Abstract
In a previous study, we found that ERGIC3 was a novel lung cancer-related gene by screening libraries of differentially expressed genes. In this study, we developed a new murine monoclonal antibody (mAb) against ERGIC3. This avid antibody (6-C4) is well suited for immunohistochemistry, immunoblotting and solid-phase immunoassays. Furthermore, we systematically investigated expressions of ERGIC3 in a broad variety of normal human tissues and various types of tumors by immunohistochemistry. In normal human tissues, 6-C4 reacted only in some epithelial cells, such as hepatocytes, gastrointestinal epithelium, ducts and acini of the pancreas, proximal and distal tubules of the kidney, and mammary epithelial cells; however, most normal human tissues were not stained. Moreover, almost all carcinomas that originated from the epithelial cells were positive for 6-C4, whereas all sarcomas were negative. Notably, 6-C4 strongly stained non-small cell lung cancer (NSCLC) cells but did not react with normal lung tissues. Hence, ERGIC3 mAb could be used in histopathological diagnosis and cytopathological testing to detect early-stage NSCLC. We also studied the mechanisms of ERGIC3 regulation in vitro and in vivo by means of bioinformatics analysis, luciferase reporter assay, miRNA expression profiling and miRNA transfection. Results showed that miR-203a downregulation induced ERGIC3 overexpression in NSCLC cells.
Keywords: Biomarker, ERGIC3, miR-203a, monoclonal antibody, non-small cell lung cancer
Cancer is a major cause of mortality worldwide.1 Lung cancer is the leading cause of cancer-related deaths, accounting for 20% of all cancer deaths.2 Lung cancer is divided into small-cell lung cancer and non-small cell lung cancer (NSCLC), and the latter accounts for more than 80% of all lung cancer types, including adenocarcinoma (AC), squamous cell carcinoma (SCC) and large-cell carcinoma.3 The overall 5-year survival rate of lung cancer has merely improved from 12% to 16% over the recent three decades, as observed in 80% of patients diagnosed with metastatic disease and more than half of patients with distant metastases.4 Early detection and improved monitoring of lung cancer are urgently needed. Sputum cytology and examination of blood biomarkers, including circulating DNA and RNA, exosomal microRNA, circulating tumor cells, and antigens, are potential approaches for the early detection of lung cancer.5 However, although some biomarkers, such as carcinoembryonic antigen (CEA) and cytokeratin 19 fragment antigen 21-1 (CYFRA 21-1), have been used to screen for lung cancer, the specificity and sensitivity remain unsatisfactory.6,7 Therefore, novel tumor biomarkers and tests need to be developed.8 In a previous study, we used NSCLC and normal lung tissues to construct libraries of differentially expressed genes, and found that endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3) is strongly overexpressed in NSCLC, and this overexpression promotes proliferation and migration of NSCLC cells.9
ERGIC3 (Erv46 and ERp43) is located in the cis face of the Golgi apparatus and vesicular tubular structures between the transitional endoplasmic reticulum (ER) and cis-Golgi.10 ERGIC3 significantly affects cell growth and causes ER stress-induced cell death, and is involved in the invasion and metastasis in hepatocellular carcinomas (HCC).11,12 Considering the high sensitivity and specificity of ERGIC3 expression in NSCLC, and the roles of ERGIC3 in cancer development and progression, in embarking on this study we believed that ERGIC3 may be a potential biomarker of NSCLC.13 In this study, we developed a new murine monoclonal antibody (mAb) against ERGIC3, and systematically investigated ERGIC3 expressions in a broad variety of normal human tissues and various types of human tumors. The mechanisms of ERGIC3 regulation were also studied.
Materials and Methods
Cell lines and tissue samples
Lung cancer cell lines, including A549 (AC), 801-D (large cell carcinoma), EPLC-32M1 (SCC), NCI-H292 (mucoepidermoid carcinoma), and XLA-07 (AC),14 were cultured with RPMI 1640 containing 10% FBS. 16HBE (an immortalized human bronchial epithelial cell line) was maintained in DMEM containing 10% FBS.
Normal adult human tissues were obtained from surgical specimens or following autopsies performed <10 h after death. Only specimens maintaining good histological preservation were used. Among these, 23 lung tissue samples obtained from patients with bullous lung disease and inflammatory pseudotumors through surgical operation were used as “normal lung tissues.” A total of 110 resected tumors, including 15 various types, were studied. The tumor types were identified based on microscopic examination according to the international classification.15 All tissue samples were fixed in 10% buffered formalin and embedded in paraffin for immunohistochemistry. Seven cases of fresh NSCLC and adjacent nonmalignant lung tissues were used for isolation of RNA and proteins. The study was approved by the local research ethics committee.
Preparation of mAb
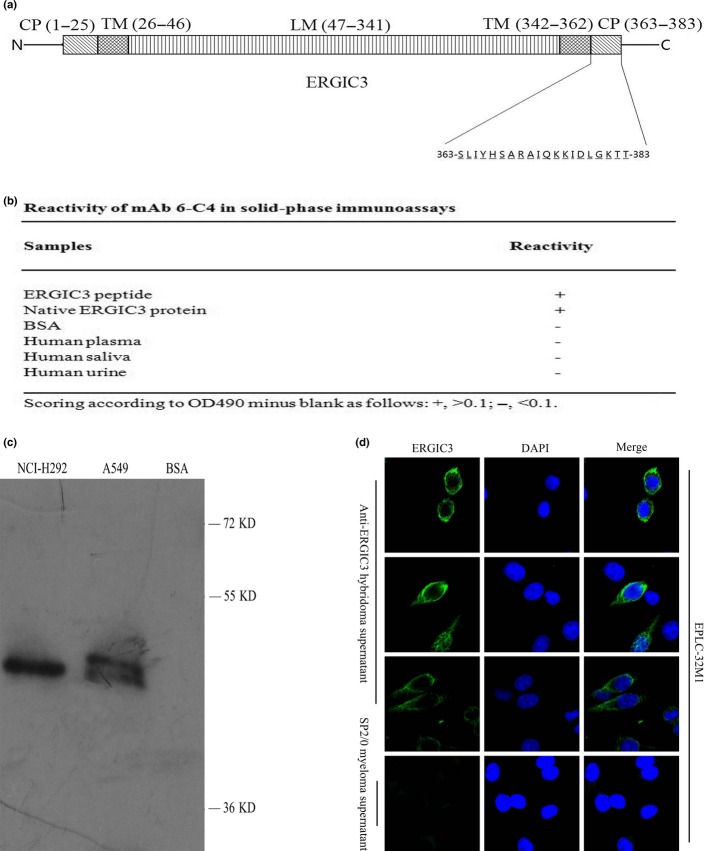
For generation of mAb to ERGIC3, a peptide of ERGIC3 (Gene ID: 51614) coupled to KLH via an N-terminal cysteine was synthesized by GL Biochem, Shanghai, China (Fig.1a). Eight-week-old BALB/c mice were immunized subcutaneously with the ERGIC3 peptide emulsified using complete Freund’s adjuvant (Sigma-Aldrich, St. Louis, MO, USA). A booster injection was given intraperitoneally with the peptide emulsified using incomplete Freund’s adjuvant (Sigma-Aldrich) 3 days before the animals were killed. Titers of anti-serums to ERGIC3 were assayed by solid-phase ELISA. Western blot analysis was used to evaluate whether the anti-serum could recognize the native ERGIC3 protein. When titers of ERGIC3-specific antibodies reached 1:10 000, the immunized mice were killed and the spleen was removed for cell fusion.16 All experiments were carried out according to the regulations for animal experimentation and approved by the appropriate authorities.
Figure 1.
Generation and identification of the monoclonal antibody (mAb) against ERGIC3. (a) The tertiary structure of ERGIC3 protein and the sequence of the synthetic peptide used as antigen. (b) The reactivity of 6-C4 in solid-phase immunoassays. (c) 6-C4 recognized the protein extracted from cultured cells at a band of approximately 50 kD in immunoblot. (d) Immunofluorescence staining of 6-C4 around the Golgi apparatus and the endoplasmic reticulum (ER) of non-small cell lung cancer (NSCLC) cells. SP2/0 cell supernatant was used as a negative control. CP, cytoplasmic domain; LM, luminal domain; TM, transmembrane domain.
The spleen cells were fused with SP2/0 myeloma using 50% polyethylene glycol solution, and then cultivated in hypoxanthine, aminopterin and thymidine medium. The candidate hybridomas that secreted the antibody with high titer and desired specificity were screened and identified by ELISA. To ensure that the mAb was secreted by the progeny of a single cell, sub-cloning was performed by limiting dilution.
ELISA assay
Antigens were coated on microplates. The plates were blocked with 2% BSA, and then incubated with antibodies. Subsequently, peroxidase-labeled anti-mouse antibody was added, followed by color reaction developed with o-phenylenediamine dihydrochloride. After the reaction was stopped with 2.5 M sulfuric acid, the optical density (OD) was determined using a microplate reader (BioRad, Hercules, CA, USA) at 490 nm.
Western blot
Total protein was prepared from cultured cells and tissues through protein lysis. The lysate was centrifuged. Supernatants were boiled in sodium dodecyl sulfate-polyacrylamide gel reducing buffer, separated by electrophoresis, and then transferred to polyvinylidene difluoride transfer membrane. After blocking with 2% BSA, the membrane was incubated with antibodies, and then treated with peroxidase-conjugated anti-mouse antibody, followed by the addition of chemiluminescence reagents (Thermo Fisher Scientific, Waltham, MA, USA). GAPDH was used as an internal control.
Immunofluorescence staining
Cells were seeded on coverslips. The slides were fixed with ice-cold acetone and the permeations were performed using digitonin for 10 min. The cells were blocked with 2% BSA and incubated with antibodies. Subsequently, the slides were incubated with FITC-conjugated goat anti-mouse antibody. The nuclei were counterstained with 4,6-diamidino-2-phenylindole. Finally, the slides were analyzed under confocal laser scanning microscopy.
Immunohistochemistry
The paraffin sections were deparaffinized, and the antigen retrieval was performed using 10 mM sodium citrate buffer through pressure cooker processing for 5 min. Endogenous peroxidase activity was eliminated with 3% H2O2. The sections were blocked using 2% BSA, incubated with antibodies, and then treated with peroxidase-labeled goat anti-mouse antibody. The color development was performed with 3,3′-diaminobenzidine. Hematoxylin was used for counterstaining. Negative controls were performed using SP2/0 cell supernatant instead of the antibody.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from the cultured cells and tissues using TRIzol (Invitrogen, Carlsbad, CA, USA). The cDNA was synthesized with M-MLV Reverse Transcriptase (Promega, Madison, WI, USA) using RNA samples as the template. The quantitative RT-PCR (qRT-PCR) assay was performed for ERGIC3 and internal control (β-actin) as described previously.9 All samples were run in triplicate to ensure quantitative accuracy, and the threshold cycle numbers (Ct) were averaged. Upregulation or downregulation of target genes was determined using the 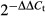 method.17
method.17
For examination of miRNA, total RNA were used in 5× miScript HiSpec Buffer (QIAGEN, Suzhou, China) according to the manufacturer’s protocol. The cDNA was prepared using the miScript II RT Kit (QIAGEN). The qRT-PCR assay was performed using a target-specific primer and the miScript SYBR Green PCR Kit (QIAGEN), which contains the miScript Universal Primer (reverse primer) and QuantiTect SYBR Green PCR Master Mix. The U6 was used as an internal control. The Ct of target miRNA were determined as described above.
All primers used in the study are listed in Supplementary Table S1.
Bioinformatics prediction
The miRNA that may target ERGIC3 were predicted through bioinformatics analysis using algorithms, RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) and miRecords (http://mirecords.biolead.org/prediction_query.php).
MiRNA expression profiling
MiRNA expression profiling was determined in cultured cells through miRNA sequencing using Hiseq 2000 platform by BGI Tech (Shenzhen, China: http://bgitechsolutions.com). Differentially expressed miRNA were found through comparing miRNA expression profiling of NSCLC cells (A549, 801D, EPLC-32M1) with that of 16HBE.
Transient transfection of miRNA
Cultured cells were seeded in 12-well plates at a density of 30–50%, and then transfected, respectively, with 150 nM has-miRNA mimics or inhibitors, as well as the same amount of mimic or inhibitor controls (RiboBio, Guangzhou, China) using lipofectamine 2000 (Invitrogen) according to the instruction manual.
Luciferase reporter assay
The predicted target site of miR-203a was amplified from the genomic DNA of EPLC-32M1 cells (primers in Suppl. Table S1) and was inserted into the KpnI and HindIII restriction sites of the pGL-3 basic vector (Promega). The cells were co-transfected with the pGL-3 luciferase construct and Renilla luciferase plasmid (pRL-TK) along with the miR-203a mimic or its negative control using Lipofectamine 2000. The pRL-TK plasmid was used as an internal control. After transfection, the luciferase activities were measured using Dual-Luciferase Reporter System (Promega) according to the manufacturer’s instructions.
Cell viability assay
Cell viability and proliferation was analyzed by using WST-1 assay. At 48, 72 and 96 h after the treatments, the WST-1 reagent (Roche Molecular Biochemicals, Rotkreuz, Switzerland) was added and incubated for 2–3 h at 37°C. The absorbance of converted dye was measured at 490 nm by microplate reader (BioRad).
Statistical analysis
Data of mRNA and protein levels, as well as cellular proliferation were analyzed using the paired t-test. All of the values were evaluated using IBM SPSS 19 (SPSS, Chicago, IL, USA). Differences were considered significant if P < 0.05.
Results
A new monoclonal antibody to ERGIC3 was developed
After cell fusion was performed, six strongly positive clones were obtained (Suppl. Fig. S1a). A monoclonal hybridoma was established from 06-C4 after three rounds of sub-cloning (Suppl. Fig. S1b). The mAb secreted by the monoclonal hybridoma was named “6-C4.” The isotype of mAb 6-C4 was IgG2b (κ light chain).
6-C4 reacted with the ERGIC3 peptide and the native protein extracted from NSCLC cells, but not with BSA, and plasma, saliva and urine samples from three normal adults, as revealed by ELISA (Fig.1b). A clear single band was detected at approximately 50 kD using 6-C4 by western blot analysis with the native protein, similar to the preliminary result using the sera of immunized mice (Fig.1c and Suppl. Fig. S1c). The immunofluorescence staining of 6-C4 was localized around the Golgi apparatus and the ER (Fig.1d). NSCLC and HCC tissues were strongly stained by immunohistochemistry using 6-C4 (Suppl. Fig. S1d). These observations are consistent with our previous finding in which anti-ERGIC3 serum (Abcam, Cambridge, UK) was used, and a previous study.9,12 These results indicated that 6-C4 specifically recognizes ERGIC3.
ERGIC3 expression was determined in normal adult human tissues
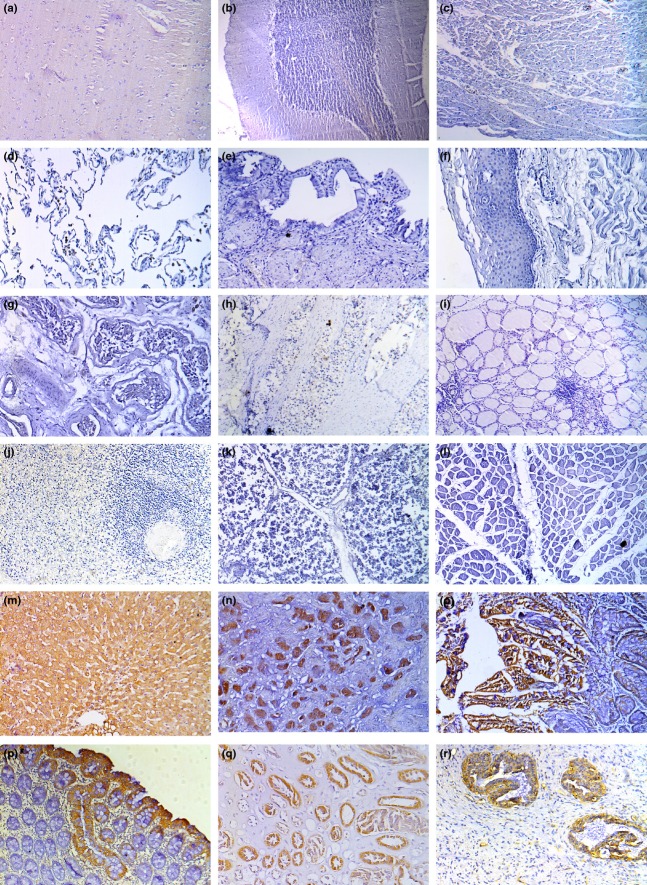
Studies have not been previously conducted on ERGIC3 expression in a broad variety of normal human tissues. Therefore, we examined the expressions of ERGIC3 in various normal human tissues using 6-C4; the results are shown in Table1 and Figure2. Most normal tissues were not stained with 6-C4. However, the cytoplasm of some epithelial cells was positively stained. By contrast, all non-malignant lung tissues were negative for 6-C4 staining.
Table 1.
Immunohistochemical analysis of ERGIC3 in normal human tissues by using mAb 6-C4
| Tissue | ERGIC3 staining |
|---|---|
| Cerebral cortex | 0/3 |
| Cerebellum | 0/1 |
| Heart | 0/4 |
| Lung | 0/23 |
| Liver | 10/11 |
| Gallbladder | 0/2 |
| Pancreas | 5/6 |
| Esophagus | 0/3 |
| Stomach | 5/5 |
| Intestine | 2/2 |
| Colon | 3/3 |
| Kidney | 6/6 |
| Urinary bladder | 0/3 |
| Testis | 0/3 |
| Prostate | 1/5 |
| Breast | 3/4 |
| Ovary | 0/3 |
| Uterus | 0/2 |
| Thyroid gland | 0/5 |
| Spleen | 0/3 |
| Thymus | 0/4 |
| Muscle | 0/3 |
| Skin | 0/3 |
Figure 2.
Immunohistochemical analysis of ERGIC3 in normal human tissues using 6-C4: (a) brain; (b) cerebellum; (c) heart; (d) lung; (e) gallbladder; (f) esophagus; (g) testis; (h) prostate; (i) thyroid gland; (j) spleen; (k) thymus; (l) skeletal muscle; (m) liver; (n) stomach; (o) intestine; (p) colon; (q) kidney; and (r) breast.
ERGIC3 expression was determined in various tumor tissues
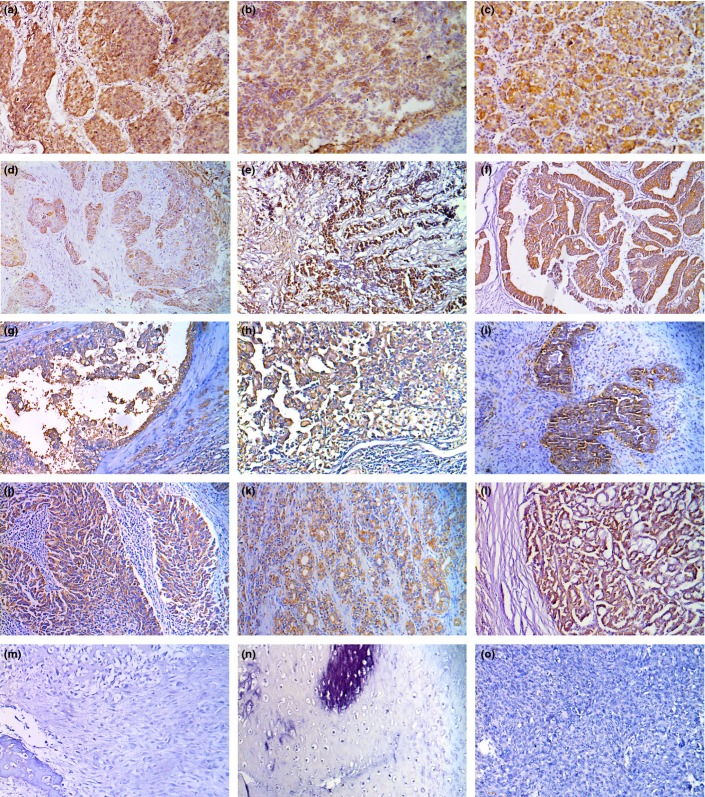
Immunohistochemical results of ERGIC3 in 15 types of human tumors using 6-C4 are shown in Table2 and Figure3. ERGIC3 was strongly expressed in all carcinomas originating from the epithelial cells, but all sarcomas were negative for 6-C4.
Table 2.
Immunohistochemical analysis of ERGIC3 in various tumor tissues by using mAb 6-C4
| Cancer | ERGIC3 staining |
|---|---|
| Non-small cell lung cancer (NSCLC) | 21/22 |
| Pancreatic carcinoma | 4/4 |
| Hepatocellular carcinoma (HCC) | 4/4 |
| Gastric carcinoma | 4/4 |
| Esophagus carcinoma | 10/10 |
| Colorectal carcinoma | 5/5 |
| Renal cell carcinoma | 9/9 |
| Bladder carcinoma | 11/12 |
| Mammary carcinoma | 5/6 |
| Cervical carcinoma | 9/10 |
| Prostate cancer | 9/11 |
| Thyroid carcinoma | 5/5 |
| Osteosarcoma | 0/3 |
| Chondrosarcoma | 0/2 |
| Fibrosarcoma | 0/3 |
Figure 3.
Immunohistochemical analysis of ERGIC3 in various tumor tissues using 6-C4. (a) non-small cell lung cancer (NSCLC); (b) pancreatic carcinoma; (c) hepatocellular carcinoma (HCC); (d) esophagus carcinoma; (e) gastric carcinoma; (f) colon carcinoma; (g) renal cell carcinoma; (h) bladder carcinoma; (i) mammary carcinoma; (j) cervical carcinoma; (k) prostate carcinoma; (l) thyroid carcinoma; (m) osteosarcoma; (n) chondrosarcoma; (o) fibrosarcoma.
Using western blot with 6-C4, all cultured NSCLC cells expressed higher levels of ERGLC3 than 16HBE. (Fig.4a,b). This expression pattern of ERGIC3 protein is consistent with that of ERGIC3 mRNA (Fig.4d).
Figure 4.
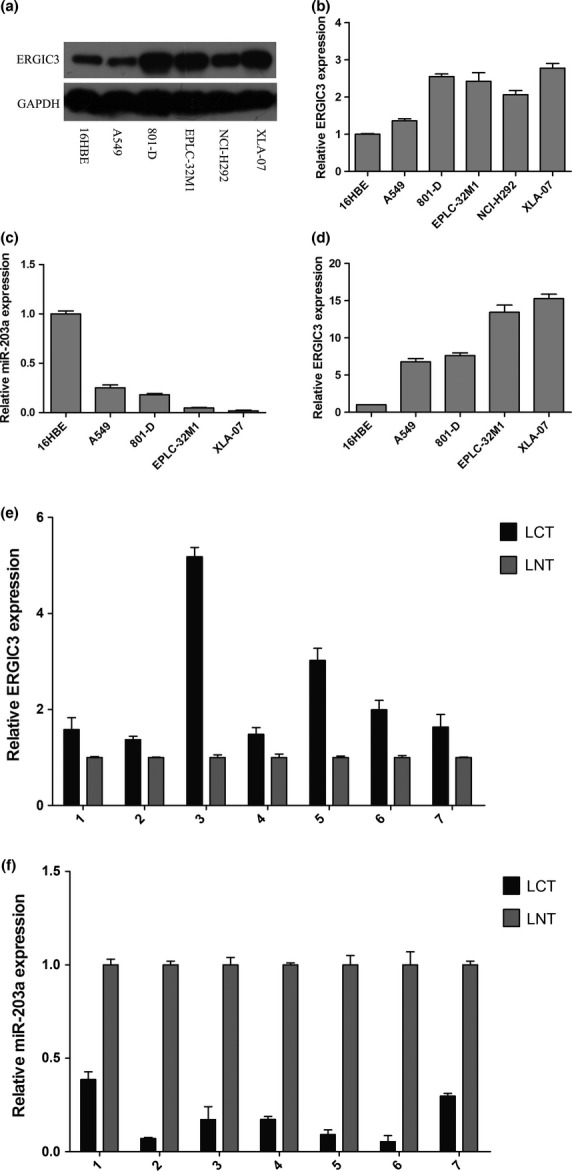
Expressions of ERGIC3 and miR-203a in cultured cells and tissues. Expressions of ERGIC3 protein (a and b) in cultured cells by western blot analysis. Expressions of miR-203a (c) and ERGIC3 mRNA (d) in cultured cells by quantitative RT-PCR. Expressions of ERGIC3 mRNA (e) and miR-203a (f) in seven pairs of non-small cell lung cancer (LCT) and their adjacent lung tissues (LNT) by quantitative RT-PCR.
ERGIC3-related miRNA were identified in non-small cell lung cancer cells
Many cancer-related genes are regulated by miRNA.18 By bioinformatics analysis we predicted that 398 miRNA may bind to ERGIC3 mRNA. Moreover, we found 87 consensus differentially expressed miRNA by comparing the miRNA profiles of NSCLC cells (A549, 801D and EPLC-32M1) with those of 16HBE. Integrating the candidate miRNA predicted by bioinformatics analysis and the differentially expressed miRNA detected by the miRNA expression profiling, we found that miR-140-3p and miR-203a may target ERGIC3 and that they were differentially expressed in NSCLC cells. Therefore, the two miRNA were selected for further research. MiR-490-3p participates in ERGIC3 regulation in HCC;12 as such, miR-490-3p was also investigated.
Through qRT-PCR, it was found that MiR-140-3p expression was higher in NSCLC cells than in 16HBE (Suppl. Fig. S2a); by contrast, miR-203a expression was lowed in NSCLC cells than in 16HBE (Fig.4c). The qRT-PCR results of miR-140-3p and miR-203a are also consistent with those of miRNA expression profiling. No significant difference in miR-490-3p expression was observed between NSCLC cells and 16HBE (Suppl. Fig. S2c). Interestingly, miR-203a expression levels were negatively correlated with ERGIC3 expression (Fig.4c,d).
ERGIC3 levels were negatively correlated with miR-203a in non-small cell lung cancer
MiR-203a was downexpressed in NSCLC tissues compared with adjacent non-malignant tissues (Fig.4f); by contrast, ERGIC3 was upregulated in NSCLC tissues (Fig.4e). ERGIC3 and miR-203a expression were negatively correlated in patients’ tissues, similar to that in cultured cells.
ERGIC3 expression was regulated by miR-203a in non-small cell lung cancer cells
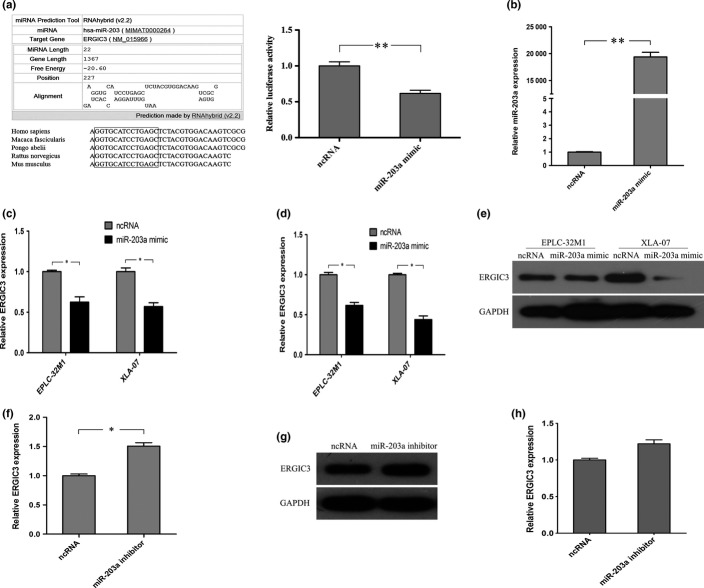
MiRNA can downregulated gene expression through either translation inhibition or mRNA degradation via binding to mRNA.18 In bioinformatic analysis, miR-203a can bind to ERGIC3, and the sequence alignment of the miR-203a binding site is conservative among primates, even in mammals (Fig.5a). A luciferase reporter assay conducted using ERGIC3 construct that contained the potential binding site of miR-203a showed that the luminescence intensities were significantly reduced by the miR-203a mimic (Fig.5a). Furthermore, the miR-203a mimic treatment increased miR-203a expression (Fig.5b) and decreased ERGIC3 expression (Fig.5c–e). However, ERGIC3 expression was increased by the miR-203a inhibitor (Fig.5f–h). Unfortunately, ERGIC3 expressions were not significantly changed by miR-140-3p and miR-490-3p mimic treatments (Suppl. Fig. S2b,d).
Figure 5.
ERGIC3 expression was regulated by miR-203a. (a) Left: The potential binding site of miR-203a on ERGIC3 predicted by RNAhybrid (v2.2) and comparison of the sequence alignment among several species. Right: Validation of the direct targeting of ERGIC31 by miR-203a using a luciferase reporter assay. (b) miR-203a level was significantly increased in EPLC-32M1 by the miR-203a mimic treatment. (c–e) ERGIC3 expression was reduced at mRNA (c) and protein (d,e) levels in non-small cell lung cancer (NSCLC) cells by the miR-203a mimic treatment. (f–h) ERGIC3 expressions were increased at mRNA (f) and protein (g,h) levels in 16HBE by miR-203a inhibitor treatment. Student’s t-test, *P < 0.05; **P < 0.01. ncRNA, negative control miRNA.
MiR-203a expression affected cellular morphology and proliferation
Considering the novel discovery of miR-203a in NSCLC, we further investigated the possible functions of miR-203a. XLA-07 (with low endogenous miR-203a level) and 16HBE (with high endogenous miR-203a level) were treated with the miR-203a mimic and inhibitor, respectively. The MiR-203a mimic caused an evident morphological change (from the spindle-like appearance to the round shape, Fig.6a) and significantly reduced the cell proliferation in XLA-07 (Fig.6c). Notably, changes induced by the miR-203a mimic were similar to the alterations caused by ERGIC3 RNA interference in NSCLC cells.9 However, significant alterations in cellular morphology and proliferation were not observed in 16HBE after miR-203a inhibitor treatment was administered (Fig.6a,b).
Figure 6.
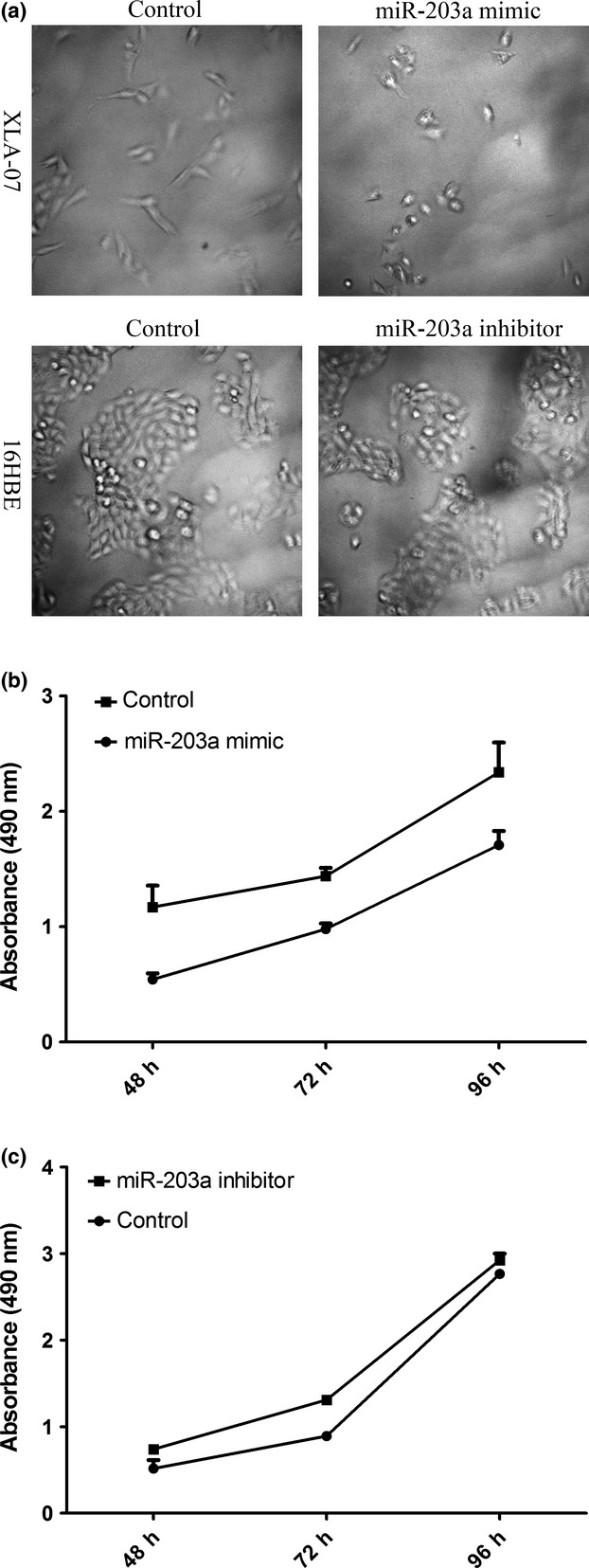
MiR-203a expression affected cellular morphology and proliferation. Changes in the morphology (a, top; 200×) and proliferation (b) were observed in XLA-07 with the miR-203a mimic treatment. However, miR-203a inhibitor treatment did not significantly affect cellular morphology (a, bottom; 200×) and proliferation (c) of 16HBE (Student’s t-test, P > 0.05). Control, negative control miRNA.
Discussion
Cancer is a disease caused by the accumulation of genetic and epigenetic alterations. As such, expressions and functions of cancer-related genes should be investigated to understand carcinogenesis and to identify cancer biomarkers. In a previous study, ERGIC3 was found to be a novel lung cancer-related gene. In addition, overexpressed ERGIC3 promotes cellular proliferation and migration.9 ERGIC3 is coupled with Erv41p to form an integral membrane protein complex as components of COPII vesicles, which play an important role in protein transportation via the secretory pathway.19–21 Abnormal ERGIC3 expression can affect cell growth and ER stress-induced cell death, and contribute to epithelial to mesenchymal transition.11,12 Thus, ERGIC3 may be a potential cancer biomarker. In this study, a new mAb (6-C4) against ERGIC3 was developed. The proposed antibody is well suited for immunohistochemistry, immunoblotting and ELISA. To our knowledge, 6-C4 is the first murine mAb towards ERGIC3. 6-C4 could be used to investigate the expression and functions of ERGIC3.
We systematically studied expressions of ERGIC3 in normal human tissues and tumors by immunohistochemistry using 6-C4. The staining was not observed in the most normal human tissues. However, 6-C4 reacted with some epithelial cells, such as hepatocytes, gastrointestinal epithelium, ducts and acini of pancreas, proximal and distal tubules of kidney, and mammary epithelial cells. The expression of ERGIC3 in these epithelial cells may be closely related with its functions. ERGIC3 participates to form vesicles of the protein transport and secretion.20,21 These epithelial cells stained by the ERGIC3 mAb are very active in protein synthesis and secretion. We found that almost all carcinomas that originated from the epithelial cells were positively stained with 6-C4; by contrast, all sarcomas were negative for 6-C4. Therefore, we suggested that ERGIC3 may be a biomarker to distinguish between carcinomas and sarcomas in histopathological diagnosis. However, a larger number of cases should be investigated to draw final conclusions. We also observed that NSCLC cells were strongly stained with 6-C4; however, all of the cells (including bronchial epithelial cells and alveolar cells) in non-malignant lung tissues were negative for 6-C4. These findings were similar to our previous observation in which anti-ERGIC3 serum purchased from Abcam was used.9 These results showed that ERGIC3 has higher specificity and sensitivity for NSCLC than CEA and CYFRA 21-1 in immunostaining.6,7 Hence, ERGIC3 may be used to distinguish NSCLC from benign lesions of the lung. More importantly, ERGIC3 mAb might be applied in the sputum test to detect early-stage lung cancer. Early diagnosis and treatment have a decisive influence on lung cancer prognosis. Thus, new methods should be developed to detect early-stage lung cancer. This study is continuing in our laboratory.
ERGIC3 overexpression has been observed in NSCLC. However, molecular mechanisms underlying ERGIC3 alteration remain elusive. MiRNA are a family of small non-coding RNA, approximately 21 to 25 nucleotides in length,22,23 that target mRNA at complementary sites in the 3′-UTR and coding domain sequence to regulate gene expression at a post-transcriptional level.24 MiR-203a (also named miR-203), which was first assigned as a skin-related miRNA,25,26 is involved in cancer development and progression by targeting different genes.27–30 In this study, reduced miR-203a expression was one of the mechanisms found to underlie ERGIC3 overexpression in NSCLC. Our finding was supported by the following evidence: (i) miR-203a can bind to ERGIC3 mRNA by bioinformatic prediction; (ii) a luciferase reporter assay verified that ERGIC3 was targeted by miR-203a directly; (iii) ERGIC3 expressions were negatively correlated with miR-203a in cultured cells and tissues obtained from patients with NSCLC; (iv) ERGIC3 was downregulated at mRNA and protein levels by the increased miR-203a expression in NSCLC cells; (v) ERGIC3 was upregulated at mRNA and protein levels by the reduced miR-203a expression in human bronchial epithelial cells; and (vi) the forced expression of miR-203a led to changes in cellular morphology and proliferation of cultured NSCLC cells, which were similar to the effects induced by the decreased ERGIC3 expression.9 MiR-203a has been regarded as a tumor suppressor miRNA that participates in cancer development and progression.27–30 Here, miR-203a regulated the cellular proliferation and the expression of functional gene in NSCLC, suggesting that miR-203a might be a tumor suppressor miRNA in lung cancer. Surprisingly, significant alterations in cellular morphology and proliferation were not observed in immortalized human bronchial epithelial cells after miR-203a inhibitor treatment. We cannot explain this phenomenon. We speculated that cancer cells showed higher sensitivity to the change of miR-203a than normal cells, because normal cells have better regulation and balanced systems than cancer cells. Taken together, miR-203a might be a reasonable biomarker and therapeutic target for lung cancer. However, further research is necessary. In a previous study, miR-490-3p increased ERGIC3 mRNA and protein levels by binding to the 3′-UTR of ERGIC3 in HCC cells; this interaction is in contrast to most miRNA–mRNA interactions.12 However, we did not find that miR-490-3p significantly affected ERGIC3 expression in NSCLC cells. The mechanisms of ERGIC3 regulation may differ in various cancers.
In conclusion, a new murine mAb against ERGIC3 (6-C4) was developed, and the avid antibody is well suited for immunohistochemistry, immunoblotting and ELISA. 6-C4 strongly stained NSCLC cells but did not react with normal lung tissues. Thus, this mAb might be used in the histopathological diagnosis and cytopathological testing to detect early-stage lung cancer. Furthermore, miR-203a downregulation induced ERGIC3 overexpression in NSCLC cells.
Acknowledgments
This work was supported by the Natural Science Foundation of China (81272617) and the 973 Program (2011CB510104).
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1. Establishment and identification of monoclonal antibodies (mAb) to ERGIC3. ELISA analysis demonstrated positive hybridomas of anti-ERGIC3 mAbs after cell fusion was completed (a) and the positive rate of approximately 95% after rounds of sub-cloning was performed by limiting dilution (b). Sera of mice immunized with the ERGIC3 peptide that recognized the protein extracted from cultured cells at approximately 50 kD in immunoblot (c). 6-C4 strongly stained non-small cell lung cancer (NSCLC) and hepatocellular carcinoma (HCC) tissues in immunohistochemistry, and SP2/0 supernatant was used as a negative control (d).
Fig. S2. Effects of miR-140-3p and miR-490-3p on the ERGIC3 expression in non-small cell lung cancer (NSCLC) cells. (a) miR-140-3p was highly expressed in cultured NSCLC cells. (b) miR-140-3p mimic treatment did not significantly affect ERGIC3 expression in 16HBE. (c) miR-490-3p was expressed at varying degrees in NSCLC cells. (d) miR-490-3p mimic treatment likely reduce the ERGIC3 expression in A549; however, no significant difference was observed between miR-490-3p mimic treatment and negative control (Student’s t-test, P > 0.05). ncRNA, negative control miRNA.
Table S1. Primers used in this study.
References
- Ferlay J, Soerjomataram II, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Travis WD World Health Organization; International Agency for Research on Cancer; International Association for the Study of Lung Cancer; International Academy of Pathology. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon Oxford: IARC Press Oxford University Press; 2004. [Google Scholar]
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- Xiang D, Zhang B, Doll D, Shen K, Kloecker G, Freter C. Lung cancer screening: from imaging to biomarker. Biomark Res. 2013;1:4. doi: 10.1186/2050-7771-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa J, Wojcik E, Reinfuss M, Kolodziejski L. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21-1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem. 2002;48:1931–7. [PubMed] [Google Scholar]
- Tomita M, Shimizu T, Ayabe T, Yonei A, Onitsuka T. Prognostic significance of tumour marker index based on preoperative CEA and CYFRA 21-1 in non-small cell lung cancer. Anticancer Res. 2010;30:3099–102. [PubMed] [Google Scholar]
- Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–43. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- Wu M, Tu T, Huang Y, Cao Y. Suppression subtractive hybridization identified differentially expressed genes in lung adenocarcinoma: ERGIC3 as a novel lung cancer-related gene. BMC Cancer. 2013;13:44. doi: 10.1186/1471-2407-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Mack GJ, Barlowe C, Otte S. Mammalian Erv46 localizes to the endoplasmic reticulum-Golgi intermediate compartment and to cis-Golgi cisternae. Proc Natl Acad Sci U S A. 2003;100:4586–91. doi: 10.1073/pnas.0730885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa M, Kira Y, Yabunaka Y, Inoue M. Identification and characterization of endoplasmic reticulum-associated protein, ERp43. Gene. 2007;386:42–51. doi: 10.1016/j.gene.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Liu M, Li X, Tang H. miR-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3) J Biol Chem. 2013;288:4035–47. doi: 10.1074/jbc.M112.410506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MS, Cao Y. ERGIC3 (20pte-q12) Atlas Genet Cytogenet Oncol Haematol. 2014 ; September. [Google Scholar]
- Ma LJ, Wang HZ, Bian L, et al. Establishment and characterization of lung adenocarcinoma cell line XLA-07. Chin J Pathol. 2012;41:335–9. doi: 10.3760/cma.j.issn.0529-5807.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059–68. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- Cao Y, Merling A, Crocker PR, Schwartz-Albiez R. Differential expression of β-galactoside α2,6 sialyltransferase (ST6Gal. I) and sialoglycans in normal and cirrhotic liver and hepatocellular carcinomas. Lab Invest. 2002;82:1515–24. doi: 10.1097/01.lab.0000038503.34655.98. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–15. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuza L, Halbeisen R, Jeno P, et al. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem. 2004;279:47242–53. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]
- Otte S, Barlowe C. The Erv41p-Erv46p complex: multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J. 2002;21:6095–104. doi: 10.1093/emboj/cdf598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh LM, Tong AH, Boone C, Jensen ON, Otte S. Genetic and molecular interactions of the Erv41p-Erv46p complex involved in transport between the endoplasmic reticulum and Golgi complex. J Cell Sci. 2006;119:4730–40. doi: 10.1242/jcs.03250. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Banerjee J, Sen CK. MicroRNAs in skin and wound healing. Methods Mol Biol. 2013;936:343–56. doi: 10.1007/978-1-62703-083-0_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Wei T, Janson PC, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang X, Liang H, et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKC alpha. PLoS One. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. MiR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- Bo J, Yang G, Huo K, et al. MicroRNA-203 suppresses bladder cancer development by repressing bcl-w expression. FEBS J. 2011;278:786–92. doi: 10.1111/j.1742-4658.2010.07997.x. [DOI] [PubMed] [Google Scholar]
- Viticchie G, Lena AM, Latina A, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–31. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Establishment and identification of monoclonal antibodies (mAb) to ERGIC3. ELISA analysis demonstrated positive hybridomas of anti-ERGIC3 mAbs after cell fusion was completed (a) and the positive rate of approximately 95% after rounds of sub-cloning was performed by limiting dilution (b). Sera of mice immunized with the ERGIC3 peptide that recognized the protein extracted from cultured cells at approximately 50 kD in immunoblot (c). 6-C4 strongly stained non-small cell lung cancer (NSCLC) and hepatocellular carcinoma (HCC) tissues in immunohistochemistry, and SP2/0 supernatant was used as a negative control (d).
Fig. S2. Effects of miR-140-3p and miR-490-3p on the ERGIC3 expression in non-small cell lung cancer (NSCLC) cells. (a) miR-140-3p was highly expressed in cultured NSCLC cells. (b) miR-140-3p mimic treatment did not significantly affect ERGIC3 expression in 16HBE. (c) miR-490-3p was expressed at varying degrees in NSCLC cells. (d) miR-490-3p mimic treatment likely reduce the ERGIC3 expression in A549; however, no significant difference was observed between miR-490-3p mimic treatment and negative control (Student’s t-test, P > 0.05). ncRNA, negative control miRNA.
Table S1. Primers used in this study.