Abstract
We investigated the role of human leukocyte antigen (HLA) class II alleles in multistage cervical carcinogenesis. Cross-sectional analysis for HLA association with cervical cancer included 1253 Japanese women: normal cytology (NL, n = 341), cervical intraepithelial neoplasia grade 1 (CIN1, n = 505), CIN grade 2 or 3 (CIN2/3, n = 96), or invasive cervical cancer (ICC, n = 311). The HLA class II allele frequencies were compared by Fisher’s exact test or the χ2-test. The Bonferroni adjustment corrected for multiple comparisons. Among the study subjects, 454 women with low-grade squamous intraepithelial lesion cytology were prospectively monitored by cytology and colposcopy every 3–4 months to analyze cumulative risk of CIN3 within the next 10 years in relation to HLA class II alleles. HLA class II DRB1*1302 allele frequency was similar between women with NL (11.7%) and CIN1 (11.9%), but significantly decreased to 5.2% for CIN2/3 and 5.8% for ICC (P = 0.0003). Correction for multiple testing did not change this finding. In women with low-grade squamous intraepithelial lesion cytology, the cumulative risk of CIN3 diagnosed within 10 years was significantly reduced among DRB1*1302-positive women (3.2% vs. 23.7%, P = 0.03). In conclusion, the two different types of analysis in this single study showed the protective effect of the DRB1*1302 allele against progression from CIN1 to CIN2/3.
Keywords: cervical cancer, cervical intraepithelial neoplasia, human leukocyte antigen, human papillomavirus, low-grade squamous intraepithelial lesion
Cervical cancer is the third most common cancer in women worldwide, with approximately 530 000 women developing the disease every year. 1 Virtually all cases of cervical cancer are caused by persistent infection with carcinogenic HPVs, specifically, HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68.2 However, although carcinogenic HPV infection is common among young women of reproductive age, only a small subset of infected women develop cervical cancer, implying the involvement of additional risk factors.3 Environmental factors such as smoking, parity, and OC use have been suggested as relevant to cervical carcinogenesis in numerous case–control studies.4
Genetic linkage with cervical cancer has also been implied.5 There is a priori biological plausibility supporting HLA involvement in the development of HPV-related cancer.6 HLA molecules are responsible for the presentation of viral antigens to the host immune system, thus playing a central role in immune recognition and subsequent clearance of virus-infected cells. Therefore, genetic variations in HLA regions may influence the efficiency of HPV antigen presentation and condition the immune responsiveness to HPV infections.7
It is known that rabbit MHC class II genes are associated with malignant conversion of cottontail rabbit papillomavirus-induced tumors.8 To date, epidemiological studies have suggested that HLA class II DRB1*1501 and DQB1*03 may be associated with an increased risk of cervical cancer, whereas DRB1*13 may protect against cervical carcinogenesis.7 However, results from these studies have not been entirely consistent. In addition, very little is known about the step at which HLA class II alleles may play a central role in multistep cervical cancer pathogenesis from HPV infection to cancer development because most HLA data on cervical cancer are based on case–control studies comparing HLA class II allele frequencies between cancer patients and healthy controls. There are few prospective studies addressing HLA association with the development of cervical cancer and precancer.
In the present study, we analyzed HLA class II data from 1253 women with normal cytology, cervical precursor lesions, or invasive cervical cancer by using a cross-sectional study design. To reduce the risk of chance findings, putative links between cervical cancer and specific HLA class II alleles were also examined in a prospective cohort study of 454 women with low-grade cervical lesions. By using cross-sectional and prospective study designs, we investigated the protective effect of the HLA class II DRB1*1302 allele against progression to cervical precancer.
Materials and Methods
Study design
Our study subjects consisted of 1253 Japanese women (341 with NL, 505 with CIN1, 86 with CIN2, 10 with CIN3, and 311 with ICC) who visited 10 hospitals for cervical cancer screening, treatment of cervical diseases, or other reasons between April 1998 and March 2011. These women were included in the cross-sectional HLA class II analysis. Non-Japanese women were excluded from the present study based on self-reported ethnicity. At enrolment, blood samples were collected for HLA genotyping. Histological specimens were stained with H&E and reviewed by two pathologists (R. F. [author] and Tomoyuki Kitagawa [Department of Pathology, Cancer Institute Hospital, Japanese Foundation of Cancer Research, Tokyo, Japan]).
Of the study subjects included in the cross-sectional analysis, 591 women with CIN1 or CIN2 were followed at 3–4-month intervals and received cytology and colposcopic examinations at each visit. Of these, 454 women with evident LSIL cytology at baseline were included in the prospective analysis. This prospective cohort study has been described elsewhere.9 Briefly, baseline cytology was reviewed by two cytopathologists (Y. H. [author] and Masafumi Tsuzuku [Department of Cytopathology, Cancer Institute Hospital, Japanese Foundation of Cancer Research]). At the time of study entry, cervical HPV DNA was determined by PCR-based methodology. In addition, information about smoking, contraceptive and reproductive history, and sexual behavior was also obtained from a self-administered questionnaire. During follow-up, a cervical biopsy was carried out only when Pap smears and colposcopic findings were suggestive of progression to CIN3 or worse.
For women whose condition was regarded as progressing, based on cytology and histology examinations carried out in the participating hospitals, the two cytopathologists and two pathologists reviewed all cytological and histological specimens collected for diagnosis of disease progression. In the prospective study, the primary endpoint was histological CIN3 lesions or worse diagnosed after rigorous pathological review. Occasionally a few difficult cases were adjudicated by joint review with consideration of cytology as well as histology. We used the primary endpoint of CIN3 or worse because CIN3 is a more certain, rigorous histological diagnosis of precancer than CIN2.3
Women entered the study voluntarily only after giving their signed informed consent. The study protocol was approved by the ethical and research review boards of the participating institutions.
Genotyping of HLA class II
Blood leukocytes were used for HLA genotyping. Total cellular DNA was extracted from these specimens and amplified by PCR using locus-specific primers. All samples were initially typed at the HLA-DRB1 and DQB1 loci using a commercially available reverse sequence-specific oligonucleotide probe typing kit (Dynal RELI SSO; Dynal Biotech, Oslo, Norway). For subtyping, group-specific amplifications were carried out as previously described.10 DRB1 and DQB1 alleles were identified by SSCP and RFLP using the PCR products. Laboratory staff who carried out the HLA class II typing were blinded to the clinical data collected from the study subjects.
Genotyping of HPV
We detected HPV DNA in cervical samples by PCR-based methodology as described previously.11 In brief, HPV DNA was amplified by PCR using consensus primers (L1C1 / L1C2 + L1C2M) for the HPV L1 region. The HPV types were identified by RFLP that has been shown to identify at least 26 types of genital HPVs.12 Laboratory staff who carried out HPV genotyping were blinded to the clinical data collected from the study subjects.
Statistical analysis
In the cross-sectional study, HLA class II allele frequencies were compared by Fisher’s exact test or the χ2-test. When an expected cell value in the 2 × 2 tables was <5, Fisher’s exact test was used. We also analyzed the data using the Bonferroni adjustment, a conventional method of correcting for multiple comparisons.
In the prospective cohort study, time to event was measured from the date of the index visit (i.e., the first instance of an abnormal cytology result) to the date of the visit at which cytological transition to CIN3 was first detected. Women whose lesions did not progress to CIN3 were censored at their last recorded return visit dates. The cumulative probability of progression to CIN3 was estimated using the Kaplan–Meier method and compared with a log–rank test, and the Cox regression model was used for statistical adjustments. Patient age, CIN grade, at the time of entry, HPV risk category (HPV16/18/31/33/35/45/52/58, other carcinogenic types, or carcinogenic HPV negative), smoking status, parity, OC use, number of lifetime sexual partners, and age at first sexual intercourse were included in the multivariate model for adjustments. As the results did not differ among the 10 hospitals, the study sites were not included in the multivariate models.
All analyses were carried out using the STATA 9 (StataCorp LP, College Station, TX, USA) statistics package. Two-sided P-values were calculated throughout and considered to be significant at <0.05.
Results
Cross-sectional analysis
The distribution of HLA-DRB1 and HLA-DQB1 alleles in 1253 Japanese women with normal cytology or cervical diseases is shown in Table1. To examine the step at which HLA class II alleles may contribute to cervical carcinogenesis, the HLA data were compared among four groups: NL (n = 341), CIN1 (n = 505), CIN2/3 (n = 96), and ICC (n = 311).
Table 1.
Correlation between human leukocyte antigen class II carrier frequencies and cervical diseases in Japanese women (n = 1253) at the commencement of the study
| NL | CIN1 | CIN2/3 | ICC | Relative risk of CIN2 or worse† | Correction for multiple comparison | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 341 | % | n = 505 | % | n = 96 | % | n = 311 | % | OR (95% CI) | P-value | Corrected P-value‡ | |
| DRB1* | |||||||||||
| 0101 | 30 | 9 | 57 | 11 | 8 | 8 | 33 | 11 | 0.98 (0.66–1.45) | 0.91 | |
| 0401 | 9 | 3 | 5 | 1 | 3 | 3 | 12 | 4 | 2.27 (1.09–4.76) | 0.03 | 0.72 |
| 0403 | 25 | 7 | 27 | 5 | 8 | 8 | 18 | 6 | 1.04 (0.64–1.69) | 0.87 | |
| 0405 | 91 | 27 | 107 | 21 | 18 | 19 | 68 | 22 | 0.87 (0.66–1.17) | 0.37 | |
| 0406 | 17 | 5 | 25 | 5 | 4 | 4 | 10 | 3 | 0.68 (0.37–1.26) | 0.21 | |
| 0407 | 5 | 1 | 6 | 1 | 0 | 0 | 1 | 0 | 0.19 (0.02–1.45) | 0.12 | |
| 0410 | 16 | 5 | 32 | 6 | 4 | 4 | 18 | 6 | 0.95 (0.57–1.60) | 0.85 | |
| 0701 | 4 | 1 | 3 | 1 | 2 | 2 | 5 | 2 | 2.10 (0.73–6.02) | 0.17 | |
| 0802 | 28 | 8 | 44 | 9 | 9 | 9 | 24 | 8 | 0.95 (0.62–1.49) | 0.81 | |
| 0803 | 45 | 13 | 92 | 18 | 16 | 17 | 48 | 15 | 0.97 (0.70–1.33) | 0.83 | |
| 0901 | 90 | 26 | 136 | 27 | 27 | 28 | 45 | 14 | 0.59 (0.44–0.79) | 0.0003§ | 0.007 |
| 1001 | 3 | 1 | 10 | 2 | 1 | 1 | 5 | 2 | 0.96 (0.36–2.54) | 0.93 | |
| 1101 | 18 | 5 | 26 | 5 | 6 | 6 | 14 | 5 | 0.94 (0.54–1.62) | 0.83 | |
| 1201 | 22 | 6 | 22 | 4 | 4 | 4 | 17 | 5 | 0.99 (0.58–1.69) | 0.98 | |
| 1202 | 9 | 3 | 13 | 3 | 1 | 1 | 4 | 1 | 0.47 (0.18–1.24) | 0.15 | |
| 1301 | 2 | 1 | 5 | 1 | 3 | 3 | 1 | 0 | 1.19 (0.35–4.09) | 0.75 | |
| 1302 | 40 | 12 | 60 | 12 | 5 | 5 | 18 | 6 | 0.44 (0.28–0.71) | 0.0003§ | 0.007 |
| 1401 | 33 | 10 | 36 | 7 | 9 | 9 | 22 | 7 | 0.92 (0.60–1.44) | 0.74 | |
| 1403 | 12 | 4 | 18 | 4 | 4 | 4 | 10 | 3 | 0.97 (0.51–1.85) | 0.92 | |
| 1405 | 11 | 3 | 18 | 4 | 7 | 7 | 14 | 5 | 1.52 (0.86–2.72) | 0.15 | |
| 1406 | 10 | 3 | 16 | 3 | 3 | 3 | 11 | 4 | 1.12 (0.58–2.18) | 0.73 | |
| 1501 | 46 | 13 | 84 | 17 | 18 | 19 | 48 | 15 | 1.07 (0.77–1.47) | 0.70 | |
| 1502 | 76 | 22 | 115 | 23 | 25 | 26 | 91 | 29 | 1.37 (1.04–1.79) | 0.02 | 0.48 |
| 1602 | 5 | 1 | 7 | 1 | 2 | 2 | 1 | 0 | 0.52 (0.14–1.84) | 0.41 | |
| DQB1* | |||||||||||
| 0202 | 2 | 1 | 2 | 0 | 1 | 1 | 3 | 1 | 2.09 (0.51–8.40) | 0.28 | |
| 03 | 203 | 60 | 282 | 56 | 58 | 60 | 176 | 57 | 1.01 (0.79–1.28) | 0.96 | |
| 0301 | 77 | 23 | 94 | 19 | 21 | 22 | 56 | 18 | 0.92 (0.68–1.24) | 0.59 | |
| 0302 | 60 | 18 | 86 | 17 | 15 | 16 | 50 | 16 | 0.91 (0.66–1.25) | 0.57 | |
| 03032 | 96 | 28 | 138 | 27 | 31 | 32 | 86 | 28 | 1.06 (0.81–1.37) | 0.69 | |
| 0401 | 89 | 26 | 105 | 21 | 18 | 19 | 66 | 21 | 0.87 (0.65–1.17) | 0.35 | |
| 0402 | 29 | 9 | 55 | 11 | 10 | 10 | 29 | 9 | 0.96 (0.64–1.43) | 0.85 | |
| 0501 | 33 | 10 | 66 | 13 | 9 | 9 | 37 | 12 | 0.96 (0.66–1.39) | 0.84 | |
| 0502 | 24 | 7 | 24 | 5 | 6 | 6 | 14 | 5 | 0.86 (0.52–1.47) | 0.58 | |
| 05031 | 26 | 8 | 41 | 8 | 8 | 8 | 29 | 9 | 1.16 (0.76–1.77) | 0.48 | |
| 0601 | 110 | 32 | 189 | 37 | 40 | 42 | 134 | 43 | 1.37 (1.07–1.74) | 0.01 | 0.24 |
| 0602 | 42 | 12 | 80 | 16 | 17 | 18 | 46 | 15 | 1.09 (0.78–1.51) | 0.62 | |
| 0603 | 2 | 1 | 5 | 1 | 3 | 3 | 1 | 0 | 1.19 (0.35–4.09) | 0.78 | |
| 0604 | 39 | 11 | 59 | 12 | 5 | 5 | 16 | 5 | 0.41 (0.26–0.68) | 0.0001§ | 0.001 |
Relative risks of cervical intraepithelial neoplasia grade 2/3 (CIN2/3)/invasive cervical cancer (ICC) were analyzed in comparison with <CIN2 (normal cytology or CIN1, n = 846).
P-values were calculated by Bonferroni adjustment to correct for multiple comparison.
Bold letters indicate statistical significance. CI, confidence interval; NL, normal cytology; OR, odds ratio.
We could not find any association between HLA class II alleles and development of CIN1, because HLA class II allele frequencies were very similar between women with NL and CIN1 (Table1). DRB1*0901 frequency was significantly decreased among women with CIN2 or worse (CIN2+) compared with women with NL or CIN1 (<CIN2) (15.0% vs. 26.2%; P = 0.0003; Pc = 0.007). In addition, DRB1*1302 frequency was also significantly lower among women with CIN2 + (5.9% vs. 11.2%, P = 0.0003, Pc = 0.007). Although the number of women with CIN2/3 was small (n = 96), DRB1*0901 frequency was significantly decreased in women with ICC compared with women with CIN2/3 (14.5% vs. 28.1%; P = 0.003). DRB1*1302 frequency was similar between women with ICC and CIN2/3 (5.8% vs. 5.2%; P = 0.99).
DQB1*0604 frequency was also significantly decreased among women with CIN2+ compared to those with <CIN2 (5.2% vs. 11.6%; P = 0.0001; Pc = 0.001).
In the cross-sectional analysis, we could not detect any significant associations between other HLA class II alleles and development of CIN2 + (Table1). The DRB1*0401, DRB1*1502, and DQB1*0601 allele frequencies were significantly associated with an increased risk of CIN2 + (P = 0.03, P = 0.02, and P = 0.01, respectively), but these associations lost significance after correction for multiple comparison. Although DRB1*1501 and DQB1*03 alleles have been suggested as relevant to an increased risk of cervical cancer in previous HLA studies,7 the frequencies of these alleles were similar between women with <CIN2 and CIN2 + (P = 0.99 and P = 0.95, respectively). Another analysis comparing HLA allele frequencies between women with and without ICC did not change the findings (data not shown).
Prospective analysis
The clinical outcomes of 454 women with LSIL cytology were monitored by cytologic and colposcopic testing at intervals of 3–4 months. Table2 shows the characteristics of the study subjects included in the prospective analysis. At baseline, 407 women had biopsy-proven CIN1 and 47 had biopsy results of CIN2. The mean age of the study subjects was 35.9 years (range, 19–54 years). In the current study, we updated the previous cohort data and extended the mean follow-up period from 39.0 months (range, 6.8–84.9 months) to 62.7 months (range, 6.8–155.9 months). By updating the data, the number of women diagnosed as progressing to CIN3 over the period of follow-up increased from 39 to 61.
Table 2.
Characteristics of Japanese women with low-grade squamous intraepithelial lesion cytology (n = 454) included in prospective analysis
| All study subjects (n = 454), n (%) | DRB1*1302 status | P-value | ||
|---|---|---|---|---|
| Positive (n = 47), n (%) | Negative (n = 407), n (%) | |||
| Age, years | ||||
| 18–29 | 86 (18.9) | 7 (14.9) | 79 (19.4) | 0.13 |
| 30–39 | 217 (47.8) | 18 (38.3) | 199 (48.9) | |
| 40+ | 151 (33.3) | 22 (46.8) | 129 (31.7) | |
| Histology at entry | ||||
| CIN grade 1 | 389 (85.7) | 44 (94) | 345 (85) | 0.10 |
| CIN grade 2 | 65 (14.3) | 3 (6) | 62 (15) | |
| HPV genotypes | ||||
| HPV16, 18, 31, 33, 35, 45, 52, 58 | 201 (44.3) | 19 (40.4) | 182 (44.7) | 0.49 |
| HPV39, 51, 56, 59, 68 | 110 (24.2) | 17 (36.2) | 93 (22.9) | |
| Low-risk types or negative | 67 (14.8) | 4 (8.5) | 63 (15.5) | |
| Undetermined | 32 (7.0) | 5 (10.6) | 27 (6.6) | |
| Multiple infection | 38 (8.4) | 2 (4.2) | 36 (8.8) | |
| Parity | ||||
| 0 | 151 (33.3) | 15 (31.9) | 136 (30.4) | 0.50 |
| 1–2 | 243 (53.5) | 28 (59.6) | 215 (52.8) | |
| 3+ | 60 (13.2) | 4 (8.5) | 56 (13.8) | |
| Use of oral contraceptives | ||||
| Yes | 39 (8.6) | 3 (6.7) | 36 (8.8) | 0.49 |
| No | 378 (90.6) | 42 (93.3) | 336 (82.6) | |
| Unknown | 37 (8.1) | 4 (8.5) | 33 (8.1) | |
| Smoking | ||||
| Never smokers | 209 (46.0) | 15 (31.9) | 185 (45.5) | 0.89 |
| Smokers | 210 (46.3) | 30 (63.8) | 189 (46.4) | |
| Current smokers | 151 (33.3) | 24 (51.1) | 136 (33.4) | |
| Former smokers | 59 (13.0) | 6 (12.8) | 53 (14.2) | |
| Unknown | 35 (7.8) | 2 (4.3) | 33 (8.1) | |
| Number of lifetime sexual partners | ||||
| 1 | 68 (15.0) | 6 (12.8) | 62 (15.2) | 0.84 |
| 2–3 | 115 (25.3) | 13 (27.7) | 102 (25.1) | |
| 4 | 235 (51.8) | 26 (55.3) | 209 (51.4) | |
| Unknown | 36 (7.9) | 2 (4.3) | 34 (8.4) | |
| Age at first sexual intercourse, years | ||||
| ≤20 | 150 (33.0) | 16 (34.0) | 134 (32.9) | 0.72 |
| 21–23 | 176 (38.8) | 17 (36.2) | 159 (39.1) | |
| ≥24 | 93 (20.5) | 12 (25.5) | 81 (19.9) | |
| Unknown | 35 (7.7) | 2 (4.3) | 33 (8.1) | |
CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus.
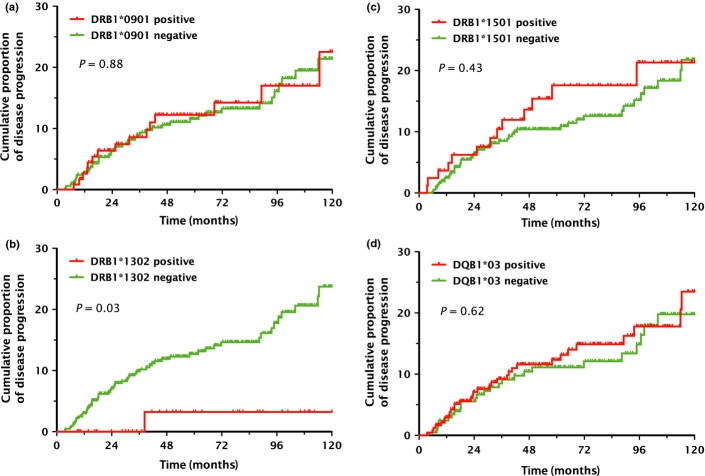
Unlike the cross-sectional analysis results, DRB1*0901 was not associated with progression to CIN3 in the prospective analysis (P = 0.88; Fig.1a). Adjustment for age, CIN grade at the time of entry, HPV risk category, parity, smoking status, OC use, number of lifetime sexual partners, and age at first sexual intercourse did not change this finding (P = 0.71; Table3).
Figure 1.
Cumulative risks of cervical intraepithelial neoplasia grade 3 within 10 years in relation to human leukocyte antigen class II polymorphisms. A Kaplan–Meier plot was used to estimate the cumulative 10-year probabilities of progression to cervical intraepithelial neoplasia grade 3 among women with low-grade squamous intraepithelial lesion cytology for selected human leukocyte antigen class II alleles: DRB1*0901 (a), DRB1*1302 (b), DRB1*1501 (c), and DQB1*03 alleles (d). P-values were calculated using the log–rank test.
Table 3.
Effect of selected human leukocyte antigen (HLA) class II alleles on progression to cervical intraepithelial neoplasia grade 3 in Japanese women with low-grade squamous intraepithelial lesion (n=454)
| HLA class II alleles | n | Person-months | Events | 10-year progression rate (95% CI) | Log–rank test | Adjusted analysis† | |
|---|---|---|---|---|---|---|---|
| P-value | Hazard ratio (95% CI) | P-value | |||||
| DRB1*0901 | |||||||
| Positive | 118 | 7274.9 | 15 | 22.5 (11.9–38.6) | 0.88 | 1.15 (0.53–2.33) | 0.71 |
| Negative | 336 | 20 715.1 | 43 | 21.4 (15.2–29.4) | 1.00 | ||
| DRB1*1302 | |||||||
| Positive | 47 | 2916.7 | 1 | 3.2 (0.5–20.8) | 0.03 | 0.13 (0.02–0.94) | 0.04 |
| Negative | 407 | 25 073.3 | 57 | 23.7 (17.6–31.5) | 1.00 | ||
| DRB1*1501 | |||||||
| Positive | 82 | 5163.5 | 13 | 21.3 (12.3–35.5) | 0.43 | 1.12 (0.56–2.425 | 0.76 |
| Negative | 372 | 22 826.5 | 45 | 21.8 (15.5–30.1) | 1.00 | ||
| DQB1*03 | |||||||
| Positive | 244 | 14 438.9 | 32 | 23.5 (15.3–35.0) | 0.62 | 1.26 (0.72–2.20) | 0.41 |
| Negative | 210 | 13 551.1 | 26 | 19.8 (13.1–29.2) | 1.00 | ||
| DQB1*0604 | |||||||
| Positive | 46 | 2879.6 | 1 | 3.2 (0.5–20.8) | 0.03 | 0.13 (0.02–0.95) | 0.04 |
| Negative | 408 | 25 110.4 | 57 | 23.7 (17.6–31.4) | 1.00 | ||
Cox regression model was used for statistical adjustments. Patient age, histological grade at the time of entry, human papillomavirus (HPV) risk category (HPV16/18/31/33/35/45/52/58, other carcinogenic types, or carcinogenic HPV negative), smoking status, parity, use of oral contraceptives, number of lifetime sexual partners, and age at first sexual intercourse were included in the multivariate model for adjustments. CI, confidence interval.
The risk of progression to CIN3 was significantly reduced among DRB1*1302-positive women (P = 0.03, log–rank test; Fig.1b), which was consistent with the cross-sectional analysis results. This protective effect of DRB1*1302 against progression to CIN3 remained statistically significant, even after adjustment for possible risk factors of cervical cancer (adjusted P = 0.04; Table3). Distributions of baseline characteristics among the DRB1*1302-positive and DRB1*1302-negative women are presented in Table2. Because of the poor reproducibility of cervical histologic interpretations,(13,14) some CIN3 lesions may have been classified incorrectly as CIN2 at baseline. Therefore, we also analyzed the follow-up data for CIN1 and CIN2 separately. In women with CIN1 histology (n = 407), the cumulative risk of CIN3 diagnosed within 10 years was lower among DRB1*1302-positive women (3.5% vs. 17.6%; P = 0.09). In women with CIN2 histology (n = 47), progression to CIN3 did not occur in DRB1*1302-positive women (cumulative probability of histological CIN3 diagnosis within 10 years, 0.0% vs. 48.9%; P = 0.28). Although these associations did not reach statistical significance due to limitations imposed by the small sample size, similar patterns were observed in separate analyses for CIN1 and CIN2.
We analyzed the protective effect of DRB1*1302 allele in relation to HPV type-specific risk category. Among DRB1*1302-positive women, only one case positive for HPV33 progressed to CIN3. Therefore, in women positive for HPV16, 18, 31, 33, 35, 45, 52, or 58, the cumulative risk of CIN3 diagnosed within 10 years was 5.9% for those who were DRB1*1302-positive and 39.6% for those who were DRB1*1302-negative (P = 0.04, log–rank test). In women positive for HPV39, 51, 56, 59, or 68, the CIN3 risk within 10 years was 0.0% for those who were DRB1*1302-positive and 9.6% for those who were DRB1*1302-negative (P = 0.29, log–rank test). In women negative for high-risk HPVs, the CIN3 risk was 0.0% for DRB1*1302-positives and 1.9% for DRB1*1302-negatives (P = 0.78).
The DQB1*0604 allele was closely linked to the DRB1*1302 allele, indicating a strong correlation (r2 = 0.96). As the DRB1*1302 and DQB1*0604 alleles were in linkage disequilibrium, similar findings were observed for the DRB1*1302 and DQB1*0604 alleles (Table3). Therefore, only DRB1*1302 data are shown in Fig.1.
We could not find any significant association between other HLA class II alleles and progression to CIN3. Data are only shown on two representative HLA class II alleles (DRB1*1501 and DQB1*03) that have been reported to be a risk factor for cervical cancer in previous HLA studies.7 In the present study, these alleles did not affect the risk of progression to CIN3 within the next 10 years (Fig.1c,d). Adjustment for possible cervical cancer risk factors did not change these findings (Table3).
Discussion
By using cross-sectional and prospective study designs, we indicated the protective effect of the DRB1*1302 allele against progression to cervical cancer and precancer. The consistent results obtained by two different analyses appear to provide stronger evidence for the protective effect of the DRB1*1302 allele. In addition, negative associations between the DRB1*13 alleles and cervical cancer have been consistently reported in case–control studies.7 In a small prospective study of French women with CIN1 (n = 86), a relationship between the DRB1*13 alleles and cytological regression was observed.15 These observations also support the protective effect of the DRB1*13 alleles. However, some studies have reported a protective effect of DRB1*1301,(16,17) and other studies have suggested that DRB1*1302 decreases the risk of cervical cancer.(18,19) In the present study, we confirmed only the protective effect of the DRB1*1302 allele because the DRB1*1301 allele is rarely detected in Japanese populations.
Cervical cancer arises by way of several carcinogenic steps: HPV acquisition, HPV persistence (development of low-grade cervical precursor lesion), progression of a persisting infection to cervical precancer, and invasion through the basement membrane of the epithelium.(3,20) In multistage cervical carcinogenesis, however, the step at which DRB1*13 alleles exert their protective effect is not fully understood. Two prospective studies of HPV infections have shown that DRB1*13 alleles do not play a protective role against the acquisition and persistence of viral infections.(21,22) This is consistent with our finding that the DRB1*1302 frequency was similar between women with NL (12%) and CIN1 (11%). In the present study, the DRB1*1302 frequency significantly decreased to 5.2% for CIN2/3 and 5.8% for ICC. In the prospective analysis of women with cytological LSIL and histological CIN1/2, the cumulative probability of CIN3 diagnosed within the next 10 years was significantly low among DRB1*1302-positive women. These observations suggest that the DRB1*1302 allele may act protectively against progression from CIN1 to CIN2/3. Several studies have reported the poor reproducibility of CIN grading, even among well-trained observers.(13,14) In particular, CIN2 is an equivocal diagnosis of precancer (a heterologous borderline category between CIN1 and CIN3).3 Therefore, it may be difficult to strictly determine whether the DRB1*1302 allele protects against progression from CIN1 to CIN2, or from CIN2 to CIN3. The protective effect of the DRB1*1302 allele against progression to cervical precancer suggests that DRB1*13 alleles could contribute to the immunological recognition of viral antigens such as the E7 protein, which is increasingly expressed during progression to cervical precancer.(23,24) However, very little is known about the binding of DRB1*13 molecules to HPV-related antigens.
As the DRB1*1302 and DQB1*0604 alleles were in linkage disequilibrium, very similar findings were observed for the DRB1*1302 and DQB1*0604 alleles in both cross-sectional and prospective analyses. Although the protective effect of DRB1*13 alleles is the most consistent HLA finding in published reports, the question is still open as to whether DRB1*1302 alone, DQB1*0604 alone, or both are associated with reduced risk of cervical diseases, or whether other MHC complex genes in linkage disequilibrium with these alleles are more important. To evaluate the independent effect of the DRB1*1302 and DQB1*0604 alleles, larger studies will be required.
The cross-sectional analysis suggested that the DRB1*0901 allele may act protectively against cervical cancer pathogenesis. A meta-analysis also reported the protective effect of DRB1*0901 against invasive cervical squamous cell carcinoma in Caucasian populations.25 Interestingly, this allele was not associated with progression to CIN3 in the prospective analysis, suggesting that the DRB1*0901 allele could exert a protective effect on progression from CIN2/3 to ICC. The DRB1*0901 allele frequency was significantly lower in women with ICC than in those with CIN2/3. Alternatively or additionally, the discrepancy may be explained by the HPV type-specific effect of the DRB1*0901 allele. When the analysis was confined to HPV16-positive women with LSIL cytology, the cumulative probability of progression to CIN3 was lower among DRB1*0901-positive women compared with DRB1*0901-negative women (data not shown), but this effect was not statistically significant (P = 0.12) due to limitations imposed by the small sample size.
We could not find any significant association between DRB1*1501 or DQB1*03 and risk of developing CIN2/3 or ICC. Effects of the DRB1*1501 and DQB1*03 alleles have not been found in prospective cohort studies so far,(15,21,22) and the results from case–control studies have not been entirely consistent.(7,16–19,25–27) Although several groups have suggested a significant association of DRB1*1501 or DQB1*0602 (or the corresponding haplotype DRB1*1501-DQB1*0602) with HPV16-positive cervical cancer,(17,26,27) an increased risk of progression to CIN3 was not observed for either the DRB1*1501 or DQB1*0602 allele, even among HPV16-positive women in the present study (data not shown). The inconsistent results regarding the DRB1*1501 and DQB1*0602 alleles may be explained by two studies suggesting that cancer risks associated with these alleles may vary according to HPV16 E6 variations.(27,28)
One may speculate that HLA class II DRB1*13 testing, alone or in combination with genotype-specific HPV testing, for women with low-grade cervical abnormalities might be useful for identifying populations at decreased risk of disease progression. Interestingly, the protective effect of the DRB1*1302 allele appears to be non-specific to the HPV genotype. However, HLA class II testing may not be recommended in clinical practice because only 10–20% of women among various ethnic populations have the DRB1*13 alleles.7 In addition, DRB1*13 does not prevent all cervical cancer; some women with cervical cancer are positive for DRB1*13.
The present study had several limitations. First, possible misclassification of CIN lesions may have affected the results. Recently, p16INK4a immunohistochemistry has been shown to increase the sensitivity and specificity of CIN2–3 detection in cervical biopsies.29 Although two pathologists reviewed all histological specimens, the histological diagnosis of cervical specimens was obtained by H&E examination alone. Second, the present study may have missed several HLA associations with cervical diseases due to limitations imposed by the small sample size. Although this is the first large-scale study on HLA association with cervical cancer in Japan, larger studies will be required to further evaluate the risk of cervical cancer and precancer in relation to HLA polymorphism. Finally, the present study did not clarify the mechanism by which the DRB1*1302 allele protects against cervical precancer. To address this, immunological studies on the binding of DRB1*13 molecules to HPV-related antigens will be required. Analyses of other genes in the MHC complex found in linkage disequilibrium with the DRB1*13 alleles may also give insight to this protective effect.
In conclusion, our results confirmed the protective effects of DRB1*1302 through two different types of analysis in a single study. Our data also suggested that the DRB1*1302 allele may function protectively against progression from CIN1 to CIN2/3. However, the mechanism underlying the protective effect of DRB1*1302 allele is not fully understood. To enhance our biological understanding of this effect, identification of specific viral epitopes presented by the DRB1*1302 allele will be needed.
Acknowledgments
The authors thank Dr. Tadahito Kanda (Center for Pathogen Genomics, National Institute of Infectious Diseases, Tokyo, Japan) for his comments on the study design, Dr. Tomoyuki Kitagawa (Department of Pathology, Cancer Institute Hospital, Japanese Foundation of Cancer Research, Tokyo, Japan) for his histological review, Mr. Masafumi Tsuzuku (Department of Cytopathology, Cancer Institute Hospital, Japanese Foundation of Cancer Research) for his cytological review, many other researchers who facilitated this study, and all the women who participated in the study. This work was supported by a grant from the Ministry of Education, Science, Sports and Culture of Japan (grant nos. 12218102 and 25462585).
Glossary
- CIN1–3
cervical intraepithelial neoplasia grade 1–3
- HLA
human leukocyte antigen
- ICC
invasive cervical cancer
- HPV
human papillomavirus
- LSIL
low-grade squamous intraepithelial lesion
- NL
normal cytology
- OC
oral contraceptive
- Pc
P corrected by the Bonferroni method
Disclosure Statement
The authors have no conflict of interest.
References
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2008;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, et al. WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009;10:321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- Castellsagué X, Muñoz N. Chapter 3: cofactors in human papillomavirus carcinogenesis–role of parity, oral contraceptives, and tobacco smoking. J Natl Cancer Inst Monogr. 2003;31:20–8. [PubMed] [Google Scholar]
- Magnusson PK, Sparén P, Gyllensten UB. Genetic link to cervical tumours. Nature. 1999;400:29–30. doi: 10.1038/21801. [DOI] [PubMed] [Google Scholar]
- Breitburd F, Ramoz N, Salmon J, Orth G. HLA control in the progression of human papillomavirus infections. Semin Cancer Biol. 1996;7:359–71. doi: 10.1006/scbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- Hildesheim A, Wang SS. Host and viral genetics and risk of cervical cancer: a review. Virus Res. 2002;89:229–40. doi: 10.1016/s0168-1702(02)00191-0. [DOI] [PubMed] [Google Scholar]
- Han R, Breitburd F, Marche PN, Orth G. Linkage of regression and malignant conversion of rabbit viral papillomas to MHC class II genes. Nature. 1992;356:66–8. doi: 10.1038/356066a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Maeda H, Oki A, et al. for Japan HPV And Cervical Cancer (JHACC) Study Group. HLA class II DRB1*1302 allele protects against progression to cervical intraepithelial neoplasia grade 3: a multicenter prospective cohort study. Int J Gynecol Cancer. 2012;22:471–8. doi: 10.1097/IGC.0b013e3182439500. [DOI] [PubMed] [Google Scholar]
- Konno Y, Numaga J, Tsuchiya N, et al. HLA-B27 subtypes and HLA class II alleles in Japanese patients with anterior uveitis. Invest Ophthalmol Vis Sci. 1999;40:1838–44. [PubMed] [Google Scholar]
- Yoshikawa H, Kawana T, Kitagawa K, et al. Detection and typing of multiple genital human papillomaviruses by DNA amplification with consensus primers. Jpn J Cancer Res. 1991;82:524–31. doi: 10.1111/j.1349-7006.1991.tb01882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H, Yoshikawa H, Kawana T, et al. Association of multiple human papillomavirus types with vulvar neoplasias. J Obstet Gynaecol Res. 1996;22:1–8. doi: 10.1111/j.1447-0756.1996.tb00927.x. [DOI] [PubMed] [Google Scholar]
- Stoler MH, Schiffman M Atypical Squamous Cells of Undetermined Significance-Low-grade Squamous Intraepithelial Lesion Triage Study (ALTS) Group. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA. 2001;285:1500–5. doi: 10.1001/jama.285.11.1500. [DOI] [PubMed] [Google Scholar]
- Carreon JD, Sherman ME, Guillén D, et al. CIN2 is a much less reproducible and less valid diagnosis than CIN3: results from a histological review of population-based cervical samples. Int J Gynecol Pathol. 2007;26:441–6. doi: 10.1097/pgp.0b013e31805152ab. [DOI] [PubMed] [Google Scholar]
- Sastre-Garau X, Cartier I, Jourdan-Da Silva N, et al. Regression of low-grade cervical intraepithelial neoplasia in patients with HLA-DRB1*13 genotype. Obstet Gynecol. 2004;104:751–5. doi: 10.1097/01.AOG.0000139834.84628.61. [DOI] [PubMed] [Google Scholar]
- Wang SS, Wheeler CM, Hildesheim A, et al. Human leukocyte antigen class I and II alleles and risk of cervical neoplasia: results from a population-based study in Costa Rica. J Infect Dis. 2001;184:1310–14. doi: 10.1086/324209. [DOI] [PubMed] [Google Scholar]
- Beskow AH, Josefsson AM, Gyllensten UB. HLA class II alleles associated with infection by HPV16 in cervical cancer in situ. Int J Cancer. 2001;93:817–22. doi: 10.1002/ijc.1412. [DOI] [PubMed] [Google Scholar]
- Maciag PC, Schlecht NF, Souza PS, et al. Major histocompatibility complex class II polymorphisms and risk of cervical cancer and human papillomavirus infection in Brazilian women. Cancer Epidemiol Biomarkers Prev. 2000;9:1183–91. [PubMed] [Google Scholar]
- Madeleine MM, Johnson LG, Smith AG, et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68:3532–9. doi: 10.1158/0008-5472.CAN-07-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarinci IC, Garcia FA, Kobetz E, et al. Cervical cancer prevention: new tools and old barriers. Cancer. 2010;116:2531–42. doi: 10.1002/cncr.25065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag PC, Schlecht NF, Souza PS, et al. Polymorphisms of the human leukocyte antigen DRB1 and DQB1 genes and the natural history of human papillomavirus infection. J Infect Dis. 2002;186:164–72. doi: 10.1086/341080. [DOI] [PubMed] [Google Scholar]
- Mahmud SM, Robinson K, Richardson H, et al. HLA polymorphisms and cervical human Papillomavirus infection in a cohort of Montreal University students. J Infect Dis. 2007;196:82–90. doi: 10.1086/518612. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Yoshikawa H, Yasugi T, et al. Ubiquitous presence of E6 and E7 transcripts in human papillomavirus-positive cervical carcinomas regardless of its type. J Med Virol. 2000;62:251–8. doi: 10.1002/1096-9071(200010)62:2<251::aid-jmv18>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Coquillard G, Palao B, Patterson BK. Quantification of intracellular HPV E6/E7 mRNA expression increases the specificity and positive predictive value of cervical cancer screening compared to HPV DNA. Gynecol Oncol. 2011;120:89–93. doi: 10.1016/j.ygyno.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Yang YC, Chang TY, Lee YJ, et al. HLA-DRB1 alleles and cervical squamous cell carcinoma: experimental study and meta-analysis. Hum Immunol. 2006;67:331–40. doi: 10.1016/j.humimm.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Apple RJ, Erlich HA, Klitz W, et al. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat Genet. 1994;6:157–62. doi: 10.1038/ng0294-157. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Terry G, Ho L, et al. Association between high-risk HPV types, HLA DRB1* and DQB1* alleles and cervical cancer in British women. Br J Cancer. 2000;82:1348–55. doi: 10.1054/bjoc.1999.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Yasugi T, Nakagawa S, et al. Human papillomavirus type 16 E6 variants and HLA class II alleles among Japanese women with cervical cancer. Int J Cancer. 2003;106:919–22. doi: 10.1002/ijc.11332. [DOI] [PubMed] [Google Scholar]
- Darragh TM, Colgan TJ, Cox JT, et al. Members of LAST Project Work Groups. The lower anogenital squamous terminology standardization Project for HPV-Associated Lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Arch Pathol Lab Med. 2012;136:1266–97. doi: 10.5858/arpa.LGT200570. [DOI] [PubMed] [Google Scholar]