichment of primary small-cell lung cancer tumors
| Gene symbol | Gene description | FISH location | Copy number aberration (average log2 copy number) |
|---|---|---|---|
| (a) For genes associated with the focal adhesion pathway | |||
| PIK3CA | Phosphatidylinositol 3-kinase catalytic subunit α | 3q26.32 | Gain (0.32) |
| MAPK10 | Mitogen-activated protein kinase 10 | 4q21.3 | Loss (−0.34) |
| LAMB1 | Laminin β1 | 7q31.1 | Gain (0.54) |
| LAMB4 | Laminin β4 | 7q31.1 | Gain (0.54) |
| FAK | Focal adhesion kinase 1 | 8q24.3 | Gain (0.30) |
| HRAS | Ras family small GTP binding protein H-Ras | 11p15.5 | Gain (0.30) |
| ACTC1 | α-cardiac actin | 15q14 | Loss (−0.45) |
| THBS1 | Thrombospondin-1 | 15q14 | Loss (−0.33) |
| ITGB4 | Integrin β4 | 17q25.1 | Gain (0.32) |
| PIK3R2 | Phosphatidylinositol 3-kinase regulatory subunit β | 19p13.11 | Gain (0.33) |
| VASP | Vasodilator-stimulated phosphoprotein | 19q13.2–q13.3 | Gain (0.32) |
| (b) For genes associated with the neuroactive ligand–receptor interaction pathway | |||
| THRB | Thyroid hormone receptor β1 | 3q24.2 | Loss (−0.34) |
| DRD4 | Dopamine receptor D4 | 11p15.5 | Gain (0.31) |
| SCT | Secretin | 11p15.5 | Gain (0.31) |
| HTR2A | Serotonin receptor 2A | 13q14.2 | Gain (0.47) |
| EDG6 | Endothelial differentiation G-protein coupled receptor 6 | 19p13.3 | Gain (0.34) |
| GRIN3B | Glutamate receptor subunit 3B | 19p13.3 | Gain (0.40) |
| PTGIR | Prostacyclin receptor | 19p13.3 | Gain (0.35) |
| GIPR | Gastric inhibitory polypeptide receptor | 19p13.3 | Gain (0.34) |
Verification of gene copy number alterations in the focal adhesion pathway
Genomic level
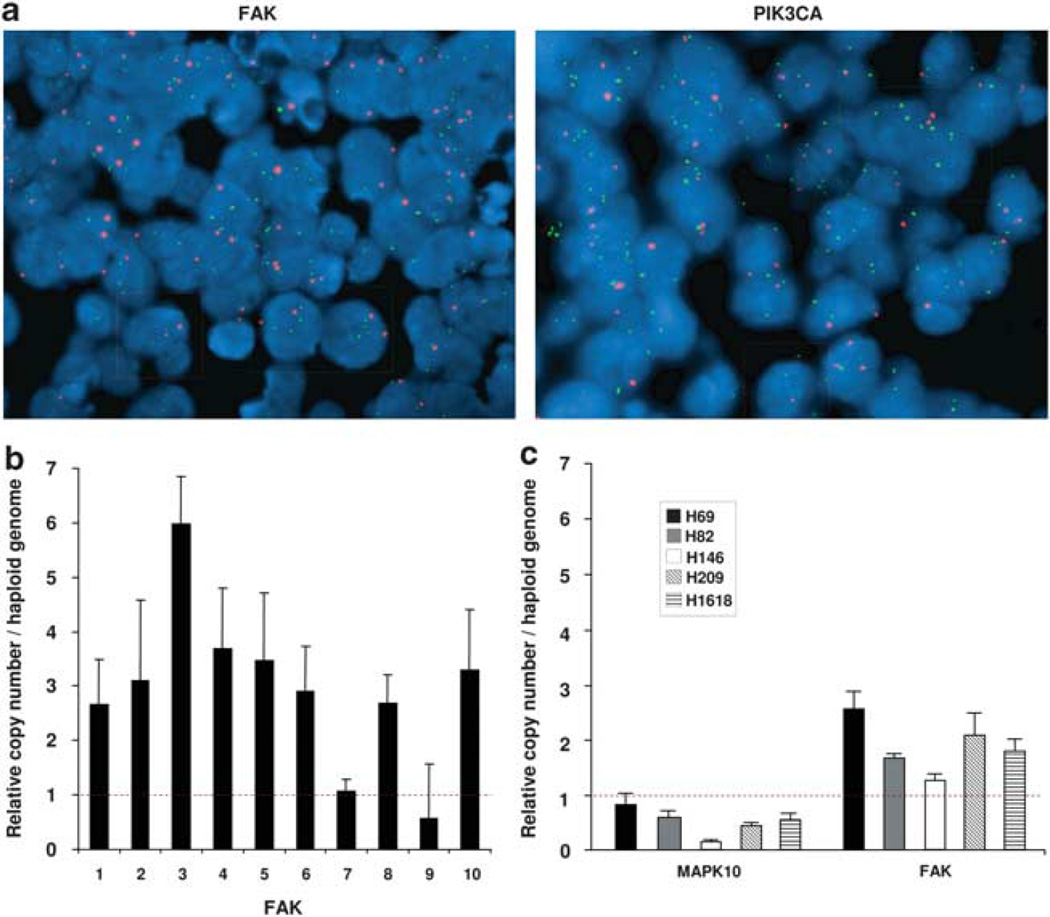
Gene copy number of FAK and PIK3CA, genes selected for their central role in the pathway, was evaluated by fluorescent in situ hybridization (FISH) in 46 SCLCs. Defining gene copy number gain as a relative copy number (test gene/corresponding centromeric probe) ≥ 1.8 in ≥ 15% nuclei, we found that FAK displayed copy number gain in 7 of 42 (17%) and PIK3CA in 10 of 46 (22%) SCLCs (Figure 2a). Among the SCLCs studied by array CGH, 12 were also tested by FISH, demonstrating concordant copy number alterations in 83% of them for FAK and in 69% for PIK3CA. FAK copy number was also verified by quantitative real-time polymerase chain reaction in 10 of the 46 SCLCs studied by array CGH. Copy number gains were confirmed in 8 of 10 (80%) of them (Figure 2b). Finally, FAK gene copy number was evaluated in five SCLC cell lines. As control, we tested MAPK10 copy number status, a gene that showed strong copy number loss by array CGH. Copy number gain of FAK and loss of MAPK10 were found in all tested cell lines (Figure 2c), in striking concordance with results obtained by array CGH in primary SCLCs.
Figure 2.
Verification of FAK and PIK3CA increased gene copy number by FISH in primary SCLCs and by quantitative RT–PCR in SCLC cell lines. (a) Dual color fluorescence in situ hybridization of FAK gene (8q24.3, green spots) and CEP8 probe (red spots) or of PIK3CA gene (3q26.3, green spots) and CEP3 probe (red spots) on the centromeric region of the same chromosome in representative SCLC tissue samples. Interphase nuclei (stained in blue with DAPI) with increased copy number of FAK (left panel) and PIK3CA (right panel) are shown in green boxes. Interphase nuclei with normal gene copy number are shown in red boxes. Pictures magnification: ×100. (b) Quantitative RT–PCR of FAK in primary SCLCs. Copy number gain of FAK was confirmed in 8 of 10 tested tumors. (c) Quantitative RT–PCR of FAK and MAPK10 genes in SCLC cell lines. Copy number gain of FAK and copy number loss of MAPK10 were observed in all tested cell lines. Copy number changes per haploid genome were calculated using the formula 2(Nt–NRNase P)–(Dt–DRNase P), where Nt is the average threshold cycle number observed for an experimental primer in the normal DNA sample, NRNase P is the threshold cycle number observed for an RNase P primer in the normal DNA sample, Dt is the average threshold cycle number observed for the experimental primer in the SCLC DNA sample and DRNase P is the average threshold cycle number observed for an RNase P primer in the SCLC DNA sample. Data represent averages and error bars ± 1 s.d. (n = 3).
Gene expression level
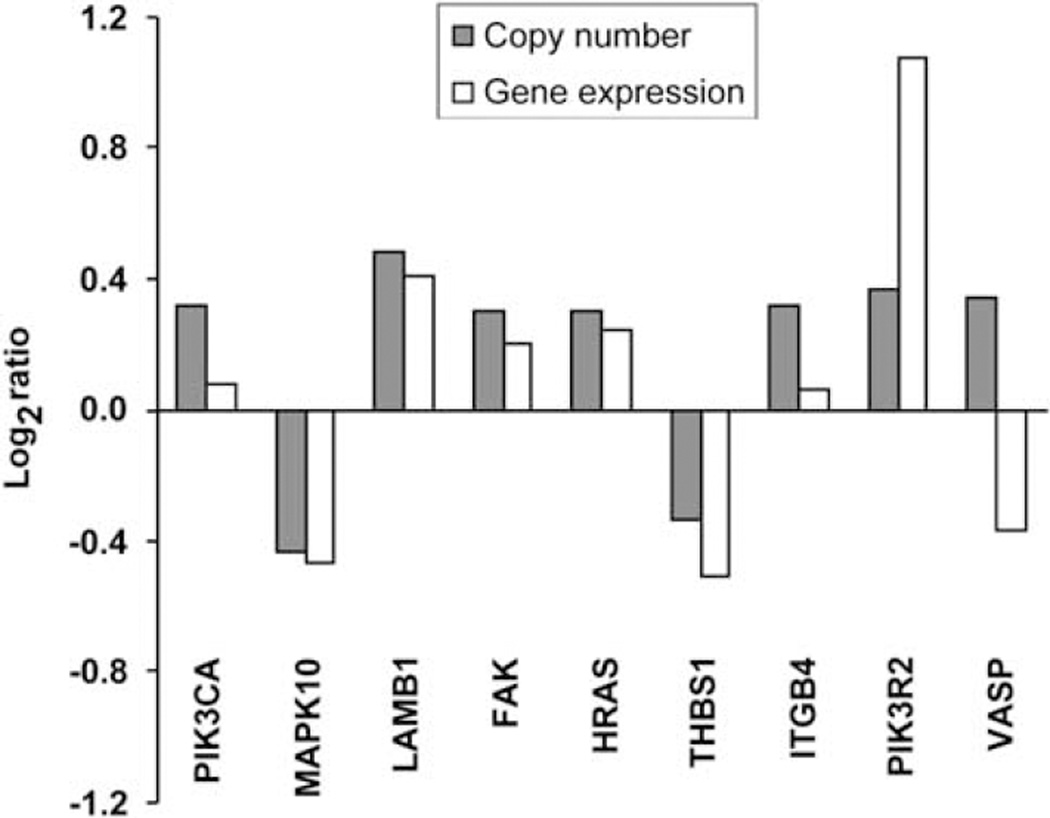
Significant and recurrent gene copy number alterations were correlated to gene expression data found in a publicly available data set (Garber et al., 2001), in which five SCLC and six normal lung tissues were analyzed by complementary DNA microarrays. We tested the correlation between median log2 gene expression ratios (SCLC/normal) from this data set and copy number ratios (SCLC/normal) from our array CGH data set. There was a significant correlation between gene copy number and expression levels (Pearson = 0.68) for genes represented in both the focal adhesion pathway and the gene expression data sets (9 of 11) (Figure 3). This further supports the potential importance of the observations made at the genomic level.
Figure 3.
Positive correlation between the focal adhesion pathway gene copy number data and gene expression level. Array CGH gene copy number of 46 SCLC tumors was compared with gene expression level of the same genes represented in Garber et al.’s public gene expression data set, based on the analysis of five primary SCLC tumors. A positive correlation between the median log2 gene expression ratio (SCLC/normal) and the median log2 gene copy number ratio (SCLC/normal) was found (Pearson: 0.68) for 9 of 11 genes represented in the focal adhesion pathway.
Protein expression level
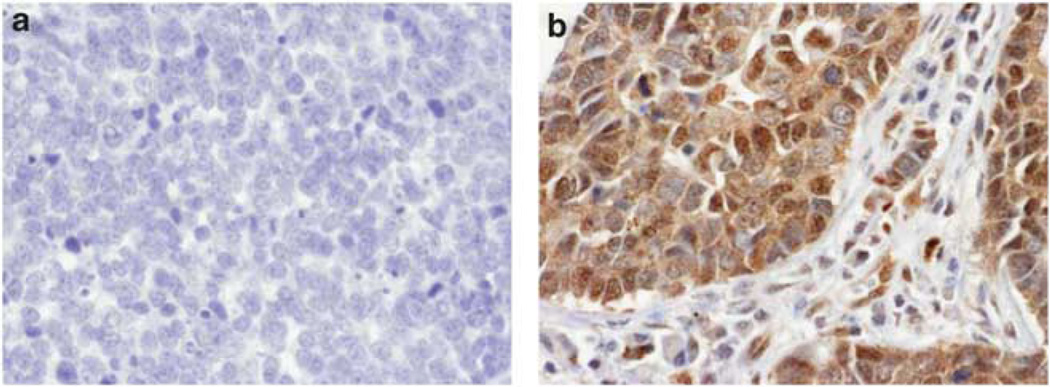
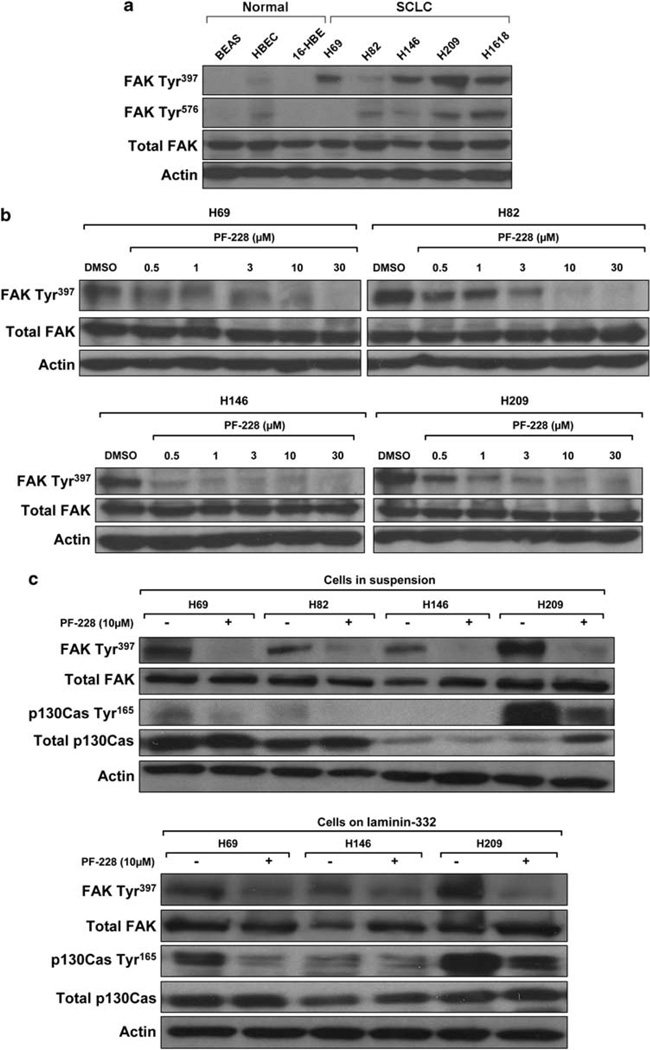
FAK protein expression was then evaluated by immunohistochemistry (IHC) in five normal lung and 52 SCLCs. FAK expression was absent in one, low in two, moderate in one and strong in one of the tested normal lung tissues. FAK was highly expressed in the cytoplasm of 47 of 52 (90%) SCLCs. Average staining score was ≥2 for 29 (56%), ≥1 and <2 for 14 (27%), and >0 and <1 for 4 (7%) of these SCLCs. Representative images of tumors with positive and negative staining are displayed in Figure 4. Among tumors tested by IHC, 14 were also tested by array CGH and 35 by FISH. FAK gene copy number and protein expression were correlated (copy number gain and average IHC score ≥2, or no copy number gain and average IHC score <2) in 8 of 14 (57%) cases for array CGH and 18 of 35 (51%) cases for FISH. FAK expression was also evaluated by western blotting in cell lines. Similar levels of total FAK expression were observed in three immortalized normal bronchial epithelial and five SCLC cell lines. However, phosphorylation of FAK Tyr397/576 was mainly observed in SCLC cell lines, demonstrating activation of FAK in SCLC (Figure 5a).
Figure 4.
FAK expression by immunohistochemistry: a representative example of 52 studied SCLCs. Tissue sections were incubated with an antibody against FAK (A-17). All immunostains were independently scored by two independent observers (SO and PPM) as follows: 0—no staining, 1—weak staining, 2—moderate staining and 3—strong staining. FAK displayed a cytoplasmic staining. (a) Representative image of SCLC with absence of total FAK expression. (b) Representative image of SCLC tumor with maximum staining score for total FAK. It is noteworthy that positively stained cancer cells are infiltrating the stroma. Pictures magnification: ×40.
Figure 5.
FAK is activated in SCLC cell lines and inhibited by PF-228 in a dose-dependent manner. (a) FAK is phosphorylated in SCLC cell lines. Whole-cell lysates were resolved with SDS-PAGE and blots were incubated with antiphospho-FAK Tyr397/576, total FAK or actin antibodies. FAK Tyr397/576 and total FAK levels were normalized to actin. FAK phosphorylation at tyrosine residues was mainly observed in SCLC cell lines, demonstrating an activation of FAK in SCLC. (b) Dose-dependent inhibition of FAK Tyr397 phosphorylation by PF-228 in SCLC cell lines. SCLC cell lines in suspension were treated with the indicated concentrations of PF-228 for 90 min. Whole-cell lysates were resolved with SDS-PAGE and blots were incubated with antiphospho-FAK Tyr397, total FAK or actin antibodies. FAK Tyr397 and total FAK levels were normalized to actin. Significant decrease of FAK phosphorylation at Tyr397 was achieved with PF-228 in a dosedependent manner. (c) PF-228 decreases p130CAS Tyr165 phosphorylation, downstream substrate of FAK, in SCLC cell lines. SCLC cell lines in suspension, as well as adhering on laminin-332, were treated with 10 µm PF-228 for 90 min. Whole-cell lysates were resolved with SDS-PAGE and blots were incubated with antiphospho-FAK Tyr397, total FAK, phospho-p130CAS Tyr165, total p130CAS or actin antibodies. FAK Tyr397 phosphorylation, total FAK, phospho-p130CAS Tyr165 and total p130CAS levels were normalized to actin. Significant decrease of p130CAS phosphorylation at Tyr165 was achieved with PF-228, parallel to the decrease of FAK phosphorylation at Tyr397.
Inhibition of FAK kinase activity by PF-573,228 (PF-228) in SCLC cell lines
Because of the expression and activation of FAK in SCLC and its central role in cellular functions contributing to cancer metastasis, we attempted to inhibit its activity in SCLC cell lines and investigate the effects on key cellular functions, including adhesion, spreading, motility and invasion. We used PF-228 (6-[(4-((3-(methanesulfonyl)benzyl)amino)-5-trifluoromethylpyrimidin- 2-yl)amino]-3,4-dihydro-1H-quinolin-2-one), a commercially available competitive inhibitor of adenosine triphosphate with specificity for FAK family proteintyrosine kinases, which prevents FAK Tyr397 autophosphorylation (Slack-Davis et al., 2007). To assess PF-228’s ability to inhibit endogenous FAK activity, FAK Tyr397 phosphorylation was evaluated by western blotting in SCLC cells treated with increasing concentrations of PF-228 for 90 min. A significant decrease in FAK Tyr397 phosphorylation was achieved with PF-228 in a dose-dependent manner (Figure 5b). FAK forms a binary complex with Src family kinases, which can phosphorylate other downstream substrates such as p130CAS and paxillin to regulate various cellular functions, including migration. Inhibition of FAK phosphorylation in SCLC by PF-228 was accompanied by inhibition of p130CAS Tyr165 phosphorylation (Figure 5c).
Effect of FAK inhibition on adhesion, spreading and migration of SCLC cell lines
FAK has been shown to promote cell adhesion, spreading and motility in different types of cell lines. However, despite their constitutive activation of FAK, SCLC cell lines grow in suspension and do not adhere on plastic substrates. We tested the ability of four SCLC cell lines (NCI-H69, NCI-H82, NCI-H146 and NCIH209) to attach on collagen I, fibronectin, vitronectin or laminin-332 (Supplementary Figure 1), and found that laminin-332 significantly promoted SCLC adhesion, except for NCI-H82 cells that did not adhere on any of these substrates. NCI-H146 displayed the highest adhesion on laminin-332 (P = 6.3 × 10−5 compared with phosphate buffered saline), followed by NCI-H69 (P = 4.2 × 10−5) and NCI-H209 (P = 2.5 × 10−5). Therefore, we decided to use, for further experiments, laminin-332-coated dishes or filters.
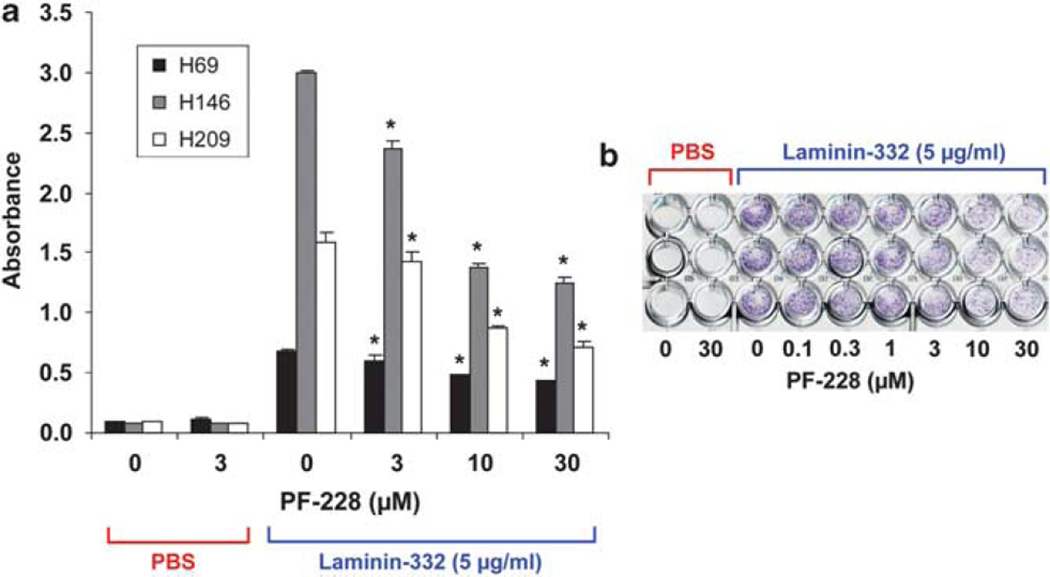
To determine the role of FAK inhibition on adhesion, NCI-H69, NCI-H146 and NCI-H209 cells were treated with increasing concentrations of PF-228 for 30 min before plating on laminin-332-coated wells and incubated for 24 h before analysis of crystal-violet staining of adherent cells. Treatment with FAK inhibitor induced a dose-dependent decrease of adhesion (Figure 6). The effect became statistically significant with 10 µm PF-228 (NCI-H69: P = 3 × 10−4, NCI-H146 and NCI-H209: P = 1 × 10−4 compared with DMSO).
Figure 6.
PF-228 induces a dose-dependent decrease in the adhesion of SCLC lines to laminin-332. Cells were treated with increasing concentrations of PF-228 for 30 min before seeding on laminin-332-coated plates and then during the entire experiment. Adhesion was evaluated by crystal violet staining 24 h after seeding. The extent of cell adhesion was determined by measuring the absorbance of wells at 595nm with an ELISA reader. (a) This figure, representative of one of the experiments performed in triplicate wells three independent times, shows that treatment with PF-228 induced a dose-dependent decrease of the adhesion of the three SCLC cell lines to laminin-332. Data represent averages and error bars ±1 s.d. (n = 3). P-values: absorbance of wells treated with PF-228 vs those treated with DMSO; *P<0.05. (b) This image, representative of one of the experiments performed with NCI-H209, shows the decrease of crystal violet staining with increasing concentrations of PF-228.
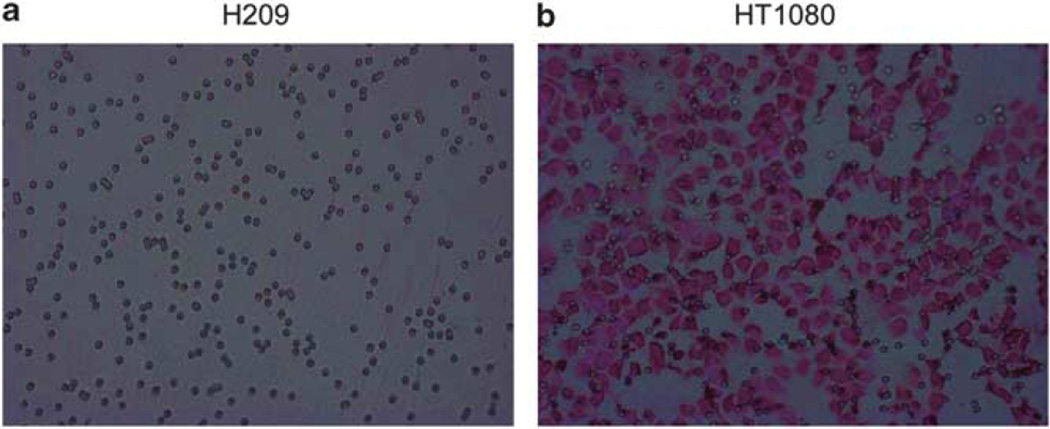
To determine whether PF-228 affects SCLC motility and invasion, we first used a transwell assay. None of the cells tested (NCI-H69, NCI-H82, NCI-H209) migrated through the porous membrane for up to 24 h in any of the conditions tested (coating the lower or upper side of the membrane with laminin-332 or basal membrane extract, PF-228 or DMSO, serum or VEGF gradient) (Figure 7a). This was in sharp contrast to the migration observed with fibrosarcoma cells HT1080, used as positive control. In the migration assay, 488 (s.d. = 24) and 402 (s.d. = 30) HT1080 cells per ×40 microscopic field were counted on the membrane’s lower side after 6 and 24 h, respectively (Figure 7b). Similarly, in the invasion assay, 206 (s.d. = 16) and 156 (s.d. = 18) HT1080 cells per ×40 microscopic field were counted on the membrane’s lower side at 6 and 24 h, respectively.
Figure 7.
Absence of motility and invasion of SCLC cells in a transwell assay. Migration and invasion were assessed for up to 24 h by counting cells on the lower side of the transwell’s membrane in five microscopic fields of high magnification (40×) under a light microscope. Cells on the membrane were fixed and stained with hematoxylin and eosin. (a) This image, representative of one of the numerous experiments performed in triplicate wells to address migration and invasion under different conditions, shows the absence of SCLC cells on the lower side of the membrane. Only pores of the membrane are observed. (b) This is in sharp contrast to the large number of HT1080 (human fibrosarcoma) cells observed on the lower side of the membrane, as demonstrated in this figure representative of one of the three independent experiments performed in triplicate wells to address migration. Pictures magnification: ×40.
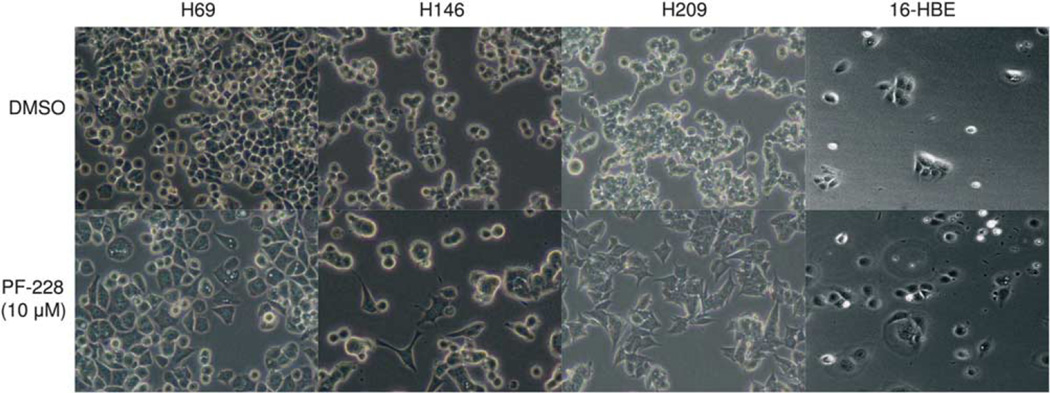
To further address migration, NCI-H209 cells were tested in a single-cell motility assay. The cells were seeded on laminin-332-coated dishes, allowed to adhere overnight, treated with increasing concentrations of PF-228 and analyzed by time-lapse microscopy for 8 h. These studies demonstrated that most SCLC cells would not move regardless of the presence of PF-228 (Supplementary Movies 1 and 2), and that a subset of SCLC cells treated with PF-228 spread on laminin-332, losing their rounded shape and presenting extended processes. Spreading was also observed on images of unstained NCI-H69, NCI-H146 and NCI-H209 cells after 90 min and 24 h treatment with 10 µm PF-228, as compared with DMSO (Figure 8). No change in morphology was observed in 16-HBE, an immortalized normal bronchial epithelial cell line constitutively lacking phospho-FAK Tyr397 expression and treated with PF-228, ruling out an off-target effect.
Figure 8.
Treatment of SCLC cells with PF-228 increases their spreading on laminin-332. NCI-H69, NCI-H146, NCI-H209 and 16-HBE cells were plated for 12 h on laminin-332-coated dishes and then treated with 10 µm PF-228 or DMSO for 24 h. Pictures of unstained cells were taken after 24 h treatment and show that SCLC cells treated with PF-228 spread on laminin-332, losing their rounded shape and presenting extended processes. No change in morphology was observed in 16-HBE, an immortalized normal bronchial epithelial cell constitutively lacking phospho-FAK Tyr397 expression. Pictures magnification: ×20.
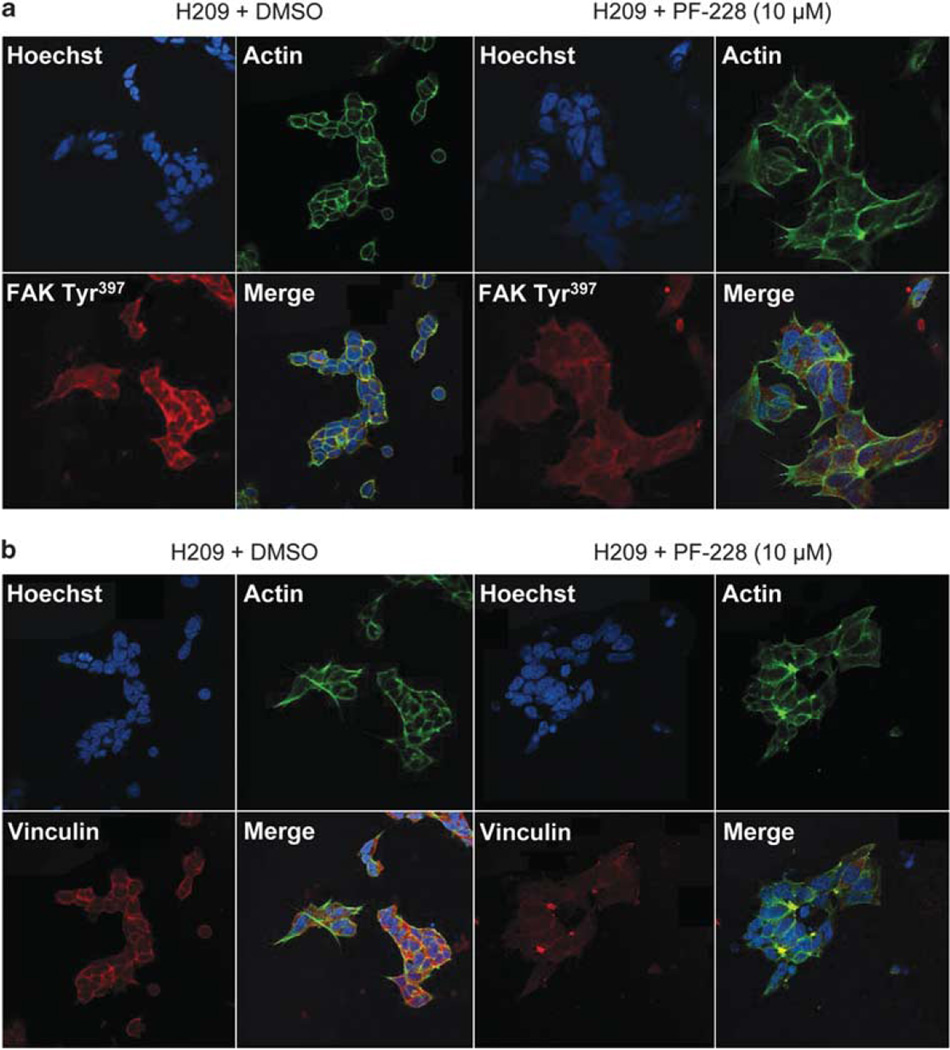
Immunostaining for phospho-FAK Tyr397 confirmed that NCI-H209 cells treated with 10 µm PF-228 were spread and displayed a lower expression of phospho-FAK, in contrast to cells treated with DMSO (Figure 9a). Moreover, PF-228 decreased focal adhesions, as demonstrated by a decrease in vinculin immunostaining in the absence of actin fiber reorganization (Figure 9b).
Figure 9.
Treatment of SCLC cells adherent to laminin-332 with PF-228 decreases immunofluorescence staining for phospho-FAK Tyr397 and vinculin. NCI-H209 cells were plated for 12 h on laminin-332-coated dishes and then treated with 10 µm PF-228 or DMSO for 12 h. After 12 h treatment, they were fixed and immunostained using Hoechst (blue, for nuclei), Phalloidin (green, for Actin) and phospho-FAK Tyr397 (red) in a or vinculin (red) in b. (a) Cells treated with PF-228 spread on laminin-332 and showed decreased phospho-FAK Tyr397 accumulation at the spreading edges and at cell–cell contact sites compared with NCI-H209 cells treated with DMSO. (b) Immunofluorescence staining of NCI-H209 cells also showed a decrease in vinculin expression in the presence of 10 µm PF-228. Pictures magnification: ×63.
Discussion
The main objective of this study was to test the hypothesis that recurrent copy number alterations in SCLCs will point toward molecular pathways relevant to tumor progression. We report the largest series of SCLCs to undergo high-throughput genomic analysis. Our array CGH analysis revealed 70 regions of copy number gain and 55 regions of copy number loss, including 329 and 99 genes, respectively, and consistent with published reports (Levin et al., 1994; Ried et al., 1994; Lindblad-Toh et al., 2000; Peng et al., 2005; Zhao et al., 2005; Coe et al., 2006; Kim et al., 2006; Olejniczak et al., 2007). Molecular pathway analysis centered on these genes led to the discovery of two enriched pathways in SCLCs, the focal adhesion and the neuroendocrine ligand–receptor interaction pathways. This is the first report of the activation of these pathways in SCLC.
We deliberately focused on the focal adhesion pathway because of the potential relevance in understanding SCLC metastatic and aggressive behavior. Among genes represented in this pathway is FAK, which has an important role in cell adhesion, spreading, migration, invasion, survival and anchorage-independent growth (Hanks et al., 2003; Parsons, 2003; Mitra et al., 2005; Siesser and Hanks, 2006). Although FAK overexpression has been reported in highly invasive cancers (Owens et al., 1995; Gabarra-Niecko et al., 2003), this is a first report of its possible implications in SCLC progression.
Once activated by integrins, G protein-coupled receptor ligands or growth factors, FAK is autophosphorylated at Tyr397 and activates proteins such as Src, p130CAS, paxillin and PI3KR2 (Hanks et al., 2003) to regulate adhesion turnover at the cell front, a process central to migration (Hanks et al., 2003; Parsons, 2003; Webb et al., 2004; Mitra et al., 2005; Siesser and Hanks, 2006). Small molecule inhibitors targeting the FAK kinase domain have been developed (NVP-TAE226, PF-562,271 and PF-573,228 (PF-228) used in our functional experiments) (Shi et al., 2007; Slack-Davis et al., 2007; Roberts et al., 2008). These inhibitors decreased FAK Tyr397 phosphorylation and induced inhibition of migration in various cell types. Preclinical studies with PF-562,271 showed tumor stasis or regression in mouse models of prostate, breast, colon, non-small-cell lung cancer, glioblastoma and pancreatic cancers (Roberts et al., 2008).
We carefully verified our findings at the DNA copy number, gene expression and protein expression levels. At the genomic level, we observed very concordant results between those obtained for FAK by array CGH and FISH. Next, We verified FAK gene copy number by quantitative real-time polymerase chain reaction on DNA extracted from SCLC primary tumors and cell lines, and found it to be amplified in 80% of tumors and in all tested cell lines. Testing the relevance of these genomic results in the focal adhesion pathway at the gene expression level, we observed a positive correlation between copy number determined by array CGH and gene expression data from a publicly available data set (Garber et al., 2001). Finally, at the protein level, we found that FAK was highly expressed in primary SCLC tumors and autophosphorylated in SCLC cell lines. Enrichment for genes associated with the focal adhesion pathway is a novel finding in SCLC and may be important because dysregulation of focal adhesions contributes to cell transformation, is observed in metastatic cancers and could explain SCLC high propensity of invasion and metastasis (Cooper and Spiro, 2006). However, we have not been able to show a direct correlation between FAK copy number and protein expression levels. Normal lung tissues adjacent to SCLC tissues tested by IHC were not available; hence, we were unable to compare FAK expression between unaffected lung and SCLC. Moreover, total FAK expression was similar in immortalized normal bronchial epithelial and SCLC cell lines. Yet, the most important observation is that FAK was preferentially phosphorylated in SCLC cell lines, even though the mechanisms of FAK activation remain unknown. Similarly, EGFR overexpression is observed more often than its amplification, and the relationship between protein expression and gene amplification is not fully understood in lung cancers (Hirsch et al., 2003). Moreover, clinical response to EGFR inhibitors is better related to activating mutations of EGFR or to its level of gene amplification than to its level of overexpression (Cappuzzo et al., 2005).
Should FAK activation be important in SCLC phenotype, inhibition-of-function studies are warranted. We therefore treated four SCLC cell lines with a FAK inhibitor, PF-228, and demonstrated a dose-dependent decrease in FAK Tyr397 phosphorylation. Inhibition of FAK phosphorylation was associated with inhibition of its downstream substrate p130CAS Tyr165 phosphorylation, providing a likely mechanism of action. FAK inhibition decreased adhesion of SCLC cells to laminin-332 in a dose-dependent manner, supporting results found in other systems (Guan et al., 1991; Burridge et al., 1992; Schwartz et al., 1995). We also studied cell migration and invasion using a transwell assay and found that none of the three tested SCLC cells migrated in the tested conditions, irrespective of FAK inhibition. Pursuing these findings, we performed a single-cell motility assay with NCI-H209 and confirmed that they did not migrate significantly, regardless of FAK inhibition. Yet, few cells that did not detach from laminin-332-coated dishes in the presence of PF-228 acquired a more spread morphology. It is important to note that this increased spreading was not associated with increased motility. In fact, PF-228 prevented focal adhesions assembly, as shown by markedly reduced vinculin immunostaining at the periphery of cells. From these experiments, we learned that FAK is constitutively activated in SCLC and may be responsible for focal adhesions turnover, which, once inhibited, may prevent cells from adhering to substrates. Our results exemplify the complexity of the focal adhesion pathway and demonstrate that FAK phosphorylation in SCLC is independent of cell–substrate and cell–cell contact. Future studies will need to clarify the role of FAK in SCLC motility and determine the potential therapeutic implications of these observations.
Further validating our approach and consistent with the literature (Taniwaki et al., 2006), our pathway analysis also implicated the neuroactive ligand–receptor interaction pathway. SCLC is thought to originate from neuroendocrine cells (Sidhu, 1979; Osada et al., 2008) and neuroendocrine mediators have been shown to activate G protein-coupled receptors that in turn activate FAK (Rozengurt, 2007). The relatively low resolution of the arrays we used leaves the possibility of other pathway activations to be discovered in SCLC.
In summary, we have established a role for FAK in SCLC. The genomic data were validated in independent data sets at the DNA, RNA and protein levels, and functionally by studying the in vitro effects of FAK inhibition on SCLC cell adhesion, spreading and motility. The results presented strengthen the validity of the approach taken and support that pathway analysis of array CGH data may uncover important molecular mechanisms of tumor progression.
Materials and methods
Primary SCLC specimens and SCLC cell lines
Primary SCLC surgical pathology blocks were obtained from the archives of the Department of Pathology at the Vanderbilt University Medical Center, the Nashville VA Medical Center and St-Thomas Hospital in Nashville, TN, USA. On the basis of the availability of surgical pathology blocks and the amount of material, 46 tumors from 46 patients were selected for array CGH. Surgical pathology specimens were obtained at the time of surgery or bronchoscopy. All tissues were obtained before medical treatment, and a lung cancer pathologist (ALG) confirmed the diagnosis. Institutional Review Boards at each medical center approved the study. Five SCLC cell lines were used, NCI-H69, NCI-H82, NCI-H146, NCI-H209 and NCIH1618 (ATCC, Manassas, VA, USA), and three immortalized normal bronchio-epithelial cell lines, BEAS-2B (ATCC), HBEC-3KT (Dr Minna’s gift, University of Texas Southwestern, TX, USA) and 16-HBE (Dr Gruenert’s gift, Children’s Hospital, Oakland Research Institute, OK, USA).
Tumor DNA extraction and array CGH
Tissue sections of tumor blocks were cut 20 µm thick and placed on glass slides. A 5 µm section was obtained in the middle of the series for pathological evaluation (ALG) and to confirm the presence of > 70% tumor cells and absence of necrosis. Other sections were stained with methyl green, tumor cells were manually microdissected under a stereomicroscope and DNA was extracted and purified as previously reported (Massion et al., 2002, 2008). Tumor DNA was random-primer labeled with Cy5 and cohybridized onto a 2464-BAC clone array CGH slide obtained from the UCSF array CGH core laboratory (http://cancer.ucsf.edu/array/services.php#human-BAC), along with Cy3-labeled sex-matched leukocyte control DNA (Invitrogen, Carlsbad, CA, USA). Array CGH hybridizations were performed as described in previous reports (Pinkel et al., 1998; Massion et al., 2002, 2008). Microarray slides were analyzed using GenePix software (Axon Inc., Darien, CT, USA). Hybridization signals were obtained from GenePix and consisted of red-to-green intensity ratios.
Pathway analysis of recurrent and significant copy number alterations
We identified recurrent (prevalence > 50% of analyzed tumors) and significant (log2 copy number ratio > 0.3 or <−0.3) regions of gene copy number alterations. From these regions, we outputted the reference sequence genes located within a 300 kb genomic distance fixed on the centroid of the BAC clone using UCSC Genome Browser (Human Mar. 2006 Assembly) following our previous protocol (Massion et al., 2008). Using WebGestalt2.0 software (http://bioinfo.vanderbilt.edu/webgestalt) (Kirov et al., 2005; Zhang et al., 2005), we determined enrichment for molecular pathways compared with a reference gene set consisting of the 7913 genes included in the array.
Fluorescent in situ hybridization
Dual-color FISH was performed on interphase nuclei in tissue sections as described before (Massion et al., 2002) on 46 primary SCLCs. Probes targeting FAK at 8q24.3 (RP11-13A18) and PIK3CA at 3q26.3 (RP11-245C23) and the reference centromeric regions of CEP3 and CEP8 (Vysis Inc., Downers Grove, IL, USA) were hybridized and spot counts were obtained in ≥50 nuclei per specimen for each probe. The ratio of mean counts for test and reference probes on the same chromosome was reported as the relative copy number for the test gene.
Quantitative real-time polymerase chain reaction
Gene copy number in 10 SCLCs and five SCLC cell lines was evaluated by quantitative real-time polymerase chain reaction following a previously reported protocol (Gallia et al., 2006). Primer sequences are reported in Supplementary Table 3. The ΔΔCt method was used for DNA quantification. DNA content was normalized to that of RNase P.
Immunohistochemistry
Tissues sections were stained following a previously reported protocol (Massion et al., 2003) with anti-FAK (A-17) (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibody. Immunostains were scored by two independent observers (SO, PPM) as follows: 0—no staining, 1—weak, 2—moderate and 3—strong staining.
Western blotting
Cell lysates were obtained following standard protocol (Qian et al., 2009). Blots were probed with antibodies against phospho- FAK (Tyr397/576) (1:1000; BioSource, Camarillo, CA, USA), FAK (C-20) (1:200; Santa Cruz Biotechnology), phosphop130CAS (Tyr165) (1:1000; Cell Signaling, Danvers, MA, USA), p130CAS (1:2000; BD Transduction Laboratories, San Jose, CA, USA) or actin (1:5000; Sigma, St Louis, MO, USA).
Cell adhesion assay
Plates of 96 wells were coated with phosphate-buffered saline, 5 µg/ml collagen I (BD, Franklin Lakes, NJ, USA), fibronectin (Chemicon, Billerica, MA, USA), vitronectin (Sigma) or laminin-332 (VUMC Protein Users Club, Nashville, TN, USA) and incubated overnight at 4 °C. Single-cell suspensions were seeded in complete medium at a density of 8×104 cells/ well and allowed to adhere for 24 h at 37 °C. Cells were treated with PF-573,228 (PF-228) (3-10-30 µm, Tocris Biosciences, Ellisville, MO,USA) or DMSO for 30 min before seeding and during the whole experiment. After 24 h, non-adherent cells were washed off, whereas adherent cells were fixed and stained with crystal violet. Absorbance was measured at 595nm and experiments performed three times in triplicate wells.
Transwell migration and invasion assay
Transwell permeable supports of 24 wells with 8-µm pores (Corning Life Sciences, Lowell, MA, USA) were coated overnight on the lower (migration) or upper (invasion) side with laminin-332 (5 µg/ml) or basal membrane extract (1×, Trevigen, Gaithersburg, MD, USA). Lower chambers were then filled with 10% fetal bovine serum medium, supplemented with PF-228 (10 µm) or DMSO. VEGF (10–50 ng/ml, R&D Systems, Minneapolis, MN, USA) was added in the lower chamber of some wells for being previously reported to induce SCLC cells migration (Tanno et al., 2004). SCLC cells were seeded at 1 × 105 cells/well in serum-free or complete medium supplemented with PF-228 (10 µm) or DMSO. Fibrosarcoma cells HT1080 were used as positive control. Cells were allowed to migrate for 24 h at 37 °C, then fixed and stained with hematoxylin–eosin. Migration and invasion were evaluated by counting cells on the membrane’s lower side in five microscopic fields of ×40 magnification under a light microscope. Experiments were performed three times in triplicate wells.
Single-cell motility assay
Six-well Corning Costar, clear plates (Sigma-Aldrich, St Louis, MO, USA) were coated overnight with 5 µg/ml laminin-332. NCI-H209 cells were seeded in complete medium at a density of 3 × 105 cells/well. PF-228 (3-10-30 µm) or DMSO was then added to the medium. Time-lapse microscopy was conducted using a Zeiss Axiovert 200M microscope (Zeiss, Thornwood, NY, USA). Ten fields were randomly selected within each well. Phase-contrast images were captured automatically every 6 min for 8 h. Following image capture, 51 individual images from each coordinate were combined using MetaMorph to produce image stacks. Cells were tracked manually using an ImageJ (NIH, Bethesda, MD, USA) plugin, MTrackJ.
Immunofluorescence
Glass-bottom microwell dishes of 35 mm (MatTek, Ashland, MA, USA) were coated with 5 µg/ml laminin-332. NCI-H209 cells were seeded at a density of 2.5 × 104 cells/dish with PF-228 or DMSO. After 12 h, adherent cells were fixed, permeabilized and incubated with Hoechst (1:1000, blue, Invitrogen), phalloidin (1:2000, green, Invitrogen) and phospho-FAK (Tyr397) (1:200, BioSource) or vinculin antibody (1:200, Sigma) for 2 h at RT. Bound primary antibody to phospho-FAK (Tyr397) or vinculin was detected using goat antirabbit IgG Alexa Fluor568 (1:200, red, Invitrogen) or goat antimouse IgG Alexa Fluor568 for 1 h at RT. Cells were observed under LSM510 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY, USA).
Statistical analyses
Array CGH analysis
Red-to-green intensity ratios were analyzed by snapCGH package in R. Data were filtered by flag and R square ≤0.49 (correlation of Cy3 versus Cy5 pixel intensities). Background was corrected by a log linear interpolation method. Normalization of background-subtracted fluorescence ratios was obtained within each array by Print-tip loess methods and between arrays by quantile normalization (Bolstad et al., 2003). All intensity ratios were log2 transformed. Threshold for determining copy number gain or loss was defined as an average log2 copy number ratio > 0.3 or < −0.3.
Webgestalt pathway analysis
Fisher’s exact test was used to evaluate enrichment of the candidate gene set in KEGG pathway categories using Webgestalt software (http://bioinfo.vanderbilt.edu/webgestalt) (Kirov et al., 2005; Zhang et al., 2005). Comparison with a public gene expression data set: mRNA expression levels of candidate genes in the format of fold changes (log2 ratio SCLC/normal) were retrieved from a public data set with a similar design, including six normal lung and five SCLC samples (Garber et al., 2001). An average fold change between SCLC and normal was calculated for each candidate gene. Integration of genetic dosages and the corresponding gene expression levels was performed by correlation plot and Pearson’s correlation analyses. Other statistical analyses were performed by Student’s t-test (two-tailed distribution).
Supplementary Material
Acknowledgements
We thank Kathy Taylor, Director of the Research Institute at St Thomas Health Services, Nashville, TN, for sharing archived SCLC tissue blocks; Dr Coe for providing array CGH data (Coe et al., 2006); and Vanderbilt’s Immunohistochemistry Core (supported by a Cancer Center Support Grant 5P30 CA068485) for their contribution. We also thank Steven K Hanks, Alissa M Weaver and Donna J Webb for their useful input in the interpretation of the paper. This work was supported by a Merit Review grant from the Department of Veterans Affairs. Dr Ocak was supported by an IASLC Young Investigator Fellowship Award and by a grant from Université Catholique de Louvain (Bourse Clinicien-Chercheur), Belgium.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Balsara BR, Testa JR. Chromosomal imbalances in human lung cancer. Oncogene. 2002;21:6877–6883. doi: 10.1038/sj.onc.1205836. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Burridge K, Turner CE, Romer LH. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrix: a role in cytoskeletal assembly. J Cell Biol. 1992;119:893–903. doi: 10.1083/jcb.119.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Coe BP, Lockwood WW, Girard L, Chari R, Macaulay C, Lam S, et al. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer. 2006;94:1927–1935. doi: 10.1038/sj.bjc.6603167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Spiro SG. Small cell lung cancer: treatment review. Respirology. 2006;11:241–248. doi: 10.1111/j.1440-1843.2006.00850.x. [DOI] [PubMed] [Google Scholar]
- Gabarra-Niecko V, Schaller MD, Dunty JM. FAK regulates biological processes important for the pathogenesis of cancer. Cancer Metastasis Rev. 2003;22:359–374. doi: 10.1023/a:1023725029589. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Rand V, Siu IM, Eberhart CG, James CD, Marie SK, et al. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res. 2006;4:709–714. doi: 10.1158/1541-7786.MCR-06-0172. [DOI] [PubMed] [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JL, Trevithick JE, Hynes RO. Fibronectin/integrin interaction induces tyrosine phosphorylation of a 120-kDa protein. Cell Regul. 1991;2:951–964. doi: 10.1091/mbc.2.11.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Ryzhova L, Shin NY, Brabek J. Focal adhesion kinase signaling activities and their implications in the control of cell survival and motility. Front Biosci. 2003;8:d982–d996. doi: 10.2741/1114. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–138. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- Kirov SA, Peng X, Baker E, Schmoyer D, Zhang B, Snoddy J. GeneKeyDB: a lightweight, gene-centric, relational database to support data mining environments. BMC Bioinformatics. 2005;6:72. doi: 10.1186/1471-2105-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin NA, Brzoska P, Gupta N, Minna JD, Gray JW, Christman MF. Identification of frequent novel genetic alterations in small cell lung carcinoma. Cancer Res. 1994;54:5086–5091. [PubMed] [Google Scholar]
- Lindblad-Toh K, Tanenbaum DM, Daly MJ, Winchester E, Lui WO, Villapakkam A, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- Massion PP, Kuo WL, Stokoe D, Olshen AB, Treseler PA, Chin K, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–3640. [PubMed] [Google Scholar]
- Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63:7113–7121. [PubMed] [Google Scholar]
- Massion PP, Zou Y, Chen H, Jiang A, Coulson P, Amos CI, et al. Smoking-related genomic signatures in non-small cell lung cancer. Am J Respir Crit Care Med. 2008;178:1164–1172. doi: 10.1164/rccm.200801-142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Miura I, Graziano SL, Cheng JQ, Doyle LA, Testa JR. Chromosome alterations in human small cell lung cancer: frequent involvement of 5q. Cancer Res. 1992;52:1322–1328. [PubMed] [Google Scholar]
- Olejniczak ET, Van Sant C, Anderson MG, Wang G, Tahir SK, Sauter G, et al. Integrative genomic analysis of small-cell lung carcinoma reveals correlates of sensitivity to bcl-2 antagonists and uncovers novel chromosomal gains. Mol Cancer Res. 2007;5:331–339. doi: 10.1158/1541-7786.MCR-06-0367. [DOI] [PubMed] [Google Scholar]
- Osada H, Tomida S, Yatabe Y, Tatematsu Y, Takeuchi T, Murakami H, et al. Roles of achaete-scute homologue 1 in DKK1 and E-cadherin repression and neuroendocrine differentiation in lung cancer. Cancer Res. 2008;68:1647–1655. doi: 10.1158/0008-5472.CAN-07-5039. [DOI] [PubMed] [Google Scholar]
- Owens LV, Xu L, Craven RJ, Dent GA, Weiner TM, Kornberg L, et al. Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Res. 1995;55:2752–2755. [PubMed] [Google Scholar]
- Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- Peng WX, Shibata T, Katoh H, Kokubu A, Matsuno Y, Asamura H, et al. Array-based comparative genomic hybridization analysis of high-grade neuroendocrine tumors of the lung. Cancer Sci. 2005;96:661–667. doi: 10.1111/j.1349-7006.2005.00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Qian J, Zou Y, Rahman JS, Lu B, Massion PP. Synergy between phosphatidylinositol 3-kinase/Akt pathway and Bcl-xL in the control of apoptosis in adenocarcinoma cells of the lung. Mol Cancer Ther. 2009;8:101–109. doi: 10.1158/1535-7163.MCT-08-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, et al. Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res. 1994;54:1801–1806. [PubMed] [Google Scholar]
- Roberts WG, Ung E, Whalen P, Cooper B, Hulford C, Autry C, et al. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res. 2008;68:1935–1944. doi: 10.1158/0008-5472.CAN-07-5155. [DOI] [PubMed] [Google Scholar]
- Rozengurt E. Mitogenic signaling pathways induced by G protein-coupled receptors. J Cell Physiol. 2007;213:589–602. doi: 10.1002/jcp.21246. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Schaller MD, Ginsberg MH. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Shi Q, Hjelmeland AB, Keir ST, Song L, Wickman S, Jackson D, et al. A novel low-molecular weight inhibitor of focal adhesion kinase, TAE226, inhibits glioma growth. Mol Carcinog. 2007;46:488–496. doi: 10.1002/mc.20297. [DOI] [PubMed] [Google Scholar]
- Sidhu GS. The endodermal origin of digestive and respiratory tract APUD cells. Histopathologic evidence and a review of the literature. Am J Pathol. 1979;96:5–20. [PMC free article] [PubMed] [Google Scholar]
- Siesser PM, Hanks SK. The signaling and biological implications of FAK overexpression in cancer. Clin Cancer Res. 2006;12:3233–3237. doi: 10.1158/1078-0432.CCR-06-0456. [DOI] [PubMed] [Google Scholar]
- Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, et al. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- Sozzi G, Bertoglio MG, Borrello MG, Giani S, Pilotti S, Pierotti M, et al. Chromosomal abnormalities in a primary small cell lung cancer. Cancer Genet Cytogenet. 1987;27:45–50. doi: 10.1016/0165-4608(87)90259-7. [DOI] [PubMed] [Google Scholar]
- Taniwaki M, Daigo Y, Ishikawa N, Takano A, Tsunoda T, Yasui W, et al. Gene expression profiles of small-cell lung cancers: molecular signatures of lung cancer. Int J Oncol. 2006;29:567–575. [PubMed] [Google Scholar]
- Tanno S, Ohsaki Y, Nakanishi K, Toyoshima E, Kikuchi K. Human small cell lung cancer cells express functional VEGF receptors, VEGFR-2 and VEGFR-3. Lung Cancer. 2004;46:11–19. doi: 10.1016/j.lungcan.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Ullmann R, Schwendel A, Klemen H, Wolf G, Petersen I, Popper HH. Unbalanced chromosomal aberrations in neuroendocrine lung tumors as detected by comparative genomic hybridization. Hum Pathol. 1998;29:1145–1149. doi: 10.1016/s0046-8177(98)90428-2. [DOI] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.