Abstract
Peripheral T-cell lymphomas (PTCL) are a heterogeneous group of non-Hodgkin lymphomas with poor prognosis. Their molecular pathogenesis has not been entirely elucidated. We previously showed that 6q24 is one of the most frequently deleted regions in primary thyroid T-cell lymphoma. In this study, we extended the analysis to other subtypes of PTCL and performed functional assays to identify the causative genes of PTCL that are located on 6q24. Genomic loss of 6q24 was observed in 14 of 232 (6%) PTCL cases. The genomic loss regions identified at 6q24 always involved only two known genes, STX11 and UTRN. The expression of STX11, but not UTRN, was substantially lower in PTCL than in normal T-cells. STX11 sequence analysis revealed mutations in two cases (one clinical sample and one T-cell line). We further analyzed the function of STX11 in 14 cell lines belonging to different lineages. STX11 expression only suppressed the proliferation of T-cell lines bearing genomic alterations at the STX11 locus. Interestingly, expression of a novel STX11 mutant (p.Arg78Cys) did not exert suppressive effects on the induced cell lines, suggesting that this mutant is a loss-of-function mutation. In addition, STX11-altered PTCL not otherwise specified cases were characterized by the presence of hemophagocytic syndrome (67% vs 8%, P = 0.04). They also tended to have a poor prognosis compared with those without STX11 alteration. These results suggest that STX11 plays an important role in the pathogenesis of PTCL and they may contribute to the future development of new drugs for the treatment of PTCL.
Keywords: Functional analyses, genomic loss of 6q24, peripheral T-cell lymphomas, STX11, tumor suppressor gene
Peripheral T-cell lymphomas (PTCL) are tumors derived from mature T-cells. They are relatively rare and account for approximately 10% of non-Hodgkin Lymphomas.1 A recent epidemiological study demonstrated that the incidence of PTCL is increasing in Japan and the USA.2 PTCL are known to be clinically, histopathologically and genetically heterogeneous.3,4 Recently, studies employing unbiased and genome-wide methods showed that the genetic alterations were involved in the pathophysiology of PTCL.5–16 Because of the heterogeneity of the diseases, additional genomic alterations are predicted to contribute to the pathophysiology of PTCL. Therefore, the molecular pathogenesis of PTCL has not been fully elucidated.
We previously analyzed the genomic alterations associated with primary T-cell lymphoma of the thyroid (primary thyroid T-cell lymphoma [PTTL]).17 In that study, we observed that 67% of PTTL cases showed genomic loss at 6q24.2. The minimal common region (MCR) lost in those cases contained STX11 and/or UTRN, suggesting the involvement of these genes in the pathophysiology of PTTL. STX11 is a member of the t-SNAP receptor (t-SNARE) family. A biallelic germline mutation in STX11 is known to cause familial hemophagocytic lymphohistiocytosis type 4 (FHL type 4).18 Although degranulation of cytotoxic T-cells (CTL) and natural killer (NK) cells is impaired by STX11 deficiency, the precise molecular function of STX11 is largely unknown.19–21 The incidence of T-cell lymphomas in patients with FHL type 4 has not been reported.18,22,23 Moreover, Stx11 knockout mice are normal in development and differentiation of T-cells.19,21 UTRN encodes utrophin, a component of cytoskeleton. Because nonsense and frameshift mutations in UTRN have been reported in a small number of cancers, UTRN is regarded as a tumor suppressor gene.24 However, alterations of UTRN have not been described in malignant lymphomas such as PTCL.
In this study, we demonstrated for the first time that STX11 functions as an important tumor suppressor gene in PTCL by using gene expression and functional analyses. In addition, we identified a loss-of-function mutation of STX11 that is associated with T-cell lymphoma.
Materials and Methods
Samples and cell lines
The Institute Review Board of the Aichi Cancer Center approved all the samples and medical records used in our study. Most patients with PTCL-NOS and PTTL were treated with anthracycline-based chemotherapy, as previously reported.6,17 Clinicopathological findings were reexamined from the results of previous our studies.6,25 CD4-positive cells were used as controls in this study and were purified as previously reported.26
Six T-cell neoplasm cell lines (ST1, KOB, Su9T01, KOB, Hut102, Hut78 and Jurkat) were used in this study. In addition to the six T-cell lines, an NK cell line (NKL), four B-cell lines (Reh, SUDHL6, Raji and Jeko1), a myeloid cell line (K562) and three epithelial cell lines (293T, MCF7 and HeLa) were also analyzed. Cell lines were cultured as previously reported.27,28 The cell lines used in the present study are summarized in Supplementary Table S1.
Array comparative genomic hybridization analysis
Focusing on chromosome 6q loss, we reexamined our previous comparative genomic hybridization (CGH) data on 430 cases of non-Hodgkin’s lymphoma. These data comprised six PTTL,17 51 PTCL-NOS,6 62 adult T-cell leukemia/lymphoma (ATL; 35 and 27 cases of the acute- and chronic-type cases, respectively),16 35 NK-cell lymphoma,27 118 diffuse large B-cell lymphoma (DLBCL),29,30 80 follicular lymphoma,31 26 Burkitt lymphoma32 21 mantle cell lymphoma33 and 31 mucosa-associated lymphoid tissue lymphoma34 cases. In addition, we evaluated the genomic loss of 6q using previous results of the genomic alterations in 39 angioimmunoblastic T-cell lymphoma (AITL)35 and 74 anaplastic large cell lymphoma (ALCL) cases.36 Platforms of array CGH used in these analyses and the accession numbers for the database are described in Supplementary Table S2.
Gene expression analysis
We evaluated the expression levels of STX11 and UTRN using published data (GSE6338 and GSE19069).5,37 Using quantitative real-time RT-PCR, we measured the gene expression levels of STX11 in 29 cases of PTCL-NOS, four cases of PTTL, and six T-cell lines, for which adequate RNA was available.
Mutation analysis of STX11
The coding region of STX11 was amplified from genomic DNA and cDNA by using PCR. The PCR primers used are detailed in a previous study.18
Western blot analysis
Western blot analyses were performed using a rabbit polyclonal antibody specific to STX11 (1:1000, HPA007992; Sigma-Aldrich, St. Louis, MO, USA), a mouse monoclonal antibody specific to Actin (1:1000, AC-40; Sigma-Aldrich) and a mouse monoclonal antibody specific to FLAG M2 (1:1000; Sigma-Aldrich).
Gene transduction, and cell proliferation, apoptosis and cell-cycle assays
We used the Retro-X Tet-Off Advanced Inducible Expression System (Clontech, Palo Alto, CA, USA).
Cell proliferation, apoptosis and cell-cycle assays were performed on the stable Tet-OFF cell lines generated for each gene. To induce the target gene, doxycycline (DOX) was removed using three washes (day 0), according to the manufacturer’s protocol. All of these experiments were performed in triplicate. Detailed methods are described in previous studies.27
Statistical analyses
All the statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University), a modified version of R commander software.38 Detailed methods can be found in Supplementary Data S1.
Results
Recurrent genomic loss of 6q24.2 found in subtypes of non-Hodgkin’s lymphoma
We investigated the genomic loss frequency of 6q24 in various subtypes of non-Hodgkin lymphoma. Heterozygous loss of 6q24 was observed in 6% (n = 232) of PTCL cases (Table1), whereas homozygous loss of this region was not observed in our analysis. The loss of 6q24 occurred most frequently in the TTCL cases (67%, n = 6). This genomic deficiency was also frequently observed in DLBCL and NK-cell lymphomas (24% and 20%, respectively), and the genes located near the 6q24 region are implicated in the pathophysiology of these lymphoma subtypes. TNFAIP3 and PRDM1 are implicated in DLBCL,39–41 and FOXO3 and PRDM1 are thought to be involved in NK-cell lymphoma.27,42 However, the genes involved in the pathophysiology of PTCL, such as PTCL-NOS, have not been identified in 6q24 locus. Therefore, we focused on identifying the genes located in 6q24 that are responsible for PTCL.
Table 1.
Frequency of STX11 loss in non-Hodgkin’s lymphoma
| Histological subgroup | n (%) |
|---|---|
| PTTL | 4/6 (67) |
| PTCL-NOS | 4/51 (8) |
| ATL | 3/62 (5) |
| Acute type | 3/35 (9) |
| Chronic type | 0/27 (0) |
| AITL† | 0/39 (0) |
| ALCL‡ | 3/74 (3) |
| PTCL | 14/232 (6) |
| NK-cell lymphoma | 7/35 (20) |
| DLBCL | 28/118 (24) |
| Mantle cell lymphoma | 3/21 (14) |
| Follicular lymphoma | 8/80 (10) |
| Burkitt lymphoma | 2/26 (8) |
| MALT lymphoma | 1/31 (3) |
| Total | 63/543 (12) |
This was determined using the published result reported by Thorns et al.35 ‡This was determined using the published result reported by Salaverria et al.36 AITL, angioimmunoblastic lymphoma; ALCL, anaplastic large cell lymphoma; ATL, adult T-cell leukemia/lymphoma; DLBCL, diffuse large B-cell lymphoma; MALT lymphoma, mucosa-associated lymphoid tissue lymphoma; PTCL, peripheral T-cell lymohomas; PTCL-NOS, peripheral T-cell lymphoma, not otherwise specified; PTTL, primary thyroid T-cell lymphoma.
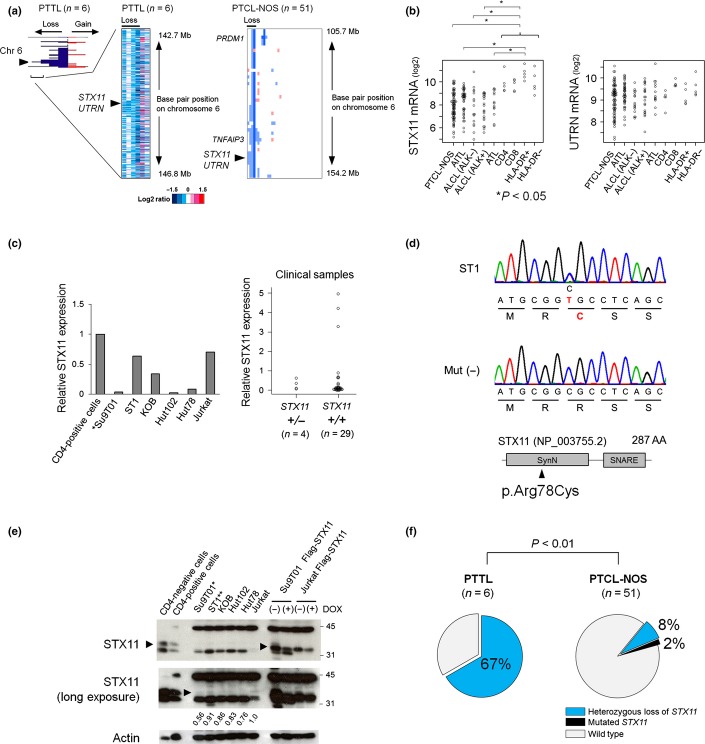
The genes that are contained within the MCR are most likely the candidate genes in the altered regions. Among the 6q24 losses observed in analyzed PTCL cases, the narrowest one was observed in a case of PTTL (Fig.1a).17 The MCR contained only two known coding genes, STX11 and UTRN. Case 39 of PTCL-NOS showed approximately 4.8-Mb loss at 6q24.1–6q24.3, which was the second narrowest loss in our analysis (Fig.1a and Suppl. Table S3). This loss involved 12 known genes, including STX11 and UTRN, but not TNFAIP3, FOXO3 and PRDM1 (Fig.1a). Thus, we regarded STX11 and UTRN as candidate genes contained within the 6q24 region that are predicted to be involved in pathogenesis of PTCL. Genomic losses of 6q24 that included STX11 and UTRN were observed in 8% (n = 51) of PTCL-NOS cases and a T-cell line (Su9T01) (Table1 and Suppl. Fig. S1a).
Figure 1.
Status of STX11 in primary thyroid T-cell lymphoma (PTTL) and peripheral T-cell lymphomas (PTCL)-NOS genomes. (a) The horizontal axis indicates the frequency of chromosome 6 alterations in PTTL cases (left). Red and blue areas represent genomic gains and losses, respectively. Heat map analyses showing log2 ratios of PTTL (middle) and PTCL-NOS (right) tumor cells relative to normal controls. White, blue and red represent genomic diploids, losses and gains, respectively. Arrowheads indicate the STX11/UTRN locus. Data of PTTL were modified from our previous study.17 (b) The expression levels of STX11 and UTRN, genes contained within the minimal common region (MCR), were determined using published data (GSE6338 and GSE19069). The data included PTCL-NOS (n = 78), angioimmunoblastic T-cell lymphoma (AITL; n = 43), anaplastic lymphoma kinase (ALK)-negative anaplastic large cell lymphoma (ALCL, n = 16), ALK-positive ALCL (n = 20) and adult T-cell leukemia/lymphoma cases (ATL; n = 13), as well as CD4-positive T-cells (n = 5), CD8-positive T-cells (n = 5), HLA-DR-positive T-cells (n = 5) and HLA-DR-negative T-cells (n = 5). Significant differences were observed in each pair (*P < 0.05, one way-analysis of variance, Tukey’s correlation). (c) STX11 expression levels in PTCL were normalized to β-actin and compared to that of healthy donor CD4-positive cells. Relative expression is shown. (Left) STX11 expression in T-cell lines was lower than in normal CD4-positive T-cells. Genomic loss of STX11 is indicated (*). (Right) Four PTCL-NOS and PTTL cases carrying genomic alterations of STX11 and 29 cases without STX11 alterations were analyzed. (d) DNA sequencing chromatogram of a T-cell line (ST1) showing a missense mutation in STX11 (p. Arg78Cys, c.232C>T; upper panel). A representative sample that did not carry the mutation is shown in the middle panel. The lower panel shows a schematic representation of the STX11 protein, depicting the location of the syntaxin N-terminus (SynN) and SNAP receptor (SNARE) domains. The arrowhead indicates the position of the mutation. (e) Western blot analysis of STX11 and β-Actin in normal CD4-positive and CD4-negative cells, and six T-cell lines. To detect STX11, we introduced Flag-STX11 into Jurkat and Su9T01 cells using the Tet-OFF system. Western blots of induced STX11 probed with anti-STX11 antibody are shown in the four lanes to the right. In this system, STX11 is expressed in response to doxycycline (DOX) removal (see also Suppl. Fig. S1c). The arrowheads mark STX11 protein signals. The middle panel shows a longer exposure of the same membrane. Signals were converted to a numerical value by using the ImageJ software. The ratio of STX11 to actin expression was calculated and displayed below each lane. The STX11/actin ratio in Jurkat cells is used as a control. Heterozygous genomic loss and heterozygous missense mutation of STX11 are indicated by * and **, respectively. (f) Genomic loss of the STX11 region was observed in 67% of PTTL cases (n = 6), whereas 10% of PTCL-NOS cases (n = 51) showed genomic losses and mutations of STX11 (P < 0.01, Fisher’s exact test). Synonymous mutations were excluded from the analysis.
Selection of the candidate gene located in minimal common region of 6q24.2
Differences in STX11 and UTRN expression levels in PTCL compared to normal T-cells were analyzed using previously published gene expression profiling data (Fig.1b). Notably, STX11 expression was significantly lower in PTCL than in the normal T-cells, but UTRN expression did not change. These data are consistent with that of our previous study reporting the reduction of STX11, but not UTRN expression, in PTTL cases missing 6q24.2.17
To accurately assess STX11 expression in PTCL-NOS and PTTL cases and T-cell lines, we performed quantitative RT-PCR. STX11 expression was lower in T-cell lymphomas and in all cases with 6q24 loss than in the normal T-cells (Fig.1c and Suppl. Table S4). The genes whose expression was affected by copy number changes were considered candidate genes in the regions of genomic alteration. Therefore, we regarded STX11 as the most likely candidate gene located in 6q24.
Mutation and protein blot analyses of STX11
Mutation analysis was performed for PTCL-NOS and PTTL cases, and for T-cell lines to further evaluate the genomic alteration of STX11. This sequence analysis revealed a missense mutation in one T-cell line (ST1); the arginine at position 78 was changed to cysteine (p.Arg78Cys, c.232C>T; Fig.1d). In addition, a PTCL-NOS case (Case 9) had a one-nucleotide substitution in the region before ATG (c.-19C>G; Suppl. Fig. S1b). Synonymous mutations were observed in a T-cell line (Jurkat, c.462C>T) and a PTCL-NOS case (Case 48, c.570C>T). The latter mutation was registered as a single-nucleotide polymorphism in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/snp/, rs148354227).
Western blots performed using anti-STX11 antibodies indicated that STX11 expression was significantly lower in T-cell lines than in the normal CD4-positive and CD4-negative cells (Fig.1e and Suppl. Fig. S1c). Longer exposure of the membranes allowed us to evaluate STX11 expression in the T-cell lines. In Su9T01 cells that have a genomic loss of STX11, STX11 expression was reduced, unlike that in the other T-cell lines that have not lost STX11.
In total, 17% (11/63; 51 PTCL-NOS cases, six PTTL cases, and six T-cell lines) of the analyzed cases had genomic loss and mutation of STX11. Notably, genomic alterations of STX11 were significantly more prevalent in PTTL (4/6; 67%) than in PTCL-NOS (5/51; 10%) (P < 0.01; Fig.1f). No apparent correlations were observed between the distribution of STX11 alterations and the alterations that were frequently observed in PTCL-NOS cases (Suppl. Fig. S1d). A RHOA mutation (p.Gly17Val) is reportedly observed in PTCL.12–14 Using Sanger sequencing, we analyzed this mutation in the cases with STX11 alterations, and no mutations were observed (data not shown).
Functional analyses of STX11
We speculated that STX11 plays a key role as a tumor suppressor gene in PTCL because genetic alteration of STX11 and its reduced expression were detected in PTCL samples. Therefore, we introduced STX11 into T-cell lines by using an established Tet-OFF system to investigate its function.27 Because PTTL and PTCL-NOS cell lines were not available, we used ATL and T-cell lineage acute lymphoblastic leukemia cell lines.
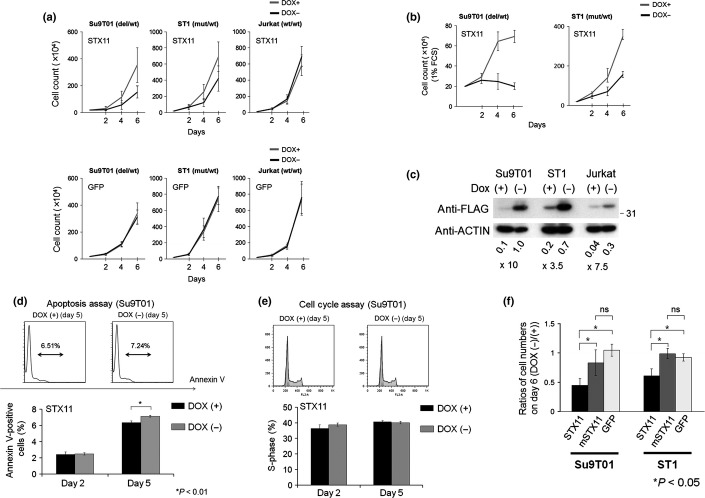
In T-cell lines with genomic alterations of STX11 (Su9T01 and ST1), induction of STX11 suppressed proliferation (Fig.2a). Induced STX11 expression had no effect on Jurkat cells that did not carry an alteration of STX11. Although proliferation of Su9T01 and ST1 cell lines was suppressed by the expression of STX11 under standard serum conditions (10% FCS), this suppression was more apparent under low-serum conditions (1% FCS; Fig.2b). As shown in Figure2c, STX11 was successfully induced in each cell line by the removal of DOX. To further characterize the suppressive effect of STX11, we performed apoptosis and cell cycle assays in Su9T01 cells (Fig.2d,e). Expression of STX11 induced cellular apoptosis in the cell line (Figs.2d and Suppl. Fig. S2a), although the number of apoptotic cells induced by STX11 was relatively small.
Figure 2.
Functional characteristics of STX11 in T-cell lymphomas. (a) The effect of reestablishing STX11 expression in Su9T01 (STX11-deficient), ST1 (STX11 mutant) and Jurkat cells (wild-type STX11). The horizontal axis indicates the time elapsed after doxycycline (DOX) removal. The vertical axis indicates the average cell numbers with standard deviation. Cell numbers were counted using the trypan blue exclusion assay on days 2, 4 and 6 post-DOX removal. Experiments were performed in triplicate. The upper and lower panels show the result of reestablishing STX11 expression and GFP expression, respectively. (b) Cell proliferation analysis of ST1 and Su9T01 cells under low-serum conditions (1% FCS). (c) Using anti-FLAG and anti-β-actin antibodies, the induction efficiency of Flag-STX11 was evaluated in the cells. Signals were converted to a numerical value by using the ImageJ software. The expression ratio relative to actin was calculated and is shown below each lane. (d) Annexin V-positive STX11-induced Su9T01 cells were quantified using flow cytometry on days 2 and 5, following the removal of DOX. Experiments were performed in triplicate, and averages with standard deviations are shown. Significant differences were observed between DOX (−) and DOX (+) samples (*P < 0.01, t-test). (e) Cell cycle assays were conducted by staining cells with propidium iodide (PI) on days 2 and 5. Induction of STX11 did not alter the cell cycle of Su9T01 cells. (f) Arg78Cys mutant STX11 (mSTX11) was introduced into Su9T01 and ST1 cells. The ratio of proliferating cells in the absence versus presence of DOX (DOX [−]/DOX [+]) on day 6 is shown on the vertical axis. Experiments were performed in triplicate. Standard deviations and average values are shown. Significant differences were observed in each pair (*P < 0.05, one-way analysis of variance with Bonferroni adjustment). ns, no significance.
The STX11 Arg78Cys mutant allele we identified in a T-cell line (ST1) by using capillary sequencing (Fig.1d and Suppl. Fig. S2b) was also transduced into the ST1 and Su9T01 cells. Induction of STX11 Arg78Cys did not have a suppressive effect on these cell lines (Fig.2f and Suppl. S2c). These results indicate that STX11 Arg78Cys is a loss-of-function mutation.
Suppressive effect of STX11 induction only in T-cell lines
To determine whether the suppressive effect is specific to PTCL with STX11 alteration, we next introduced STX11 and GFP into cell lines of various lineages by using a Tet-Off system and performed cell proliferation assays. Forced STX11 expression suppressed the proliferation of ST1 and Su9T01 cells, but did not affect the other cell lines analyzed in this study (Fig.3a and Suppl. Fig. S3). Although Jeko1 and NKL also had genomic losses of STX11, induction of STX11 did not have a suppressive effect on these cell lines.
Figure 3.
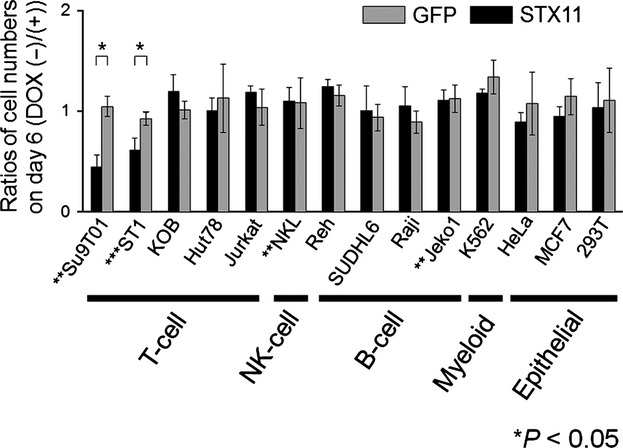
Reestablished STX11 expression in cells of various lineages. STX11 or GFP was transduced into 14 cell lines using the Tet-OFF system. The average number of proliferating cells expressed as a ratio (DOX [−]/DOX [+]) on day 6; the standard deviation is shown. Experiments were performed in triplicate. Significant differences were observed in each pair (*P < 0.05, t-test). Genomic loss and mutation of STX11 are indicated by ** and ***, respectively.
Clinicopathological findings of STX11 alteration
Previously, we showed that PTTL is a distinct entity from PTCL-NOS.17 Therefore, we separately analyzed the clinicopathological characteristics of STX11 alteration in the PTTL and PTCL-NOS cases (Tables2 and 3 and Suppl. Tables S5 and S6). Among PTCL-NOS cases, patients with STX11 alteration tended to have poorer prognoses than those without the alteration (P = 0.07). In addition, STX11-altered PTCL-NOS cases were characterized by the presence of hemophagocytic syndrome at diagnosis (67% vs 8%, P = 0.04). STX11 alteration was frequently observed in PTTL characterized by past histories of autoimmune diseases and extra-nodal lesions.17 However, no significant differences in these findings were observed among PTCL-NOS cases based on STX11 alteration. Morphologically, STX11-altered PTCL-NOS cases were associated with anaplastic nuclei compared with the STX11-wild type cases (60% vs 9%, P = 0.016), although the numbers of the analyzed cases were small. No significant differences were seen in other immunohistochemical findings. In PTTL cases, there were no differences in clinicopathological findings based on STX11 alteration.
Table 2.
Clinical characteristics of PTCL-NOS with STX11 alteration
| Case† | Age (years) | Sex | CS | B-symptom | Extra-nodal lesion | LDH >ULN | Auto-immune diseases | Hemophagocytic syndrome | OS (months) | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Case 9 | 4 | F | IVS | + | + (Liver) | 0 | 0 | + | 15 | Death |
| Case 31 | 3 | M | NA | NA | NA | NA | NA | NA | 3 | Death |
| Case 39 | 48 | M | NA | NA | NA | NA | NA | NA | 11 | Death |
| Case 42 | 80 | F | IV | + | + (Soft tissue) | 0 | 0 | + | 29 | Survival |
| Case 46 | 56 | M | III | + | − | 0 | 0 | 0 | 1 | Death |
Representations in previous Nakagawa et al. paper were used.6 CS, clinical stage; F, female; LDH, lactate dehydrogenase; M, male; NA, not available; OS, overall survival; ULN, upper limit of normal.
Table 3.
Pathological characteristics of PTCL-NOS with STX11 alteration
| Case* | Nuclear size* | Capillary proliferation* | Eosinophils/plasma cells proliferation* | Lymphoepitheloid cells proliferation* | CD3 | CD4 | CD8 | TIA-1 | CCR4* | CCR3* | CXCR3* | EBER | Ki-67 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 9 | Anaplastic | − | − | − | + | + | − | − | − | + | − | + | 20 |
| Case 31 | Large | − | − | − | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Case 39 | Anaplastic | − | − | − | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Case 42 | Pleomorphic | − | − | − | + | + | − | − | + | − | − | − | 30 |
| Case 46 | Anaplastic | − | − | − | + | + | − | − | − | − | + | − | 80 |
“−” means negative, and “+” means positive. NA, not available.
Representations in previous Nakagawa et al.(6) paper were used.
Discussion
Genomic alterations of STX11 in T-cell lymphomas
In the present study, we examined the MCR of 6q24 loss and showed that STX11 plays an important role in PTCL. Genomic alteration of STX11 in malignancies had not been previously reported. Therefore, our study is the first to investigate STX11 in this context. The loss of 6q occurred frequently in DLBCL and NK-cell lymphomas, but our functional analysis indicated that STX11 does not act as a tumor suppressor gene in these types of lymphomas (Fig.3).
Interestingly, overexpression of STX11 had suppressive effects on T-cell lines, but only on those with STX11 alteration. These results indicate that the genetic events of STX11 are involved in the pathophysiology of PTCL. STX11 mRNA and protein levels were reduced in PTCL with or without STX11 alterations compared with normal cells. These results suggested that epigenetic events modify the expression of STX11. Because of the limited availability of samples, we could not perform epigenetic analyses for STX11. However, this should be addressed in future studies.
Haploinsufficiency of STX11 in T-cell lymphoma
Syntaxin 11, encoded by STX11, is a t-SNARE protein that plays a role in binding vesicles to cell membranes.43 Homozygous loss and mutation of STX11 in the germline causes FHL type 4.18 The genomic alterations of STX11 in PTCL were all heterozygous, suggesting that PTCL is associated with STX11 haploinsufficiency. The loss-of-function STX11 mutation (p.Arg78Cys, c.232C>T) identified in this study was not observed in FHL type 4 patients. Therefore, the role of STX11 alteration in PTCL might differ from that in FHL type 4. We speculate that the STX11 Arg78Cys mutant will provide further insight into the pathobiology of STX11 in PTCL and will reveal novel roles of STX11 in normal T-cells.
Contribution of STX11 alteration to peripheral T-cell lymphoma pathophysiology
Until now, T-cell lymphoma has not been found to occur in Stx11 knockout mice or in patients with FHL type 4. Therefore, we surmised that STX11 alteration alone does not cause lymphoma. In fact, we found that the cases with STX11 alterations also had genomic alterations that were typically found in PTCL-NOS samples (Suppl. Fig. S1d).
We also found that PTCL-NOS cases with STX11 alterations tended to have poor prognoses, show a hemophagocytosis, and possess anaplastic nuclei (Tables2 and 3). A previous study reported that expression of cytotoxic molecules tended to be associated with hemophagocytosis in PTCL-NOS,44 but STX11-altered PTCL-NOS with hemophagocytosis lacked an expression of cytotoxic molecules. It is speculated that additional genomic alteration(s) are required for the pathophysiology of PTCL having STX11 alteration. To reveal such genes that act synergistically with STX11, future studies using the appropriate T-cell lymphoma model mice are needed.45
In conclusion, we identified that STX11 loss occurred in a portion of PTCL cases. STX11 mutations, including a loss-of function mutation, were also observed in PTCL. Overexpression of STX11 only suppressed proliferation of T-cell lines with STX11 alterations, indicating that STX11 acts as a T-cell lineage-specific tumor suppressor gene. We believe that these findings provide a novel approach to understand the molecular mechanisms involved in PTCL pathogenesis.
Acknowledgments
This work was supported in part by Grants-in-Aid from the Ministry of Health, Labor, and Welfare of Japan; the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Society for the Promotion of Science; and the Takeda Science Foundation. The authors thank Drs Tohru Izumi, Daisuke Niino, Kotaro Arita and Tatsuo Kakiuchi for their discussions and encouragement throughout this study. We also thank Yumiko Kasugai, Seiko Sato and Kyoko Hirano for their technical assistance.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1. Genetic alterations of STX11, related to Figure 1.
Fig. S2. Functional analysis for STX11 in cell lines, related to Figure 2.
Fig. S3. Western blot analysis of STX11 in cell lines belonging to various cell lineages, related to Figure 3.
Table S1. The characteristics of each cell line.
Table S2. Platforms of array CGH in each histological subgroup.
Table S3. Genetic lesion of STX11 in T-cell lymphomas.
Table S4. Characteristics of patients and cell lines analyzed in this study.
Table S5. Clinicopathological characteristics of PTCL-NOS cases according to STX11 status.
Table S6. Clinicopathological characteristics of PTTL cases according to STX11 status.
Data S1. Supplementary methods.
References
- Piccaluga PP, Tabanelli V, Pileri SA. Molecular genetics of peripheral T-cell lymphomas. Int J Hematol. 2014;99:219–26. doi: 10.1007/s12185-014-1522-1. [DOI] [PubMed] [Google Scholar]
- Chihara D, Ito H, Matsuda T, et al. Differences in incidence and trends of haematological malignancies in Japan and the United States. Br J Haematol. 2014;164:536–45. doi: 10.1111/bjh.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edn. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- Pileri SA, Piccaluga PP. New molecular insights into peripheral T cell lymphomas. J Clin Invest. 2012;122:3448–55. doi: 10.1172/JCI61205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccaluga PP, Agostinelli C, Califano A, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117:823–34. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Nakagawa-Oshiro A, Karnan S, et al. Array comparative genomic hybridization analysis of PTCL-U reveals a distinct subgroup with genetic alterations similar to lymphoma-type adult T-cell leukemia/lymphoma. Clin Cancer Res. 2009;15:30–8. doi: 10.1158/1078-0432.CCR-08-1808. [DOI] [PubMed] [Google Scholar]
- Braun FC, Grabarczyk P, Mobs M, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sezary syndrome. Leukemia. 2011;25:1494–501. doi: 10.1038/leu.2011.101. [DOI] [PubMed] [Google Scholar]
- Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–13. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boi M, Rinaldi A, Kwee I, et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. 2013;122:2683–93. doi: 10.1182/blood-2013-04-497933. [DOI] [PubMed] [Google Scholar]
- Odejide O, Weigert O, Lane AA, et al. A targeted mutational landscape of angioimmunoblastic T cell lymphoma. Blood. 2014;123:1293–6. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odqvist L, Sanchez-Beato M, Montes-Moreno S, et al. NIK controls classical and alternative NF-kappaB activation and is necessary for the survival of human T-cell lymphoma cells. Clin Cancer Res. 2013;19:2319–30. doi: 10.1158/1078-0432.CCR-12-3151. [DOI] [PubMed] [Google Scholar]
- Palomero T, Couronne L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–70. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Sung MK, Lee SH, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–5. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- Vaque JP, Gomez-Lopez G, Monsalvez V, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014;123:2034–43. doi: 10.1182/blood-2013-05-504308. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Karube K, Utsunomiya A, et al. Molecular characterization of chronic-type adult T-cell leukemia/lymphoma. Cancer Res. 2014;74:6129–38. doi: 10.1158/0008-5472.CAN-14-0643. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Nishikori M, Izumi T, et al. Primary peripheral T-cell lymphoma, not otherwise specified of the thyroid with autoimmune thyroiditis. Br J Haematol. 2013;161:214–23. doi: 10.1111/bjh.12255. [DOI] [PubMed] [Google Scholar]
- zur Stadt U, Schmidt S, Kasper B, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet. 2005;14:827–34. doi: 10.1093/hmg/ddi076. [DOI] [PubMed] [Google Scholar]
- Sepulveda FE, Debeurme F, Menasche G, et al. Distinct severity of HLH in both human and murine mutants with complete loss of cytotoxic effector PRF1, RAB27A, and STX11. Blood. 2013;121:595–603. doi: 10.1182/blood-2012-07-440339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson YT, Rudd E, Zheng C, et al. Defective cytotoxic lymphocyte degranulation in syntaxin-11 deficient familial hemophagocytic lymphohistiocytosis 4 (FHL4) patients. Blood. 2007;110:1906–15. doi: 10.1182/blood-2007-02-074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogl T, Muller J, Jessen B, et al. Hemophagocytic lymphohistiocytosis in syntaxin-11-deficient mice: T-cell exhaustion limits fatal disease. Blood. 2013;121:604–13. doi: 10.1182/blood-2012-07-441139. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ishii E, Horiuchi H, et al. Mutations of syntaxin 11 and SNAP23 genes as causes of familial hemophagocytic lymphohistiocytosis were not found in Japanese people. J Hum Genet. 2005;50:600–3. doi: 10.1007/s10038-005-0293-1. [DOI] [PubMed] [Google Scholar]
- Rudd E, Goransdotter Ericson K, Zheng C, et al. Spectrum and clinical implications of syntaxin 11 gene mutations in familial haemophagocytic lymphohistiocytosis: association with disease-free remissions and haematopoietic malignancies. J Med Genet. 2006;43:e14. doi: 10.1136/jmg.2005.035253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang J, Zhao YL, et al. UTRN on chromosome 6q24 is mutated in multiple tumors. Oncogene. 2007;26:6220–8. doi: 10.1038/sj.onc.1210432. [DOI] [PubMed] [Google Scholar]
- Vose J, Armitage J, Weisenburger D International TCLP. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–30. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- Umino A, Nakagawa M, Utsunomiya A, et al. Clonal evolution of adult T-cell leukemia/lymphoma takes place in the lymph nodes. Blood. 2011;117:5473–8. doi: 10.1182/blood-2010-12-327791. [DOI] [PubMed] [Google Scholar]
- Karube K, Nakagawa M, Tsuzuki S, et al. Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood. 2011;118:3195–204. doi: 10.1182/blood-2011-04-346890. [DOI] [PubMed] [Google Scholar]
- Karube K, Tsuzuki S, Yoshida N, et al. Lineage-specific growth inhibition of NK cell lines by FOXO3 in association with Akt activation status. Exp Hematol. 2012;40:1005–15. doi: 10.1016/j.exphem.2012.08.005. e6. [DOI] [PubMed] [Google Scholar]
- Zhang X, Karnan S, Tagawa H, et al. Comparison of genetic aberrations in CD10+ diffused large B-cell lymphoma and follicular lymphoma by comparative genomic hybridization and tissue-fluorescence in situ hybridization. Cancer Sci. 2004;95:809–14. doi: 10.1111/j.1349-7006.2004.tb02186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa H, Suguro M, Tsuzuki S, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770–7. doi: 10.1182/blood-2005-02-0542. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Karube K, Guo Y, et al. Trisomy 3 is a specific genomic aberration of t(14;18) negative follicular lymphoma. Leukemia. 2007;21:2549–51. doi: 10.1038/sj.leu.2404817. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 2007;98:1482–90. doi: 10.1111/j.1349-7006.2007.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Yoshida N, Suguro M, et al. Clonal heterogeneity of mantle cell lymphoma revealed by array comparative genomic hybridization. Eur J Haematol. 2013;90:51–8. doi: 10.1111/ejh.12030. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Nakamura T, Nakagawa M, et al. Chromosomal imbalances are associated with outcome of Helicobacter pylori eradication in t(11;18)(q21;q21) negative gastric mucosa-associated lymphoid tissue lymphomas. Genes Chromosom Cancer. 2007;46:784–90. doi: 10.1002/gcc.20464. [DOI] [PubMed] [Google Scholar]
- Thorns C, Bastian B, Pinkel D, et al. Chromosomal aberrations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma unspecified: a matrix-based CGH approach. Genes Chromosom Cancer. 2007;46:37–44. doi: 10.1002/gcc.20386. [DOI] [PubMed] [Google Scholar]
- Salaverria I, Bea S, Lopez-Guillermo A, et al. Genomic profiling reveals different genetic aberrations in systemic ALK-positive and ALK-negative anaplastic large cell lymphomas. Br J Haematol. 2008;140:516–26. doi: 10.1111/j.1365-2141.2007.06924.x. [DOI] [PubMed] [Google Scholar]
- Iqbal J, Weisenburger DD, Greiner TC, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115:1026–36. doi: 10.1182/blood-2009-06-227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–8. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–75. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–6. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Mandelbaum J, Bhagat G, Tang H, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–79. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucuk C, Iqbal J, Hu X, et al. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Nat Acad Sci USA. 2011;108:20119–24. doi: 10.1073/pnas.1115128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–53. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Asano N, Suzuki R, Kagami Y, et al. Clinicopathologic and prognostic significance of cytotoxic molecule expression in nodal peripheral T-cell lymphoma, unspecified. Am J Surg Pathol. 2005;29:1284–93. doi: 10.1097/01.pas.0000173238.17331.6b. [DOI] [PubMed] [Google Scholar]
- Arita K, Maeda-Kasugai Y, Ohshima K, et al. Generation of mouse models of lymphoid neoplasm using retroviral gene transduction of in vitro-induced germinal center B and T cells. Exp Hematol. 2013;41:731–41. doi: 10.1016/j.exphem.2013.04.001. e9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Genetic alterations of STX11, related to Figure 1.
Fig. S2. Functional analysis for STX11 in cell lines, related to Figure 2.
Fig. S3. Western blot analysis of STX11 in cell lines belonging to various cell lineages, related to Figure 3.
Table S1. The characteristics of each cell line.
Table S2. Platforms of array CGH in each histological subgroup.
Table S3. Genetic lesion of STX11 in T-cell lymphomas.
Table S4. Characteristics of patients and cell lines analyzed in this study.
Table S5. Clinicopathological characteristics of PTCL-NOS cases according to STX11 status.
Table S6. Clinicopathological characteristics of PTTL cases according to STX11 status.
Data S1. Supplementary methods.