GLABRA2 targets and suppresses five basic helix-loop-helix transcription factor genes with diverse functions in root hair development during root hair pattern formation of Arabidopsis thaliana.
Abstract
The Arabidopsis thaliana GLABRA2 (GL2) gene encodes a transcription factor involved in the cell differentiation of various epidermal tissues. During root hair pattern formation, GL2 suppresses root hair development in non-hair cells, acting as a node between the gene regulatory networks for cell fate determination and cell differentiation. Despite the importance of GL2 function, its molecular basis remains obscure because the GL2 target genes leading to the network for cell differentiation are unknown. We identified five basic helix-loop-helix (bHLH) transcription factor genes (ROOT HAIR DEFECTIVE6 [RHD6], RHD6-LIKE1 [RSL1], RSL2, Lj-RHL1-LIKE1 [LRL1], and LRL2) as GL2 direct targets using transcriptional and posttranslational induction systems. Chromatin immunoprecipitation analysis confirmed GL2 binding to upstream regions of these genes in planta. Reporter gene analyses showed that these genes are expressed in various stages of root hair development and are suppressed by GL2 in non-hair cells. GL2 promoter-driven GFP fusions of LRL1 and LRL2, but not those of the other bHLH proteins, conferred root hair development on non-hair cells. These results indicate that GL2 directly suppresses bHLH genes with diverse functions in root hair development.
INTRODUCTION
The morphological differentiation and deposition patterns of cells are crucial determinants of functional plant structures. Among the plant tissues composed of multiple types of cells, the root epidermis of Arabidopsis thaliana has served as an excellent model system for studying the morphological differentiation and pattern formation of plant cells (Dolan et al., 1993; Schiefelbein, 2000; Tominaga-Wada et al., 2011). In Arabidopsis, the root epidermis is composed of non-hair (N) and hair (H) cell files that are in contact with either one or two underlying cortical cell file(s), respectively, and only the cells in H cell files develop root hairs (Dolan et al., 1994; Galway et al., 1994; Clowes, 2000; Schiefelbein et al., 2009). A mechanism that converts a positional cue from the underlying structure to a signal directing the root hair pattern is assumed to involve a receptor-like kinase expressed in the root epidermis (Kwak et al., 2005; Kwak and Schiefelbein, 2007, 2008). Downstream of this mechanism, numerous transcription factor genes, including GLABRA2 (GL2), constitute regulatory networks responsible for cell pattern formation and subsequent cell differentiation (Schiefelbein et al., 2009, 2014; Tominaga-Wada et al., 2011; Grebe, 2012).
GL2 encodes a homeodomain-leucine-zipper transcription factor and is thought to be a negative regulator of root hair development because it is expressed preferentially in N cell files and because mutant defects in GL2 result in ectopic root hair formation in N cell files (Rerie et al., 1994; Di Cristina et al., 1996; Masucci et al., 1996). GL2 is activated in a cell file-specific manner by a transcription factor complex consisting of the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1), the basic helix-loop-helix (bHLH) protein GL3 or ENHANCER OF GLABRA3 (EGL3), and the R2R3-type MYB protein WEREWOLF (WER) or MYB23 (Galway et al., 1994; Masucci et al., 1996; Lee and Schiefelbein, 1999; Payne et al., 2000; Bernhardt et al., 2003, 2005; Zhang et al., 2003; Kang et al., 2009). The complex also transcriptionally activates the R3-type MYB protein gene CAPRICE (CPC) and its paralogs in N cell files (Wada et al., 1997; Schellmann et al., 2002; Esch et al., 2004; Kirik et al., 2004; Simon et al., 2007; Tominaga et al., 2008). However, these R3-type MYB proteins move to H cell files to negatively regulate the transcription factor complex by replacing WER and MYB23, thereby suppressing the expression of GL2 with consequent root hair development in H cell files (Schellmann et al., 2002; Bernhardt et al., 2003; Esch et al., 2004; Kirik et al., 2004; Kurata et al., 2005; Tominaga et al., 2007, 2008). Consistent with this gene regulatory network, mutant defects in the transcription factor complex and its negative regulators confer ectopic root hair and scarce root hair phenotypes, respectively, and defects in GL2 are epistatic to these mutant defects with respect to root hair development (Wada et al., 1997; Hung et al., 1998; Lee and Schiefelbein, 1999; Bernhardt et al., 2005; Wang et al., 2010). Furthermore, because the expression of a modified GL2 with a constitutive transactivating function (VP16-GL2ΔN) resulted in the formation of root hair-like structures from various epidermal cells, including leaf pavement cells (Ohashi et al., 2003), GL2 is thought to recognize a set of genes that are sufficient for root hair development. Unlike root hair development, the file-specific cell fate of the root epidermis is thought to be determined independently of GL2 function because the characteristics of H and N cell files in cell length and vacuolation timing, which are affected in ttg mutants, are normal in gl2 mutants (Galway et al., 1994; Masucci et al., 1996). This line of evidence clarifies the role of GL2 as a genetic switch that suppresses root hair development between cell fate determination and cell differentiation during root hair pattern formation.
A regulatory network that acts downstream of GL2 for root hair cell differentiation comprises many bHLH transcription factor genes (Bruex et al., 2012; Pires et al., 2013). Mutants of the ROOT HAIR DEFECTIVE6 (RHD6)/bHLH83 gene, which encodes a transcription factor belonging to the bHLH VIIIc subfamily (Heim et al., 2003), have fewer root hairs than the wild type (Masucci and Schiefelbein, 1994). RHD6 expression is normally specific to H cell files (Masucci and Schiefelbein, 1994) but extends to include N cell files in gl2 mutants (Menand et al., 2007), suggesting that GL2 suppresses RHD6 expression directly or indirectly in N cell files. RHD6 LIKE1 (RSL1)/bHLH86, the closest putative paralog of RHD6, has a function partly redundant with that of RHD6 in the initiation of root hair development (Menand et al., 2007). Other VIIIc subfamily genes, RSL2/bHLH85 and RSL4/bHLH54, are functionally redundant with each other in root hair elongation (Yi et al., 2010). Posttranslational induction experiments using a modified RHD6 protein with the glucocorticoid receptor (GR) domain showed that RHD6 activated the RSL4 and RSL2 genes directly and indirectly, respectively (Yi et al., 2010). Lotus japonicus mutants of the transcription factor gene ROOTHAIRLESS1 (Lj-RHL1) exhibited severe defects in root hair development (Karas et al., 2009). Cross-species complementation analysis showed that the Arabidopsis bHLH XI subfamily genes Lj-RHL1-LIKE1 (LRL1)/bHLH66, LRL2/bHLH69, and LRL3/bHLH82 (Heim et al., 2003) are functional equivalents of Lj-LRHL1 (Karas et al., 2009). Multiple mutants of these genes have shorter root hairs than the wild type (Karas et al., 2009), while single mutants exhibited only moderate phenotypes in root hair length and morphology (Bruex et al., 2012), suggesting redundancy of their functions in root hair elongation. The transcript levels of LRL3, but not those of LRL1 or LRL2, were affected markedly in the rhd6-3 rsl1-1 double mutant (Karas et al., 2009), indicating that LRL3 is regulated differently from LRL1 and LRL2.
Despite the pivotal role of GL2 in suppressing root hair development, the molecular basis of the suppression remains obscure because, to date, few GL2 target genes are known and none of them are connected to the gene regulatory network for root hair cell differentiation. The PHOSPHOLIPASE Dζ1 (PLDζ1) gene is suppressed directly by GL2 in N cell files, and its ectopic expression causes root hair bulges in N cell files, suggesting its involvement in promoting root hair development (Ohashi et al., 2003). CELLULOSE SYNTHASE5 and XYLOGLUCAN ENDOTRANSGLYCOSYLASE17, both of which encode enzymes involved in polysaccharide synthesis, are also GL2 target genes, but their roles in root hair development are unclear (Tominaga-Wada et al., 2009). GL2 and MYB23 constitute a positive feedback loop in the shoot epidermis where GL2 directly recognizes MYB23 (Khosla et al., 2014). Recently, GL2 was shown to negatively regulate anthocyanin biosynthesis through direct recognition of the bHLH IIIf subfamily gene TRANSPARENT TEST8 and the R2R3-type MYB genes PRODUCTION OF ANTHOCYANIN PIGMENT2 and MYB113 (Wang et al., 2015). Upstream regions of these genes typically contain the L1 box-like sequence 5′-TAAATGT-3′ (Abe et al., 2001; Tominaga-Wada et al., 2009; Lin and Aoyama, 2012; Khosla et al., 2014; Wang et al., 2015). Although GL2 physically interacted with DNA regions containing this sequence (Ohashi et al., 2003; Khosla et al., 2014; Wang et al., 2015), whether the sequence is necessary and/or sufficient for recognition by GL2 in planta remains unclear.
In this study, we identified five bHLH transcription factor genes, RHD6, RSL1, RSL2, LRL1, and LRL2, as direct targets of GL2 using transcriptional and posttranslational induction systems with the chimeric transcription factors VP16-GL2ΔN and GR-VP16-GL2ΔN, respectively. Chromatin immunoprecipitation (ChIP) analysis using a GFP-fused GL2 protein confirmed the physical interaction of GL2 with these genes in planta. Expression analyses showed that they are suppressed by GL2 in N cell files and are expressed in various developmental stages of root hair development. Moreover, the phenotypes caused by the ectopic expression of their GFP-fusion proteins differed among them. These results indicate that GL2 acts as a negative factor for root hair development via multimodal pathways by targeting bHLH genes with diverse functions.
RESULTS
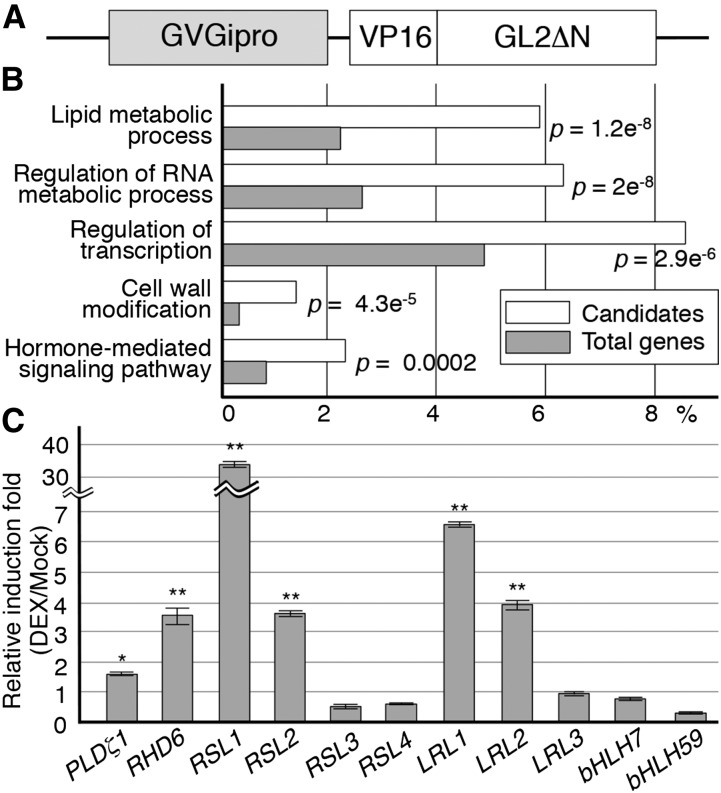
RHD6, RSL1, RSL2, LRL1, and LRL2 Are Candidate GL2 Target Genes
We performed a microarray screening to identify candidate GL2 target genes that were transcriptionally activated in response to the expression of the modified GL2 VP16-GL2ΔN, which has a constitutive transactivating function (Ohashi et al., 2003), in a glucocorticoid-inducible gene expression system using the GVG transcription factor (Figure 1A). Triplicate microarray experiments using total RNA from seedlings harboring the GVG-inducible VP16-GL2ΔN gene detected 864 genes with steady state transcript levels that were consistently increased by more than 2-fold after dexamethasone (DEX) treatment (Supplemental Data Set 1). These candidate genes were categorized by gene product functions as defined using the agriGO Web-based software tool (Du et al., 2010). The top five groups with most significant enrichment in the candidate target genes are associated with lipid metabolic processes, regulation of RNA metabolic processes, regulation of transcription, cell wall modification, and hormone-mediated signaling pathways, respectively (Figure 1B). Of these, the group involved in the regulation of transcription contained the largest number of genes, suggesting the presence of a large transcriptional network downstream of GL2. We subsequently focused on the transcription factor genes RHD6 and LRL1 because they are known to be involved in root hair development (Masucci and Schiefelbein, 1994; Menand et al., 2007; Karas et al., 2009; Bruex et al., 2012).
Figure 1.
Microarray and Individual Expression Analyses for Candidate GL2 Target Genes.
(A) The structure of the modified GL2 gene (GVGipro-VP16-GL2ΔN) used in the expression analyses for candidate GL2 target genes is schematically illustrated. The promoter is glucocorticoid inducible via the function of the GVG transcription factor.
(B) The candidate target genes obtained from the microarray analysis were categorized by gene product functions as defined using the agriGO Web-based software tool (Du et al., 2010), and the top five groups with most significant enrichment are shown with the ratios of their populations in the candidates (white box) and the total Arabidopsis genes (gray box). P values are indicated on the right of boxes.
(C) Relative induction folds (DEX/Mock) of transcript levels in the expression analysis for individual bHLH VIIIc and XI subfamily genes, and the PLDζ1 gene as a positive control, are shown (mean ± sd, n = 3). Asterisks indicate significant differences between the levels of the DEX and mock samples (*P < 0.05 and **P < 0.01, Student’s t test).
RHD6 and LRL1 belong to the bHLH VIIIc and XI subfamilies, respectively (Heim et al., 2003). Other members of these subfamilies, RSL4, LRL3, bHLH7, and bHLH59 were not induced significantly, and probes for RSL1, RSL2, LRL2, and RSL3/bHLH84 were absent from the microarray used in this study (Table 1). We examined the transcript levels of all the bHLH VIIIc and XI subfamily genes, together with that of PLDζ1 as a positive control, individually by qRT-PCR analysis using the same RNA preparations as used in the microarray experiments. The transcript levels of RHD6, RSL1, RSL2, LRL1, and LRL2, but none of the other VIIIc or XI subfamily genes, were induced significantly by the DEX treatment (Figure 1C). Therefore, we analyzed the five bHLH genes further as candidate GL2 target genes.
Table 1. Changes in Transcript Levels of Arabidopsis bHLH VIIIc and XI Subfamily Genes Caused by the Inducible Expression of VP16-GL2ΔN.
| bHLH Subfamily | Gene Name | AGI Gene ID | Average Fold Changea |
|---|---|---|---|
| VIIIc | RHD6/bHLH83 | AT1G66470 | 3.325 (<0.1) |
| VIIIc | RSL1/bHLH86 | AT5G37800 | Not representedb |
| VIIIc | RSL2/bHLH85 | AT4G33880 | Not representedb |
| VIIIc | RSL3/bHLH84 | AT2G14760 | Not representedb |
| VIIIc | RSL4/bHLH54 | AT1G27740 | 0.724 (0.83) |
| XI | LRL1/bHLH66 | AT2G24260 | 3.732 (<0.1) |
| XI | LRL2/bHLH69 | AT4G30980 | Not representedb |
| XI | LRL3/bHLH82 | AT5G58010 | 0.831 (0.67) |
| XI | bHLH7 | AT1G03040 | 0.871 (0.71) |
| XI | bHLH59 | AT4G02590 | 0.456 (1.00) |
The value is the average fold change of three independent experiments using the GeneChip ATH1 Arabidopsis microarray. False discovery rate (q-value) is shown in parentheses.
Data were not represented on the ATH1 microarray.
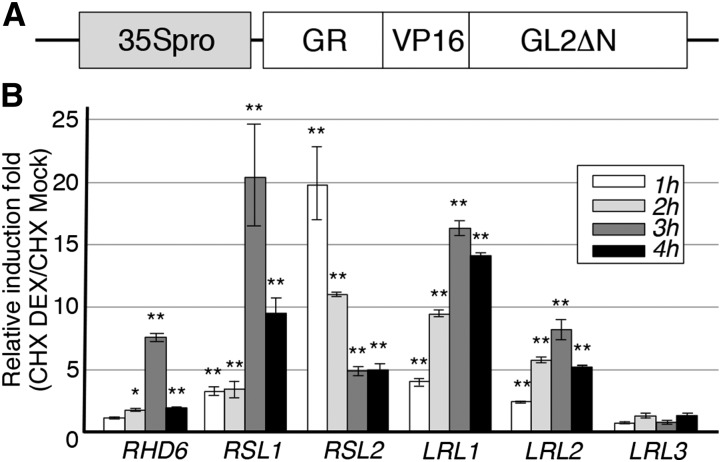
GL2 Recognizes RHD6, RSL1, RSL2, LRL1, and LRL2 Directly
To examine the direct recognition of the five bHLH genes by GL2 in planta, we used a posttranslational induction system with a GR domain-fused transcription factor. Transgenic seedlings harboring the 35S promoter-driven GR-VP16-GL2ΔN gene (Figure 2A) were treated or not treated with DEX for 1 to 4 h in the presence of cycloheximide (CHX), a protein synthesis inhibitor that prevents the secondary induction of transcription by the protein products of GL2 target genes. Following the treatment, qRT-PCR analysis using total RNA from the transgenic roots showed that the transcript levels of the five bHLH genes were increased by more than 5-fold, while the time required for the maximum induction differed for each bHLH gene (Figure 2B). By contrast, the noncandidate gene LRL3 was not activated significantly by DEX treatment (Figure 2B). These results indicate that GR-VP16-GL2ΔN directly recognized the five bHLH genes through the GL2 DNA binding domain and that GL2 can also recognize these genes directly.
Figure 2.
GL2 Direct Target Analysis of the Candidate GL2 Target Genes.
(A) The structure of the modified GL2 gene with a posttranslationally inducible function (35Spro-GR-VP16-GL2ΔN) used in the GL2 direct target analysis is schematically illustrated.
(B) Results of the inducible expression analysis using 35Spro-GR-VP16-GL2ΔN are shown for the candidate GL2 target genes RHD6, RSL1, RSL2, LRL1, and LRL2 and a negative control gene, LRL3. Relative induction folds (CHX DEX/CHX Mock) of transcript levels 1 to 4 h after induction are presented by different gray colors (mean ± sd, n = 3). Asterisks indicate significant differences between the levels of the CHX-DEX and CHX-Mock samples (*P < 0.05 and **P < 0.01, Student’s t test).
Based on previous findings suggesting that GL2 recognizes the L1 box-like sequence 5′-TAAATGT-3′ (Lin and Aoyama, 2012), we searched the 3-kb regions upstream of the initiation codons of the five bHLH genes and identified at least one L1 box-like site in each (Table 2). To test the possibility that GL2 recognizes the five bHLH genes via the L1 box-like sequence, we performed Agrobacterium tumefaciens-mediated transient expression analysis of GFP reporter genes containing the upstream regions of the five bHLH genes as promoters in tobacco (Nicotiana benthamiana) leaves (Figure 3). GFP fluorescence levels were increased for all five of the upstream regions by cointroduction with the 35S promoter-driven VP16-GL2ΔN gene (Figures 3A to 3J and 3P). Changing the L1 box-like sequence to 5′-TACATCT-3′ at all the sites in each upstream region reduced the increment in all instances (Figures 3F to 3P). These results suggest that GL2 recognized the five bHLH genes via the L1 box-like sequence.
Table 2. Candidate Binding Sites of GL2 within the 3-kb Upstream DNA Regions of PLDζ1, RHD6, RSL1, RSL2, LRL1, and LRL2.
| Site Name | Sequencea | Positionb |
|---|---|---|
| PLDζ1-S | 5′-ATTAATAAATGTTAAGA-3′ | 164 bp |
| RHD6-S | 5′-ATGAGTAAATGTTAACT-3′ | 2187 bp |
| RSL1-S1 | 5′-ACATCTAAATGTCATCA-3′ | 206 bp |
| RSL1-S2 | 5′-CCAAGTAAATGTGGTAT-3′ | 2324 bp |
| RSL2-S1 | 5′-ATAAATAAATGTCAACA-3′ | 146 bp c |
| RSL2-S2 | 5′-ATTTTTAAATGTTTCAT-3′ | 1850 bp |
| RSL2-S3-1 | 5′-TCAGTTAAATGTGTATG -3′ | 2283 bp c |
| RSL2-S3-2 | 5′-AGAAGTAAATGTCATAC-3′ | 2295 bp |
| LRL1-S1-1 | 5′-AGAAGTAAATGTCATAC-3′ | 506 bp |
| LRL1-S1-2 | 5′-GTAAATAAATGTGTTTT -3′ | 579 bp c |
| LRL1-S2 | 5′-AACCTTAAATGTATTAG-3′ | 2187 bp |
| LRL2-S | 5′-TTCCTTAAATGTAATTT -3′ | 2139 bp c |
The sequence of the candidate GL2-binding site is presented. The L1 box-like sequence is indicated in bold.
Base-pair length from the initiation codon to the L1 box-like sequence is presented. The “c” represents the complementary strand.
Figure 3.
Transient Expression Analysis for the Promoter Activity Directed by VP16-GL2ΔN.
(A) to (O) Representative fluorescence images of the N. benthamiana leaf epidermis transfected with each bHLH promoter-driven GFP gene, bHLHpro-GFP ([A] to [E]), each bHLHpro-GFP and the transfactor gene 35Spro-VP16-GL2ΔN ([F] to [J]), or each mutant bHLH promoter-driven GFP gene, bHLH-mL1pro-GFP, and 35Spro-VP16-GL2ΔN ([K] to [O]) are shown. The structures of the genes are schematically illustrated on the left of the images. The L1 box-like sequence 5′-TAAATGT-3′ was altered to 5′-TACATCT-3′ at all sites in each mutant bHLH promoter. Bars = 100 μm.
(P) Intensities of the GFP fluorescence signal were quantified, and the relative intensity of the signal was calculated with the mean value for each bHLHpro-GFP arbitrarily set as 1 (mean ± sd, n = 3). Asterisks indicate that the values are significantly different from their corresponding bHLHpro-GFP values (*P < 0.05 and **P < 0.01, Student’s t test). Hashes indicate that the values are significantly different from their corresponding bHLHpro-GFP/35Spro-VP16-GL2ΔN values (#P < 0.05 and ##P < 0.01, Student’s t test).
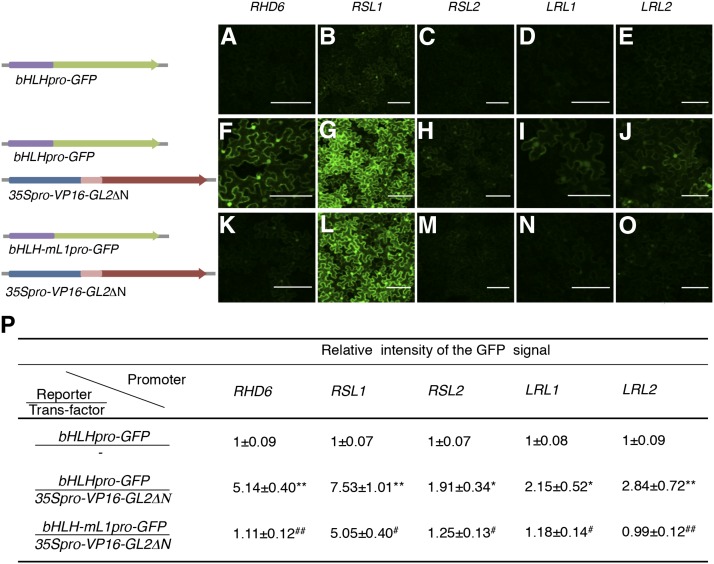
GL2 Binds to Upstream Regions of the bHLH Transcription Factor Genes in Planta
To examine the physical interaction of GL2 with the five bHLH genes in planta, we performed ChIP analysis using an anti-GFP antibody and a transgenic line harboring the GL2 promoter-driven the GFP-GL2 fusion gene GL2pro-GFP-GL2 in the gl2-5 mutant background (Figure 4A). To ensure that the fusion protein gene was functionally equivalent to the endogenous GL2 gene, we confirmed that the transgene complemented gl2-5 mutant phenotypes in the root and leaf epidermis (Figures 4B to 4E) and was expressed in the same manner as reported previously for GL2 in the root epidermis (Figure 4F; Galway et al., 1994; Masucci et al., 1996).
Figure 4.
ChIP Analysis for the Binding of GL2 to the GL2 Target Genes.
(A) The structure of the GL2 promoter-driven GFP-GL2 gene (GL2pro-GFP-GL2) used in the ChIP analysis is schematically illustrated.
(B) to (E) Phenotypes in the root ([B] and [C]) and leaf ([D] and [E]) epidermis of the gl2-5 mutant ([B] and [D]) and GL2pro-GFP-GL2/gl2-5 transgenic ([C] and [E]) plants are shown.
(F) The GFP fluorescence signal in a root tip region of the GL2pro-GFP-GL2/gl2-5 transgenic plant is shown. Bars = 100 μm in (B), (C), and (F) and 500 μm in (D) and (E).
(G) Results of the ChIP analysis are shown. The quantified DNA regions PLDζ1-S, RHD6-S, RSL1-S1, RSL1-S2, RSL2-S1, RSL2-S2, RSL2-S3, LRL1-S1, LRL1-S2, and LRL2-S correspond to the sites of the L1 box-like sequence listed in Table 2. The regions RSL2-S3 and LRL2-S1 contain two sites of the L1 box-like sequence for each. An upstream DNA region of ACT7 was used as a negative control. Relative enrichment folds (anti-GFP/IgG) of GL2pro-GFP-GL2/gl2-5 roots and the wild-type roots are shown in white and gray boxes, respectively (mean ± sd, n = 3). Asterisks indicate significant differences between the enrichment folds in GL2-pro-GFP-GL2/gl2-5 and in the wild type (*P < 0.05 and **P < 0.01, Student’s t test).
A chromosome fraction was prepared from the roots of the transgenic line and fragmented DNA bound to GFP-GL2 was coimmunoprecipitated using the anti-GFP antibody. We examined the coimmunoprecipitates by quantitative real-time PCR to detect the short 100- to 150-bp DNA regions containing the L1 box-like sequence upstream of the bHLH genes. The short DNA regions of all five bHLH genes and the PLDζ1 positive control gene were enriched significantly in the coimmunoprecipitates, whereas either a negative control region of LRL1 or upstream regions of the negative control genes ACT7 and LRL3 were not (Figure 4G; Supplemental Figure 1). These results indicated that GL2 physically interacts with the upstream regions of the five bHLH genes in roots, thereby confirming that they are GL2 target genes.
GL2 Suppresses the Expression of the bHLH Transcription Factor Genes in N Cells
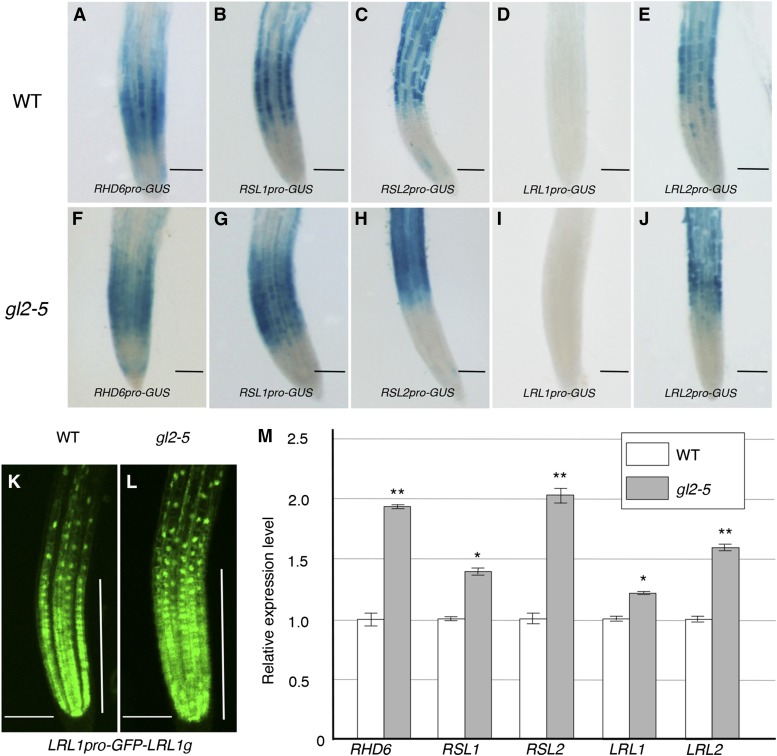
To investigate how GL2 modulates the expression of the five bHLH genes via binding to their upstream regions, we analyzed the promoter activity of each upstream region in wild-type and gl2-5 mutant roots using the histochemical GUS reporter system. The upstream regions of RHD6, RSL1, RSL2, and LRL2 exhibited promoter activity preferentially in the H cell files of the wild-type root epidermis (Figures 5A to 5C and 5E), but their activities extended to all cell files in the gl2-5 mutant (Figures 5F to 5H and 5J), indicating that the promoter activities of these upstream regions were suppressed by GL2 gene function in N cell files. The LRL1 upstream region exhibited little promoter activity in the root epidermis based on the GUS reporter analysis (Figures 5D and 5I); however, a GFP reporter gene containing the LRL1 upstream region and a genomic region encompassing the entire protein-coding sequence produced GFP florescence preferentially in H cell files in the wild type (Figure 5K). This preference disappeared in the gl2-5 mutant (Figure 5L), indicating that LRL1 is also suppressed by GL2 in N cell files. Consistent with these results, qRT-PCR analysis showed that the transcript levels of the five bHLH genes were significantly higher in gl2-5 mutant roots than in wild-type roots (Figure 5M). These results, together with the evidence for GL2 binding, indicate that the transcription of the five bHLH genes is suppressed directly by GL2 in N cell files.
Figure 5.
Expression Analyses of the GL2 Target Genes.
(A) to (J) Histochemical GUS expression patterns in root tips at 7 d after germination (DAG) in the wild-type ([A] to [E]) and gl2-5 ([F] to [J]) genetic backgrounds are shown for the upstream regions of RHD6 ([A] and [F]), RSL1 ([B] and [G]), RSL2 ([C] and [H]), LRL1 ([D] and [I]), and LRL2 ([E] and [J]).
(K) and (L) The GFP fluorescence signal directed by the GFP reporter gene containing the upstream and total genomic regions of LRL1 (LRL1pro-GFP-LRL1g) is shown. Root tips of the 7-DAG transgenic plants harboring LRL1pro-GFP-LRL1g in the wild-type (K) and gl2-5 (L) genetic backgrounds were observed. Vertical bars indicate the root tip region containing proliferating cells. Bars = 100 μm in (A) to (L).
(M) Relative transcript levels of RHD6, RSL1, RSL2, LRL1, and LRL2 in 7-DAG wild-type (white box) and gl2-5 (gray box) roots are shown. Transcript levels were determined by qRT-PCR, and their relative values were calculated by setting each wild-type mean value arbitrarily as 1 (mean ± sd, n = 3). Asterisks indicate significant differences between the transcript levels in gl2-5 and in the wild type (*P < 0.05 and **P < 0.01, Student’s t test).
The upstream regions of RHD6 and RSL1 exhibited promoter activities at earlier stages in root hair development than those of RSL2 and LRL2 (Figures 5A to 5C and 5E). The GFP reporter gene for LRL1 was active in the root tip epidermis, including the cell proliferation zone (Figure 5K). These results strongly suggest that the five bHLH genes are expressed differentially during the various stages of root hair development.
LRL1 and LRL2 Function Redundantly in Root Hair Elongation Downstream of GL2
RHD6 functions downstream of GL2 in root hair initiation redundantly with RSL1 (Masucci and Schiefelbein, 1994; Menand et al., 2007), and RSL2 functions further downstream in root hair elongation (Yi et al., 2010). To examine the functional relationship of LRL1 and LRL2 with GL2 in root hair development, the epistasis of their mutant effects was analyzed. Although the lines SALK_006430 (Atlrl1-2 in Karas et al., 2009, and bhlh66-1 in Bruex et al., 2012) and SALK_029317c (bhlh69-1 in Bruex et al., 2012), which are designated as lrl1-2 and lrl2-2, respectively, contain T-DNA insertions in introns (Supplemental Figure 2A), transcripts from both mutant genes were only faintly detected by the RT-PCR analysis that produced intense signals for the wild-type genes (Supplemental Figure 2B). This indicates that the expression of LRL1 and LRL2 is severely affected in lrl1-2 and lrl2-2, respectively.
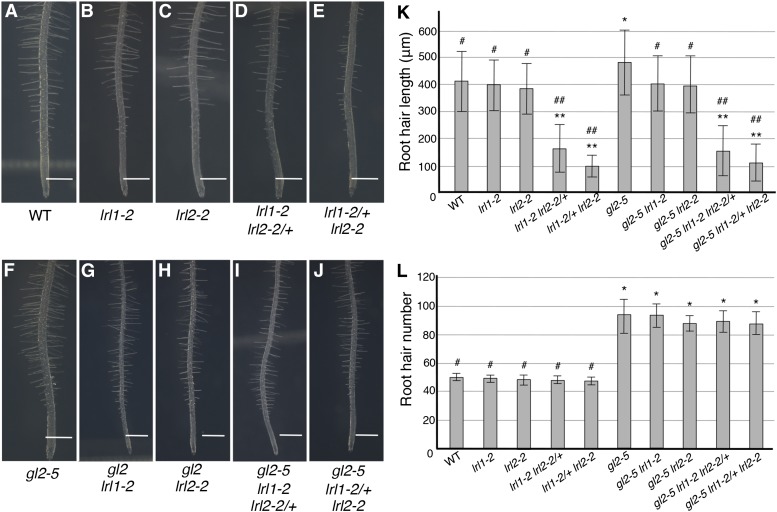
Under the conditions used in this study, neither lrl1-2 nor lrl2-2 exhibited significant difference from the wild type in root hair length (Figures 6A to 6C and 6K). Whereas the homozygous double mutant plants could not be obtained, the partially homozygous double mutants lrl1-2 lrl2-2/+ and lrl-12/+ lrl2-2 exhibited short root hair phenotypes (Figures 6D, 6E, and 6K), as reported previously (Karas et al., 2009). Regarding root hair density, none of the single or partially homozygous double mutants exhibited significant differences from the wild type (Figure 6L), as also reported previously (Karas et al., 2009). The gl2-5 mutant exhibited significantly longer root hairs than the wild type (Figure 6F). However, root hairs of the gl2-5 lrl1-2 and gl2-5 lrl2-2 double mutants were significantly shorter than those of gl2-5 and similar in length to those of the wild type (Figures 6F to 6H and 6K). Moreover, the partially homozygous triple mutants gl2-5 lrl1-2 lrl2-2/+ and gl2-5 lrl-12/+ lrl2-2 exhibited root hairs of similar lengths to those of lrl1-2 lrl2-2/+ and lrl-12/+ lrl2-2, respectively (Figures 6D, 6E, and 6I to 6K). These results provide genetic evidence that LRL1 and LRL2 function redundantly as positive factors in root hair elongation downstream of GL2. By contrast, the gl2-5 phenotype of higher root hair density due to ectopic root hair development in N cell files was not affected by the lrl mutations (Figure 6L).
Figure 6.
Root Hair Phenotypes of gl2-5, lrl1-2, lrl2-2, and Their Multiple Mutants.
(A) to (J) Main roots of the wild type (A), lrl1-2 (B), lrl2-2 (C), lrl1-2 lrl2-2/+ (D), lrl1-2/+ lrl2-2 (E), gl2-5 (F), gl2-5 lrl1-2 (G), gl2-5 lrl2-2 (H), gl2-5 lrl1-2 lrl2-2/+ (I), and gl2-5 lrl1-2/+ lrl2-2 (J) at 7 DAG are shown. Bars = 500 μm in (A) to (J).
(K) Root hair lengths of the wild type, gl2-5, lrl1-2, lrl2-2, and their multiple mutants at 7 DAG are shown. Root hairs in the region 5 to 7 mm away from the root tip were measured (mean ± sd, n = 400). Asterisks indicate that the root hair lengths are significantly different from those of the wild type (*P < 0.05 and **P < 0.01, Student’s t test). Hashes indicate that the root hair lengths are significantly different from those of gl2-5 (#P < 0.05 and ##P < 0.01, Student’s t test).
(L) Root hair numbers of the wild type, gl2-5, lrl1-2, lrl2-2, and their multiple mutants at 7 DAG are shown. Root hairs and bulges in the visible side in the region 5 to 7 mm away from the root tip were counted (mean ± sd, n = 10). Asterisks indicate that the root hair numbers are significantly different from those of the wild type (P < 0.01, Student’s t test). Hashes indicate that the root hair numbers are significantly different from those of gl2-5 (P < 0.01, Student’s t test).
The bHLH Transcription Factors Have Diverse Functions in Cell Differentiation
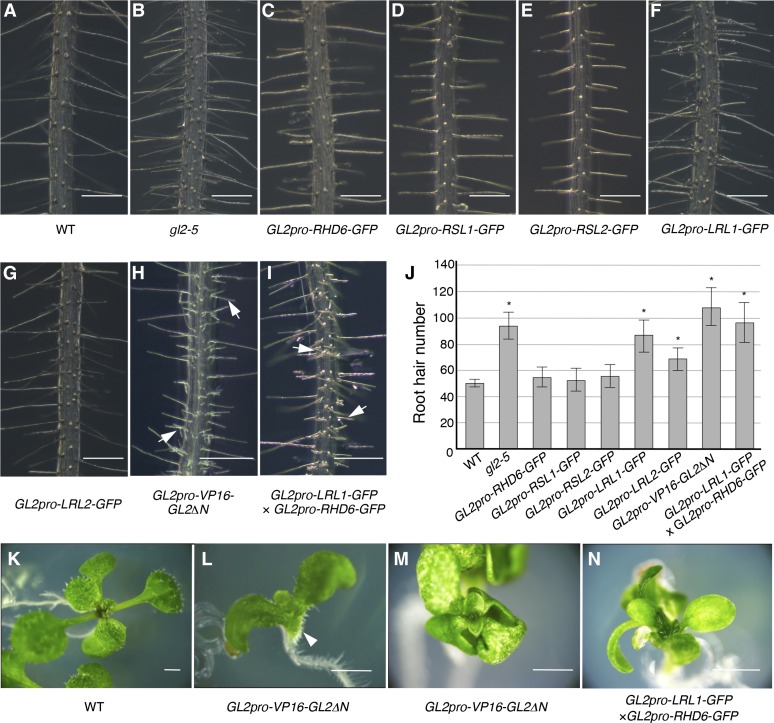
To investigate the biological functions of the five bHLH transcription factors, we examined the effects of ectopic expression of their GFP-fusion proteins directed by the GL2 promoter. Of transgenic plants that expressed the five fusion proteins at comparable levels (Supplemental Figure 3), those harboring GL2pro-LRL1-GFP and GL2pro-LRL2-GFP exhibited severe and moderate phenotypes, respectively, characterized by ectopic root hairs in N cell files and an increase in the number of root hairs (Figures 7F, 7G, and 7J; Supplemental Figure 4). By contrast, none of the other fusion genes exhibited significant effects on the root hair pattern (Figures 7C to 7E and 7J).
Figure 7.
Phenotypes Caused by the GL2 Promoter-Driven bHLH-GFP Genes.
(A) to (I) Main roots of the wild-type (A) and gl2-5 (B) plants and the transgenic plants harboring GL2pro-RHD6-GFP (C), GL2pro-RSL1-GFP (D), GL2pro-RSL2-GFP (E), GL2pro-LRL1-GFP (F), GL2pro-LRL2-GFP (G), GL2pro-VP16-GL2ΔN (H), and both GL2pro-LRL1-GFP and GL2pro-RHD6-GFP (I) at 7 DAG are shown. Typical branching root hair structures are indicated by arrows in (H) and (I).
(J) Root hair numbers of the wild-type and gl2-5 roots and the transgenic roots harboring GL2pro-RHD6-GFP, GL2pro-RSL1-GFP, GL2pro-RSL2-GFP, GL2pro-LRL1-GFP, GL2pro-LRL2-GFP, GL2pro-VP16-GL2ΔN, and both GL2pro-LRL1-GFP and GL2pro-RHD6-GFP at 7 DAG are shown (mean ± sd, n = 10). Asterisks indicate that the root hair numbers are significantly different from those of the wild type (P < 0.01, Student’s t test).
(K) to (N) Phenotypes of the transgenic seedlings harboring GL2pro-VP16-GL2ΔN and both GL2pro-LRL1-GFP and GL2pro-RHD6-GFP in aerial organs. Seedlings of the wild type (K) and transgenic lines harboring GL2pro-VP16-GL2ΔN ([L] and [M]) and both GL2pro-LRL1-GFP and GL2pro-RHD6-GFP (N) are shown. The root hair-like structures on the abaxial surface of the hypocotyl/cotyledon junction are indicated by an arrowhead in (L).
Bars = 250 μm in (A) to (I) and 1 cm in (K) to (N).
Because it was suggested that ectopic expression of a set of GL2 target genes caused severe phenotypes in cell differentiation including root hair-like structures produced from aerial epidermal cells (Ohashi et al., 2003), the cooperative effects of the fusion genes were examined in the F1 progeny of homozygous transgenic plants. Among the F1 plants, those that harbored both GL2pro-RHD6-GFP and GL2pro-LRL1-GFP exhibited a phenotype of ectopic and branching root hairs similar to the GL2pro-VP16-GL2ΔN plants (Figures 7H and 7I), while they did not develop aerial protuberant structures like those observed in the GL2pro-VP16-GL2ΔN plants (Figure 7L). Their leaf epidermis exhibited various degrees of abnormalities in the initiation and branching pattern of trichomes. Some plants had no trichomes, similar to the GL2pro-VP16-GL2ΔN plants (Figures 7M and 7N), and others had fewer trichomes with abnormal spatial patterns and divergent branch numbers (Supplemental Figure 5 and Supplemental Table 1). These results from single- and double-transgenic plants indicate that the five bHLH transcription factors have diverse functions in cell differentiation.
DISCUSSION
In this study, we showed that GL2 directly suppresses the five bHLH transcription factor genes RHD6, RSL1, RSL2, LRL1, and LRL2 in N cell files. These genes have been identified as positive factors at various stages in root hair development. RHD6 and RSL1 promote root hair initiation in a partly redundant manner and RSL2 subsequently promotes root hair elongation redundantly with RSL4 (Menand et al., 2007; Yi et al., 2010; Pires et al., 2013). LRL1 and LRL2, together with LRL3, are thought to function redundantly in root hair elongation based on the root hair phenotypes of their single and multiple mutants (Karas et al., 2009; Bruex et al., 2012). This diversity in function is supported by the result that the upstream regions of RHD6 and RSL1 exhibited promoter activities at earlier stages in root hair development than those of RSL2 and LRL2.
We confirmed that the partially homozygous double mutants of LRL1 and LRL2, lrl1-2 lrl2-2/+ and lrl1-2/+ lrl2-2, exhibited shorter root hairs than the wild type and found that their phenotypes in root hair lengths were epistatic to those of gl2-5. The possibility that they are also involved in processes other than root hair elongation cannot be excluded, however, because lrl1 lrl2 double homozygous mutants could not be obtained. LRL1 and LRL2 are functional homologs of L. japonicus RHL1, a mutant that exhibited no indication of root hair initiation on almost all parts of the root surface (Karas et al., 2005). Moreover, the GL2pro-LRL1-GFP and GL2pro-LRL2-GFP transgenes caused ectopic root hairs in N cell files where RHD6 and RLS1 are normally inactive (Menand et al., 2007). These findings suggest that LRL1 and LRL2 are also involved in root hair initiation. This idea is supported, especially for LRL1, by the result that the GFP reporter gene for LRL1 was expressed in root epidermal cells including those in the cell proliferation zone.
Different sets of transcription factor genes are known to be responsible for root hair development under various conditions. While RHD6 is predominantly responsible for root hair initiation under normal growth conditions on agar media, RSL1 contributes to an extent comparable to RHD6 when roots are grown on cellophane disks (Menand et al., 2007). Moreover, in the presence of exogenous auxin, root hair initiation does not require either RHD6 or RSL1 (Yi et al., 2010) suggesting that other transcription factor genes are responsible for the root hair initiation. Despite such diversity of transcription factor genes for root hair initiation, the file-specific pattern of root hair formation is sustained robustly (Masucci and Schiefelbein, 1994). The mechanism ensuring this robustness might involve the targeting of multiple bHLH transcription factor genes by GL2. GL2 may also act as a negative factor for root hair elongation by suppressing RSL2, LRL1, and LRL2. Because GL2 is expressed at a low level in H cells (Lee and Schiefelbein, 2002), GL2 might modulate root hair elongation in H cells via these bHLH genes. This idea is supported by the result that gl2-5 exhibited significantly longer root hairs than the wild type. These findings suggest that for the fine and robust regulation of root hair development, GL2 suppresses multimodal pathways by targeting bHLH genes with diverse functions.
Transgenic plants harboring both the GL2pro-RHD6-GFP and GL2pro-LRL1-GFP transgenes exhibited abnormalities in root hair morphology and the initiation frequency and branching pattern of trichomes. Because these abnormalities were not observed in GL2pro-RHD6-GFP or GL2pro-LRL1-GFP homozygous transgenic plants, but were observed in the F1 progeny of crosses between them, the phenotype is thought to be due to a cooperative effect of the transgenes. The ectopic expression of GL2 target genes directed by VP16-GL2ΔN resulted in the development of root hair-like structures from aerial organs (Ohashi et al., 2003). In a related finding, ectopic coexpression of the Physcomitrella patens RHD6-like transcription factors RSL1 and RSL2 converted developing leafy shoot axes into rhizoids, which are protuberances similar in structure and function to root hairs (Jang et al., 2011). However, plants harboring both the GL2pro-RHD6-GFP and GL2pro-LRL1-GFP genes did not develop aerial protuberant structures like those observed in the GL2pro-VP16-GL2ΔN plants, while they exhibited root hair and trichome phenotypes similar to those of the GL2pro-VP16-GL2ΔN plants. GL2 target genes in addition to those encoding bHLH transcription factors may be required for such protuberant cell morphogenesis from aerial organs.
The GL2 DNA binding domain has been shown to recognize DNA regions containing the L1 box-like sequence 5′-TAAATGT-3′ in in vitro DNA footprinting analysis (Ohashi et al., 2003) and yeast one-hybrid analysis (Tominaga-Wada et al., 2009; Khosla et al., 2014). The GL2 target genes identified in this study contain at least one copy of the L1 box-like sequence in each 3-kb upstream region, and we confirmed the binding of GFP-GL2 in the close proximity of each L1 box-like sequence by ChIP analysis. To date, more than 10 L1 box-like sites, including those from previous studies (Khosla et al., 2014; Wang et al., 2015), have been confirmed individually by ChIP analysis. Alignment of their sequences does not show strong conservation in the sequences surrounding the L1 box-like sites except for a relatively high A/T content (Supplemental Figure 6). Although this reflects a critical contribution of the L1 box-like sequence to the recognition by GL2, some imperfect L1 box-like sequences might be recognized by GL2 because the mutated RSL1 upstream region that lacked all of the L1 box-like sequence sites still functioned as a promoter activated by VP16-GL2ΔN (Figures 3L and 3P).
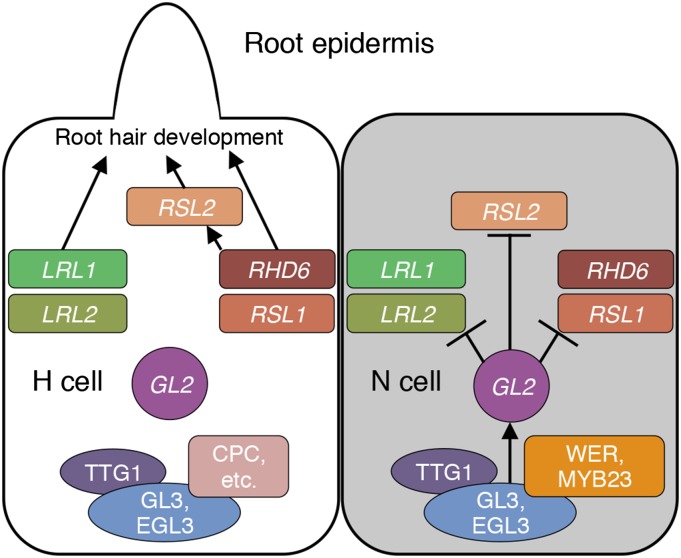
The gene regulatory networks for the cell fate determination in the root epidermis and for the morphological differentiation of root hair cells are the best studied mechanisms of cell pattern formation and cell morphogenesis, respectively, in plants. This study integrates these two elements into a regulatory cascade via the direct interaction between GL2 and its target genes as illustrated in Figure 8. In N cells, a transcription factor complex composed of WER/MYB23, GL3/EGL3, and TTG1 activates GL2 expression, and GL2 negatively regulates the bHLH transcription factor genes RHD6, RSL1, RSL2, LRL1, and LRL2 to suppress root hair development. In H cells, GL2 is not activated due to the inhibitory function of CPC and its paralogs, and the bHLH genes remain active to promote root hair development. In addition to the bHLH genes, GL2 may recognize a large number of target genes with various functions, as shown in the list of candidate GL2 target genes in Supplemental Table 1. The enrichment for genes related to signal transduction (i.e., those for lipid metabolic processes and hormone-mediated signaling pathways) and gene expression (i.e., those for regulation of RNA metabolic processes and regulation of transcription) among the candidate genes is notable. Systematic approaches to identify GL2 target genes will help to elucidate the roles of GL2 in the differentiation of various types of epidermal cells.
Figure 8.
A Model of the Transcriptional Network Surrounding GL2 in Root Epidermal Cells.
In N cells (gray), the WER/MYB23-GL3/EGL3-TTG1 complex activates GL2. GL2 directly suppresses the bHLH transcription factor genes RHD6, RSL1, RSL2, LRL1, and LRL2. In H cells (white), WER and MYB23 in the complex are replaced by CPC and its paralogs. Consequently, GL2 is not activated and the bHLH genes remain active to promote various stages of root hair development. Arrows indicate activation of genes or promotion of root hair development. T-bars indicate suppression of genes.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana, ecotype Columbia-0, served as the wild type. The mutant line gl2-5 (the Columbia genetic background; Ohashi et al., 2003) was donated by G. Morelli (Food and Nutrition Research Centre, Agricultural Research Council, Rome, Italy). lrl1-2 (SALK_006430) and lrl2-2 (SALK_029317c) were ordered from the ABRC (Alonso et al., 2003). Sequences of the primers used in genotyping are presented in Supplemental Table 2. Plants were grown as described previously (Qin et al., 2005) unless otherwise noted.
Construction of Transgenes and Transformants
The GVG-inducible and GL2 promoter-driven VP16-GL2ΔN genes were constructed previously (Ohashi et al., 2003). To construct the 35S promoter-driven GR-VP16-GL2ΔN gene, the promoter of the GL2 promoter-driven VP16-GL2ΔN gene was replaced with the 35S promoter of pBI121 (Bevan et al., 1983), and a GR-coding fragment amplified by PCR from the GVG gene (Aoyama and Chua, 1997) was inserted at the 5′-end of the VP16-coding region in an in-frame manner. The GL2 promoter-driven GFP-GL2 gene was constructed by assembling PCR products of the 2.1-kb GL2 promoter fragment (Ohashi et al., 2003) and fragments encoding GFP and GL2 in the three-fragment recombination vector pK7m34GW (Invitrogen). To construct reporter genes for transient expression analysis, upstream fragments of bHLH genes were amplified by PCR from Arabidopsis genomic DNA and assembled with the GFP-coding fragment in the vector pK7FWG0 (Invitrogen). To make mutant fragments, in which the L1 box-like sequence 5′-TAAATGT-3′ was altered to 5′-TACATCT-3′ at all the sites, the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) was used. To construct the 35S promoter-driven VP16-GL2ΔN gene, the DNA region encoding VP16-GL2ΔN was cloned into pENTR-D-TOPO (Invitrogen) and transferred into the pK7WG2 expression vector (Invitrogen). To construct reporter genes for histochemical GUS analysis, upstream fragments of bHLH genes were amplified by PCR from Arabidopsis genomic DNA and cloned into the vector pKGWFS7 (Invitrogen). To construct GL2 promoter-driven bHLH-GFP fusion genes, bHLH-coding fragments were amplified by PCR from Arabidopsis cDNA and assembled with the GL2 promoter fragment and the GFP-coding fragment in the vector pK7m34GW. The DNA was cloned into the vectors pK7m34GW, pK7FWG0, pK7WG2, and pKGWFS7 using the Gateway system (Invitrogen) as described previously (Tao et al., 2013). Constructs were transformed into Agrobacterium tumefaciens GV3101/pMP90 using a freeze-thaw procedure. Arabidopsis transformation and transgenic plant screening were conducted as described previously (Qin et al., 2005). PCR primers used in DNA construction are shown in Supplemental Table 2.
Analyses for Transcript Levels
Total RNA was extracted using TRIzol reagent (Invitrogen) and treated with RNase-free DNase (TaKaRa). cDNA was synthesized from total RNA using the Superscript II RT kit (Invitrogen). Microarray analysis was performed using GeneChip ATH1 Arabidopsis (Affymetrix) according to Affymetrix protocols. qRT-PCR was performed as described previously (Wu et al., 2011) using SYBR Green Real-time PCR Master Mix (Toyobo) on an ABI7500 FAST (Applied Biosystems). Gene expression levels were standardized to the constitutive expression level of reference. At least three replications were performed for each sample in one experiment. The relative expression level of each gene was calculated based on the cycle threshold (CT) 2−ΔΔCT method (Livak and Schmittgen, 2001). PCR primers used in qRT-PCR are shown in Supplemental Table 2.
Conditions of Induction Experiments
Seedlings grown on agar medium (Murashige and Skoog [MS] salts, 1% sucrose, and 0.8% agar) around the four-leaf stage (about 2 weeks after germination) were transferred onto filter paper on agar medium (MS salts, 0% sucrose, and 2% agar) and adapted to the open-air condition gradually for 3 d. Then, the seedlings were sprayed with induction or control solution and kept for appropriate induction periods. For the microarray analysis, seedlings were treated with the induction solution containing 30 μM DEX and 0.01% Silwet L-77 for 5 h. The induction solution for the DEX/CHX analysis contained 30 μM DEX, 30 μM CHX, and 0.01% Silwet L-77. For both analyses, the control solution was the same as the induction solution except for DEX.
Transient Expression Analysis in Tobacco Leaves
The reporter and effector constructs were introduced into abaxial leaf epidermal cells of 2- to 4-week-old Nicotiana benthamiana plants by Agrobacterium (GV3101/pMP90)-mediated transfection, as described previously (Sparkes et al., 2006). Fluorescence images were captured using a TCS SPE confocal laser scanning microscope (Leica) 3 d after the transfection. The experiment was replicated three times for each effector-reporter combination. The intensity of the GFP signal was quantified on the image using ImageJ (http://rsbweb.nih.gov/ij/).
ChIP Analysis
ChIP analysis was performed using total root tissues of the wild-type and GL2pro-GFP-GL2/gl2-5 plants as described previously (Gendrel et al., 2005). Immunoprecipitation was performed using Protein A beads (Millipore), anti-GFP antibody (Abcam), and IgG serum (Millipore). Coimmunoprecipitated DNA containing a particular region was quantified by real-time PCR with primers shown in Supplemental Table 2. The quantified value from the coimmunoprecipitated DNA was first normalized to that from the input DNA. The relative enrichment fold was then calculated by dividing the normalized value using the anti-GFP antibody with that using the IgG sample.
GUS and GFP Reporter Analyses
The histochemical GUS reporter analysis was performed as described previously (Jefferson et al., 1987). Tissues were submerged in 90% acetone for 30 min at −20°C. After several washes with 0.1 M Na2HPO4 (pH 7.0), tissues were infiltrated under vacuum for 30 min and then incubated at 37°C for 2 to 4 h in a solution of 0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide, 0.1 M Na2HPO4 (pH 7.0), 10 mM Na2EDTA, 0.5 mM potassium ferricyanide/ferrocyanide, and 0.06% Triton X-100. The staining solution was removed, and tissues were cleared in 70% ethanol. The GFP fluorescence signal was observed and images were acquired using a confocal laser scanning microscope TCS SPE (Leica).
Observation and Evaluation of Root Hair and Trichome Phenotypes
To evaluate root hair phenotypes, plants were germinated and grown on the surface of vertically positioned agar medium containing MS salts, 1% sucrose, and 1% Phytagel (Sigma-Aldrich). Images of the primary root on the agar medium were captured with a CCD camera DFC425 C (LEICA) coupled to a stereomicroscope M205 FA (Leica). The observation and image capture of root surface structures were performed using an Axioplan 2 microscope (Zeiss) equipped with a CSU-X1 confocal laser scanning unit (Yokogawa) after staining with 5 μg/mL propidium iodide. For root hair lengths, root hairs elongated horizontally to the agar surface in the region 5 to 7 mm from the root tip were measured on the image with the assistance of ImageJ. Immature root hairs or bulges with lengths of <5 μm were omitted. For root hair numbers, both mature and immature root hairs including bulges in the visible side in the region 5 to 7 mm from the root tip were counted. Trichomes were observed by an S-3500 N variable-pressure scanning electron microscope (Hitachi) equipped with a cooling stage. The 10th to 12th leaves were cut from the plants and directly observed in variable-pressure mode (100 Pa) at −20°C. To evaluate trichome and trichome branch numbers, 2-week-old seedlings germinated and grown on the surface of agar medium containing MS salts, 1% sucrose, and 1% Phytagel (Sigma-Aldrich) were used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: GL2 (At1g79840), PLDζ1 (At3g16785), RHD6 (At1g66470), RSL1 (At5g37800), RSL2 (At4g33880), RSL3 (At2g14760), RSL4 (At1g27740), LRL1 (At2g24260), LRL2 (At4g30980), LRL3 (At5g58010), bHLH7 (At1g03040), bHLH59 (At4g02590), ACT7 (At5g09810), and TUB2 (At5g62690).
Supplemental Data
Supplemental Figure 1. ChIP Analysis for the GL2 Binding to the LRL1 and LRL3 Upstream Regions.
Supplemental Figure 2. Expression Analysis of the lrl1-2 and lrl2-2 Mutant Genes.
Supplemental Figure 3. Expression Patterns and Intensities of bHLH-GFP Fusion Proteins Driven by the GL2 Promoter.
Supplemental Figure 4. Root Hair Development Patterns of the Transgenic Plants Harboring the GL2 Promoter-Driven bHLH-GFP Genes.
Supplemental Figure 5. Trichome Phenotypes of Transgenic Plants Harboring Both GL2pro-LRL1-GFP and GL2pro-RHD6-GFP.
Supplemental Figure 6. Sequences Surrounding the L1 Box-Like Sites Recognized by GL2.
Supplemental Table 1. Trichome and Trichome Branch Number of Transgenic Plants Harboring GL2 Promoter-Driven bHLH-GFP Genes.
Supplemental Table 2. List of Primers Used in This Study.
Supplemental Data Set 1. List of Candidate GL2 Target Genes.
Supplementary Material
Acknowledgments
We thank K. Yasuda for technical assistance and G. Morelli for providing gl2-5 mutant seeds. This work was supported by the National Transgenic Research Project (Grant 2013ZX08009-002), by the Natural Science Foundation of China and the Japanese Society of Promotion of Science (NSFC-JSPS 31211140041 to L.-J.Q. and T.A.), by the Japanese Society for the Promotion of Science (Grants-in-Aid for Research Activity Start-up; 26891013) to M.K., and partially by the 111 Project.
AUTHOR CONTRIBUTIONS
Q.L. conceived the study, designed the experiments, performed the research, analyzed the data, and prepared the article. Y.O. performed the experiments and contributed to experimental design. M.K. and T.T. contributed to experimental design and analyzed the data. H.G. and L.-J.Q. contributed to experimental design, analyzed the data, and prepared the article. T.A. contributed to experimental design, data analysis, and article preparation and submitted the article.
Glossary
- N
non-hair
- H
hair
- bHLH
basic helix-loop-helix
- ChIP
chromatin immunoprecipitation
- DEX
dexamethasone
- CHX
cycloheximide
- MS
Murashige and Skoog
- DAG
days after germination
References
- Abe M., Takahashi T., Komeda Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26: 487–494. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Aoyama T., Chua N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298. [DOI] [PubMed] [Google Scholar]
- Bevan M., Barnes W.M., Chilton M.D. (1983). Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 11: 369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes F.A.L. (2000). Pattern in root meristem development in angiosperms. New Phytol. 146: 83–94. [Google Scholar]
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10: 393–402. [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C.M., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S., Roberts K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474. [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch J.J., Chen M.A., Hillestad M., Marks M.D. (2004). Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 40: 860–869. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Grebe M. (2012). The patterning of epidermal hairs in Arabidopsis--updated. Curr. Opin. Plant Biol. 15: 31–37. [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Hung C.-Y., Lin Y., Zhang M., Pollock S., Marks M.D., Schiefelbein J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G., Yi K., Pires N.D., Menand B., Dolan L. (2011). RSL genes are sufficient for rhizoid system development in early diverging land plants. Development 138: 2273–2281. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker inhigher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B., Amyot L., Johansen C., Sato S., Tabata S., Kawaguchi M., Szczyglowski K. (2009). Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 151: 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B., Murray J., Gorzelak M., Smith A., Sato S., Tabata S., Szczyglowski K. (2005). Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs. Plant Physiol. 137: 1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A., Paper J.M., Boehler A.P., Bradley A.M., Neumann T.R., Schrick K. (2014). HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26: 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J., Hulskamp M. (2004). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55: 389–398. [DOI] [PubMed] [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2007). The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 302: 118–131. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 18: 1949–1954. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Aoyama T. (2012). Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. J. Integr. Plant Biol. 54: 729–737. Erratum. J. Integr. Plant Biol. 55: 485. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ ΔC(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430. [DOI] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N.D., Yi K., Breuninger H., Catarino B., Menand B., Dolan L. (2013). Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 110: 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Huang L., Zheng X. (2014). Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loop. Front. Plant Sci. 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Kwak S.H., Wieckowski Y., Barron C., Bruex A. (2009). The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J.W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124: 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311: 566–578. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., Zhang J., Wei T., Gu H., Qu L.J., Qin G. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Sano R., Inoue K., Okada K., Wada T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Ishida T., Wada T. (2011). New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286: 67–106. [DOI] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Okada K., Wada T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19: 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R., Iwata M., Sugiyama J., Kotake T., Ishida T., Yokoyama R., Nishitani K., Okada K., Wada T. (2009). The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J. 60: 564–574. [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wang S., Barron C., Schiefelbein J., Chen J.G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185: 387–400. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang X., Hu Q., Dai X., Tian H., Zheng K., Wang X., Mao T., Chen J.-G., Wang S. (2015). Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 83: 300–311. [DOI] [PubMed] [Google Scholar]
- Wu R., Li S., He S., Wassmann F., Yu C., Qin G., Schreiber L., Qu L.J., Gu H. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23: 3392–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42: 264–267. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.