Abstract
Afatinib is an irreversible epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI) that is known to be effective against the EGFR T790M variant, which accounts for half of the mechanisms of acquired resistance to reversible EGFR-TKIs. However, acquired resistance to afatinib was also observed in clinical use. Thus, elucidating and overcoming the mechanisms of resistance are important issues in the treatment of non-small cell lung cancer. In this study, we established various afatinib-resistant cell lines and investigated the resistance mechanisms. EGFR T790M mutations were not detected using direct sequencing in established resistant cells. Several afatinib-resistant cell lines displayed MET amplification, and these cells were sensitive to the combination of afatinib plus crizotinib. As a further investigation, a cell line that acquired resistance to afatinib plus crizotinib, HCC827-ACR, was established from one of the MET amplified-cell lines. Several afatinib-resistant cell lines including HCC827-ACR displayed epithelial-to-mesenchymal transition (EMT) features and epigenetic silencing of miR-200c, which is a suppresser of EMT. In addition, these cell lines also exhibited overexpression of ALDH1A1 and ABCB1, which are putative stem cell markers, and resistance to docetaxel. In conclusion, we established afatinib-resistant cells and found that MET amplification, EMT, and stem cell-like features are observed in cells with acquired resistance to EGFR-TKIs. This finding may provide clues to overcoming resistance to EGFR-TKIs.
Keywords: Afatinib, cancer stem cells, drug resistance, EGFR-TKI, non-small cell lung cancer
Lung cancer is the leading cause of cancer-related death worldwide.1 To improve the clinical outcomes of lung cancer, new therapeutic agents have been developed including epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs). EGFR-TKIs display significant efficacy against EGFR-mutated non-small cell lung cancers (NSCLCs) by inhibiting EGFR-AKT signaling.2–4 However, most of these malignancies eventually acquire resistance to EGFR-TKIs.5,6 Several mechanisms of acquired resistance to EGFR-TKIs have been elucidated, such as secondary EGFR T790M5 and minor mutations,6 MET amplification,7 activation of the MET/hepatocyte growth factor axis,8 AXL upregulation,9 and the acquisition of epithelial-to-mesenchymal transition (EMT) features.10,11 In addition, our group previously reported that stem cell-like properties were present in EGFR-TKI-resistant cells.12
Afatinib is an irreversible TKI for EGFR and HER2 that was approved by the United States Food and Drug Administration in 201313; it exhibited in vitro and in vivo activity against the T790M variant14 and suppressed the progression of NSCLC harboring the T790M mutation in clinical use.15 In a randomized phase III trial, afatinib improved PFS compared with placebo in patients with NSCLC who experienced disease progression after reversible EGFR-TKI treatment.16 However, acquired resistance to afatinib was also observed in most of the patients.13,16 Therefore, it remains a critical issue to elucidate and overcome the mechanisms of acquired resistance to irreversible EGFR-TKIs.
In this study, we established various NSCLC cell lines with acquired resistance to afatinib and investigated the molecular profile of resistant cells to uncover the mechanisms of resistance to irreversible EGFR-TKIs.
Materials and Methods
Cell lines and reagents
EGFR-mutant HCC827 (exon 19 del. E746-A750), PC-9 (exon 19 del. E746-A750), HCC4006 (exon 19 del. L747-A750, P ins), and HCC4011 (L858R) cells were used in this study. These cell lines excluding PC-9 were kindly provided by Dr Adi F. Gazdar (The University of Texas Southwestern Medical Center at Dallas, Dallas, TX, USA), who established these lines with Dr John D. Minna.17,18 PC-9 cells were obtained from Immuno-Biological Laboratories (Takasaki, Gunma, Japan). All cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and grown in a humidified incubator with 5% CO2 at 37°C. Acquired afatinib-resistant sublines were established using the following procedures: (i) cells were exposed to afatinib with stepwise escalation from 10 nM to 2 μM over 6 months; (ii) cells were intermittently and briefly exposed to the drug at 2 μM for 6 months, (iii) cells were subjected to the drug with stepwise escalation from 1 nM to 2 μM for 3 months; and (iv) cells were continuously exposed to the drug at 2 μM for up to 3 months. The afatinib-resistant sublines established using method #1 were designated as the name of the parental cell lines followed by “-AR1,” e.g., HCC827-AR1. Similarly, the sublines established using methods #2, #3, and #4 were designated as the name of parental cell lines followed by “-AR2,” “-AR3,” and “-AR4,” respectively (Table1). Of note, acquired resistance to drugs was defined as exhibiting a higher IC50 value than the parental cell line.
Table 1.
Afatinib-resistant cell lines and resistant mechanisms
| Cell lines | Method of drug exposure | EGFR T790M mutation | MET amplification | EMT | Stem cell markers |
|---|---|---|---|---|---|
| HCC827 | N/A | – | – | – | – |
| HCC827-AR1 | Stepwise, 10 nM | – | – | + | + |
| HCC827-AR2 | Intermittent (briefly), 2 μM | – | + | – | – |
| HCC827-ACR | Continuous, Crizotinib 0.2 μM | – | + | + | + |
| HCC827-AR3 | Stepwise, 1 nM | – | + | – | – |
| HCC827-AR4 | Intermittent (as long as possible), 2 μM | – | – | + | + |
| PC-9 | N/A | – | – | – | – |
| PC-9-AR1 | Stepwise, 10 nM | – | – | – | – |
| PC-9-AR2 | Intermittent (briefly), 2 μM | – | – | – | – |
| HCC4006 | N/A | – | – | – | – |
| HCC4006-AR1 | Stepwise, 10 nM | – | – | + | + |
| HCC4006-AR2 | Intermittent (briefly), 2 μM | – | – | + | – |
| HCC4011 | N/A | – | – | – | – |
| HCC4011-AR1 | Stepwise, 10 nM | – | + | – | – |
| HCC4011-AR2 | Intermittent (briefly), 2 μM | – | – | – | – |
EGFR, epidermal growth factor receptor; EMT, epithelial to mesenchymal transition; N/A, not applicable.
Afatinib and crizotinib were purchased from Synkinase Pty ltd. (San Diego, CA, USA) and Sigma-Aldrich (St. Louis, MO, USA), respectively. Docetaxel was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Bortezomib was purchased from LC Laboratories (Woburn, MA, USA).
The primary antibodies used for western blot analysis were as follows: anti-EGFR, phospho-EGFR (Tyr1068), HER-2, phospho-HER2 (Tyr877), Akt, phospho-Akt (Ser473), p44/p42 mitogen-activated protein kinase (MAPK), phospho-p44/p42 MAPK, Met, phospho-Met (Tyr1234/1235), TCF8/ZEB1, E-cadherin, vimentin (Cell Signaling Technology, Danvers, MA, USA), and β-actin (Merck Millipore, Billerica, MA, USA). The following secondary antibodies were used: goat anti-rabbit or anti-mouse immunoglobulin G-conjugated horseradish peroxidase (Santa Cruz Biotechnology, Dallas, TX, USA).
Western blot analysis
Cells were harvested at 80–90% confluence, and cellular proteins were extracted with lysis buffer [RIPA buffer, phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich), and Complete Mini (Roche, Basel, Switzerland)]. Total proteins were separated with electrophoresis and electroblotted using the Trans-Blot Turbo Transfer system (Mini-PROTEAN TGX Precast Gel and Trans-Blot Turbo Mini PVDF Transfer Pack) (Bio-Rad, Hercules, CA, USA). After blocking with 5% non-fat dry milk and 0.1% Tween 20 in Tris-buffered saline, the membranes were incubated at 4°C overnight with primary antibodies. The membranes then were developed with secondary antibodies. Monoclonal anti-actin antibody was used as an equal loading control. To detect specific signals, the membranes were examined using ECL Prime Western Blotting Detection System (GE Healthcare, Amersham, UK) and LAS-3000 (Fujifilm, Tokyo, Japan).
Fluorescence immunocytochemistry
Cells were cultured and fixed by 4% formaldehyde on chamber slide, and blocked in 5% normal goat serum (Sigma-Aldrich) in PBS containing 0.3% Triton X-100 (Sigma-Aldrich) for 1 h at room temperature. The blocking solution was aspirated and sections were incubated overnight at 4°C in primary antibodies diluted in PBS with 0.3% Triton and 1% bovine serum albumin (BSA) (Sigma-Aldrich). Primary and secondary antibody were used PathScan EMT Duplex IF Kit (Cell Signaling Technology), and chromatin was counterstained using ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific, Waltham, MA, USA).
DNA and RNA extraction
Genomic DNAs were extracted from cell lines using a DNeasy Blood and Tissue Kit (Qiagen, Venlo, the Netherlands). Total RNAs were extracted from cell lines using an RNeasy Mini Kit (Qiagen). The complementary DNA (cDNA) was synthesized from total RNA using High-Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific), according to the manufacturer’s instructions.
DNA analyses
EGFR exon 20 mutation was examined using direct sequencing as previously reported.19 The primer sequences are shown in Supplementary Table S1A. Copy number gains (CNGs) of EGFR and MET were determined by a quantitative real-time PCR assay using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) as previously reported.20 LINE-1 gene was used as a reference gene. The relative copy number of each sample was determined by comparing the ratio of the target gene to LINE-1 in each sample with the ratio of these genes in human genomic DNA (Merck Millipore). The primer sequences are shown in Supplementary Table S1B.
mRNA and microRNA expression analysis by quantitative reverse transcription-PCR
The gene expression of the putative stem cell markers ALDH1A1 and ABCB1 were analyzed by quantitative reverse transcription-PCR using cDNAs, TaqMan Gene Expression Assays, and the ABI StepOnePlus Real-Time PCR Instrument (Thermo Fisher Scientific). mRNA and microRNA (miR) expression was calculated using delta-delta-CT method. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene and miR-374 were used as the endogenous control for mRNA and miR expression analyses, respectively. The catalog numbers of TaqMan assays are shown in Supplementary Tables S1C,D.
Cell proliferation assay
Cell proliferative ability was determined by a modified MTS assay using CellTiter 96 AQueous One Solution Reagent (Promega, Fitchburg, WI, USA) as previously reported.21 The antiproliferative effects are shown as the 50% inhibitory concentration (IC50).
DNA methylation analysis
The DNAs were subjected to bisulfite treatment using an Epitect Bisulfite Kit (Qiagen). The DNA methylation statuses were analyzed using these bisulfite DNAs and methylation-specific PCR as previously reported.21,22 The primer sequences are shown in Supplementary Table S1E.
Cell growth assay
Cells were plated in 96-well plate at a density of 2.0 × 103 cells per well and cultured for 120 h at drug-free condition. Four images per well were taken every 6 h using IncuCyteZOOM. (Essen BioScience, Ann Arbor, MI, USA), and cell confluence was evaluated using IncuCyte ZOOM 2015A software (Essen BioScience) as previously reported.23
Soft agar colony formation assay
Cells were plated in 24-well plate with Ultra-Low Attachment surface (Corning Inc., Corning, NY, USA) at a density of 2.5 × 103 cells per well and cultured for 21 days in RPMI1640 with 10% FBS and 0.3% agarose. RPMI1640 with 10% FBS were added to the top of agar layer and exchanged every 3 days. Nine images per well were taken every day using IncuCyteZOOM. The colony formation was defined as the cell aggregates occupying an area at least 8000 μm2 (about 100 μm diameter) and the occupied area were measured using IncuCyte ZOOM 2015A software.24
Antibody array
Cells were cultured with or without 2 μM afatinib for 6 h. After that, they were harvested at 80–90% confluence and cellular proteins were extracted. Antibody array for these proteins were performed using PathScan RTK Signaling Antibody Array Kit (Cell Signaling Technology) as manufacturer’s recommendation.
Next generation sequencing
The targeted mRNAseq for 100 stem cell-related markers was performed. The libraries were prepared using TruSeq Targeted RNA Expression Kit (Illumina, San Diego, CA, USA) and sequencing were performed using MiSeq Reagent Kit v2 (Illumina) and MiSeq Desktop Sequencer (Illumina) as manufacturer’s recommendation. Data analysis was performed using Illumina MiSeq Reporter version 2.5.1 (Illumina).
Xenograft model and immunohistochemical staining
All experimental animals were housed under specific pathogen-free conditions and handled in accordance with the Policy on the Care and Use of Laboratory Animals, Okayama University. HCC827 and HCC827-AR1 cells were inoculated into 5–8-week-old female non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice that were purchased from Charles River Laboratories Japan (Yokohama, Japan). Each suspension of cell lines was subcutaneously injected at 1 × 106 cells in 300 μL of serum-free RPMI 1640 medium with Matrigel Basement Membrane Matrix (Corning). After tumor formation, mice were sacrificed, and tumors were harvested. The formalin-fixed and paraffin-embedded tissues of these tumors were stained immunohistochemically using anti-ALDH1 antibody (Becton Dickinson, Franklin Lakes, NJ, USA) as previously reported.25
Statistical analyses
Statistical analysis was performed using GraphPad Prism, version 6.0.3, J (GraphPad Software, San Diego, CA, USA) and R statistical software version 3.2.026 with Bioconductor packages ‘Heatplus’.27,28 Values of P < 0.05 were considered as statistically significant.
Results
EGFR-mutated cell lines that acquired resistance to afatinib
We established 10 afatinib-resistant cell lines from four parental NSCLC cell lines harboring activating EGFR mutations. Initially, we tried to establish afatinib-resistant cell lines via continuous exposure to a high concentration of afatinib (2 μM), but this was unsuccessful. Therefore, we used two exposing procedures, namely intermittent high-dose exposure and stepwise dose escalation from 10 nM, which is higher than the IC50 of the parental cell lines and similar to the in vitro IC50 for EGFR with L858R/T790M mutations.14 The characteristics of these cell lines including the IC50 values of afatinib are shown in Table1. The representative examples of the expression of EGFR, HER2, and their downstream targets are shown in Supplementary Figure S1.
EGFR T790M mutation and MET amplification in afatinib-resistant cell lines
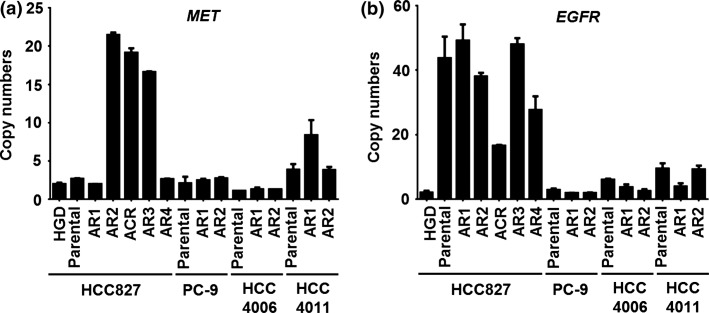
The T790M mutation was not detected in all 10 afatinib-resistant sublines by direct sequencing. The CNGs of MET were detected in HCC827-AR2 and HCC827-AR3 significantly and in HCC4011-AR1 slightly (Fig. 1).
Figure 1.
Copy number gains of MET and EGFR in EGFR-mutant lung cancer cell lines and their afatinib-resistant sublines. (a) The copy numbers of MET examined by real-time PCR were significantly amplified in HCC827-AR2, -AR3 and -ACR cells and slightly amplified in HCC4011-AR1 cells. (b) The copy number of EGFR in HCC827-ACR cells was approximately half of that in the parental cell line. All experiments were performed at least three times, and error bars indicate standard deviations.
Acquired resistance to afatinib plus crizotinib in MET-amplified cell line
HCC827-AR2 with MET amplification was sensitive to combination treatment with afatinib and crizotinib, which is a MET inhibitor (Table2). We had an interest in whether this combined treatment completely overcomes resistance to afatinib in HCC827-AR2 cells. Therefore, HCC827-AR2 cells were treated with 2 μM afatinib and 0.2 μM crizotinib via continuous exposure. Finally, a subline resistant to afatinib plus crizotinib treatment was established from HCC827-AR2 cells within 2 months of drug exposure and was named HCC827-ACR. The CNG of MET was retained in HCC827-ACR cells, although copy number of EGFR was reduced to approximately half of that in the parental cell line (Fig. 1).
Table 2.
IC50 values (μM) against various agents in EGFR-mutant NSCLC cell lines
| Cell Lines | EGFR-TKI | Chemotherapeutic agent | MET inhibitor | Protease inhibitor | |
|---|---|---|---|---|---|
| Afatinib | DOC | Crizotinib | Afatinib with Crizotinib (0.2 μM) | Bortezomib | |
| HCC827 | 0.0019 | 0.0031 | 6.9 | 0.0018 | 0.0082 |
| HCC827-AR1 | >10† | >1† | 7.9 | 4.8† | 0.0094 |
| HCC827-AR2 | 4.1† | 0.0042 | 4.6 | 0.00019 | 0.0018 |
| HCC827-ACR | >10† | >1† | N/A | 6.3† | 0.0084 |
| HCC827-AR3 | 4.3† | 0.0022 | N/A | N/A | 0.0068 |
| HCC827-AR4 | 4.8† | 0.003 | N/A | N/A | 0.0066 |
| PC-9 | 0.0023 | 0.0022 | N/A | N/A | 0.014 |
| PC-9-AR1 | 2† | 0.0023 | N/A | N/A | 0.041 |
| PC-9-AR2 | 2.2† | 0.0056 | N/A | N/A | 0.0093 |
| HCC4006 | 0.0031 | 0.0062 | N/A | N/A | 0.019 |
| HCC4006-AR1 | 3.7† | 0.062† | N/A | N/A | 0.036 |
| HCC4006-AR2 | 5.4† | 0.0011 | N/A | N/A | 0.038 |
| HCC4011 | 0.0052 | 0.0012 | 1.4 | 0.0041 | 0.001 |
| HCC4011-AR1 | 3.3† | 0.0018 | 1.5 | 7.1† | 0.0042 |
| HCC4011-AR2 | 4.1† | 0.0019 | 1.8 | 4.4† | 0.0049 |
DOC, docetaxel; EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitor; N/A, not applicable.
The ratio of the IC50 value in each resistant line to that in the parental line is higher than five-fold.
Acquisition of EMT or other features in afatinib-resistant cell lines
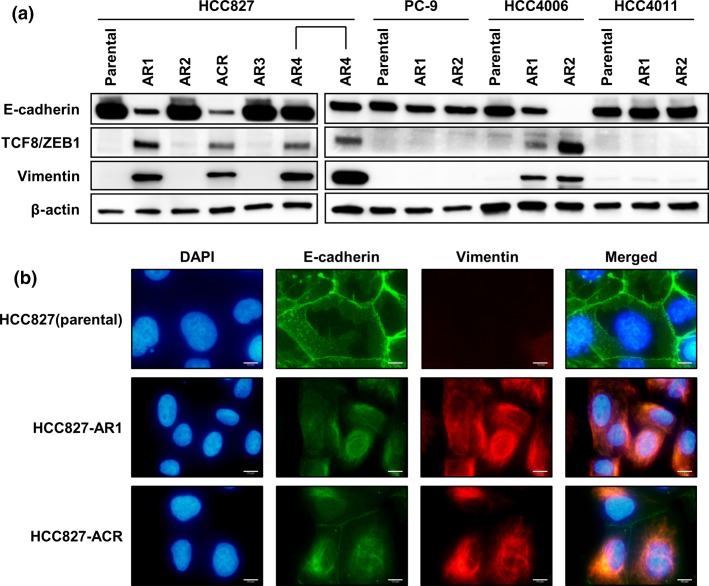
HCC827-AR1, HCC827-ACR, HCC827-AR4, and all HCC4006 sublines exhibited downregulation of E-cadherin, which is the epithelial marker, and upregulation of vimentin, which is the mesenchymal marker (Fig. 2a,b). ZEB-1, one of the regulators of EMT and a target of the miR-200c, was also expressed in cells with acquired resistance and EMT features. In fact, the expression of miR-200c was downregulated, and the methylation of the promoter regions in miR-200c was observed in these cell lines (Suppl. Fig. S2). Of note, although HCC827-AR2 that was a MET-amplified cell line did not display EMT features, HCC827-ACR, which was derived from HCC827-AR2 by afatinib plus crizotinib exposure, acquired EMT features. For PC-9 and it resistant sublines, antibody array was performed because the mechanisms of resistance to afatinib in PC-9 sublines were not identified with other assays mentioned above. In the result, phospho-stat1 upregulation by afatinib exposure was shown in PC-9 sublines although not shown in parental cell line,
Figure 2.
Epithelial to mesenchymal transition in afatinib-resistant cell lines. (a) HCC827-AR1, -ACR, -AR4, HCC4006-AR1, and -AR2 displayed downregulation of E-cadherin and upregulation of vimentin. (b) Immunofluorescence cytochemistry of E-cadherin and vimentin on HCC827 and its several sublines are shown. HCC827 displayed epithelial phenotype although HCC827-AR1 and ACR displayed mesenchymal phenotype. Scale bars, 10 μm.
Acquisition of stem cell-like properties in afatinib-resistant cell lines
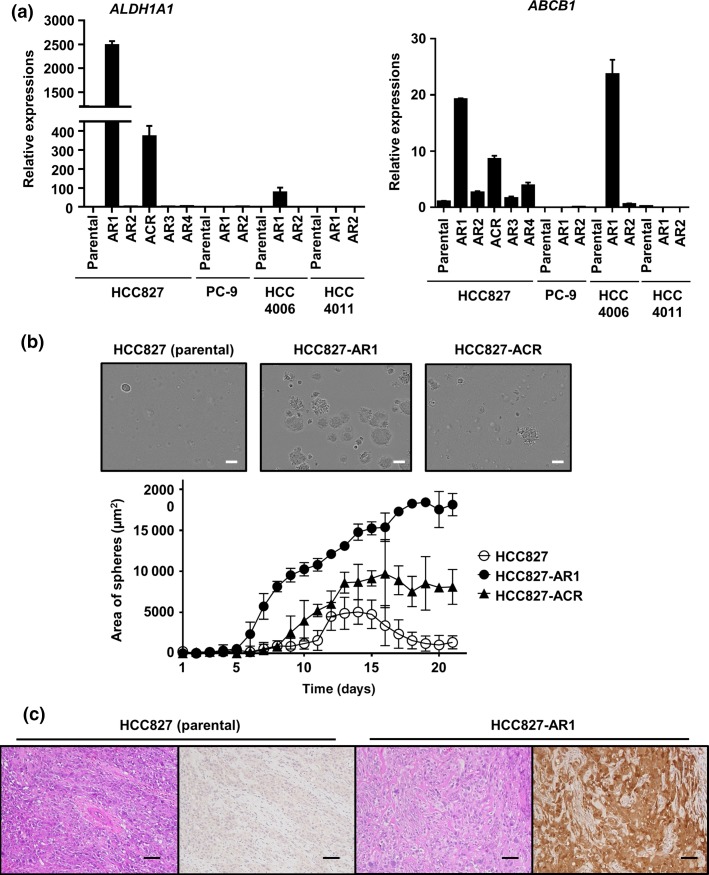
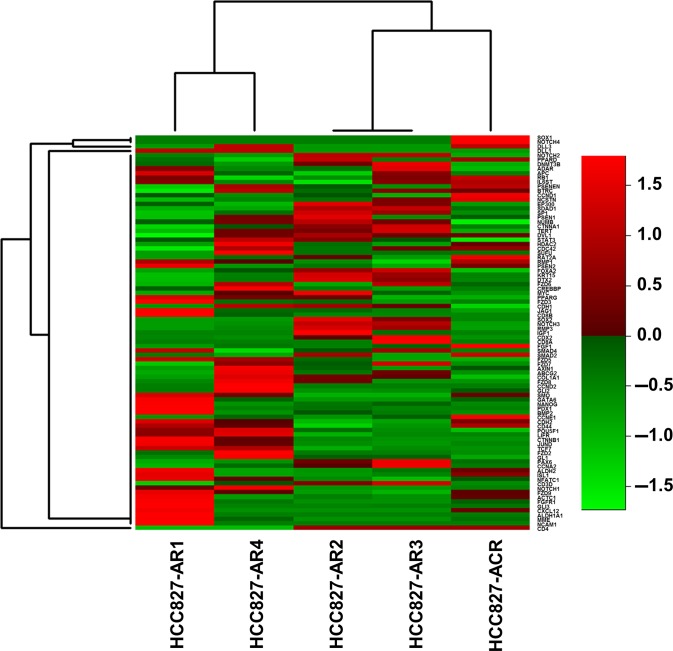
ALDH1A1 and ABCB1, which are putative stem cell markers, were overexpressed in HCC827-AR1, HCC827-ACR, and HCC4006-AR1 cells (Fig. 3a). These cell lines also exhibited EMT features. HCC827-AR1 and HCC827-ACR were displayed higher colony formation ability than HCC827 in soft agar assay (Fig. 3b). The mouse xenograft model of HCC827-AR1 cells displayed significant ALDH1 expression, although the parental HCC827 cell line did not exhibit ALDH1 expression by immunohistochemical staining (Fig. 3c). The fold changes of expressions of stem cell-related markers in HCC827 sublines from its parental cell line, which were explored using next generation sequencing, were shown in Figure 4. Among the 100 stem cell-related markers examined, the expressions of fibroblast growth factor receptor 1 (FGFR1), membrane metallo-endopeptidase (MME), Notch1 and CD44 in HCC827-AR1, HCC827-ACR, and HCC827-AR4 were significantly increased from parental cell lines (P < 0.05), but not increased in MET-amplified sublines. Additionally, HCC827-AR1 and -ACR displayed higher cell growth ability than their respective parental cell lines (Suppl. Fig. S3). These findings above supported that the established cell lines had stem cell-like properties.
Figure 3.
Stem cell-like properties of afatinib-resistant cell lines. (a) The putative stem cell markers, ALDHA1 and ABCB1 were overexpressed in HCC827-AR1, HCC827-ACR, and HCC4006-AR1 cells. The error bars indicate standard deviations. (b) The images of colonies of HCC827, -AR1 and –ACR were represented and time-lapse change of occupied area of the colonies in soft agar colony formation assays were shown. Scale bars, 100 μm. (c) In immunohistochemical staining of mouse xenograft models, HCC827 cells were negative for ALDH1 expression, but HCC827-AR1 cells displayed ALDH1 expression. Scale bars, 100 μm.
Figure 4.
The changes of expressions of stem cell-related markers in HCC827 sublines. The result of target mRNAseq for 100 stem cell-related markers in in HCC827 sublines was shown. The fold changes of gene expressions from parental cell line (HCC827) were displayed as heatmap.
Afatinib-resistant cell lines with stem cell-like properties displayed the resistance to chemotherapeutic agents
HCC827-AR1, HCC827-ACR, and HCC4006-AR1 cells harboring EMT feature and expressing stem cell markers acquired resistance to both afatinib and docetaxel. Additionally, all cell lines were sensitive to bortezomib, which is a proteasome inhibitor. The IC50 values of these drugs in all cell lines are shown in Table2.
Discussion
In this study, we established nine cell lines that acquired resistance to afatinib and one cell line that acquired resistance to afatinib plus crizotinib, and examined these resistant mechanisms. In the result, T790M mutation was not detected in all 10 cell lines, although acquisition of MET amplification, EMT, and stem cell-like properties were observed. The mechanisms of resistance to afatinib in PC-9 sublines were still unclear, although upregulation of phospho-stat1 by afatinib exposure were shown in PC-9 sublines. Further study is necessary.
Afatinib was effective against the NSCLC cell line harboring EGFR L858R/T790M mutations, and a transgenic mouse model of de novo EGFR L858R/T790M-driven lung cancer.14 In the clinical environment, there are some patients with NSCLC harboring EGFR exon 19 deletion and T790M mutation, who were sensitive to afatinib.29 However, Kim et al.30 reported acquired resistance to afatinib associated with T790M mutation. Although the reason for this discrepancy is uncertain, it may be caused by the initial concentration of afatinib, as our results along with previous reports suggest that the drug concentration affects the establishment of the mechanism of drug resistance. We previously reported that gefitinib-resistant cell lines established via step-wise exposure to the drug acquired EGFR T790M mutation or MET amplification, although those established via high-dose exposure to the drug did not acquire the T790M mutation.21 In this study, T790M mutation was not detected in our resistant sublines, and the initial concentrations of afatinib were higher than the IC50 for cells featuring EGFR L858R/T790M mutation14 in nine of the 10 cell lines. On the contrary, Kim et al.30 started afatinib exposure using a dose of < 10 nM. These facts suggest that the first-line use of afatinib with sufficient dosage may tend to avoid acquired resistance associated with T790M mutation. Regarding the MET amplification observed in the sublines derived from HCC827 and HCC4011 in the current study, it had also been identified in the resistant sublines from same parental cell lines established with gefitinib exposure in the previous reports.7,21 This fact suggests that some cell lines may have preferred resistant mechanism to EGFR-TKIs.
It is uncertain whether preventing T790M mutation contributes to prolonging the overall survival of patients. NSCLC harboring activating EGFR mutation and T790M mutation exhibit indolent progression compared with NSCLC harboring only activating EGFR mutations,31 and the prognosis of patients with T790M after EGFR-TKI failure was better than that of patients lacking this mutation.32 In addition, T790M-mutant EGFR-selective EGFR-TKIs were recently developed.33,34 In the current and our previous studies,12 the strict selection of cancer cells with a sufficient concentration of EGFR-TKIs may prevent the acquisition of conventional resistance mechanisms, such as T790M or MET amplification, and may cause cancer cells to acquire stem cell-like properties. As these cell lines displayed higher growth rate than their respective parental cell lines (Suppl. Fig. S3) and resistance to both EGFR-TKIs and chemotherapeutic agents, the acquisition of stem cell-like properties may have an unfavorable influence on overall survival.
The HCC827-ACR cell line, the afatinib and crizotinib-resistant cell line, acquired resistance associated with stem cell-like properties, although HCC827-AR2 cells, the parental cell line of HCC827-ACR cells, harbored MET amplification but did not display EMT or stem cell-like properties. Suda et al.35 also observed the acquisition of EMT features beyond T790M mutation and MET amplification. Previous studies attempted to overcome the conventional mechanisms of resistance to EGFR-TKIs, such as a change of conformation of EGFR5,6 or the dependence on another receptor tyrosine kinase (RTK),7,8,36 by targeting RTKs.33,34,37,38 However, our findings suggest that this approach may result in resistance associated with stem cell-like properties. Therefore, strategies targeting other hallmarks of cancer39 may be required to overcome these mechanisms of resistance. In the current study along with our previous study,12 a proteasome inhibitor, bortezomib displayed antitumor effect in EGFR-TKI-resistant cell lines with stem cell-like properties in our study.
In conclusion, we established cell lines with acquired resistance to afatinib and one cell line that acquired resistance to the combination of afatinib plus crizotinib. As several cell lines displayed stem cell-like properties, overcoming resistance to EGFR-TKIs associated with stem cell-like properties may be a critical component of the treatment of NSCLCs.
Acknowledgments
The authors thank Ms Fumiko Isobe (Department of Thoracic, Breast and Endocrinological Surgery, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan), Dr Takehiro Matsubara, Ms Yuko Hanafusa and Ms Yayoi Kubota (Okayama University Hospital Biobank, Okayama University Hospital, Okayama, Japan) for their technical support. Assays using IncuCyteZOOM (Essen bioscience) and Miseq (Illumina) were performed in Okayama University Hospital Biobank. This study was supported by a Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (JSPS KAKENHI Grant Number: 25293302 to ST).
Glossary
- CNG
Copy number gain
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- NSCLC
non-small cell cancer
- RTK
receptor tyrosine kinase
- TKI
tyrosine kinase inhibitor
Disclosure Statement
None of the authors have any potential conflicts of interest to disclose.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1. Receptor tyrosine kinases and these downstream signaling in HCC827 and its afatinib-resistant sublines (685 KB)
Fig. S2. The expressions and epigenetic silencing of miR-200c in EGFR-mutant lung cancer cell lines and their afatinib-resistant sublines (146 KB)
Fig. S3. Cell growth assay for HCC827 and its afatinib-resistant sublines (148 KB)
Table S1. Detailed information of primers and TaqMan® assays (11 KB)
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–87. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K, Tomizawa K, Fujii M, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol. 2011;6:1152–61. doi: 10.1097/JTO.0b013e318216ee52. [DOI] [PubMed] [Google Scholar]
- Chung JH, Rho JK, Xu X, et al. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer. 2011;73:176–82. doi: 10.1016/j.lungcan.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Shien K, Toyooka S, Yamamoto H, et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell-like properties in cancer cells. Cancer Res. 2013;73:3051–61. doi: 10.1158/0008-5472.CAN-12-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–34. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–11. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakami N, Atagi S, Goto K, et al. LUX-Lung 4: a phase II trial of afatinib in patients with advanced non-small-cell lung cancer who progressed during prior treatment with erlotinib, gefitinib, or both. J Clin Oncol. 2013;31:3335–41. doi: 10.1200/JCO.2012.45.0981. [DOI] [PubMed] [Google Scholar]
- Miller VA, Hirsh V, Cadranel J, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528–38. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- Gandhi J, Zhang J, Xie Y, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–906. [PubMed] [Google Scholar]
- Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–73. [PubMed] [Google Scholar]
- Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shien K, Toyooka S, Yamamoto H, et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell-like properties in cancer cells. Cancer Res. 2013;73:3051–3061. doi: 10.1158/0008-5472.CAN-12-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Moutinho C, Villanueva A, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062–74. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craveiro V, Yang-Hartwich Y, Holmberg JC, et al. Phenotypic modifications in ovarian cancer stem cells following Paclitaxel treatment. Cancer Med. 2013;2:751–62. doi: 10.1002/cam4.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumwalde NA, Domae E, Mescher MF, Shimizu Y. ICAM-1-dependent homotypic aggregates regulate CD8 T cell effector function and differentiation during T cell activation. J Immunol. 2013;191:3681–93. doi: 10.4049/jimmunol.1201954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shien K, Toyooka S, Ichimura K, et al. Prognostic impact of cancer stem cell-related markers in non-small cell lung cancer patients treated with induction chemoradiotherapy. Lung Cancer. 2012;77:162–7. doi: 10.1016/j.lungcan.2012.02.006. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Cited 2013 April 21] Available from URL: http://www.R-project.org/ [Google Scholar]
- Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Tamura T, Takahashi T, et al. Phase I study of continuous afatinib (BIBW 2992) in patients with advanced non-small cell lung cancer after prior chemotherapy/erlotinib/gefitinib (LUX-Lung 4) Cancer Chemother Pharmacol. 2012;69:891–9. doi: 10.1007/s00280-011-1738-1. [DOI] [PubMed] [Google Scholar]
- Kim Y, Ko J, Cui Z, et al. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Ther. 2012;11:784–91. doi: 10.1158/1535-7163.MCT-11-0750. [DOI] [PubMed] [Google Scholar]
- Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Katakami N, Yoshioka H, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: comparison between T790M mutation-positive and mutation-negative populations. Cancer. 2013;119:4325–32. doi: 10.1002/cncr.28364. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K, Tomizawa K, Osada H, et al. Conversion from the “oncogene addiction” to “drug addiction” by intensive inhibition of the EGFR and MET in lung cancer with activating EGFR mutation. Lung Cancer. 2012;76:292–9. doi: 10.1016/j.lungcan.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Ware KE, Hinz TK, Kleczko E, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regales L, Gong Y, Shen R, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–10. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Receptor tyrosine kinases and these downstream signaling in HCC827 and its afatinib-resistant sublines (685 KB)
Fig. S2. The expressions and epigenetic silencing of miR-200c in EGFR-mutant lung cancer cell lines and their afatinib-resistant sublines (146 KB)
Fig. S3. Cell growth assay for HCC827 and its afatinib-resistant sublines (148 KB)
Table S1. Detailed information of primers and TaqMan® assays (11 KB)