Abstract
Commensal microorganisms colonize the nasal mucosa without inducing inflammation. Pathogens perturbing the commensal flora often invade evading immune defenses. The different types of adaptive responses that drive the distinct behaviors of commensals and pathogens, allowing one to persist at mucosal surfaces and the other to survive within tissues, are not yet clear. In the present work we demonstrate that although crossing epithelial barriers, the commensal Lactobacillus murinus stimulates epitope-specific CD4+ T cells in nasal-associated lymphoid tissue (NALT) less efficiently than the pathogen Streptococcus pyogenes. In NALT antigen-presenting cells other than CCR6+ CD11c+ dendritic cells process and present the microbial antigens. Effector/memory CD4+ T cells generated after intranasal priming with L. murinus and S. pyogenes surprisingly express similar proinflammatory cytokines and are not CD25+/FoxP3+ T-regulatory cells when recirculating in the spleen. These findings suggest that when a commensal crosses the nasal epithelial barrier it induces a proinflammatory response similar to a pathogen but without causing disease.
INTRODUCTION
Infections initiated at upper respiratory tract are one of the most prevalent worldwide,1 particularly affecting the young and the immunocompromised.2,3 Invasive pathogens like Streptococcus pyogenes, often persist and are difficult to eradicate because they can acquire intracellular location avoiding antibiotics4,5 and ultimately residing within normal commensal microbial communities in healthy individuals.6 Alternatively, commensal microbes permanently reside in such ecological niches and at least in the gastrointestinal tract, appear to be readily eliminated once they cross the epithelial barriers.7
The B-cell response against T-independent antigens (carbohydrates and lipopolysaccharides), provides low affinity, isotype switched secretory immunoglobulins8 that limit microbial translocation across epithelial barriers.9 Alternatively, the induction of protective, high affinity, isotype switched, secretory immunoglobulins against T-cell-dependent antigens is subordinate to T-cell activation,10 cognate T/B-cell interaction,11 and cytokine production.12–15 The phenotype of memory T-cell and the subsequent adaptive secretory humoral immune response are therefore central to shaping the microbial community on mucosal tissues and protecting against virulent pathogens.
Despite the presence of a substantial microbiota on the surface of the epithelium,16 the absence of inflammation within the submucosa of the upper respiratory tract contrasted with the sustained proinflammatory response elicited by an invading pathogen17 suggest two possible hypotheses. First, commensal microorganisms do not cross the epithelial barrier efficiently, compared to pathogens, or there is an inherent substantial difference between commensals and pathogens in their potentials to activate the innate and consequently the adaptive mucosal immune response.
In this study we hypothesized that a commensal Lactobacillus induces the differentiation of T-helper cells with regulatory and/or immunosuppressive phenotype, whereas S. pyogenes would induce T cells expressing proinflammatory cytokines. To test this hypothesis we tracked the antigen-specific T-cell response against Lactobacillus murinus and a strain of S. pyogenes, both tagged with a chicken ovalbumin peptide 323–339 (OVA). Specific for OVA on I-Ad molecules, CD4+ T cells from the T-cell receptor (TCR)-transgenic mouse line DO11.1018 were injected into normal syngeneic mice and tracked with flow cytometry by staining cells in the recipient’s lymphoid organs with anti-CD4 and the anticlonotypic KJ1-26 mAb.19,20 This strategy is extremely powerful because it allows studying the antigen-specific memory T-cell response at the single-cell level after tagged bacteria are delivered to the upper respiratory tract.
RESULTS
Viable L. murinus and S. pyogenes are recovered in NALT tissues
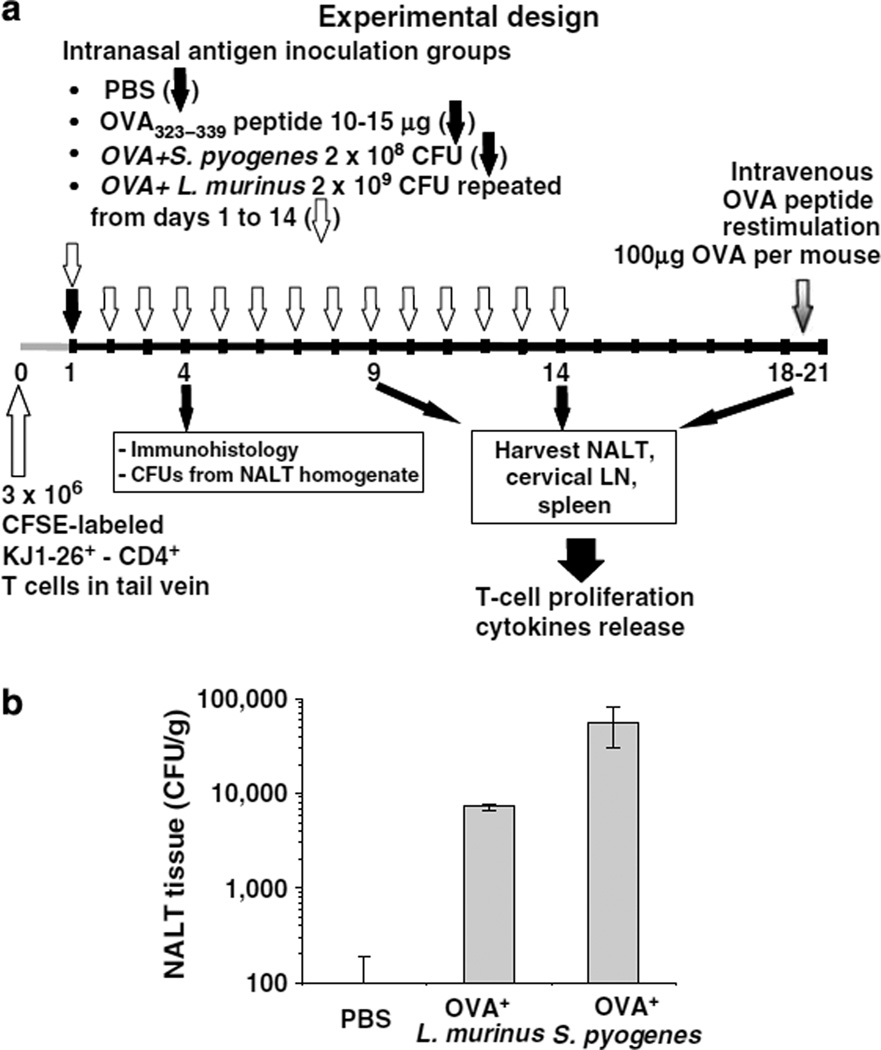
After a single intranasal inoculation (Figure 1a), we recovered seven times more S. pyogenes CFUs per gram of nasal-associated lymphoid tissue (NALT) tissue than three consecutive inocula of L. murinus (Figure 1b). Despite delivering 10 times more L. murinus CFUs per inoculation over 3-day period, more S. pyogenes was recovered from NALT 4 days after a single inoculation. This indicates that both bacteria translocate across the nasal follicle-associated epithelium (NFAE) but S. pyogenes either translocates by microfold-cells (M cells)21 more efficiently or it persists in higher numbers within tissues.
Figure 1.
L. murinus and S. pyogenes are recovered from nasal-associated lymphoid tissue (NALT) tissues. (a) Summary of experimental design. (b) Bacterial recovery from NALT tissues after anesthetized mice were inoculated intranasally once with 2×108 CFU of OVA+ S. pyogenes or with 1×109 CFU of OVA+ L. murinus for 3 consecutive days in two aliquots of 7.5 µl each in phosphate-buffered saline (PBS) delivered at in front of each nostril. At 4 days after S. pyogenes and 24 h after L. murinus last inoculations NALT tissues were dissected, weighed, rinsed in PBS, mashed, and plated in serial dilutions on Lactobacilli deMan–Rogosa–Sharpe (MRS) agar or blood–Todd–Hewitt yeast extract (THY) agar plates containing 5 µg/ml erythromycin for 24 h at 37°C.
As L. murinus and S. pyogenes translocate the NFAE into NALT we tested whether the physical location of CD11c+ dendritic cells (DCs) is affected by the presence of bacteria in the nasal passages.
Macro- and microscopic architecture of the nasal-associated lymphoid tissue
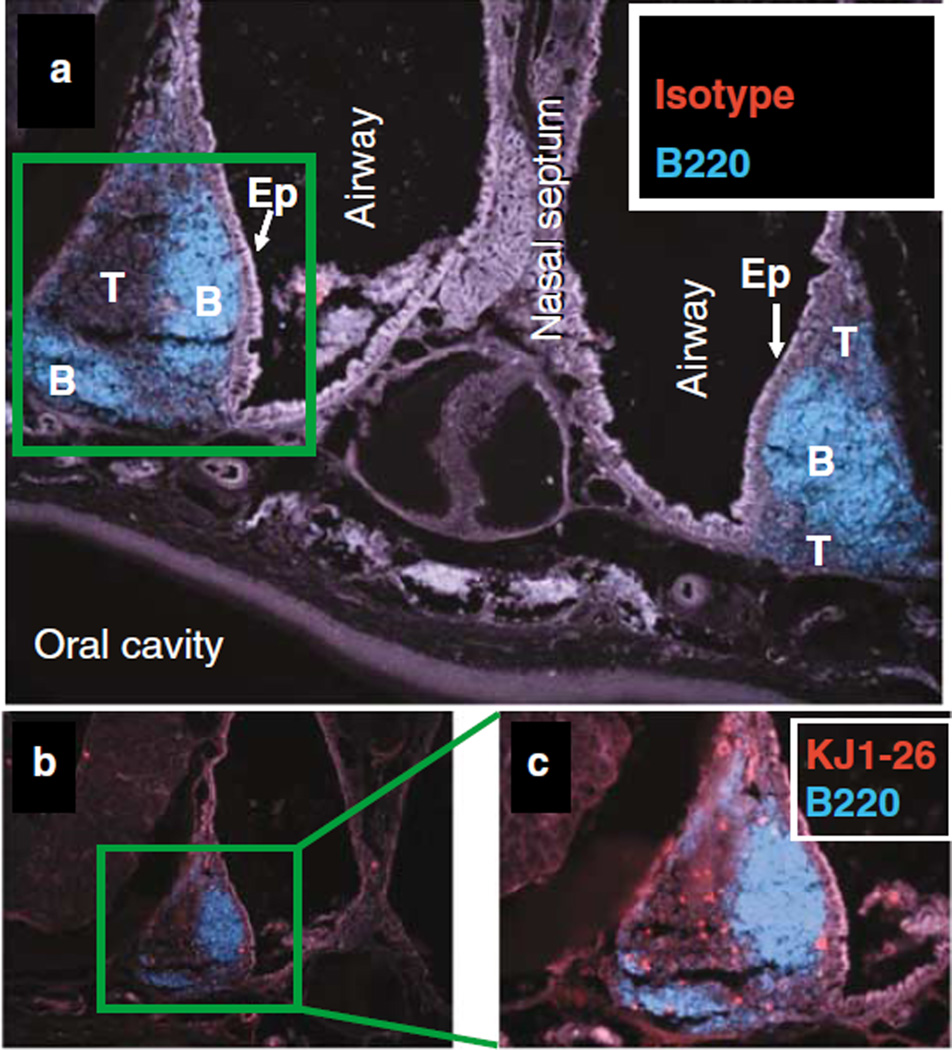
A frontal immunohistological section of the mouse maxilla and nasal passages performed between the upper incisors and the first molar teeth reveals two bilateral triangular lymphoid structures cranial to the roof of the oral cavity, located at the junction between the lateral walls and the floor of the nasal passages (Figure 2). These lymphoid structures are covered by columnar ciliated epithelium and M-cells as previously described21 (Figure 2a, light gray stain “Ep”). In the sham-infected mouse the B-cell-rich region containing B220 + follicular cells are generally underlying the NFAE (Figure 2a, blue stain “B”). Lateral to the B-cell region, away from the epithelium lays the T-cell-rich region (Figure 2a, dark gray area “T”). In OVA+ S. pyogenes-infected mice, OVA-specific T cells were detected within the T-cell-rich region of NALT with the anticlonotypic KJ1-26 mAb (Figure 2b and c).
Figure 2.
T cells are located in the T-cell rich region and B cells are facing the nasal-associated follicular epithelium (NFAE). Nasal-associated lymphoid tissue (NALT) of a BALB/c mouse 4 days after intranasal infection with OVA+ S. pyogenes. Immunohistology at ×20 magnification of NALT tissues double stained with Cy3-amplified (a) IgG2a isotypes or (b, c) anti-OVA T-cell receptor (TCR) (KJ1-26 mAb) and fluorescein isothiocyanate (FITC)-conjugated anti-B220 and recolored in Adobe Photoshop.
CD11c+ cells localize under the subepithelial dome of the NALT after bacterial inoculation
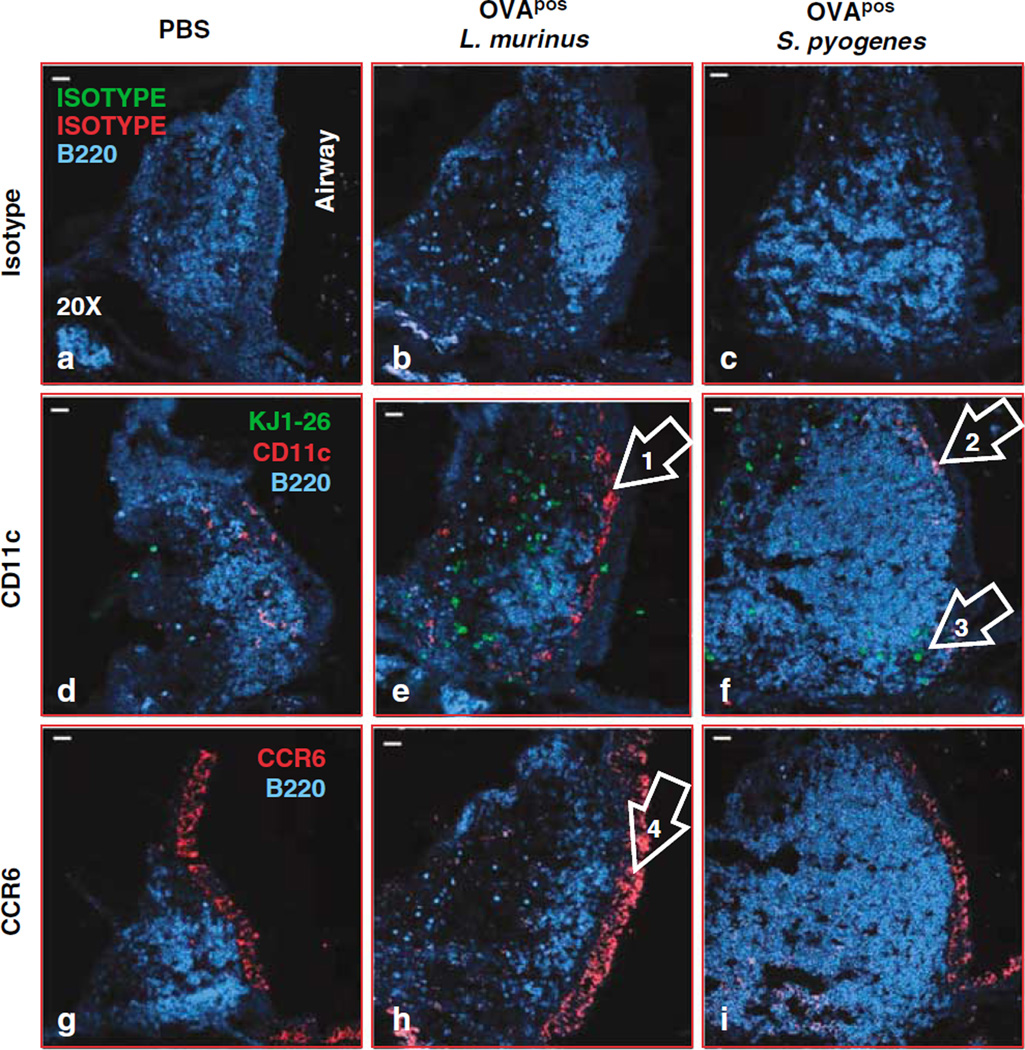
CD11c+ cells were distributed within the epithelium and in the T-cell-rich region around the B cell follicles in sham control mice (Figure 3a). At 4 days after repeated inoculation of L. murinus or single inoculation of S. pyogenes, CD11c+ cells (red stain) were consistently located just below the NFAE (Figure 3e, arrow no. 1; Figure 3f, arrow no. 2). In mice infected with S. pyogenes the extent of CD11c staining was consistently weaker than in mice infected with L. murinus. Consistent with the behavior of Peyer’s patches and lamina propria DCs,22,23 or even skin DCs,24 we speculate that S. pyogenes activates CD11c+ DCs to migrate out of the NALT into the blood stream or toward draining cervical lymph nodes (LNs). CD11c+ cells do not accumulate under the NFAE after delivery of phosphate-buffered saline (PBS) (negative control) but they were uniformly distributed in T-cell-rich region where OVA-specific T cells were located (Figure 3d). CD11c+ and OVA-specific T cells were also detected in the T-cell-rich region of NALT of mice inoculated with L. murinus (Figure 3e, red and green dots). In mice inoculated with S. pyogenes fewer CD11c+ cells were detected in the T-cell-rich region of NALT (Figure 3f, arrow no. 3).
Figure 3.
CD11c cells localize under the epithelium of nasal-associated lymphoid tissue (NALT) 4 days after bacterial inoculation. Immunohistology at ×20 magnification of triple-stained NALT tissue oriented with the luminal side is on the right side of the images. The three columns display mice treated with phosphate-buffered saline (PBS) (a, d, g), OVA+ L. murinus (b, e, h), or S. pyogenes (c, f, i). The first row displays NALT tissues stained with IgG1 and IgG2a isotypes and anti-B220 as shown in a and b or polyclonal rabbit IgG and anti-B220 as shown in c. The tissues in the second row as shown in d–f were stained with anti-CD11c, KJ1-26, or anti B220 displaying B-cell follicles (blue), OVA-specific T cells (green), and CD11c+ cells (red). Images in the third row display tissues double stained with rabbit IgG anti-mouse CCR6 and anti-B220 as shown in g–i. Mice were inoculated intranasally once with 2×108 CFU of OVA+ S. pyogenes or with 1×109 CFU of OVA+ L. murinus for 3 consecutive days in two aliquots of 7.5 µl each in PBS delivered at in front of each nostril. At 4 days after S. pyogenes and 24 h after L. murinus last inoculations the maxillae containing NALT tissues were dissected, demineralized, and embedded in OCT for cryosection.
As in the Peyer’s patches chemokine receptor 6 (CCR6)+ CD11c+ DCs activate antigen-specific T cells after encountering intestinal pathogens25 we tested in NALT tissues whether CCR6+ and CD11c+ staining colocalize. The NFAE and the epithelium lining the nasal passages expressed significant amounts of CCR6. In infected mice, CD11c+ cells were adjacent to CCR6+ epithelial cells in NFAE, but did not precisely colocalize when assessed in adjacent sections (Figure 3e and h, arrows nos. 1 and 4). We evaluated adjacent sections because CCR6 and CD11c staining protocols are incompatible with sequential staining on the same slide. These data suggest that in NALT a CD11c+ DC population that is not CCR6+ migrates under the NFAE when either L. murinus or S. pyogenes are present at the epithelial surface.
L. murinus and S. pyogenes prime epitope-specific CD4+ T cells in the NALT and cervical lymph nodes
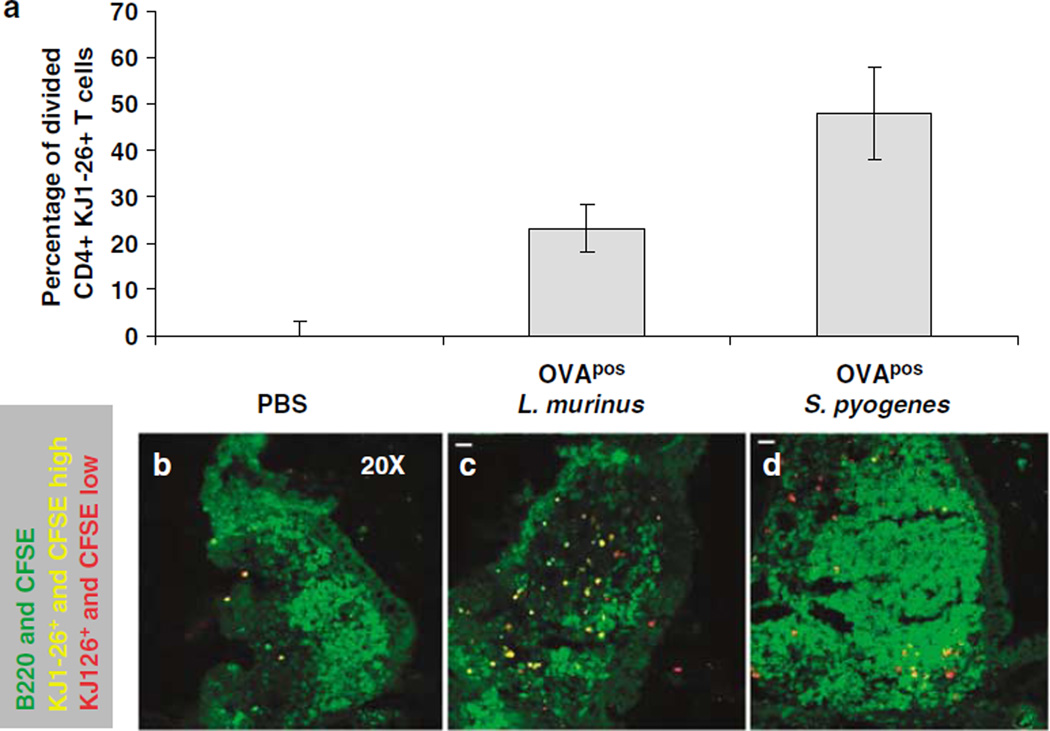
Using immunohistology we determined that 4 days after initial intranasal priming a higher number of dividing OVA-specific T cells were observed in NALT from mice inoculated with L. murinus or S. pyogenes than NALT from mice treated with PBS (Figure 4a). In standardized digital images, naive OVA-specific T cells were unequivocally identified because of their yellow secondary color resulting from merging of green 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) fluorescence and red fluorescence signal of anti-clonotypic KJ1-26 mAb (Figure 4b). After a few rounds of division, primed OVA-specific T cells dilute their CFSE content appear primarily red, due to fluorescence from the anticlonotypic KJ1-26 mAb signal. When normalized for the total NALT area, the number of red pixels within NALT is directly proportional to the level of antigen-specific priming occurring in the NALT tissue. Calculations revealed that S. pyogenes and L murinus had induced the proliferation of 48 and 23% more OVA-specific T cells, respectively, compared to mice treated with PBS just 4 days after bacterial inoculation.
Figure 4.
Direct visualization of proliferating T cells in nasal-associated lymphoid tissue (NALT) by immunohistology 4 days after bacterial inoculation. (a) Histogram estimating the fraction of activated antigen-specific T cells when normalized against the background phosphate-buffered saline (PBS) group 4 days after initial priming. The percentage of dividing cells is estimated by calculating the number of yellow pixels (CFSEhigh (green) and KJ1-26+ cells (red)) minus the number of red pixels (all KJ1-26+ cells) pixels divided by the total number of red pixels measured from mice inoculated with (b) PBS, (c) OVA+ L. murinus, or (d) OVA+ S. pyogenes. 5-(and-6)-Carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled cells (green channel) that are also KJ1-26+ (red dots) will appear yellow after merging of the colors. Cells that divide lose their CFSE and become red because they are now stained only with the KJ1-26 mAb.
L. murinus primes OVA-specific T cells less efficiently than S. pyogenes
Immunological protection depends on the increased frequency of effector/memory T cells recirculating between tissue and secondary lymphoid organs over time.26 Intranasal priming with L. murinus or S. pyogenes for 2 days induced proliferation of OVA-specific T cells. Primed OVA-specific T cells were detected in the spleen 21 days after initial immunization using flow cytometry (Figure 5A). Significantly fewer OVA-specific T cells had proliferated after L murinus intranasal priming for 2 days than after S. pyogenes (Figure 5B, gray bars). These data suggest that L. murinus does not penetrate the epithelial barrier as efficiently as S. pyogenes to be presented by antigen presenting cells (APCs) to OVA-specific T cells. Such hypothesis is corroborated by the lower number of L. murinus recovered from NALT (Figure 1b).
Figure 5.
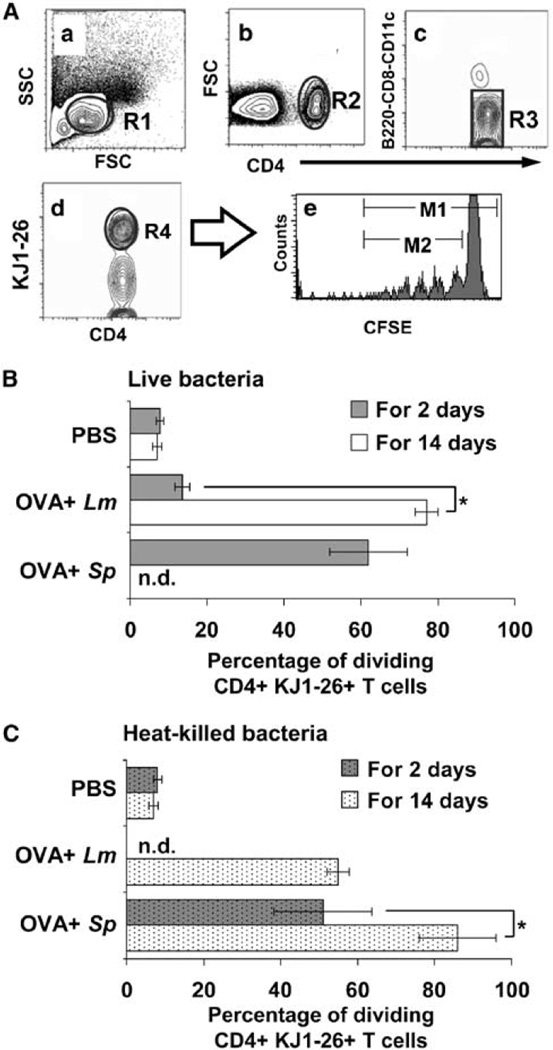
L. murinus primes OVA-specific T cells less efficiently than S. pyogenes. (A) Flow cytometry example of contour plots of splenocytes from mice inoculated intranasally with L. murinus for 9 days. (a) R1 identifies lymphocytes in forward and side scatter plot. (b) Subgate of R1 identifies CD4+ leukocytes (R2). (c) Subgate of R2 excludes B cells, CD8 T cells, CD11c+ CD4+ DCs, and potential circulating double-positive lymphocytes. (d) Subgate of R3 identifies the percentage of OVA-specific T cells (R4) out of total lymphocytes (R1). (e) Subgate of R4 identifies the percentage of dividing OVA-specific T cells when the ratio of cells diluting their 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) in the M2/M1 gates×100 is calculated. (B, C) Percentage of dividing CD4+ KJ1-26+ T cells in spleen 21 days after initial intranasal inoculation with phosphate-buffered saline (PBS), 2×109 OVA+ L. murinu s or 2×108 OVA+ S. pyogenes for 2 days (gray bars) or 14 days (white bars). To avoid septicemia, S. pyogenes was heat killed at 65 °C for 1 h before the inoculation for 14 days. Data shown are averages±s.e.m. from three experiments (n = 6 mice per data point). T-cell proliferation was significantly greater (P < 0.05) in each treatment group when compared to PBS control (* P < 0.05; Student’s t-test).
To mimic the persistent colonization of commensal bacteria on mucosal surfaces, 2×109 L. murinus were inoculated intranasally daily for 14 days. Repeated inoculation of L. murinus significantly increased the frequency of dividing OVA-specific T cells recovered in the spleen (P < 0.05) to a level similar to the one induced by S. pyogenes inoculated for only 2 days (Figure 5B, Lm white bar VS. Sp gray bar).
To determine whether bacteria actively cross the epithelial barrier of NALT S. pyogenes and L. murinus were heat-killed and inoculated for 2 or 14 days (Figure 5C). Heat-killed bacteria induced significant T-cell proliferation when delivered multiple times. A total of 2×108 S. pyogenes inoculated for 2 consecutive days were sufficient to induce T-cell proliferation similar to the one induced by L. murinus delivered in 14 consecutive inoculations (Figure 5C). These results collectively indicate that S. pyogenes and L. murinus need not to be viable to move across the epithelial barrier and activate CD4+ T cells in NALT. S. pyogenes crosses the epithelial barrier and/or persists in NALT more efficiently than L. murinus resulting in a greater percentage of proliferating antigen-specific T cells. However when L. murinus persists on the epithelium of the nasal passages dead or live, it is processed by NALT APCs and it does activate antigen-specific T cells to a comparable level induced by the episodic encounter with S. pyogenes.
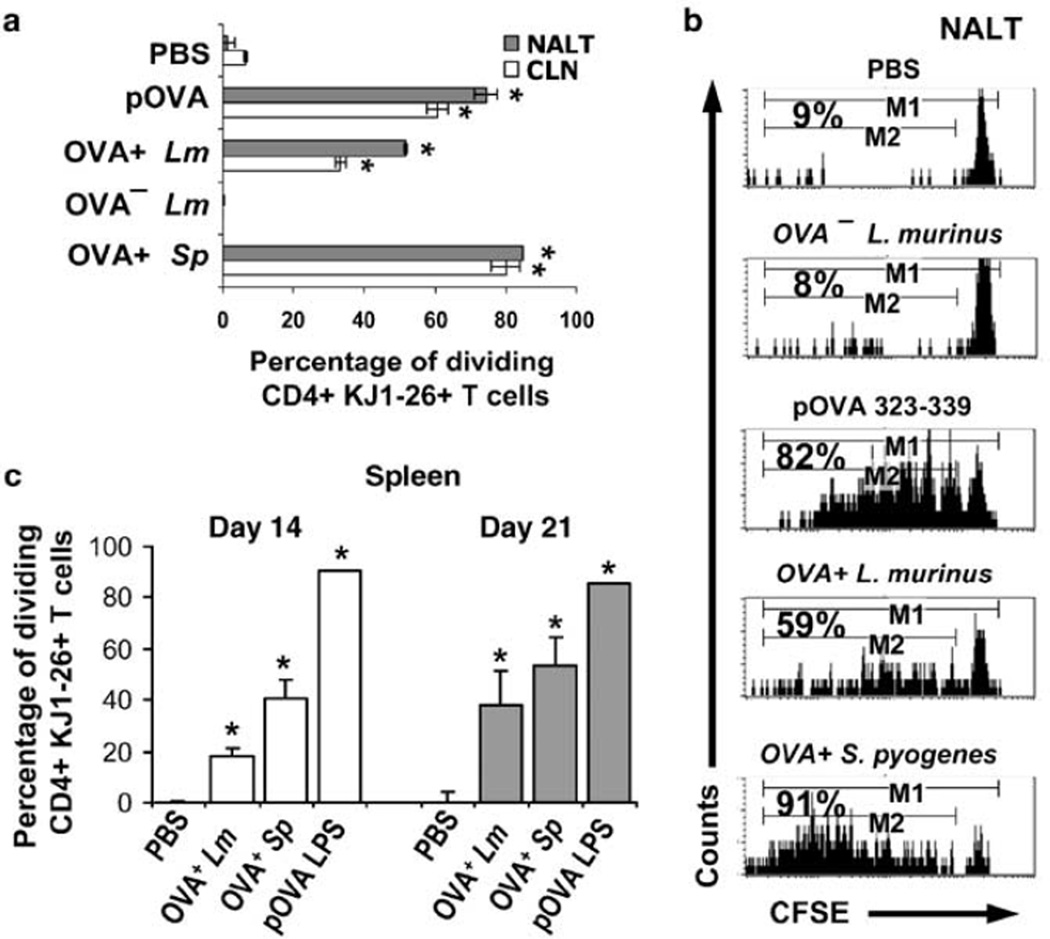
The effects of sustained colonization with L. murinus were measurable in the NALT 9 days after initial inoculation. OVA-specific T cells within the NALT had undergone significantly more divisions after intranasal priming with OVA+ L. murinus, OVA+ S. pyogenes or OVA peptide, than those in mice primed with OVA− L. murinus or PBS control (Figure 6a,—gray bars; Figure 6b). A greater frequency of T cells division in NALT than in the cervical LN (CLN) indicated that NALT is the initial priming site (Figure 6a) confirming our previous findings.27 Nonetheless, as the frequency of OVA-specific T cells in the CLN is approximately six times greater than that in NALT (Supplementary Figure S2), the total number OVA-specific T cells that had proliferated is greater in CLN than in NALT. This effect was similar across the treatment groups. As predicted OVA+ S. pyogenes induced a greater clonal expansion than OVA+ L murinus by 9 days after initial inoculation.
Figure 6.
Nasal inoculum of L. murinus and S. pyogenes induce proliferation first in nasal-associated lymphoid tissue (NALT) and cervical lymph nodes. (a) Summary data showing the percentage of dividing CD4+ KJ1-26+ T cells in NALT and cervical LN at day 9 after initial intranasal inoculation with phosphate-buffered saline (PBS), 10–15 µg of OVA 323–339 peptide (pOVA), single inoculation of OVA+ S. pyogenes or daily inoculation of OVA− or OVA+ L. murinus for 9 consecutive days. Data shown are averages±s.e.m. from three experiments (n = 6 mice per data point). (b) Representative flow cytometry histograms showing CFSE dilution profiles and percentage dividing CD4+ KJ1-26+ T cells isolated from NALT of mice 9 days after inoculation with antigens. (c) Summary data from from experiments showing the percentage of divided CD4+ KJ1-26+ T cells in the spleen 14 and 21 days after initial intranasal inoculation with PBS, single inoculation of OVA+ S. pyogenes, or daily inoculation of OVA+ L. murinus for 14 consecutive days or single intravenous inoculation of 100 µg of OVA peptide and 50 µg E. coli LPS. Data shown are averages±s.e.m. from four experiments (n = 8 mice per data point). * = Student’s t-test P < 0.05 when compared to PBS group.
Among all treatment groups the total number of OVA-specific T cells isolated from NALT 9 days after priming ranged between 3.2×104 and 1×105 cells. After L. murinus inoculation half of the total OVA-specific T cells had proliferated (Figure 6b). As only a fraction of OVA-specific T cells that had proliferated is expected to express a detectable amount of cytokine after intranasal restimulation with OVA peptide, our system was not sufficiently sensitive to detect a reproducible and statistically meaningful number of effector/memory T cells within the NALT of each mouse.
Nasally primed antigen-specific T cells recirculate through the spleen as effector/memory T cells
The limitations mentioned above led us to study the phenotype of OVA-specific effector/memory T cells in the spleen, compared to NALT, as they recirculate between blood, tissues, and lymphoid organs (reviewed in26). Recirculating effector/memory OVA-specific T cells were detected in the spleen at increasing frequency at 14 and 21 days for each priming bacterium (Figure 6c). Only T cell that proliferated the most (i.e., with the lowest CFSE content) recirculate in significant numbers in the spleen. As we previously reported,27 this indicates that S. pyogenes or L. murinus does not prime T cells directly in the spleen, even when S. pyogenes can potentially be recovered the spleen.
L. murinus and S. pyogenes stimulate differentiation into effector/memory T cells that are not T-regulatory cells
The working hypothesis for this set of experiments was that the commensal L. murinus induces the differentiation of regulatory T cells or effector T cells capable of regulating the inflammatory response, whereas the invasive pathogen S. pyogenes elicits proinflammatory T-cell clones. This hypothesis was tested by priming intranasally with L. murinus and S. pyogenes and then assaying regulatory antigen-specific T cell in the spleen.
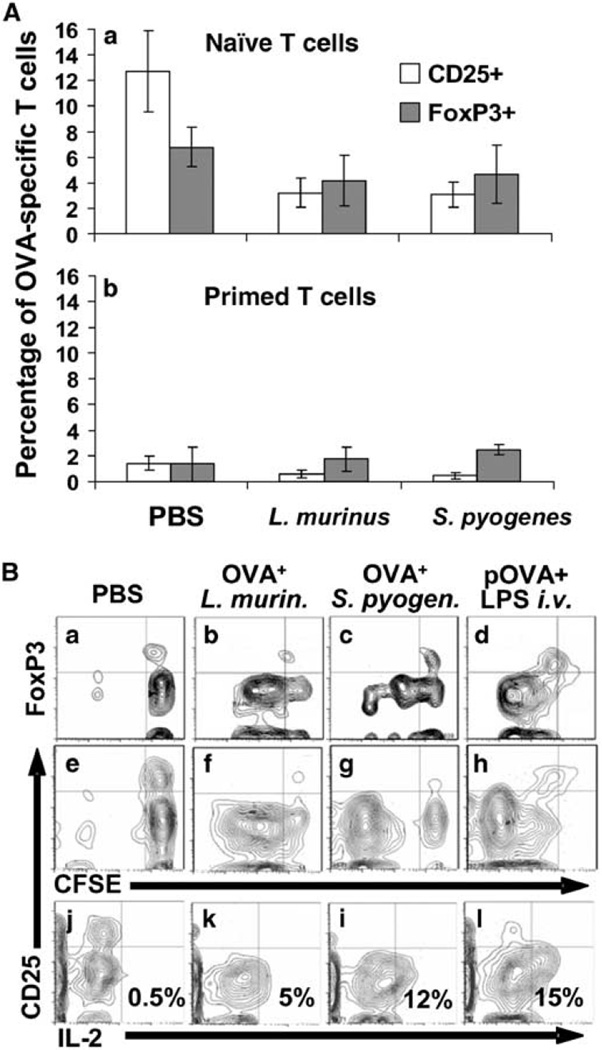
With surprise L. murinus or S. pyogenes do not induce the differentiation of central T-regulatory cells (Tregs) (Figure 7A). To the contrary, we observed that a fraction of naive OVA-specific T cells do express the two markers identifying Tregs, FoxP3, and CD25 (Figure 7B, panels a and e upper right quadrants). These two markers disappear in dividing OVA-specific T cells after priming with either bacteria (panels b, c and f, g upper left quadrant) or intravenous soluble positive control antigen (Figure 7B, panels d and h upper left quadrant). Nonetheless, dividing OVA-specific T cells do show intracellular interleukin (IL)-2 2.5 h after intravenous rechallenge with OVA peptide (Figure 7B, panels k, i, l lower right quadrant). None of the OVA-specific T cells primed with either bacterium show CD25+ cells releasing IL-2 (Figure 7B, panels k and i upper right quadrants) indicating that the low-affinity IL-2 receptor (CD25) characteristic of regulatory T cells, probably shifts to an high-affinity receptor complex (CD25–CD122–CD132) typical of effector T cells.
Figure 7.
L. murinus or S. pyogenes do not induce the maturation central T-regulatory (Treg) cells. (A) Using the gating strategy outlined in Figure 5A, the fraction of naive OVA-specific Treg cells that is generated 21 days after adoptive transfer disappears after priming with L. murinus or S. pyogenes. Data represent averages of five experiments; total mice per treatment group n = 10. (B) Representative contour plots from six-color flow cytometry indicating the expression of FoxP3 (a–d) and CD25 (e–h) of cells labeled with CFSE stained intracellularly with anti-IL-2 and anti-CD25 mAbs (j–l). Panels “a–d” and “e–l” are representative of two independent experiments.
L. murinus and S. pyogenes drive the differentiation of central effector/memory T cells that express inflammatory cytokines
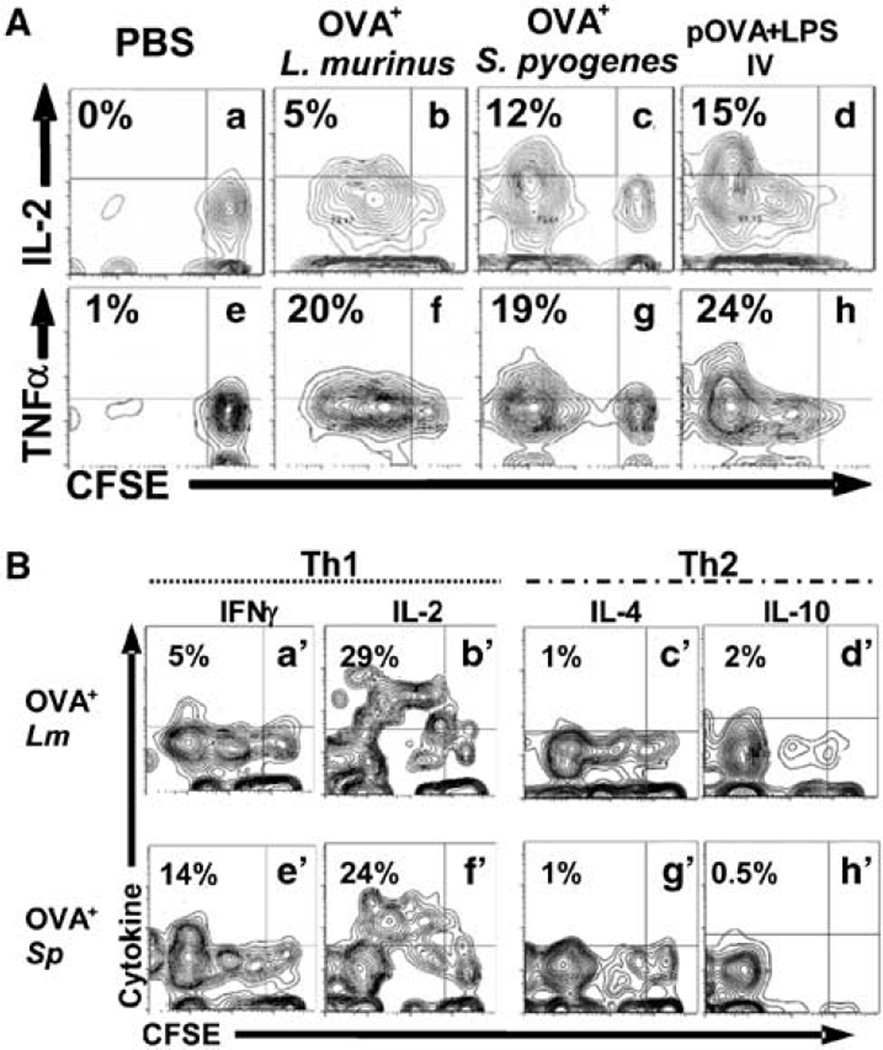
Both microorganisms induce effector T cells that when residing in the spleen, express primarily T helper 1 (Th1) cytokines 2.5–3.5 h after intravenous rechallenge with OVA peptide. IL-2 is expressed intracellularly primarily by CD4+ KJ1-26+ T cells that have diluted their CFSE (Figure 8A, panels b, c, d upper left quadrant). In this experimental design, intravenous rechallenge with OVA peptide primes residual naive T cells to release IL-2 between 2 and 4 h as previously demonstrated.28 To compare the frequency of cytokine positive cells in the effector and naive populations the cytokine gates were set at the upper level of the naive population. There were more effectors than naive T cells expressing intracellular IL-2 3.5 h after in vivo rechallenge with OVA peptide (Figure 7B, panels j–l, left vs. right quadrants). The same is true for tumor necrosis factor-α (TNFα) (Figure 8A, panels f–h). The most divided cells produce the most cytokine in all treatment groups. This indicates that T cells primed by bacteria intranasally differentiate into effector T cells that produce inflammatory cytokines.
Figure 8.
Proinflammatory cytokines are expressed and released in effector/memory antigen-specific T cell after rechallege in vivo with OVA peptide. (A) Using the gating strategy outlined in Figure 5A we report contour plots of splenocytes of T cell adoptively transferred mice 18–21 days after initial priming and 2.5 h after OVA IV peptide restimulation. Cells were permeabilized and stained for intracellular cytokines interleukin-2 (IL-2) and tumor-necrosis factor-α (TNFα). Mice were primed intranasally with phosphate-buffered saline (PBS) (a, e), daily inoculations of OVA+ L. murinus for 14 days (b, f), single inoculation of OVA+ S. pyogenes (c, g), or intravenous inoculation of OVA peptide plus adjuvant E. coli LPS (d, h). (B) Flow cytometry contour plots of spleens of T cell adoptively transferred mice 21 days earlier and primed intranasally as described in A. Mice were restimulated with OVA peptide 100 µg intravenously. Splenocytes were harvested 2.5 h later, incubated with cytokine-capture mAb (Miltenyi Biotech) for 60 min secretion period at 37°C. Cytokines were revealed with PE-labeled anticytokine sandwich mAb and analyzed in flow cytometry. Priming with OVA+ L. murinus for 14 days induces T cells to release of interferon (IFN)γ and IL-2 (a′, b′ upper left quadrants) but not of IL-4 or IL-10 (c′, d′ upper left quadrants) after 2.5 h rechallenge with OVA peptide. Single inoculation of OVA+ S. pyogenes parallels the IFNγ and IL-2 (e′, f′ upper left quadrants) and IL-4 and IL-10 release (g′, h′ upper left quadrants).
L. murinus and S. pyogenes generated effector T cells that produce and release interferon-γ (IFNγ) and IL-2 (Figure 8B) and TNFα (data not shown) 2.5 h after in vivo rechallenge with intravenous OVA peptide. Priming with S. pyogenes induced a greater percentage of divided T cells to release IFNγ than L murinus (Figure 8B, panel e′ vs. a′), whereas IL-2 release was comparable (Figure 8B, panel b′ vs. f′). Surprisingly, neither IL-4 and the immunosuppressive IL-10 were detected intracellularly (data not shown), nor released (Figure 8B, panels e′ vs. g′ and d′ vs. h′) after in vivo rechallenge with OVA peptide in any of the priming condition, except after intravenous priming with OVA and pertussis toxin (positive control).
DISCUSSION
Infections of mucosal surfaces, such as pharyngitis or gastroenteritis affect millions of people worldwide, with extremely high costs to society.1,29 Millions of bacteria coat the surfaces of the upper respiratory tract16 and intestine,30 making such mucosal surfaces potential portals of entry for diverse microorganisms. To develop efficacious mucosal vaccines, the mechanisms that regulate a protective response at mucosal sites must be better understood (reviewed in31). Like the lymphoepithelial structures of the Waldeyer’s ring for primates, in rodents NALT is strategically located to sample potential microbes in the air stream coming from the nares32 and in fluids from the oral cavity accessing the nose through the nasopalatine canal.33 Therefore, the characterization of memory T cells primed in upper respiratory mucosa is critical to the understanding of a protective adaptive response. In this study we compared whether a virulent pathogen or a commensal generate memory T cell of different phenotype.
The data show that despite being inoculated with high numbers, significantly fewer L. murinus were recovered from NALT when compared to S. pyogenes recovery. S. pyogenes translocates, forms microcolonies,21 and persists into the NALT more efficiently than the commensal L. murinus. These data are consistent with the capacity of S. pyogenes to invade the NALT tissues21 and provides new information regarding the potential that commensal bacteria have to move across nasal epithelial barriers of the upper respiratory tract, and to stimulate antigen-specific T cells.
At 4 days after inoculation with either bacterium, CD11c+ DCs locate at the periphery the B-cell follicles in the subepithelial dome (SED) just below the epithelium of NALT. Surprisingly, naive antigen-specific T cells (CFSE high) did not colocalize with the CD11c+ cells in the SED, but were primarily located near CD11c+ cells in the T-cell-rich region of NALT. This localization of T cells suggests that antigen presentation is performed by mature myeloid CD11b+ CD11c+ DCs and possibly by NALT macrophages. Within NALT, CD11b+ I-Ad+ represent approximately 4% of total cells.34 We corroborated this observation with F4/80 and CD11c staining in flow cytometry (data not shown).
After the bacterial inoculation, antigen-specific T-cell proliferation initiates primarily in NALT and later in draining CLN as we have previously shown for S. pyogenes.27 We demonstrate here that the antigen-specific T-cell proliferate (CFSE low cells) in the T-cell-rich region of the NALT as soon as 3 days after mice were infected with either L. murinus or S. pyogenes. This localization of proliferating T cells in secondary lymphoid organs is consistent with our study of mesenteric LN during Salmonella typhimurium infections.35
By 9 days after initial inoculation there has been significant T-cell proliferation in NALT and in CLN in mice inoculated with either bacterium. L. murinus induced significantly lower T-cell proliferation than S. pyogenes, particularly in the CLN. This difference can be explained by a lower number of L. murinus crossing the epithelial barrier and/or more rapid clearance from NALT. Nonetheless, as both bacteria induce T-cell proliferation we concluded that both microorganisms are processed by APCs and presented locally to antigen-specific T cells after translocating or invading across the epithelium.
After activation, antigen-experienced T cells recirculate between blood, tissues, and secondary lymphoid organs36 and at anyone time a fraction of such T cells can be recovered as central effector/memory T cells recirculating to the spleen. We show that by 21 days after initial inoculation, OVA-specific T cells recirculate to the spleen as memory T cells, capable of releasing cytokines upon restimulation with the OVA epitope.
The data indicate that persistent intranasal colonization by commensals or episodic infections by pathogens induce nasal APCs to stimulate antigen-specific T cells to proliferate in NALT and CLN and then recirculate as antigen-experienced T cells in the spleen.
Mucosal surfaces are surprisingly free of inflammatory infiltrate despite the proximity with high numbers of microbes.16,30 We tested the hypothesis that commensal microorganisms induce the differentiation of regulatory T cells capable of dampening inflammation of the submucosal and detrimental cell-mediated responses during infection.37 Expression of CD25 and FoxP3 is absent on T cells from naive transgenic DO11.10 donor mice, but become significant 21 days after T-cell-adoptive transfer into uninfected recipient BALB/c mice (Figure 7B, panels a and e). Preliminary evidence suggests that this change is related to low-affinity interactions with cross-reactive T-cell epitopes present in the intestine of BALB/c mice (data not shown). Surprisingly, however, when T cells were efficiently primed and begin to divide in the NALT, neither S. pyogenes, nor L. murinus induce the differentiation of antigen-specific CD25+ FoxP3+ regulatory T cells that can be recovered as recirculating T cells in the spleen. Although unlikely, it is possible that regulatory T cells are generated locally and remain in the NALT.
Commensal and probiotic microorganisms residing within mucosal microbiota are hypothesized to induce immunosuppressive and/or regulatory cytokines that support the generation of secretory immunoglobulins. Consistent with the work of Mohamadzadeh et al.,38 we demonstrate that the commensal L. murinus and the pathogen S. pyogenes induce antigen-experienced T cells that when residing in the spleen can be restimulated to release primarily proinflammatory. The dividing antigen-specific T cells recovered in the spleen were surprisingly all CD25 negative but expressed IL-2, TNFα, and IFNγ. Unlike IL-2, the expression of TNFα and IFNγ in particular is attributable to T cells that have divided the most. Surprisingly, none of the T helper 2 (Th2) cytokines, IL-5 (data not shown), IL-4 were expressed intracellularly or released after intravenous OVA peptide restimulation. The frequency of T cells expressing IL-10 was inconsistent across repeated experiments and extremely low at best. Collectively, these findings are important because they suggest that the commensal L. murinus as well as the pathogen S. pyogenes induce the differentiation of effector/memory T cells of similar Th1 phenotype rather than T cells with regulatory or suppressive phenotype. This finding becomes important in light of potential use of commensal microorganisms to treat atopic diseases.39,40 L. murinus crosses mucosal barriers, it remains localized in mucosal lymphoid organs, does not spread to distant sites inducing disease, but rather elicits an antigen-specific CD4 T-cell response potentially capable of inhibiting Th2-mediated atopic responses. Although the mechanisms of antiallergic effects elicited by probiotic microorganisms remains unclear,41,42 our study provides data in favor of a Th1 response induced by multiple inoculations of Lactobacilli that could be potentially exploited to counteract atopic responses.
After S. pyogenes infection the protective role of proinflammatory cytokines like IFNγ or TNFα is controversial depending on whether the innate or adaptive response is considered. IFNγ released by NK cells during acute intravenous Group A Streptococcus (GAS) infection induces rapid shock and death of C3H/HeN mice, whereas lack of NK cells and low IFNγ is protective.43 Similarly, after intranasal infection with S. pyogenes, IFNγ prevents clearance of GAS in NALT and increases dissemination to CLN of C57BL/6 mice (K. Highland, personal communication). Alternatively, in a skin infection model S. pyogenes inoculation is lethal for 50% of C57BL/6 mice within 4 days when IFNγ is completely absent or neutralized with circulating mAbs.44 It is possible that these contrasting results are related to the mouse strains nonetheless the release of IFNγ in a secondary adaptive response may have a differential protective impact against a L. murinus or S. pyogenes. Therefore, it appears that the despite the characteristics of the microbe, commensal or invasive pathogen, once the microorganism crosses the nasal mucosa in sufficient numbers the Th1 adaptive response that is generated is effectively controlling L. murinus but is unable to protect against the invasive S. pyogenes.
To conclude, we demonstrated that L. murinus and S. pyogenes translocate and persist across the epithelium with different efficiency, are processed locally possibly by CD11c+ DCs that are not CCR6+, and presented to antigen-specific T cells. After priming the antigen-specific T cells divide locally and in the CLN then migrate to the spleen as central effector/memory T cells. In the spleen regardless of the priming microorganism, these antigen-experienced T cells are not regulatory T cells, but after antigen restimulation release proinflammatory rather than immunosuppressive cytokines. These are important findings particularly for L. murinus where the proinflammatory response could be potentially used not only in antiallergy protocols41 but also as a safe carrier of immunodominant epitopes in intranasal vaccines.45,46
METHODS
Bacteria
S. pyogenes strain 90–226 was engineered to express the chicken ovalbumin 323–339 peptide by chromosomal allelic replacement on the bacterial surface as a fusion protein with streptococcal M1 protein as described in ref.27 L. murinus was isolated from the small intestine of a BALB/c mouse and speciated through 16sRNA sequencing (accession no. AF157049). In L. murinus the OVA sequence was engineered on a truncated nonpathogenic form of M1 protein and expressed by a derivative of plasmid vector, pIL252, named pLM1Δ54–215:OVA. Expression of the fusion proteins M1-OVA on the surface of S. pyogenes or M1Δ54–215-OVA on L. murinus was best in log phase as confirmed by fluorescence-activated cell sorting (FACS) using the anti-M1 C-repeats, mAb 10B6, kindly provided by Vincent A. Fischetti, Rockefeller University, NYC, NY (Supplementary Figure S1A and S1B). As L. murinus expresses approximately 10 times less M1-OVA fusion protein during log phase than S. pyogenes (Supplementary Figure S1B), to approximate similar delivery of OVA epitope we inoculated one log more L. murinus CFUs at each single inoculation.
Mice
DO11.10 TCR transgenic mice were bred in a specific pathogen-free facility according to the National Institutes of Health guidelines and screened as previously described.20 Recipient BALB/c mice were purchased from Taconic Farms and housed after infection in a biosafety level 2 facility.
Adoptive transfer of DO11.10 T cells
Spleen and LN cells were harvested from DO11.10 transgenic mice and labeled in Hank’s buffer salt solution (HBSS) containing 7.5 µM CFSE at 37°C for 10 min as described in ref.47 A total of 3×106 TCR transgenic T cells were adoptively transferred into recipient BALB/c mice through the lateral tail vein. The frequency of TCR transgenic T cells was determined with anti-CD4 mAb and antitransgenic TCR mAb KJ1-26 (Caltag Laboratories; Invitrogen, Carlsbad, CA) as previously described.19
Intranasal bacterial priming and antigenic rechallenge
At 24 h after adoptive transfer of T cells, mice were anesthetized with a 1:1 oxygen–isofluorane mixture for 2 min and inoculated intranasally with either 2×108 OVA+ S. pyogenes, or wild-type S. pyogenes, 1–2×109 OVA+ L. murinus or OVA− L. murinus or 10–15 µg of OVA peptide or PBS. To mimic persistent colonization, L. murinus was inoculated every day to a maximum of 14 consecutive days whereas S. pyogenes was inoculated once or for 2 consecutive days. According to the specific experiment L. murinus was inoculated for 3, 9, or 14 days. In selected experiments OVA+ S. pyogenes was heat-killed at 65°C for 1 h before intranasal inoculation (Figure 5C). All antigens were delivered in 7.5 µl of PBS per nostril to avoid inoculation of the lungs. In experiments assessing cytokine expression, positive control animals were primed intravenously with 100 µg of OVA peptide plus 50 µg of LPS from E. coli J5 (List Biological Labs, Campbell, CA). On days 18–21 after initial inoculation effector/memory OVA-specific T cells were restimulated intravenously 100 µg of OVA peptide in 0.25 ml of PBS. Spleen cells were harvested 2.5–3.5 h after intravenous restimulation for assessment of intracellular expression and release of cytokines. The kinetics of T-cell priming and proliferation was determined by harvesting NALT, CLN, and spleen at 3, 9, 14–17, and 18–21 days after initial intranasal inoculation (Figure 1).
Recovery of bacteria from NALT tissues
At 4 days after S. pyogenes and 24 h after the last of three L. murinus inocula, NALT tissues were harvested as previously described.34, 48 Briefly, the hard palate was dissected as part of the tissues forming the base and the lateral walls of the nasal cavities (Figure 2a). Specimens were weighed, rinsed in PBS to remove contaminating blood, and disrupted over a nylon mesh. Single-cell suspensions serially diluted and plated on sheep blood–Todd–Hewitt yeast or lactobacilli-selective deMan–Rogosa–Sharpe (MRS) agar plates containing 5 µg/ml erythromycin for 24 h at 37°C.
T-cell proliferation assessment
Mononuclear cells from NALT, cervical LNs, and spleens containing CFSE-labeled DO11.10 cells were washed in HBSS containing 2% fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA) and 0.05% sodium azide and incubated on ice for 30 min in the presence of Fc block (spent 2.4G2 culture supernatant, 2% rat serum, 2% mouse serum, and 0.01% sodium azide) and primary antibodies: PerCP/Cyanin (Cy) 5.5-conjugated anti-CD4 (eBioscience, San Diego, CA), biotin-conjugated KJ1-26 mAb, followed by allophycocyanin (APC)-conjugated streptavidin. All samples were fixed with 1% formaldehyde in PBS. Single-cell data were collected using a FACScalibur (BD Biosciences, San Jose, CA) and CFSE dilution profiles of CD4+ KJ1-26+ cells were analyzed using CellQuest (BD Biosciences) or FlowJo software (TreeStar, Ashland, OR). We designed a subgating strategy to detect adoptively transferred antigen-specific T cells (Figure 5A), where CD4+ cells were identified within a lymphocyte gate (Figure 5A, panel a). CD11b+ CD11c+ DCs, CD8+ T cells, or CD45R + (B220) B cells were excluded from the CD4+ cells population (Figure 5A, panel c). In the CD4+ lymphocyte population, adoptively transferred OVA-specific T cells, were identified using the KJ1-26 mAb (Figure 5A, panel d). Within the CD4+, KJ1-26+ T-cell population we determined the percentage of cells that had proliferated by analyzing their CFSE dilution profile (Figure 5A, panel e). The fraction of proliferating T cells (M1 gate/M2 gate ×100) defines the extent of antigen-specific T-cell activation induced by the antigens delivered intranasally.
Immunomagnetic positive selection and intracellular cytokine staining
Erythrocytes were removed from spleen’s single-cell suspensions with ACK lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM Na2 EDTA, pH 7.2) or centrifugation over a density gradient (Histopaque 8003; Sigma, St. Louis, MO). Approximately 3×107 mononuclear cells per spleen were incubated in MACS buffer (PBS containing 0.5% biotin-free BSA (fraction V), 2 mM EDTA, 0.09% sodium azide) (Miltenyi Biotec,Bergisch Gladbach, Germany). After preincubation with unlabeled anti-CD16/32 (Fc Block, mAb 2.4G2), surface staining with biotin-conjugated KJ1-26 mAb, PerCP-conjugated anti-CD4 (BD Biosciences), Pacific orange-conjugated anti-CD8 and anti-CD45R, and APC-conjugated anti-CD25 (eBioscience), cells were washed and resuspended at 107 cells in 100 µl MACS buffer containing 10 µl streptavidin-conjugated paramagnetic beads (Miltenyi Biotec). Immunomagnetically labeled cells were loaded on positive selection MS columns at 2×107 cells/ml following manufacturer’s instructions (Miltenyi Biotec). Magnetically retained cells (positively selected) were eluted when removed from magnetic stand (QuadroMACS; Miltenyi Biotec) and stained with phycoerythrin (PE)-cyanin7 (Cy7)-conjugated streptavidin. Samples were fixed with an ethanol based (“FoxP3 Staining Buffer Set” cat. no. 00-5523; eBioscience) or a formaldehyde-based fixation buffer (eBioscience; cat. no. 00-8222). Buffers used for intacellular staining of FoxP3 are incompatible with intracellular staining of cytokines. Cells were treated with FoxP3-permeabilization buffer (eBioscience; cat. no. 8222) or a 2% saponin permeabilization buffer (eBioscience; cat. no. 8333) and incubated with APC- or PE-conjugated anti-FoxP3 (mAb FJK-16s) or PE-conjugated anticytokine mAbs. Proliferation, intracellular cytokines, and FoxP3 expression data were collected by using an LSR II cytometer (BD Biosciences) using the gating strategy shown in Figure 5A.
Cytokines secretion assay
At 2.5–3.5 h after intravenous restimulation with 100 µg of OVA peptide, spleen mononuclear cells were concentrated to 108 cells/ml in complete Eagle’s–Ham’s amino acids (EHAAs) (EHAA medium supplemented with 5% murine serum, 2 mM L-glutamine, 100 U/ml penicillin G, 100 µg/ml streptomycin, 20 µg/ml gentamicin, and 0.05 mM 2-mercaptoethanol) containing a dimeric antibody molecule (“Catch Reagent”; Miltenyi Biotec). The dimeric antibody is tetravalent. Half of this dimeric antibody molecule binds to CD45 on the surface of each bone marrow-derived cell and the other half of the molecule is specific for the cytokine of interest: IL-2, IL-4, IL-5, IL-10, IFNγ, or TNFα. After 10 min incubation on ice, cells were washed, diluted 100 times to prevent cytokine cross-capture between cells and incubated at 37°C in prewarmed complete EHAA for 60 min with gentle rocking to allow secretion and capture of cytokines by the dimeric antibody according to manufacturer’s instructions (Miltenyi Biotec). Cell suspensions were chilled on ice, concentrated to 108 cells/ml, preincubated with unlabeled anti-CD16/32 (Fc Block) for 15 min, and subsequently with a PE-conjugated rat IgG isotypes or anti-IL-2, IL-4, IL-5, IL-10, IFNγ, or TNFα secondary detection mAb, biotin-conjugated mAb KJ1-26, PerCP-conjugated anti-CD4, Pacific orange-conjugated anti-CD8, anti-CD45R and anti-CD11c, and APC-conjugated anti-CD25. All samples were then processed for immunomagnetic-positive selection and detection of KJ1-26+ cells by streptavidin-conjugated paramagnetic beads followed by PE-Cy7-conjugated streptavidin as described above. All cytokine release samples were fixed with 1% formaldehyde in PBS and data collected an LSR II cytometer.
Immunohistology
At 4 days after initial intranasal-antigen inoculation of adoptively transferred BALB/c mice, the anterior maxilla and nasal cavities were dissected, fixed overnight in 1% paraformaldehyde, decalcified in 10% EDTA for 7 days, washed with 30% sucrose in PBS, and flash-frozen in OCT embedding medium (Tissue-Tek, Hatfield, PA). Tissue sections (10 µm) were cut with a cryostat (Leica CM3050), and stored at −20°C. Sections were rehydrated in PBS, treated with 3% H2O2 to inhibit endogenous peroxidases, and blocked with anti-CD16/32 (mAb 2.4G2) and Avidin/Biotin Blocking (Vector Laboratories, Burlingame, CA) for 15 min each as described in ref.49 DCs were detected in tissue sections by sequential addition of biotin labeled anti-mouse CD11c or rat IgG1 isotype control (Caltag Laboratories) in Tris-NaCl2 buffer (TNB) for 1 h, washed in Tris-NaCl2 Tween 20 (TNT) buffer, incubation with streptavidin–horseradish peroxidase (Perkin-Elmer, Waltham, MA) for 30 min. The signal was finally amplified with tyramide-conjugated cyanine 3 (tyramide-cyanin3 (Cy3)) (Perkin-Elmer) for 5 min. Slides were subsequently incubated again in 3% H2O2 for 1 h to quench horseradish peroxidase activity. DO11.10 T cells were detected on the same slide by sequential addition of dioxygenin-labeled KJ1-26 mAb (courtesy of Marc Jenkins, University of Minnesota) or IgG2a isotype control (eBioscience), peroxidase (POD)-conjugated antidioxygenin (Roche Applied Science, Indianapolis, INA) and finally the signal was amplified with tyramide-conjugated cyanine 5 (tyramide-cyanin5 (Cy5)) (Perkins-Elmer) for 5 min.
To detect CCR6+ cells, adjacent sections were blocked with 5% goat serum in PBS for 20 min and incubated with rabbit anti-mouse CCR6 (Santa Cruz Biotechnology, SantaCruz, CA) for 2 h. Signal was revealed by biotin-labeled goat anti-rabbit IgG, followed by streptavidin-conjugated Cy3 for 15 min. Lastly, B-cell follicles were identified in all tissue sections by FITC-conjugated anti-CD45R. Slides were mounted by using Vectashield (Vector Laboratories) to preserve fluorescence and the images acquired by using a Bio-Rad (Hercules, CA) MRC-1000 confocal microscope equipped with a krypton/argon laser (Bio-Rad). Separate green (FITC), blue (Cy3), and far-red (Cy5) images were collected for each section analyzed. Final image processing was performed by using ImageJ software (www.nih.gov) and Photoshop (Adobe, San Jose, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was entirely supported by Public Health Service grants DE014371 (M. Costalonga) from the National Institute of Dental and Craniofacial Research. We thank Dr Khoruts and Dr Jenkins for the support, insightful discussions, and feedback on the article.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Saidi SM, et al. Epidemiological study on infectious diarrheal diseases in children in a coastal rural area of Kenya. Microbiol. Immunol. 1997;41:773–778. doi: 10.1111/j.1348-0421.1997.tb01925.x. [DOI] [PubMed] [Google Scholar]
- 3.Snyder JD, Merson MH. The magnitude of the global problem of acute diarrhoeal disease: a review of active surveillance data. Bull. World Health Organ. 1982;60:605–613. [PMC free article] [PubMed] [Google Scholar]
- 4.Phelps HA, Neely MN. SalY of the Streptococcus pyogenes lantibiotic locus is required for full virulence and intracellular survival in macrophages. Infect. Immun. 2007;75:4541–4551. doi: 10.1128/IAI.00518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medina E, Rohde M, Chhatwal GS. Intracellular survival of Streptococcus pyogenes in polymorphonuclear cells results in increased bacterial virulence. Infect. Immun. 2003;71:5376–5380. doi: 10.1128/IAI.71.9.5376-5380.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin JM, Green M, Barbadora KA, Wald ER. Group A streptococci among school-aged children: clinical characteristics and the carrier state. Pediatrics. 2004;114:1212–1219. doi: 10.1542/peds.2004-0133. [DOI] [PubMed] [Google Scholar]
- 7.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 8.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qadri F, et al. Comparison of immune responses in patients infected with Vibrio cholerae O139 and O1. Infect. Immun. 1997;65:3571–3576. doi: 10.1128/iai.65.9.3571-3576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins MK, Taylor PS, Norton SD, Urdahl KB. CD28 delivers a costimulatory signal involved in antigen-specific IL-2 production by human T cells. J. Immunol. 1991;147:2461–2466. [PubMed] [Google Scholar]
- 11.Garside P, et al. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 1998;281:96–99. doi: 10.1126/science.281.5373.96. [DOI] [PubMed] [Google Scholar]
- 12.Beagley KW, et al. Recombinant murine IL-5 induces high rate IgA synthesis in cycling IgA-positive Peyer’s patch B cells. J. Immunol. 1988;141:2035–2042. [PubMed] [Google Scholar]
- 13.Shockett P, Stavnezer J. Effect of cytokines on switching to IgA and alpha germline transcripts in the B lymphoma 1.29 mu. Transforming growth factor-beta activates transcription of the unrearranged C alpha gene. J. Immunol. 1991;147:4374–4383. [PubMed] [Google Scholar]
- 14.Ehrhardt RO, Strober W, Harriman GR. Effect of transforming growth factor (TGF)-beta 1 on IgA isotype expression. TGF-beta 1 induces a small increase in sIgA+ B cells regardless of the method of B cell activation. J. Immunol. 1992;148:3830–3836. [PubMed] [Google Scholar]
- 15.Sonoda E, et al. Differential regulation of IgA production by TGF-beta and IL-5: TGF-beta induces surface IgA-positive cells bearing IL-5 receptor, whereas IL-5 promotes their survival and maturation into IgA-secreting cells. Cell Immunol. 1992;140:158–172. doi: 10.1016/0008-8749(92)90184-q. [DOI] [PubMed] [Google Scholar]
- 16.Hull MW, Chow AW. Indigenous microflora and innate immunity of the head and neck. Infect. Dis. Clin. North Am. 2007;21:265–282. doi: 10.1016/j.idc.2007.03.015. v. [DOI] [PubMed] [Google Scholar]
- 17.Nimishikavi S, Stead L. Images in clinical medicine. Streptococcal pharyngitis. N. Engl. J. Med. 2005;352:e10. doi: 10.1056/NEJMicm040442. [DOI] [PubMed] [Google Scholar]
- 18.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 19.Kearney ER, Pape KA, Loh DY, Jenkins MK. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1994;1:327–339. doi: 10.1016/1074-7613(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 20.Haskins K, et al. The major histocompatibility complex-restricted antigen receptor on T cells. I. Isolation with a monoclonal antibody. J. Exp. Med. 1983;157:1149–1169. doi: 10.1084/jem.157.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park HS, Francis KP, Yu J, Cleary PP. Membranous cells in nasal-associated lymphoid tissue: a portal of entry for the respiratory mucosal pathogen group A streptococcus. J. Immunol. 2003;171:2532–2537. doi: 10.4049/jimmunol.171.5.2532. [DOI] [PubMed] [Google Scholar]
- 22.Iwasaki A, Kelsall BL. Localization of distinct Peyer’s patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J. Exp. Med. 2000;191:1381–1394. doi: 10.1084/jem.191.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang MH, et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J. Immunol. 2006;176:803–810. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- 24.Ohl L, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Salazar-Gonzalez RM, et al. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity. 2006;24:623. doi: 10.1016/j.immuni.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins MK, et al. In vivo activation of antigen-specific CD4 T cells. Annu. Rev. Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 27.Park HS, et al. Primary induction of CD4 T cell responses in nasal associated lymphoid tissue during group A streptococcal infection. Eur. J. Immunol. 2004;34:2843–2853. doi: 10.1002/eji.200425242. [DOI] [PubMed] [Google Scholar]
- 28.Merica R, Khoruts A, Pape KA, Reinhardt RL, Jenkins MK. Antigen-experienced CD4 T cells display a reduced capacity for clonal expansion in vivo that is imposed by factors present in the immune host. J. Immunol. 2000;164:4551–4557. doi: 10.4049/jimmunol.164.9.4551. [DOI] [PubMed] [Google Scholar]
- 29.Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. World Health Organ. 2003;81:197. [PMC free article] [PubMed] [Google Scholar]
- 30.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Brandtzaeg P. Induction of secretory immunity and memory at mucosal surfaces. Vaccine. 2007;25:5467. doi: 10.1016/j.vaccine.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Kimbell JS, et al. Computer simulation of inspiratory airflow in all regions of the F344 rat nasal passages. Toxicol. Appl. Pharmacol. 1997;145:388. doi: 10.1006/taap.1997.8206. [DOI] [PubMed] [Google Scholar]
- 33.Donald RA. Olfactory and non-olfactory epithelia in the nasal cavity of the mouse. Peromyscus. Am. J. Anat. 1972;133:37–49. doi: 10.1002/aja.1001330104. [DOI] [PubMed] [Google Scholar]
- 34.Heritage PL, Underdown BJ, Arsenault AL, Snider DP, McDermott MR. Comparison of murine nasal-associated lymphoid tissue and Peyer’s patches. Am. J. Respir. Crit. Care Med. 1997;156:1256–1262. doi: 10.1164/ajrccm.156.4.97-03017. [DOI] [PubMed] [Google Scholar]
- 35.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 36.Reinhardt RL, Bullard DC, Weaver CT, Jenkins MK. Preferential accumulation of antigen-specific effector CD4 T cells at an antigen injection site involves CD62E-dependent migration but not local proliferation. J. Exp. Med. 2003;197:751–762. doi: 10.1084/jem.20021690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuji NM. Antigen-specific CD4(+) regulatory T cells in the intestine. Inflamm. Allergy Drug Targets. 2006;5:191–201. doi: 10.2174/187152806778256043. [DOI] [PubMed] [Google Scholar]
- 38.Mohamadzadeh M, et al. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. USA. 2005;102:2880–2885. doi: 10.1073/pnas.0500098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalliomaki M, et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 40.Boyle RJ, Tang MLK. The role of probiotics in the management of allergic disease. Clin. Exp. Allergy. 2006;36:568–576. doi: 10.1111/j.1365-2222.2006.02472.x. [DOI] [PubMed] [Google Scholar]
- 41.Ouwehand AC. Antiallergic effects of probiotics. J. Nutr. 2007;137:794S–797S. doi: 10.1093/jn/137.3.794S. [DOI] [PubMed] [Google Scholar]
- 42.Feleszko W, et al. Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin Exp Allergy. 2007;37:498–505. doi: 10.1111/j.1365-2222.2006.02629.x. [DOI] [PubMed] [Google Scholar]
- 43.Goldmann O, Chhatwal GS, Medina E. Contribution of natural killer cells to the pathogenesis of septic shock induced by Streptococcus pyogenes in mice. J. Infect. Dis. 2005;191:1280–1286. doi: 10.1086/428501. [DOI] [PubMed] [Google Scholar]
- 44.Raeder RH, Barker-Merrill L, Lester T, Boyle MD, Metzger DW. A pivotal role for interferon-gamma in protection against group A streptococcal skin infection. J. Infect. Dis. 2000;181:639–645. doi: 10.1086/315281. [DOI] [PubMed] [Google Scholar]
- 45.Fischetti VA, Medaglini D, Oggioni M, Pozzi G. Expression of foreign proteins on gram-positive commensal bacteria for mucosal vaccine delivery. Curr. Opin. Biotechnol. 1993;4:603–610. doi: 10.1016/0958-1669(93)90084-a. [DOI] [PubMed] [Google Scholar]
- 46.De Magistris MT. Mucosal delivery of vaccine antigens and its advantages in pediatrics. Adv. Drug. Deliv. Rev. 2006;58:52–67. doi: 10.1016/j.addr.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Parish CR. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol. Cell Biol. 1999;77:499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- 48.Wu HY, Nikolova EB, Beagley KW, Russell MW. Induction of antibody-secreting cells and T-helper and memory cells in murine nasal lymphoid tissue. Immunology. 1996;88:493–500. doi: 10.1046/j.1365-2567.1996.d01-690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK. Visualizing the generation of memory CD4 T cells in the whole body. Nature. 2001;410:101–105. doi: 10.1038/35065111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.