Abstract
Neuroblastoma (NB) is the most common extracranial solid tumor that originates from multipotent neural crest cells. NB cell populations that express embryonic stem cell-associated genes have been identified and shown to retain a multipotent phenotype. However, whether somatic reprogramming of NB cells can produce similar stem-cell like populations is unknown. Here, we sought to reprogram NB cell lines using an integration-free Sendai virus vector system. Of four NB cell lines examined, only SH-IN cells formed induced pluripotent stem cell-like colonies (SH-IN 4F colonies) at approximately 6 weeks following transduction. These SH-IN 4F colonies were alkaline phosphatase-positive. Array comparative genomic hybridization analysis indicated identical genomic aberrations in the SH-IN 4F cells as in the parental cells. SH-IN 4F cells had the ability to differentiate into the three embryonic germ layers in vitro, but rather formed NBs in vivo. Furthermore, SH-IN 4F cells exhibited resistance to cisplatin treatment and differentiated into endothelial-like cells expressing CD31 in the presence of vascular endothelial growth factor. These results suggest that SH-IN 4F cells are partially reprogrammed NB cells, and could be a suitable model for investigating the plasticity of aggressive tumors.
Keywords: Induced pluripotent stem cells, neuroblastoma, plasticity, reprogramming, Sendai virus
Neuroblastomas (NBs) are solid tumors that occur during childhood. These tumors arise in the peripheral sympathetic nervous system, particularly in the adrenal medulla and sympathetic ganglia, manifesting as thoracic, paraspinal, or abdominal tumors with metastasis to other parts of the body.1 NBs originate from multipotent neural crest stem cells; these particular stem cells give rise to diverse cell lineages, including Schwann cells, melanocytes, craniofacial cartilage, peripheral neurons, and glia.2 Some human NB cell lines partially retain a multipotent phenotype and can differentiate into several cell types.3–6 Intermediate type (I-type) NB cells possess morphological and biochemical properties of both neuroblastic and substrate-adherent NB cells, and differentiate into either type upon retinoic acid or BrdU treatment, respectively.3–6 I-type NB cells show increased tumorigenic potential compared with neuroblastic or substrate-adherent NB cells,3 suggesting that epigenetic changes in NBs contribute to aggressiveness. Indeed, recent studies have revealed that abnormal epigenetic regulation is one of the driving forces underlying NB tumorigenesis.7–10
Somatic cell reprogramming by forced expression of defined transcription factors has paved the way to generate patient-specific stem cell sources.11 These induced pluripotent stem cells (iPSCs) possess key features of embryonic stem cells (ESCs), including self-renewal and pluripotency. Several cancer cell lines have been reprogrammed via ectopic expression of defined transcription factors.12–22 Reprogrammed sarcoma and glioblastoma cells retain the same genomic aberrations as parental cells and can differentiate into multiple lineages.20,21 While reprogramming of cancer cells is a useful technique for investigating how epigenetic changes affect the plasticity of tumor cells, this technique has not been reported in NB cells to date. Here, we attempted to reprogram human NB cells to promote plasticity using a Sendai virus (SeV) vector encoding Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC) for transgene-free reprogramming of iPSCs.23,24 We used this SeV vector system to reprogram NB cells to minimize the deleterious effects on the host genome and prevent subsequent gain of artificial tumorigenic activities.
Materials and methods
SeV-mediated expressions of reprogramming factors in NB cells
SeV vectors encoding the four Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC) were obtained from DNAVEC Corporation (Ibaraki, Japan) and used according to the manufacturer’s instructions.23 We plated 5 × 105 cells and incubated them overnight in the appropriate medium. The next day, cells were transduced with SeV vectors at a multiplicity of infection of three and maintained at 37°C in 5% CO2. At 24 h post-transduction, the virus-containing medium was removed and fresh medium was added every subsequent day. At 6 days post-transduction, cells were dissociated with Accutase (Stemcell Technologies, Vancouver, BC, Canada), counted, and re-plated onto mouse embryonic fibroblasts (MEFs). The next day, the media was switched to primate ES cell medium (ReproCELL) containing 4 ng/mL basic fibroblast growth factor (bFGF, ReproCell, Yokohama, Japan) and 10 μM ROCK inhibitor and incubated at 37°C in 3% CO2. Medium was changed daily.
Endothelial tube formation assay
In vitro capillary-like tube formation was studied on Matrigel-coated wells in specific culture medium (Tube Formation Kit; Trevigen, Gaithersburg, MD, USA). NB cells were seeded onto matrigel-coated wells in Endothelial Basal Medium without serum in the presence of vascular endothelial growth factor (VEGF; 5–15 ng/mL) and bFGF (20–50 ng/mL). Normal human umbilical vein endothelial cells (HUVECs) maintained in Endothelial Cell Growth Medium 2 (PromoCell GmbH, Heidelberg, Germany) served as a positive control for tube formation. Tube-like structure formation on matrigel was observed over a 6–48 h period and results were recorded. To evaluate cell differentiation, NB cells were incubated in EndoGRO-MV-VEGF complete media kit (Millipore) with VEGF (5 ng/mL) on gelatin-coated plates. The medium was changed every other day for 1 week. Cells were then stained by immunofluorescence for CD31.
More detailed descriptions of the Material & Methods are provided in Suppl. Data S1.
Results
SeV-mediated expression of reprogramming factors in NB cells
High expression levels of pluripotency-associated genes in parental cells are related to the efficiency of iPSC generation.25 To identify suitable candidate cell lines for reprogramming, we analyzed the expression levels of pluripotency-associated genes—including NANOG, OCT4, SOX2, and KLF4—in 24 NB cell lines (Suppl. Fig. S1). We selected the NB cell lines SK-N-AS and SK-N-DZ for reprogramming based on their high expression of two (NANOG and KLF4) and three (NANOG, OCT4, and KLF4) pluripotency-associated genes, respectively (Suppl. Fig. S1). We also selected the two I-type NB cell lines SH-IN and BE(2)-C, as they exhibit stem cell-like characteristics and express pluripotency-associated genes at high levels (Suppl. Figs S1B and S2).
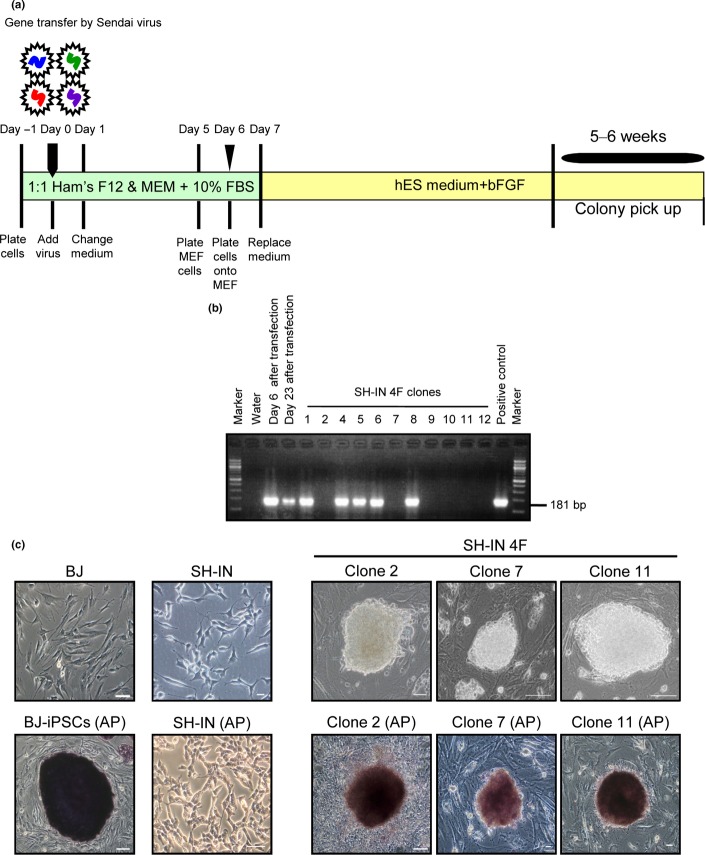
In contrast to BJ cells, all four NB cell lines proliferated rapidly in serum-free ES medium, with MEF cells dying within a week. Therefore, we prepared new MEF feeder cells weekly. SH-IN cells gave rise to iPSC-like colonies (SH-IN 4F) approximately 6 weeks after transduction (Fig.1 and Suppl. Fig. S3). Each colony was then transferred into two 96-well plates using a transfer pipet. One plate was used for AP staining and the other for passage. We selected 12 clones based on strong AP activity and cultured them for 2 months to eliminate the SeV transgenes. We were unable to detect any colonies from the SK-N-AS, BE(2)-C, or SK-N-DZ cell lines up to 7 weeks after induction. The SH-IN 4F clones could be successfully expanded and maintained in culture for at least 40 passages. To determine the presence of transgenes or a viral backbone, we examined clones using reverse transcription-polymerase chain reaction (RT-PCR). We selected three clones (2, 7, and 11) based on eliminated transgenes (Fig.1b). SH-IN 4F cells were positive for AP staining (Fig.1c). Some cells at the periphery of SH-IN 4F colonies lost AP activity 3 days after passage. Therefore, SH-IN 4F cells were passaged every 3 days.
Figure 1.
Transgene-free reprogramming of SH-IN cells. (a) Schematic outlining the transgene-free reprogramming of SH-IN cells using a SeV vector. SH-IN cells were infected with the SeV vector encoding the transcription factors OCT4, SOX2, KLF4, and c-MYC. (b) Total RNA was extracted from cells at 6 days to 4 months post-transduction and analyzed by semi-quantitative RT-PCR to verify transgene elimination. (c) Typical morphology of parental SH-IN and foreskin fibroblast BJ cells (left panels). Formation of induced pluripotent stem cells (iPSCs)-like colonies after transduction (right panels). SH-IN 4F cells stained positive for alkaline phosphatase (AP, bottom row). Scale bar: 100 μm.
Characterization of SH-IN 4F cells
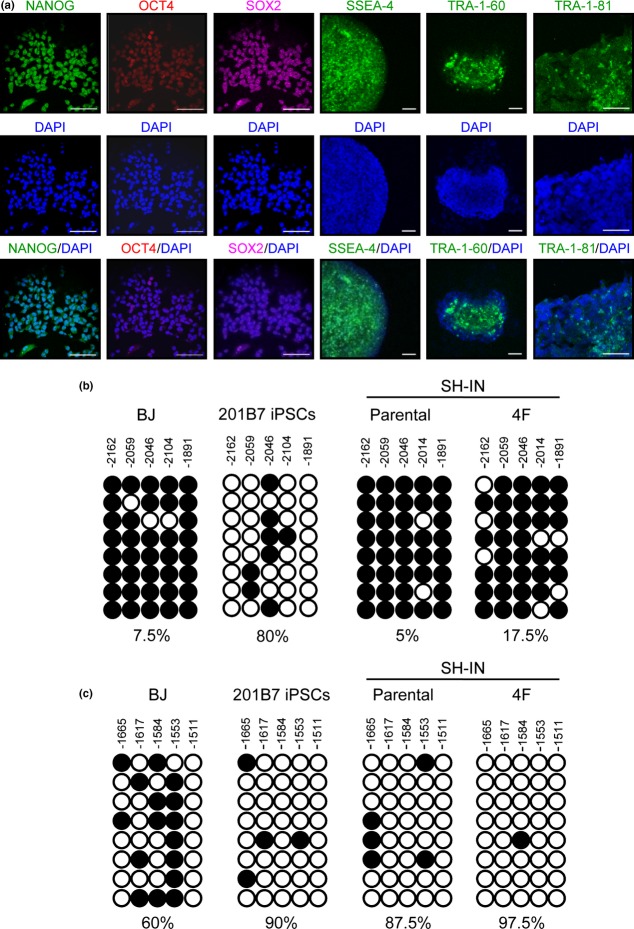
Immunocytochemistry analysis showed that SH-IN 4F cells expressed ESC-related core transcription factors, including NANOG, OCT4, and SOX2 in a manner similar to neonatal human foreskin fibroblast BJ-iPSCs and human dermal fibroblast-derived iPSCs (201B7; Fig.2a and Suppl. Fig. S4). Expression of ESC-specific surface markers, including stage specific embryonic antigen-4 (SSEA-4), tumor related antigen-1 (TRA-1-60), and tumor related antigen-1-81 (TRA-1-81), was also apparent in SH-IN 4F cells (Fig.2a). qPCR analysis revealed that expression of endogenous OCT4, SOX2, and KLF4 was induced in SH-IN 4F cells at levels comparable with those in iPSCs (Suppl. Fig. S5). NANOG, c-MYC, LIN28, and hTERT were highly expressed in SH-IN 4F cells compared with iPSCs, whereas CHD1, DNMT3B, and TDGF1 were not induced (Suppl. Fig. S5).
Figure 2.
SH-IN 4F cells express high levels of pluripotency-associated genes. (a) SH-IN 4F cells (clone 2) expressed undifferentiated embryonic stem cell (ESC) markers and surface antigens (NANOG, OCT4, SOX2, SSEA-4, TRA-1-60, and TRA-1-81) as determined by immunocytochemical analysis. Nuclei were stained with DAPI (blue). Results are representative of three independent experiments. Scale bar: 75 μm. (b) Epigenetic modification of pluripotency-related genes was examined by bisulfite genomic sequencing. (c) Reprogramming of SH-IN cells reduces NANOG promoter methylation. BJ and 201B7-iPSC lines are included as negative and positive controls, respectively. Values above each column indicate the CpG position examined from the translation initiation start codon. Each horizontal row of circles indicates the methylation status of CpG dinucleotides in one individual sequencing reaction of a bacterial clone. White circles indicate unmethylated CpGs and black circles indicate methylated CpGs. The proportion (%) of unmethylated CpGs is indicated below each cell line. Results are representatives of two independent experiments.
Reprogramming of somatic cells is accompanied by demethylation of the promoter regions of key pluripotency-associated transcription factors.11 We used bisulfite genomic sequencing to determine the degree of CpG methylation in the OCT4 and NANOG promoters of SH-IN and SH-IN 4F cells (Fig.2b,c). In contrast with 201B7 and BJ-iPSCs,23 methylation of the OCT4 promoter in SH-IN and SH-IN 4F cells remained high even after reprogramming (Fig.2b). Meanwhile, methylation of the NANOG promoter was low in both SH-IN and SH-IN 4F cells (Fig.2c).
Analysis of SH-IN 4F cells with aCGH
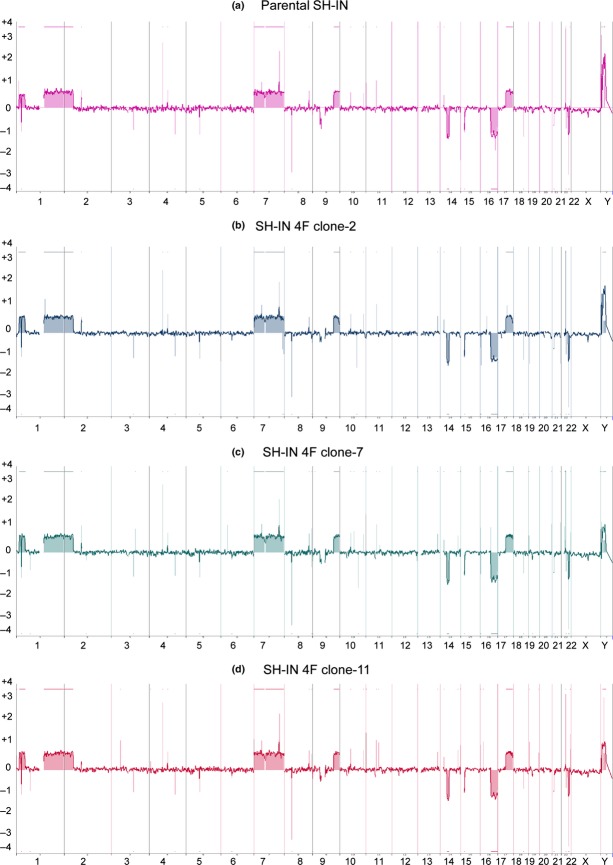
Analysis of SH-IN and SH-IN 4F cells by aCGH showed that both cells exhibited similar genomic alterations, including gains of chromosomes 1p, 1q, 2p, 7, 9q, and 17q, and losses of 14q, 16q, and 22q (Fig.3). No additional gains or losses of chromosomes were observed in SH-IN 4F cells by this array resolution.
Figure 3.
SH-IN 4F cells possess similar genomic alterations as parental SH-IN cells. Array comparative genomic hybridization. Panels show chromosomal gains and losses in parental SH-IN (a) and SH-IN 4F clones 2, 7, and 11 (b–d). A value of zero (0) indicates no loss or gain of chromosome, whereas + and − indicate gain and loss of copy number for each genomic location, respectively. SH-IN parental cells (a) exhibited ‘mixed’ genomic alterations, including gains of chromosomes 1p, 1q, 2p, 7, 9q, and 17q, and losses of 14q, 16q, and 22q. Similar genomic alterations were observed in the SH-IN 4F colonies (b–d).
Differentiation of SH-IN 4F cells into three embryonic germ layers
To determine whether SH-IN 4F cells are multipotent, we analyzed their ability to differentiate into the cells of three embryonic germ layers in vitro using embryoid body (EB) formation assays. SH-IN 4F cells readily formed EBs (Fig.4a). We observed upregulation of ectodermal (PAX6 and SOX1), mesodermal (HAND1 and FOXF1), and endodermal (AFP and GATA6) markers in SH-IN 4F cells, similar to EBs from iPSCs (Fig.4b and Suppl. Fig. S6). We also confirmed the expression of three germ layer-specific markers by immunocytochemical analyses (Fig.4c). However, in contrast with iPSCs, endogenous expression levels of pluripotency-associated genes (OCT4, SOX2, c-MYC, and KLF4) were minimally downregulated during EB formation (Suppl. Fig. S7). Furthermore, GATA6 was localized to the cytoplasm but not the nucleus (Fig.4c). We injected SH-IN 4F or parental SH-IN into the testes of NOD-SCID mice and found that SH-IN 4F formed NBs rather than teratomas (Suppl. Fig. S8). These results suggest that SH-IN 4F cells are partially reprogrammed cells, maintaining endogenous expression of reprogramming factors at high levels.
Figure 4.
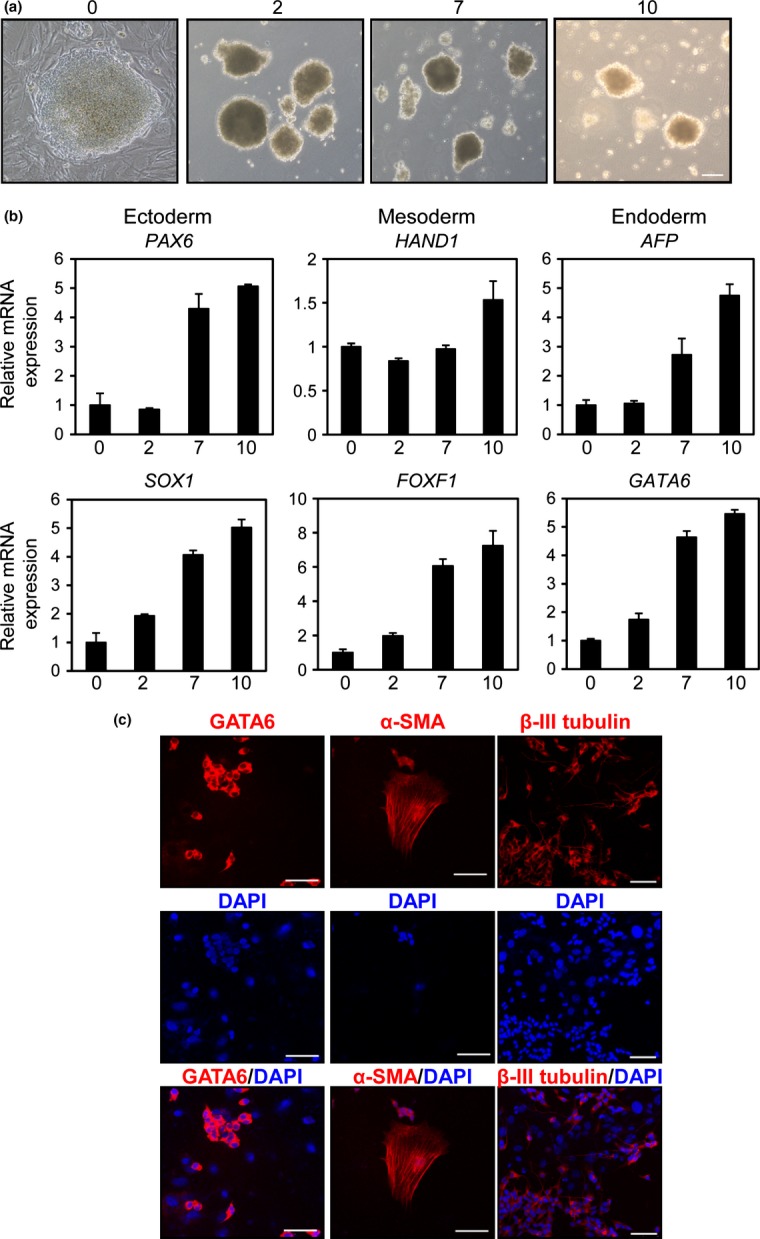
Embryoid body (EB)-mediated differentiation of SH-IN 4F cells. (a) Representative phase contrast images of EBs generated from SH-IN 4F cells. Undifferentiated cells at day 0 and differentiated EBs at days 2, 7, and 10. Scale bar: 100 μm. (b) Expression of ectodermal (PAX6 and SOX1), mesodermal (HAND1 and FOXF1) and endodermal (AFP and GATA6) markers were examined by qPCR. The x-axis represents each time point in days and the y-axis represents relative fold induction compared with day 0. All values were normalized to β-actin mRNA expression levels. (c) Immunofluorescence analysis of SH-IN 4F (clone 2) after EB differentiation: endoderm (GATA6), mesoderm (α-smooth muscle actin; [α-SMA]) and ectoderm (βIII tubulin). Nuclei were stained with DAPI (blue). Scale bars: 75 μm (GATA6 and α-SMA) and 50 μm (βIII tubulin). Results are representative of three independent experiments.
Spontaneous differentiation of SH-IN 4F cells
Cells in the center of the SH-IN 4F colonies showed similar morphology to iPSCs. However, at more than 3 days after passage, some cells at the periphery of the colonies resembled parental SH-IN cells. These cells migrated from colonies and were dispersed over the culture surface and formed monolayers (Suppl. Figs S3A and S9A). Previous reports suggest that reprogrammed cells preserve the memory of their origin and demonstrate a tendency toward spontaneous differentiation.26 We therefore examined whether cells at the periphery of the SH-IN 4F colonies had lost expression of specific markers of pluripotency (TRA-1-81 and OCT4) and recovered expression of the neuronal specific maker (βIII tubulin; Suppl. Fig. S9). The centers of SH-IN 4F colonies exhibited high levels of AP activity and expressed TRA-1-81 (Suppl. Fig. S9A,B). Meanwhile, cells at the periphery were negative for pluripotency markers (Suppl. Fig. S9A,B). The 201B7-iPSCs expressed OCT4 at high levels in the undifferentiated state (Suppl. Fig. S9C). However, expression of OCT4 was repressed in EBs from 201B7-iPS cells, and this was accompanied by the induction of βIII tubulin (Suppl. Fig. S9C). In contrast, SH-IN parental cells exhibited homogeneous staining of βIII tubulin and lower expression of OCT4 (Suppl. Fig. S9D). Most cells in SH-IN 4F colonies were OCT4-positive, with some of the peripheral cells staining positive for βIII tubulin (Suppl. Fig. S9D). At more than 3 days after passage, OCT4 localized to both the nucleus and cytoplasm of SH-IN 4F cells in colonies (Suppl. Fig. S9D). However, similar to human iPSCs (Suppl. Fig. S4), OCT4 mainly localized to the nuclei of SH-IN 4F cells when cells were passaged every 3 days (Fig.2a). Therefore, spontaneous differentiation of SH-IN 4F cells changed the subcellular localization of OCT4.
SH-IN 4F cells differentiate into vascular endothelial-like cells
OCT4 is expressed in a subpopulation of NB cells,27 and OCT4-positive NB cells serve as progenitors of tumor-derived endothelial cells.28 We therefore examined the ability of SH-IN 4F cells to form vascular endothelial-like structures. Normal HUVECs were used as a positive control. SH-IN 4F cells formed a network of cells (Fig.5a, center panel), although the tube networks were thick and less organized compared with HUVECs (Fig.5a, left panel). Parental SH-IN cells gathered together under the differentiating medium without forming tube-like structures (Fig.5a, right panel). SH-IN 4F cells expressed the endothelial-specific marker CD31 (Fig.5b) and the proportion of CD31+ cells was significantly increased compared with parental SH-IN cells (Fig.5c).
Figure 5.
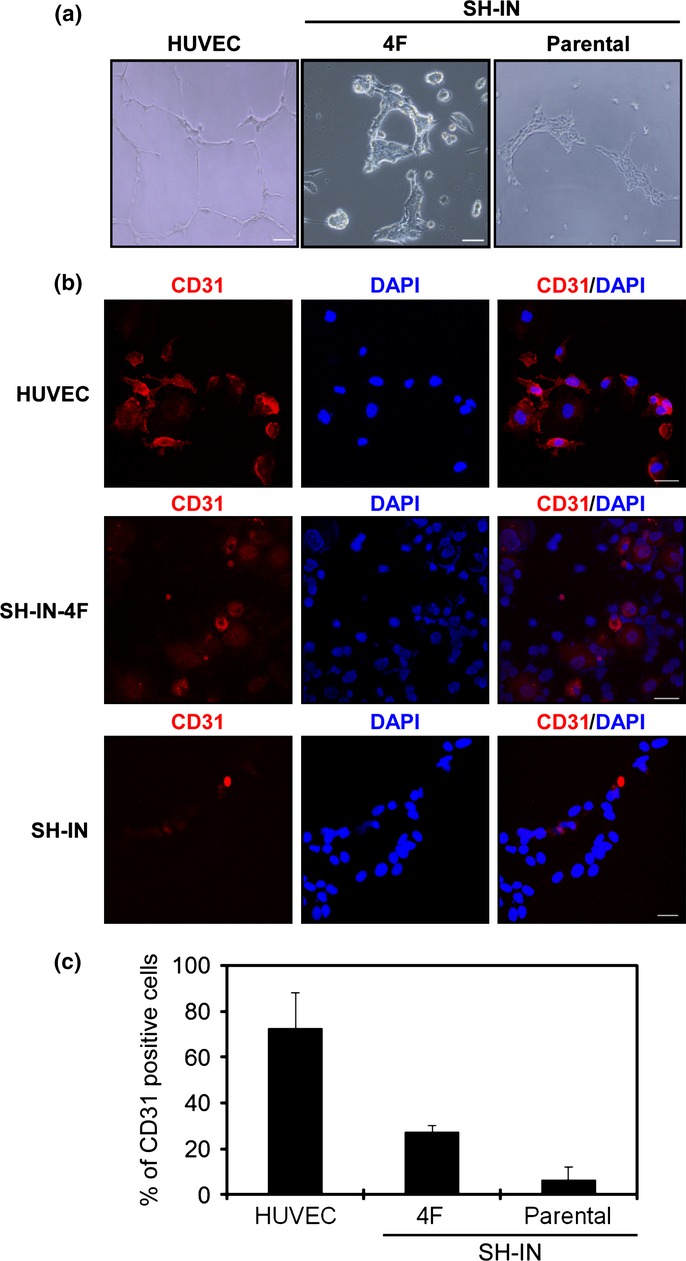
Endothelial tube formation by SH-IN 4F cells. (a) SH-IN 4F cells cultured in serum-free medium supplemented with basic fibroblast growth factor (bFGF) and EGF to examine plasticity. Representative micrograph of the tube network formed by SH-IN 4F cells and HUVEC cells. Scale bar: 300 μm. (B) HUVEC cells or SH-IN cells were cultured in differentiating medium supplemented with 2% FCS and vascular endothelial growth factor (VEGF for 7 or 10 days, respectively. Expression of the endothelial-specific marker CD31 was detected by immunofluorescence. Scale bar: 50 μm. Nuclei are stained with DAPI (blue). (c) Quantification of CD31-expressing cells as measured by immunofluorescence. Columns represent mean values from three replicate experiments, error bars represent SD. Results are representatives of three independent experiments performed using clone 7.
We next performed immunohistochemical staining for human CD31 (Suppl. Fig. S10A,B) and the immature endothelial cell maker prostate-specific membrane antigen28 (PSMA; Suppl. Fig. S10C,D), using tumor xenografts. Some spindle-shape cells were positive for human PSMA, but no specific signals for human CD31 were detected in the xenograft tumors, although western blots for CD31 suggested the induced expression of CD31 in SH-IN 4F tumors (Suppl. Fig. S10E). These results suggest that both SH-IN 4F and SH-IN cells have limited capacity for endothelial differentiation in vivo. The number of PSMA-positive cells in SH-IN 4F tumors was not significantly different from that in SH-IN tumors.
SH-IN 4F cells have a chemoresistant phenotype
We next treated SH-IN and SH-IN 4F cells with CDDP and evaluated apoptotic cell death by TUNEL assays. Apoptosis induced by CDDP was significantly reduced in SH-IN 4F cells compared with SH-IN cells (Fig.6). We then investigated the molecular mechanisms underlying chemoresistance in SH-IN 4F cells by measuring the expression of several genes known to promote chemoresistance in NB. Increased expression of CD133,29 ALDH1A1,30 ARID3B,10,31 and NCYM32 was detected in SH-IN 4F cells, while MYCN expression did not change (Suppl. Fig. S11). In contrast, reprogramming of BJ cells induced expression of MYCN, NCYM, and ARID3B (Suppl. Fig. S11C).
Figure 6.
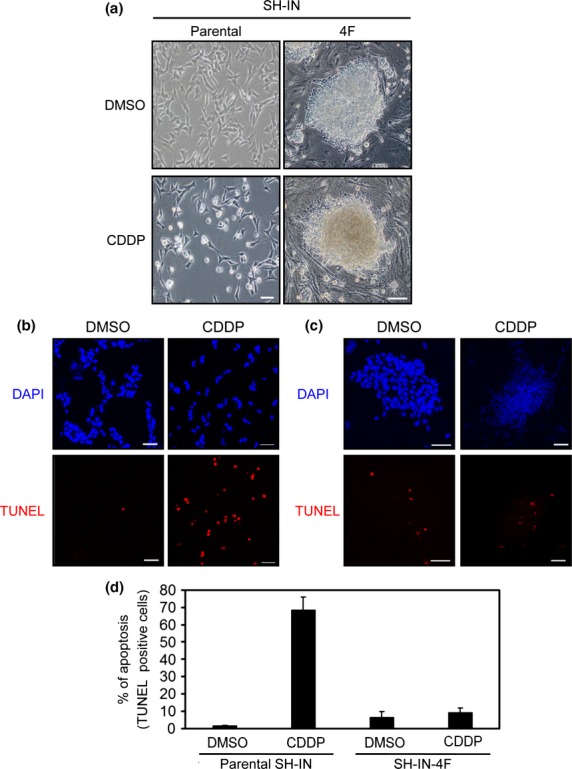
Reprogramming of SH-IN cells resulted in chemoresistance. (a) Brightfield micrographs of SH-IN cells (clone 2) treated with DMSO (top left) or 20 μM CDDP (bottom left) for 12 h. All images were acquired with an Olympus microscope using a 10 × objective lens. Scale bar: 100 μm. SH-IN (b) and reprogrammed (c) SH-IN 4F cells were treated with DMSO and CDDP (20 μM) for 12 h and subjected to TUNEL assays (red). Cell nuclei were stained with DAPI (blue). Scale bar: 50 μm. (d) TUNEL-positive cells from three independent experiments. Values are presented as the mean ± SD. Results are representatives of three independent experiments.
Discussion
We expressed reprogramming factors in NB cells using a non-integrating SeV vector. Similar to human iPSCs, SH-IN 4F cells expressed pluripotency-associated genes following reprogramming. Furthermore, markers of all three embryonic germ layers were induced during EB formation. Some cells at the periphery of SH-IN 4F colonies spontaneously differentiated into neuronal cells, with cells positive for both OCT4 and βIII tubulin detected. OCT4-positive NB cells possess the ability to differentiate into tumor cell-derived vascular-endothelial cells,28 and OCT4 is highly expressed in stem-like cells—such as side-population cells.27 Consistent with these reports, SH-IN 4F cells formed vascular endothelial-like structures in differentiating medium, and expressed the endothelial marker CD31. Taken together these findings indicate that SeV-mediated expression of reprogramming factors increased the plasticity of SH-IN cells in vitro.
Reprogramming of SH-IN cells was not sufficient to allow endothelial differentiation in vivo, though human CD31 expression was slightly induced. Given that the percentage of cells positive for PSMA in parental SH-IN cells was relatively low compared with other NB cell lines,28 endothelial differentiation of SH-IN cells in vivo may also be rare. Further experiments are required to clarify the molecular mechanisms underlining the barriers to endothelial differentiation of SH-IN cells in vivo.
Demethylation of the OCT4 promoter was limited in SH-IN 4F cells, although the expression level of OCT4 mRNA was comparable with that in iPSCs. Additionally, there was no induction of CHD1, DMNT3B, or TDGF1 expression in SH-IN 4F cells, and endogenous expression of reprogramming factors was minimally downregulated during EB formation. GATA6 localized to the cytoplasm in SH-IN 4F-derived EBs, and SH-IN 4F cells formed NBs—but not teratomas—in vivo. These results suggest that SH-IN 4F cells, while de-differentiated, were not fully reprogrammed.
Similar to SH-IN 4F cells, reprogrammed A549 human lung cancer cells highly expressed OCT4 with limited demethylation of the OCT4 promoter.33 Reprogrammed A549 cells generated invasive tumors compared with the parental cells, but the tumors were not typical teratomas, suggesting that the partial de-differentiation induced by reprogramming contributes to tumor aggressiveness. Using in vivo reprogramming mouse models, Ohnishi et al. recently reported that premature termination of reprogramming resulted in development of childhood blastomas—but not teratomas. Meanwhile long-term in vivo activation of Yamanaka factors facilitated teratoma formation.34 Ikegaki et al.9 revealed that treatment of NB cell lines with epigenetic modifiers in sphere-forming cultures enabled re-activation of reprogramming factors and induced development of poorly differentiated stem cell-like NB cells. Injection of these induced cancer stem cells into mice resulted in the formation of NBs—but not teratomas—supporting the notion that de-differentiation by partial reprogramming promotes tumor aggressiveness.
Our findings reveal that partial reprogramming of SH-IN cells facilitates CDDP resistance and enhanced differentiation into endothelial-like cells, resulting in NB heterogeneity in vitro. Therefore, de-differentiation of SH-IN cells by partial reprogramming may model the plasticity of tumor cells in NBs. Additionally, expression of CD133, ALDH1A1, ARID3B, and NCYM—but not MYCN—was induced in SH-IN 4F cells. Previous reports suggest that the expression level of MYCN is not associated with poor prognosis in MYCN non-amplified NBs, while the level of NCYM is associated with poor prognosis.32 Therefore, overexpression of reprogramming factors may contribute to chemoresistance in MYCN-non amplified SH-IN cells, possibly by induction of these stem-cell related genes.29–32,35,36
Our present findings indicate that partially reprogrammed NB cells could be a valuable in vitro model for understanding how NB cells maintain plasticity and aggressiveness.
Acknowledgments
This work was supported in part by: the Japan Agency for Medical Research and Development, AMED (AN, MO, and YN); a Grant-in-Aid from the Ministry of Health, Labor, and Welfare for the Third Term Comprehensive Control Research for Cancer, Japan (AN); a Grant-in-Aid from the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (AN); a Grant-in-Aid from the Takeda Science Foundation (AN); a Grant-in-Aid for Scientific Research on Priority Areas (Japan Society for the Promotion of Science [JSPS] KAKENHI Grant Number 17015046, AN); a Grant-in-Aid for Scientific Research (A; JSPS KAKENHI Grant Number 24249061, AN); a Grant-in-Aid for Research Activity start-up (JSPS KAKENHI Grant Number 22890241, YS); a Grant-in-Aid for Young Scientists (B; JSPS KAKENHI Grant Number 24700957, YS); and a Fund for Health and Labor Sciences Research (Grant Number 26271201, AN).
Glossary
- CDDP
Cis-diamminedichloroplatinum
- iPSCs
Induced pluripotent stem cells
- NB
Neuroblastoma
- SeV
Sendai virus
Disclosure Statement
Yasuji Ueda is an employee of DNAVEC Corporation. This does not alter the authors’ adherence to all Cancer Science policies. There are no other relevant declarations relating to employment, consultancy, patents, or products in development or marketed products.
Supporting Information
Additional supporting information may be found in the online version of this article:
Data S1. More detailed descriptions of the Material & Methods.
Fig. S1. Expression levels of pluripotency-associated genes in 24 human NB cell lines.
Fig. S2. I-type NB cells highly express pluripotency-associated genes.
Fig. S3. SH-IN 4F cells disperse over the culture surface and form monolayers.
Fig. S4. Generation of iPSCs from BJ cells by SeV vectors.
Fig. S5. Expression of pluripotency-associated genes in SH-IN 4F cells.
Fig. S6. EB-mediated differentiation of human iPSCs.
Fig. S7. SH-IN 4F cells retain expression of pluripotency-associated gene during EB formation.
Fig. S8. SH-IN 4F cells formed neuroblastomas in NOD-SCID mice.
Fig. S9. SH-IN 4F cells spontaneously differentiate towards neuronal lineages.
Fig. S10. Expression of CD31 and PSMA protein in SH-IN 4F tumors analyzed by immunohistochemistry.
Fig. S11. Expression levels of genes related to stemness or chemoresistance in SH-IN 4F cells.
References
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RA, Spengler BA, Domenech C, Porubcin M, Rettig WJ, Biedler JL. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995;6:449–56. [PubMed] [Google Scholar]
- Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res. 1989;49:219–25. [PubMed] [Google Scholar]
- Walton JD, Kattan DR, Thomas SK, et al. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838–45. doi: 10.1593/neo.04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RA, Spengler BA. Human neuroblastoma stem cells. Semin Cancer Biol. 2007;17:241–7. doi: 10.1016/j.semcancer.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Abe M, Ohira M, Kaneda A, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–34. [PubMed] [Google Scholar]
- Asada K, Watanabe N, Nakamura Y, et al. Stronger prognostic power of the CpG island methylator phenotype than methylation of individual genes in neuroblastomas. Jpn J Clin Oncol. 2013;43:641–5. doi: 10.1093/jjco/hyt058. [DOI] [PubMed] [Google Scholar]
- Ikegaki N, Shimada H, Fox AM, et al. Transient treatment with epigenetic modifiers yields stable neuroblastoma stem cells resembling aggressive large-cell neuroblastomas. Proc Natl Acad Sci U S A. 2013;110:6097–102. doi: 10.1073/pnas.1118262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Jakt LM, Nishikawa SI. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene. 2013;32:2640–8. doi: 10.1038/onc.2012.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagai K, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A. 2010;107:40–5. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Pruszak J, Varadarajan M, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–42. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Yu J, Suknuntha K, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–19. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Arai S, Hosoi M, et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–42. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cheng H, Gao S, et al. Reprogramming of MLL-AF9 leukemia cells into pluripotent stem cells. Leukemia. 2014;28:1071–80. doi: 10.1038/leu.2013.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122:3502–10. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Fan Y, Qin D, Xiaocui Bian X, Bi X. Generation and characterization of virus-free reprogrammed melanoma cells by the piggy Bac transposon. J Cancer Res Clin Oncol. 2013;139:1591–9. doi: 10.1007/s00432-013-1431-3. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32:2249–60. doi: 10.1038/onc.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engström PG, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–69. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam D, Kong CM, Lai J, Tay LL, Yang H, Wang X. Reversal of aberrant cancer methylome and transcriptome upon direct reprogramming of lung cancer cells. Sci Rep. 2012;2:592. doi: 10.1038/srep00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H, Nishishita N, Fusaki N, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108:14234–9. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–53. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Hu Q, Friedrich AM, Johnson LV, Clegg DO. Memory in induced pluripotent stem cells: reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28:1981–91. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- Das B, Tsuchida R, Malkin D, Koren G, Baruchel S, Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells. 2008;26:1818–30. doi: 10.1634/stemcells.2007-0724. [DOI] [PubMed] [Google Scholar]
- Pezzolo A, Parodi F, Marimpietri D, et al. Oct-4+/Tenascin C+ neuroblastoma cells serve as progenitors of tumor-derived endothelial cells. Cell Res. 2011;21:1470–86. doi: 10.1038/cr.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenobu H, Shimozato O, Nakamura T, et al. CD133 suppresses neuroblastoma cell differentiation via signal pathway modification. Oncogene. 2011;30:97–105. doi: 10.1038/onc.2010.383. [DOI] [PubMed] [Google Scholar]
- Cioce M, Valerio M, Casadei L, et al. Metformin-induced metabolic reprogramming of chemoresistant ALDH bright breast cancer cells. Oncotarget. 2014;30:4129–43. doi: 10.18632/oncotarget.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Era T, Takebe A, Jakt LM, Nishikawa S. ARID3B induces malignant transformation of mouse embryonic fibroblasts and is strongly associated with malignant neuroblastoma. Cancer Res. 2006;66:8331–6. doi: 10.1158/0008-5472.CAN-06-0756. [DOI] [PubMed] [Google Scholar]
- Suenaga Y, Islam SM, Alagu J, et al. NCYM, a Cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3β resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet. 2014;10:e1003996. doi: 10.1371/journal.pgen.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Zhang Z, Zhou W, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–52. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Semi K, Yamamoto T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–77. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Suenaga Y, Islam SM, et al. Functional interplay between MYCN, NCYM, and OCT4 promotes aggressiveness of human neuroblastomas. Cancer Sci. 2015;106:840–7. doi: 10.1111/cas.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji W, Suenaga Y, Kaneko Y. NCYM promotes calpain-mediated Myc-nick production in human MYCN-amplified neuroblastoma cells. Biochem Biophys Res Commun. 2015;461:501–6. doi: 10.1016/j.bbrc.2015.04.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. More detailed descriptions of the Material & Methods.
Fig. S1. Expression levels of pluripotency-associated genes in 24 human NB cell lines.
Fig. S2. I-type NB cells highly express pluripotency-associated genes.
Fig. S3. SH-IN 4F cells disperse over the culture surface and form monolayers.
Fig. S4. Generation of iPSCs from BJ cells by SeV vectors.
Fig. S5. Expression of pluripotency-associated genes in SH-IN 4F cells.
Fig. S6. EB-mediated differentiation of human iPSCs.
Fig. S7. SH-IN 4F cells retain expression of pluripotency-associated gene during EB formation.
Fig. S8. SH-IN 4F cells formed neuroblastomas in NOD-SCID mice.
Fig. S9. SH-IN 4F cells spontaneously differentiate towards neuronal lineages.
Fig. S10. Expression of CD31 and PSMA protein in SH-IN 4F tumors analyzed by immunohistochemistry.
Fig. S11. Expression levels of genes related to stemness or chemoresistance in SH-IN 4F cells.