Abstract
Tamoxifen, an estrogen receptor-α (ER) antagonist, is an important agent for the treatment of breast cancer. However, this therapy is complicated by the fact that a substantial number of patients exhibit either de novo or acquired resistance. To characterize the signaling mechanisms underlying this resistance, we treated the MCF7 breast cancer cell line with tamoxifen for over six months and showed that this cell line acquired resistance to tamoxifen in vitro and in vivo. We performed SILAC-based quantitative phosphoproteomic profiling on the tamoxifen resistant and vehicle-treated sensitive cell lines to quantify the phosphorylation alterations associated with tamoxifen resistance. From >5600 unique phosphopeptides identified, 1529 peptides exhibited hyperphosphorylation and 409 peptides showed hypophosphorylation in the tamoxifen resistant cells. Gene set enrichment analysis revealed that focal adhesion pathway was one of the most enriched signaling pathways activated in tamoxifen resistant cells. Significantly, we showed that the focal adhesion kinase FAK2 was not only hyperphosphorylated but also transcriptionally up-regulated in tamoxifen resistant cells. FAK2 suppression by specific siRNA knockdown or a small molecule inhibitor repressed cellular proliferation in vitro and tumor formation in vivo. More importantly, our survival analysis revealed that high expression of FAK2 is significantly associated with shorter metastasis-free survival in estrogen receptor-positive breast cancer patients treated with tamoxifen. Our studies suggest that FAK2 is a potential therapeutic target for the management of hormone-refractory breast cancers.
Approximately 70% of all breast tumors express estrogen receptor (ER)1 and are classified as estrogen receptor-alpha positive (ER+) breast cancers (1). Activation of ER by its ligand estrogen (E2), plays an essential role not only in regulating normal mammary gland development but also in the progression of hormone dependent breast cancer (2). Tamoxifen, a selective ER modulator (SERM), competes with E2 binding on ER and induces conformational changes leading to inactivation of ER (3, 4). Tamoxifen has been used for the treatment and prevention of breast cancer for more than three decades (5, 6). However, up to 40% of patients receiving tamoxifen adjuvant therapy develop recurrent disease within 5 years (7, 8). This resistance to tamoxifen and other endocrine therapy remains a major challenge in breast cancer management.
During the past two decades, a large battery of studies has been carried out to explore the mechanisms underlying resistance to endocrine therapy. One important mechanism for the development of resistance is a shift of tumor cells from growth dependent on estrogenic steroids to growth driven by growth factor signaling pathways which is independent of estrogenic steroids (9, 10). For example, activation of receptor tyrosine kinases, such as EGFR, HER2 and IGF-1R (11–15) and their downstream signaling pathways including PI3K/AKT and MAPK pathways have been linked to endocrine therapy resistance (16–18). However, early clinical trials combining tyrosine kinase inhibitors (TKIs), farnesyltransferase (RAS) inhibitor, or mammalian target of rapamycin (mTOR) inhibitors with endocrine therapy have been disappointing (19, 20). Therefore, there is an urgent need to further delineate the molecular mechanisms of endocrine resistance to identify novel therapeutic targets.
Recent advances in mass spectrometry have enabled researchers to identify and quantify thousands of proteins and phosphorylated peptides from in vitro and in vivo models. However, in contrast to a large number of genomic and transcriptomic studies investigating the mutational and gene expression changes involved in endocrine resistance, only a handful of proteomic-based studies have been reported (21–26) and only two of these studied the phosphoproteomic alterations in endocrine resistant cells (21, 22). In the current study, we employed stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative mass spectrometry approaches to identify the signaling pathways activated in tamoxifen resistant breast cancer cells derived from the ER+ MCF7 breast cancer cell line with long-term (>6 months) tamoxifen treatment. In order to comprehensively profile the phosphoproteome of the tamoxifen resistant cells, we employed two phosphopeptide enrichment approaches: anti-phosphotyrosine antibody to capture tyrosine phosphorylated peptides and TiO2 beads to enrich for serine/threonine phosphorylated peptides prior to LC-MS/MS analysis. Our study identified and quantified 5640 unique phosphopeptides corresponding to 2189 proteins, thereby generating the largest quantitative phosphoproteomic data set to date for tamoxifen resistant breast cancer cells. This enabled us to investigate the signaling mechanisms of tamoxifen resistance in a much more comprehensive manner compared with other published studies. We identified multiple signaling pathways activated in tamoxifen resistant cells with the focal adhesion pathway being one of the most enriched pathways. More importantly, we discovered the non-receptor tyrosine kinase, PTK2B (more widely known as FAK2 or PYK2) to be hyperphosphorylated and up-regulated in cells with tamoxifen resistance. Suppression of FAK2 with siRNA knockdown or a small molecule pharmacological inhibitor significantly inhibits the resistant cell proliferation and tumor formation in a xenograft mouse model. Thus our study demonstrates the potential of FAK2 as a novel therapeutic target for the treatment of endocrine resistant breast cancer.
EXPERIMENTAL PROCEDURES
Cell Culture and Establishment of Tamoxifen Resistant Cells
MCF7 was purchased from American Type Culture Collection (ATCC, Manassas, VA). To establish the tamoxifen resistant cell line (MCF7-TamR), MCF7 cells were grown in RPMI 1640 medium with 5% FBS and 1 μm tamoxifen (Sigma, St. Louis, MO) for more than 6 months. MCF7 control cells (MCF7-CTRL) were cultured in RPMI 1640 medium supplemented with 5% FBS and 0.1% ethanol as vehicle. In order to label cells with stable isotopic amino acids, MCF7-CTRL and MCF7-TamR cells were propagated in RPMI 1640 SILAC media (Thermo Fisher Scientific, Waltham, MA) with 5% FBS supplemented with light lysine (K) and arginine (R) for light and 13C615N2-K and 13C615N4-R for heavy state labeling (Cambridge Isotope Laboratories, Tewksbury, MA). The labeling efficiency was confirmed by mass spectrometry analysis.
Immunoblotting and siRNA Knockdown
Cells were harvested and lysed in modified RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate, and 1 mm sodium orthovanadate in the presence of protease inhibitors). Whole-cell protein extracts were denatured and separated in NuPAGE gels (Invitrogen, Grant Island, NY), transferred to nitrocellulose membranes, and probed with primary antibody followed by horseradish peroxidase-conjugated secondary antibodies. Primary antibodies used were pFAK1-Tyr576/577 (3281), FAK1 (4332), pFAK2-Tyr402 (3291), FAK2 (3480S), pPaxillin-Tyr118 (2541S), Paxillin (2542S), Claudin-1 (13255), E-Cadherin (3195), N-Cadherin (13116), Slug (9585), Snail (3879), TCF8/ZEB1 (3396), Vimentin (5741), ZO-1 (8193) and β-Catenin (8480) purchased from Cell Signaling Technology (Danvers, MA), β-ACTIN (A5316, Sigma, St. Louis, MO) and 4G10 anti-phosphotyrosine antibody (Millipore, Belerica, MA). 50 nm siRNAs (AM51331 from Ambion, Austin, TX and CACCAGGAGCAUAUCAACAUA from Dharmacon, Lafayette, CO) targeting FAK2 were used for transfections with RNAiMax (Invitrogen, Grant Island, NY). Cells were harvested 48 h post transfection for assessing knockdown efficiency or other follow-up experiments.
In-solution Trypsin Digestion
Cell lysates were prepared in urea lysis buffer containing 20 mm HEPES pH 8.0, 9 m urea, 1 mm sodium orthovanadate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate and 5 mm sodium fluoride. Protein estimation was carried out using BCA protein assays. Equal amount of protein from heavy labeled MCF7-CTRL and light labeled MCF7-TamR cell lysates was mixed, reduced with 5 mm dithiothreitol and alkylated with 10 mm iodoacetamide. Lysates were then diluted to less than 2 m urea final concentration using 20 mm HEPES (pH 8.0) and in-solution digestion was carried out using TPCK-treated trypsin. The tryptic peptides were desalted using C18 reverse phase column (Waters, Milford, MA) and eluted peptides were lyophilized and subjected to phosphopeptide enrichment.
Immunoaffinity Purification of Phosphotyrosine Peptides
The phosphotyrosine peptide enrichment was performed according to the manufacturer's protocol (Cell Signaling Technology). Briefly, 250 μg anti-phosphotyrosine antibody (pY100, Cell Signaling Technology) was used to immunoprecipitate (IP) tyrosine phosphorylated peptides in IP buffer containing 50 mm MOPS pH 7.2, 10 mm sodium phosphate, 50 mm NaCl. The enriched phosphopeptides were eluted using 0.1% TFA, and the eluted phosphopeptides were desalted using C18 STAGE tips, vacuum dried and kept at −80 °C before liquid chromatography-mass spectrometry (LC-MS) analysis.
TiO2-based Phosphopeptide Enrichment
Peptides were fractionated by strong cation exchange (SCX) chromatography as described earlier (27). Briefly, 10 mg of lyophilized peptides mixture was resuspended in 1 ml of SCX solvent A (5 mm KH2PO4 pH 2.7, 30% ACN) and separated on a PolySULPHOETHYL A column (5 μm, 200 Å, 200 × 9.4 mm; PolyLC Inc., Columbia, MD) with an increasing gradient of SCX solvent B (5 mm KH2PO4 pH 2.7, 30% ACN, 350 mm KCl) on an Agilent 1100 HPLC system. In total, 15 fractions were collected. Each fraction was subjected to TiO2-based phosphopeptide enrichment as described earlier (28). Briefly, each fraction was resuspended in DHB solution (80% ACN, 1% TFA, 3% 2,5-dihydroxybenzoic acid (DHB)) and incubated with TiO2 beads for 2 h. Phosphopeptide-bound TiO2 beads were sequentially washed with DHB solution followed by 80% ACN in 1% TFA). Peptides were eluted with 40 μl of 2% ammonia into 10 μl of 2% TFA.
Liquid Chromatography Tandem Mass Spectrometry
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of enriched phosphopeptides was carried out using a reverse-phase liquid chromatography system interfaced with an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). The peptides were loaded onto an analytical column (10 cm × 75 μm, Magic C18 AQ 5 μm, 120 Å) in 0.1% formic acid and eluted with a linear gradient from 5 to 60% ACN. For the analysis of phosphotyrosine peptides, Fourier transform mass spectrum (FTMS) was used for precursor scans in the range of 350–1800 m/z at 60,000 resolution on an Orbitrap analyzer. Ten most abundant precursor ions from a survey scan were selected for CID fragmentation (isolation width of 1.90 m/z; 30% normalized collision energy and activation time of 10 ms were allowed). For the analysis of TiO2 enriched phosphopeptides, FTMS were acquired in the range of 350–1800 m/z at 30,000 resolution on an Orbitrap analyzer. Ten most abundant precursor ions from a survey scan were selected for HCD fragmentation (isolation width of 1.90 m/z; 35% normalized collision energy and activation time of 0.1 ms were allowed) and MS2 spectra were acquired at 15,000 resolution at 400 m/z on the Orbitrap analyzer.
Mass Spectrometry Data Analysis
Proteome Discoverer (v 1.4; Thermo Fisher Scientific) suite was used for quantitation and database searches. The tandem mass spectrometry data were searched using SEQUEST HT algorithms against a Human RefSeq database (v59 containing 33,249 entries) supplemented with frequently observed contaminants. Trypsin was specified as the protease and a maximum of two missed cleavages were allowed. The search parameters included carbamidomethylation of cysteine as a static modification; oxidation at methionine, phosphorylation at serine, threonine and tyrosine and SILAC labeling 13C6,15N2-lysine; and 13C6,15N4-arginine as dynamic modifications. The MS tolerance was set at 10 ppm and MS/MS tolerance was set at 0.5 Da for the analysis using CID fragmentation method and 0.05 Da for the analysis using HCD fragmentation method. The relative abundance of phosphopeptides was quantitated based on the area under the MS peaks using the quantitation node in Proteome Discoverer. The Percolator algorithm (29) in Proteome Discoverer was used to filter peptide spectrum matches at a false discovery rate <1% using q-values. The probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated by the phosphoRS algorithm. Phosphorylation sites were assigned based on the phosphoRS probability ≥75% threshold. In the phosphotyrosine antibody enriched experiments, if the phosphoRS probabilities of ambiguous sites are equal for tyrosine and serine/threonine residues, phosphorylation was assigned onto the tyrosine residue. We averaged the intensities of phosphopeptides identified from the two biological replicates. We chose a twofold cut off to consider peptides as hyperphosphorylated and a 0.5-fold for peptides to be considered as hypophosphorylated. The relative ratio was calculated by dividing the intensity of each phosphorylated peptide over the average intensity of the corresponding peptide and used for plot. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.
Matrigel Invasion Assays
Cells were washed once with PBS, detached using trypsin (Life Technologies) and 5 × 104 cells were seeded into Biocoat matrigel invasion chambers (BD Biosciences, San Jose, CA) in RPMI 1640 media supplemented with 1% FBS. RPMI 1640 media supplemented with 10% serum was added in the lower wells as chemoattractant. After 24 h, matrigel and the cells retained in the invasion chamber were removed with cotton swab and the filter membranes were fixed with 4% formalin and stained with DAPI (Invitrogen). The number of cells that penetrated through the matrigel and membrane was counted for ten randomly selected viewing fields at 20× magnification.
MTT Cell Proliferation Assay
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays were performed to measure the cell proliferation. For validation of tamoxifen resistance of MCF7-TamR cells, both MCF7-TamR and MCF7-CTRL cells were seeded in a 96-well plate at a density of 2000 cells/well with the treatment of different concentrations of tamoxifen or 0.1% ethanol. For siRNA knockdown experiments, cells were transfected with different siRNAs in a 96-well plate. Cells were left to grow for 5–7 days before the MTT assay. 1 mg/ml MTT in growth media was added into each well and the plate was incubated for two hours in 37 °C. Media was then removed and 100 μl of DMSO and ethanol (1:1 by volume) was added into each well. The plate was then read for absorption at 530 nm on a microplate reader.
Immunofluorescence Staining
MCF7-CTRL and MCF7-TamR cells were seeded in 8-well chamber slides with 5% RPMI 1640 medium. After 2 days, cells were fixed with 5% formalin and permeabilized with 0.1% triton X-100. Fixed cells were blocked with 5% BSA in PBS-T and stained with anti-pY402 FAK2 (Thermo Fisher Scientific, 44–618A1) antibody conjugated with Alexa Fluor® 488. Nuclei were stained with DAPI. The staining was observed using Nikon Eclipse TE2000 microscope under 60x objective lens and images were taken using the EZ-C1 software. Foci were counted using 5 random fields and more than 20 cells were counted for each cell line.
In Vivo Tumor Xenograft Assays
Xenograft assays in NCr-nu/nu or NOD scid gamma (NSG) mouse were performed as previously described (30). Briefly, four to 6 week old female mice were embedded under the skin with either one estrogen pellet alone or one estrogen pellet along with one tamoxifen pellet 3 days prior to the cancer cell transplantation. MCF7-CTRL or MCF7-TamR cells were resuspended in matrigel:PBS (1:1 volume). 1 × 106 cells of each type of cell lines were injected onto the mammary gland fat pads of mice at two sites per mouse. To test the small molecule inhibitor, PF562271 (Selleckchem, Houston, TX), 1 × 106 of MCF7-CTRL or MCF7-TamR cells were transplanted onto the mammary gland fat pads of each NSG mice previously embedded with one tamoxifen pellet. When the tumor became palpable, 10 μl of 0.25 μm PF562271 or DMSO was administrated intratumorally at each tumor site. Tumor growth was measured two times a week. At the end of the experiment, mice were sacrificed for tissue analysis. All procedures were approved by Johns Hopkins University institutional Animal Care and Use Committee and were performed in accordance with the Animal Welfare Act regulations.
RESULTS
Establishment and Characterization of Tamoxifen-resistant MCF7 Breast Cancer Cells
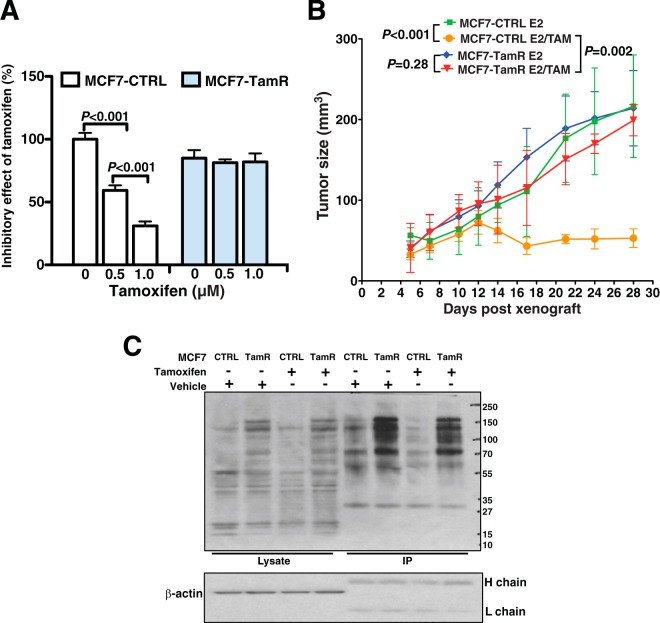
In order to identify phosphorylation regulated signaling changes in tamoxifen resistant breast cancers, we first generated tamoxifen resistant MCF7 breast cancer cells (MCF7-TamR) with long-term treatment of 1 μm tamoxifen for more than 6 months. In parallel, another set of MCF7 cells were treated with 0.1% ethanol as vehicle control cells (MCF7-CTRL). To assess the tamoxifen resistance developed in MCF7-TamR cells, we performed proliferation assays using the MCF7-TamR and MCF7-CTRL cells with treatment of different concentrations of tamoxifen. We found that MCF7-TamR cells grew ∼25% slower than MCF7-CTRL cells in the absence of tamoxifen treatment (Fig. 1A). However, inhibition induced by tamoxifen was dramatically attenuated in MCF7-TamR cells compared with MCF7-CTRL cells (Fig. 1A). We then subcutaneously transplanted MCF7-TamR cells and MCF7-CTRL cells into immunocompromised mice with the supplement of E2 pellets or E2 with tamoxifen pellets. We demonstrated that MCF7-TamR cells exhibited resistance to tamoxifen in vivo and formed significantly larger tumors than MCF7-CTRL control cells (Fig. 1B). Thus we show that we have established a tamoxifen-resistant cell line that appears to reflect hormone resistance acquired by tumors in patients treated with tamoxifen.
Fig. 1.
Tamoxifen-resistant MCF7 cell line exhibits resistance to tamoxifen and increase in global phosphotyrosine levels. A, Inhibitory effect of different concentrations of tamoxifen on the proliferation of MCF7 tamoxifen-resistant (MCF7-TamR) cell line and MCF7 sensitive control (MCF7-CTRL) cell line. Cell proliferation was assessed using MTT assay after 7 days of treatment. Quantification was done relative to vehicle-treated MCF7-CTRL. Student's t test was performed for statistical analysis. B, Tumor size of xenograft on mice grown with either MCF7-TamR or MCF7-CTRL cell lines with estrogen (E2) supplementation and treated with or without tamoxifen (TAM). Two-way Anova test was performed for statistical analysis. C, Immunoblot of tyrosine phosphorylated proteins using the anti-phosphotyrosine antibody (pTyr) on the cell lysates or after immunoprecipitation (IP) with pTyr antibody. β-actin, heavy (H) chain and light (L) chain serve as loading control.
Phosphoproteomic Analysis of Tamoxifen Resistant MCF7 Cells
Deregulation of kinase-mediated protein phosphorylation signaling pathways has been demonstrated to be involved in tumor progression and resistance to therapy (31). Given the central role of protein kinases in cell signaling networks, phosphoproteomic profiling is an ideal approach to identify activated kinase pathways and to discover therapeutic candidates for tamoxifen resistant breast cancers. To survey the phosphoproteome alterations in tamoxifen resistant cells, we first compared protein tyrosine phosphorylation levels of MCF7-CTRL with MCF7-TamR cells using anti-phosphotyrosine (anti-pTyr) antibody. As shown in Fig. 1C, the phosphotyrosine level of MCF7-TamR cells is significantly elevated compared with MCF7-CTRL cells. This suggests that activation of tyrosine kinases and their downstream signaling pathways is important for developing resistance to tamoxifen treatment.
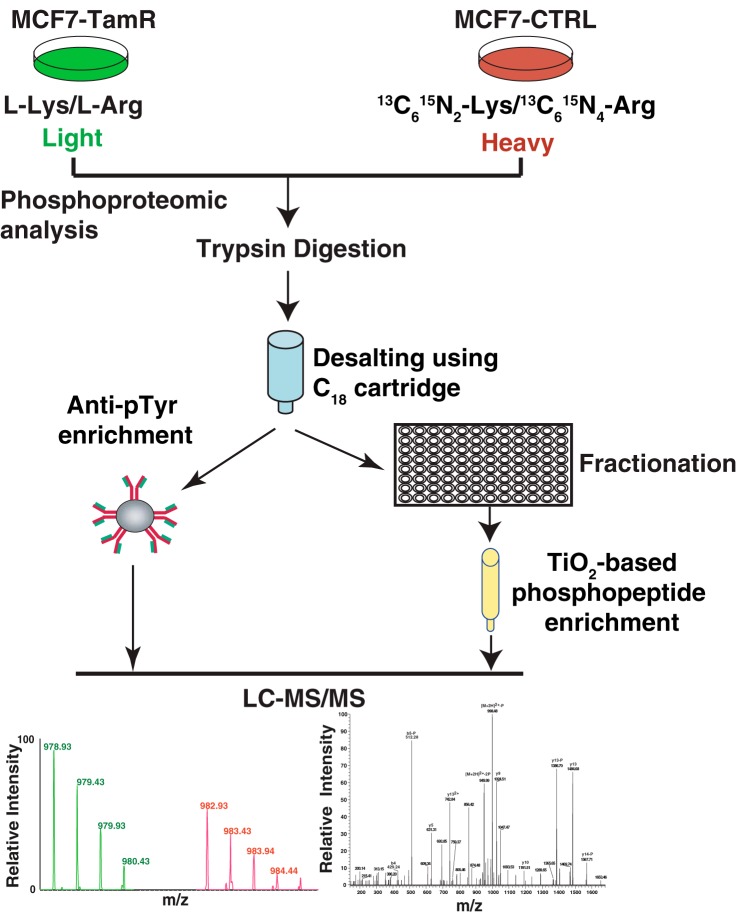
To systematically and quantitatively interrogate the phosphorylation signaling alterations that occur in the development of tamoxifen resistance, we used stable isotope labeling by amino acids in cell culture (SILAC) (32) based quantitative phosphoproteomic approach that combined two different phosphopeptide enrichment methods with high-resolution mass spectrometry (Fig. 2). Heavy-labeled MCF7-CTRL cells and light-labeled MCF7-TamR cells were lysed, mixed and digested with trypsin. In order to interrogate the observed elevation in tyrosine phosphorylation in MCF7-TamR cells, we performed antibody-based phosphotyrosine peptide enrichment. In addition, we also employed the TiO2-beads enrichment method to enrich for phosphoserine/threonine peptides in order to obtain the global phosphorylation profiles for both the resistant and sensitive cell lines. We performed biological replicate experiments to increase the reliability of our phosphoproteomic analyses. The mass spectrometry data were processed and searched against databases using SEQUEST-HT algorithms through the Proteome Discoverer platform. Using a false discovery rate (FDR) cutoff of 1%, the Percolator algorithm generated 23,798 phosphopeptide spectrum-matches. The probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated by the phosphoRS algorithm. Overall, we identified 5640 unique phosphopeptides derived from 2189 proteins. 477 of these were tyrosine phosphorylated peptides derived from 329 proteins, and ∼90% (427) of them were identified through the antibody based phosphotyrosine peptide enrichment. In addition, we identified 4723 peptides with serine phosphorylation and 599 with threonine phosphorylation (Fig. 3A). Most of the phosphopeptides were singly or doubly phosphorylated (supplemental Tables S1–S3). The SILAC ratios (MCF7-TamR cells versus MCF7-CTRL cells) of phosphopeptides obtained from the two replicate experiments showed a strong positive correlation (r = 0.80) for two independent biological replicates (Fig. 3B).
Fig. 2.
Workflow of phosphoproteomic profiling of tamoxifen-resistant and sensitive cell line. MCF7 tamoxifen-resistant (MCF7-TamR) cell line was labeled with light amino acids and MCF7 tamoxifen-sensitive control (MCF7-CTRL) cell line was labeled with heavy amino acids, mixed in equal amounts, digested with trypsin and subjected to either anti-phosphotyrosine antibody enrichment or strong cation exchange (SCX) fractionation prior to TiO2-based phosphopeptide enrichment. The enriched phosphopeptides were then subjected to LC-MS/MS analysis.
Fig. 3.
Phosphoproteomic profiling data analysis reveals global elevation of protein phosphorylation. A, Distribution of identified phosphorylated serine, threonine, and tyrosine residues in the combined data sets of anti-phosphotyrosine antibody- and TiO2- based phosphopeptide enrichments from all biological replicate samples. B, Density scatter plot of log2-transformed phosphopeptide ratios (MCF7-TamR versus MCF7-CTRL) from two biological replicates. Pearson coefficient correlation (R) is indicated. C, Distribution of phosphopeptides comparing the fold changes of phosphorylation levels of MCF7-TamR over MCF7-CTRL cells. Red dots: hyperphosphorylated peptides (>twofold), green dots: hypophosphorylated peptides (<0.5-fold). D, Top signaling pathways identified from pathway enrichment analysis of the hyperphosphorylated proteins (>twofold) in the MCF7-TamR cells. E, Cell morphology of MCF7-CTRL and MCF7-TamR viewed under phase contrast microscopy. F, Matrigel invasion assay of MCF7-CTRL and MCF7-TamR. Student's t test was performed for statistical analysis.
The relative abundance of phosphopeptides was quantitated based on the area under the MS peaks using the quantitation node in Proteome Discoverer. We found phosphorylation levels of 1529 peptides were increased (>twofold) in MCF7-TamR cells compared with MCF7-CTRL cells (Fig. 3C). Among them, 187 up-regulated phosphopeptides were identified in phosphotyrosine antibody based enrichment and 1342 were identified in TiO2 based enrichment. 409 peptides exhibited decrease in phosphorylation (<0.5-fold) in MCF7-TamR cells compared with MCF7-CTRL cells. Of these, 76 were identified from phosphotyrosine antibody enrichment and 333 were identified from TiO2 based enrichment. These differentially phosphorylated peptides correspond to 1050 proteins. Of these, 850 proteins showed up-regulation in phosphorylation levels in MCF7-TamR cells, and 233 proteins showed down-regulation in phosphorylation levels in MCF7-TamR cells. 33 proteins contained both up- and down-regulated phosphorylation sites. These results are consistent with our phosphotyrosine immunoblot observation, where we saw substantial and global elevation of protein phosphorylation in MCF7-TamR cells (Fig. 1C). This suggests that there is a robust activation of kinases in the MCF7-TamR cells which could contribute to the development of resistance.
Pathway Analysis to Identify Activated Pathways Involved in Tamoxifen Resistance
To better understand the signaling pathways involved in tamoxifen resistance, we performed a KEGG pathway analysis using an integrated online functional annotation tool, DAVID, for the proteins with increased phosphorylation in MCF7-TamR cells. This analysis revealed that several tyrosine kinase-mediated signaling pathways, such as focal adhesion kinase, ERBB, neurotrophin and insulin signaling pathways, are highly enriched in MCF7-TamR cells (Fig. 3D). Some of these signaling pathways such as focal adhesion (33, 34), MAPK (35, 36), ERBB (15, 37, 38) and insulin signaling pathways (39) have been reported to be activated and contribute to the development of resistance to tamoxifen. Significantly, multiple pathways regulating cell migration/invasion including actin cytoskeleton regulation, focal adhesion and tight junction signaling pathways, were also found to be enriched in MCF7-TamR cells (Fig. 3D). This suggests that, in addition to developing resistance to tamoxifen, MCF7-TamR cells also acquired a migration/invasion advantage. In support of this, we observed that the MCF7-TamR cells form more invadopodia-like structures compared with the MCF7-CTRL cells (Fig. 3E). We then performed matrigel invasion assay and found that MCF7-TamR cells are significantly more invasive than MCF7-CTRL cells (Fig. 3F). These observations thus support our phosphoproteomic data reflecting changes consistent with increased motility in tamoxifen resistant cells. The increase of invasiveness and morphological changes imply that the MCF7-TamR cells might have undergone epithelial-mesenchymal transition (EMT) during the development of resistance to tamoxifen. However, when we examined the key regulators and markers of EMT, we did not observe significant expression changes of most EMT markers between the two cell lines, including TCF8, vimentin, β-catenin, E-cadherin, ZO-1 and N-cadherin. We did however observe up-regulation of SLUG and CLAUDIN1 and down-regulation of SNAIL (supplemental Fig. S1). These results suggest that development of tamoxifen resistance in MCF7 cells could be accompanied by EMT-like changes but the transition was not complete.
Activation of Focal Adhesion Kinases in Tamoxifen Resistant MCF7 Cells
One of the top most enriched signaling pathways as revealed by our analysis is the focal adhesion pathway. 31 proteins in this pathway showed regulation of phosphorylation levels in the MCF7-TamR cells. Twenty-seven of these proteins were hyperphosphorylated and four were hypophosphorylated (Table I). The hyperphosphorylated proteins include both of the focal adhesion kinases, PTK2 (FAK1) and PTK2B (FAK2), their upstream kinase, SRC and multiple downstream substrates and interaction partners including Shc, p130Cas, Paxillin and Talin (Fig. 4A). The representative MS spectra of up-regulated phosphopeptides belonging to key proteins in focal adhesion pathway are depicted in Fig. 4B–4E. Our phosphoproteomic data are consistent with studies reported by different groups showing that SRC and FAK1 kinases were activated in tamoxifen resistant breast cancer cells and inhibition of these kinases could suppress the proliferation and migratory ability of tamoxifen resistant cells (33, 34). Another member of the focal adhesion complex, p130Cas, encoded by the BCAR1 gene, has been shown to be up-regulated in breast cancer with tamoxifen resistance and is associated with increased relapse and aggressiveness of the disease (40, 41). As a scaffolding protein, p130Cas can be phosphorylated by multiple kinases including SRC, FAK1 and FAK2. Phosphorylation of p130Cas regulates its interaction with many of its downstream partners (42). For instance, p130Cas can be tyrosine phosphorylated by SRC and reciprocally, p130Cas can elevate SRC kinase activity (43, 44). Tyrosine phosphorylation of p130Cas by SRC can also be enhanced by the docking of both p130Cas and SRC to focal adhesion kinases (45). Further, the interaction between p130Cas, SRC and focal adhesion kinases can promote survival and migration of breast cancer cells with tamoxifen resistance (46, 47).
Table I. A list of representative regulated proteins involved in focal adhesion pathway.
| Gene symbol | Phosphopeptide sequence | Protein name | Site | TamR/CTRL |
|---|---|---|---|---|
| PTK2B | YIEDEDYyKASVTRLPIK | focal adhesion kinase 2 (FAK2) | Y580 | 25.74 |
| RHsMREEDFIQPSSR | S778 | 8.91 | ||
| PTK2 | YMEDSTYyKASK | focal adhesion kinase 1 (FAK1) | Y662 | 2.20 |
| GSIDREDGSLQGPIGNQHIyQPVGKPDPAAPPK | Y946 | 2.74 | ||
| SRC | Y439 | |||
| FYN | WTAPEAALyGR | v-src viral oncogene | Y440 | 4.21 |
| YES | Y446 | |||
| BRAF | sPQKPIVR | v-raf oncogene homolog B | S151 | 0.45 |
| BCAR1 | RPGPGTLyDVPR | breast cancer anti-estrogen resistance 1 | Y433 | 7.41 |
| AQQGLyQVPGPSPQFQSPPAK | Y174 | 8.35 | ||
| PXN | VGEEEHVySFPNK | paxillin | Y124 | 3.42 |
| FIHQQPQSSsPVyGSSAK | Y94, S91 | 3.64 | ||
| TLN1 | STVLQQQyNR | talin 1 | Y436 | 4.61 |
| TMQFEPSTMVyDACR | Y26 | 5.94 | ||
| FYN | KLDNGGyYITTR | FYN oncogene | Y213 | 2.80 |
| GAySLSIR | Y185 | 4.14 | ||
| CRK | YRPAsASVSALIGGR | v-crk avian sarcoma virus CT10 oncogene | S194 | 5.89 |
| JUN | LAsPELER | jun proto-oncogene | S73 | 3.68 |
| MAPK8 MAPK10 | TAGTSFMMTPyVVTR | mitogen-activated protein kinase 8, 10 | Y185 | 0.37 |
| Y226 | ||||
| ACTG1 | EITALAPsTMK | actin, gamma 1 | S323 | 3.18 |
| ACTB | actin, beta | |||
| ACTN1 | HRPELIDyGK | actinin, alpha 1 | Y193 | 4.81 |
| ARHGAP5 | GGIDNPAITsDQELDDKK | Rho GTPase activating protein 5 | S1218 | 4.41 |
| RTHsDAsDDEAFTTSK | S1173, S1176 | 3.91 | ||
| BAD | RMsDEFVDSFKK | BCL2-associated agonist of cell death | S118 | 4.99 |
| FLNA | CSGPGLsPGMVR | filamin A, alpha | S1459 | 5.06 |
| APsVANVGSHCDLSLK | S2152 | 4.92 | ||
| ILK | NGtLNKHSGIDFK | integrin-linked kinase | T181 | 2.25 |
| PAK2 | FYDsNTVK | p21 protein (Cdc42/Rac)-activated kinase 2 | S132 | 2.79 |
| DGFPsGTPALNAK | S152 | 2.04 | ||
| PARVA | SPSVPKsPTPKSPPSR | parvin, alpha | S54 | 12.72 |
| PPP1CA | yGQFSGLNPGGRPITPPR | protein phosphatase 1, catalytic subunit, alpha isozyme | Y317 | 3.53 |
| PPP1CB | YQYGGLNSGRPVtPPR | protein phosphatase 1, catalytic subunit, beta isozyme | T316 | 2.49 |
| PPP1R12A | LAsTSDIEEK | protein phosphatase 1, regulatory subunit 12A | S507 | 5.55 |
| FPTTATKIsPK | S422 | 6.37 | ||
| VASP | VQIYHNPtANSFR | vasodilator-stimulated phosphoprotein | T43 | 2.13 |
| VAV2 | ASsRsPVFTPR | vav 2 guanine nucleotide exchange factor | S769, S771 | 0.50 |
| VCL | ILLRNPGNQAAyEHFETMK | vinculin | Y693 | 3.48 |
| DPSAsPGDAGEQAIR | S291 | 28.40 | ||
| ZYX | FsPGAPGGSGSQPNQK | zyxin | S341 | 4.23 |
| sPGAPGPLTLK | S404 | 7.30 |
Fig. 4.
Activation of focal adhesion pathways in tamoxifen resistance. A, Hyperphosphorylated and hypophosphorylated (MCF7-TamR versus MCF7-CTRL) proteins identified in the MCF7-TamR cells that are involved in the focal adhesion pathways. Protein phosphorylation sites and phospho-regulation patterns were specified with color-coded circles. Diagram adapted from Pathvisio, a pathway analysis tool. B-E, Representative MS spectra of hyperphosphorylated peptides of FAK2, FAK1, PXN and BCAR1.
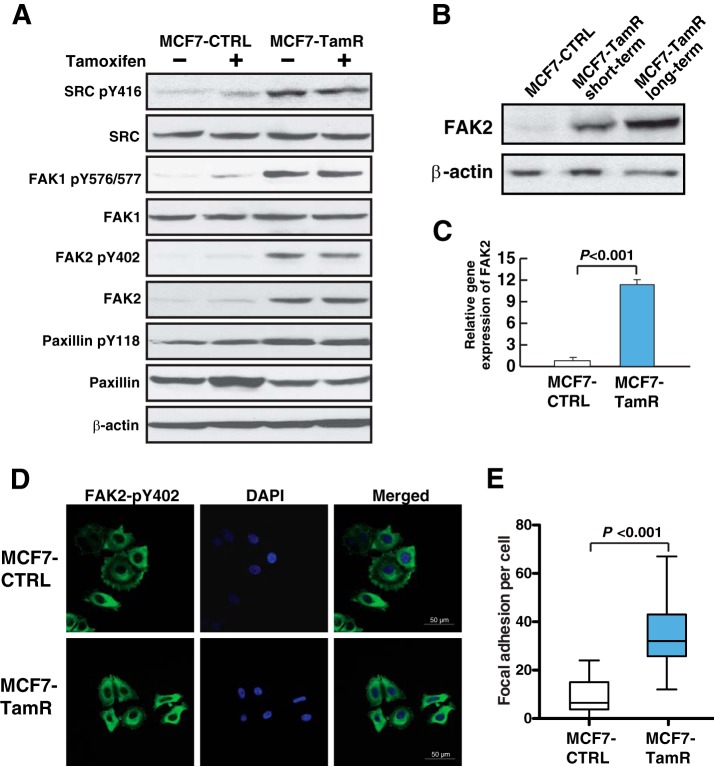
In order to confirm the activation of SRC-FAK signaling pathway in the MCF7-TamR cells, we performed Western blot analyses to examine the phosphorylation levels of the key kinases SRC, FAK1, and FAK2, and downstream protein Paxillin. We observed significant elevation of phosphorylation of SRC Y416, FAK1 Y576/Y577, FAK2 Y402, and Paxillin Y118 in tamoxifen resistant cells (Fig. 5A). Notably, short-term treatment with tamoxifen did not affect the phosphorylation levels of these signaling proteins in either MCF7-CTRL or MCF7-TamR cells, and these proteins retained hyperphosphorylation in MCF7-TamR cells even without tamoxifen treatment. This suggests that the increase in phosphorylation is stable and developed during the long-term exposure to tamoxifen. Interestingly, FAK2 also had a concomitant increase in protein expression levels in the MCF7-TamR cells compared with MCF7-CTRL cells (Fig. 5A). When we examined FAK2 expression in cells with different exposure time to tamoxifen, we observed that FAK2 expression level gradually increased during the development of tamoxifen resistance (Fig. 5B). We sought to investigate whether the up-regulation of FAK2 expression was at the transcriptional or post-transcriptional level. Our quantitative real-time RT-PCR result showed that FAK2 mRNA level increased by more than 10-fold in MCF7-TamR cells compared with MCF7-CTRL cells (Fig. 5C).
Fig. 5.
FAK2 is overexpressed and hyperphosphorylated in tamoxifen resistant cells. A, Western blot validation of phosphorylation sites of proteins involved in the focal adhesion pathway identified in phosphoproteomic profiling. β-actin serves as loading control. B, Increase in expression of FAK2 in MCF7 cells treated with tamoxifen short term or long term. C, Real-time RT-PCR analysis of FAK2 mRNA expression in MCF7-TamR and MCF7-CTRL cells. Student's t test was used for statistical analysis. D, Immunofluorescence staining of pY402 FAK2 in MCF7-CTRL and MCF7-TamR cells. Green: FAK2 pY402; Blue: DAPI staining of nuclei. E, Focal adhesions of each cell were counted and Student's t test was used for statistical analysis.
As a member of the focal adhesion kinase family, FAK2 has been reported to be mainly localized at the focal adhesions and nuclei of cells (48). Autophosphorylation at tyrosine residue Y402 activates FAK2, which then functions as a docking site for the SH2 domain of Src (49). In order to further interrogate the activation status of FAK2 in MCF7-TamR cells, we performed immunofluorescence staining of the active form of FAK2 (pY402 FAK2). As demonstrated in Fig. 5D, we observed similar cytoplasmic staining patterns of pY402 FAK2 in both MCF7-CTRL and MCF7-TamR cells. However, there was a substantial increase in the staining of foci on the periphery of MCF7-TamR cells compared with MCF7-CTRL cells. Quantification of these focal adhesions revealed an increase of ∼fivefold in MCF7-TamR cells than in MCF7-CTRL cells (p > 0.001) (Fig. 5E). Taken together, this suggests that transcriptional up-regulation and overexpression of FAK2 could play a critical role in the development of tamoxifen resistance, and suppression of FAK2 could potentially reverse the resistance to tamoxifen.
Targeting FAK2 to Suppress Proliferation of Cells with Tamoxifen Resistance
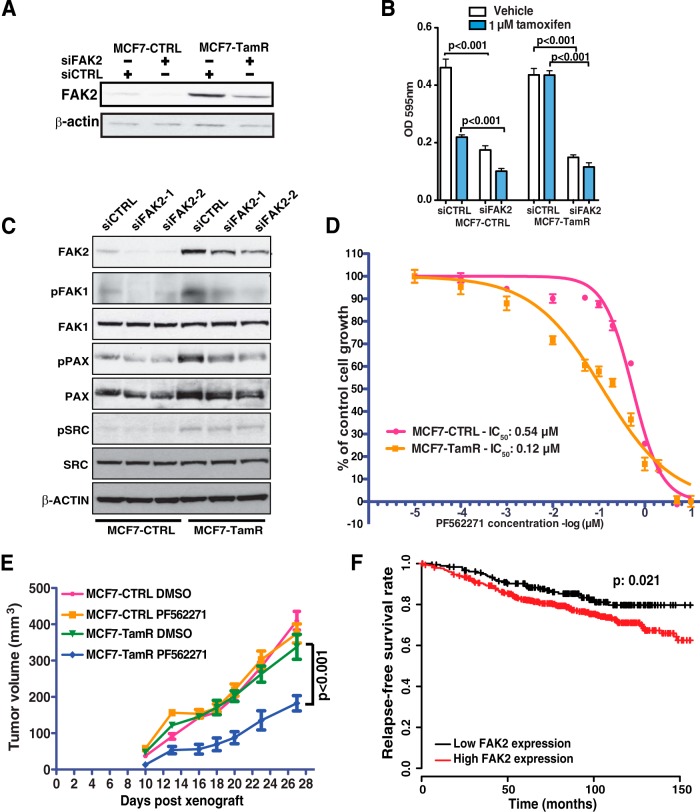
In order to evaluate the role of FAK2 in the development of tamoxifen resistance, we performed siRNA-mediated knockdown to specifically suppress FAK2 expression. Fig. 6A shows that FAK2 knockdown effectively inhibited FAK2 expression in both MCF7-CTRL and MCF7-TamR cells. We found that knockdown of FAK2 significantly reduced the proliferation of MCF7-TamR cells (Fig. 6B). Interestingly, knockdown of FAK2 also suppressed the growth of MCF7-CTRL cells and sensitized these cells to tamoxifen even further. We next examined the effect of suppressing FAK2 on signaling molecules downstream of the focal adhesion pathway. Remarkably, knockdown of FAK2 with two different sets of siRNAs reduced the phosphorylation of FAK1 and Paxillin but not SRC (Fig. 6C), suggesting that overexpression of FAK2 is pivotal in the activation of focal adhesion signaling pathway in MCF7-TamR cells.
Fig. 6.
FAK2 is a novel therapeutic target in tamoxifen-resistant breast cancer. A, Western blot analysis of siRNA knockdown of FAK2 (siFAK2) or control (siCTRL). β-actin serves as a loading control. B, Proliferation assay following siRNA knockdown of FAK2 (siFAK2) or control siRNA (siCTRL) with tamoxifen or vehicle. Student's t test was performed for statistical analysis. C, Western blot analysis of MCF7-CTRL and MCF7-TamR cells treated with two different siRNA against FAK2. Protein expression and phosphorylation levels of the proteins involved in the focal adhesion pathway were examined. D, The inhibitory effect of PF562271 on cell growth of MCF7-CTRL and MCF7-TamR cell. The IC50 curve was plotted using a nonlinear regression dose-response (variable slope) curve fit. E, Tumor volumes of mice xenograft grown with either MCF7-TamR or MCF7-CTRL cells with the treatment of PF562271 or DMSO control. Two-way Anova was performed for statistical analysis. F, Kaplan-Meier curve of metastasis-free survival of ER+ patients treated with tamoxifen. The patients were stratified based on expression of FAK2.
In order to determine the therapeutic potential of targeting focal adhesion pathway in tamoxifen resistant breast cancer, we employed a potent pan-FAK selective pharmacological inhibitor, PF562271. PF562271 has been selected for clinical trials in patients with pancreatic, head and neck, and prostatic neoplasms (50). We performed proliferation assays to assess the IC50 of PF562271 in MCF7-CTRL or MCF7-TamR cells and found that MCF7-TamR cells were four times more sensitive to PF562271 than MCF7-CTRL cells (Fig. 6D). This suggests that activation of focal adhesion pathway significantly contributes to the resistance to tamoxifen treatment. We further validated the therapeutic potential of PF562271 in suppression of tamoxifen resistant tumor growth in a xenograft tumor model. MCF7-CTRL or MCF7-TamR cells were orthotopically transplanted into the mammary gland fat pad of immunocompromised mice and treated with DMSO or PF562271. As demonstrated in Fig. 6E, the treatment with PF562271 significantly reduced the tumor formation of MCF7-TamR cells.
Finally, we sought to examine whether FAK2 expression correlated with clinical outcomes in patients with ER+ breast cancers. A publicly available gene expression database (51) of tumors from breast cancer patients was used for this survival analysis. We focused our analysis on 657 ER+ tumors treated with tamoxifen-based therapy. We found that high expression level of FAK2 is significantly associated (p = 0.021) with poorer outcomes of these patients (Fig. 6F). These clinical data suggest that FAK2 is a potential target of relevance to aggressive clinical behavior and hormone resistance in breast cancer.
DISCUSSION
Tamoxifen resistance remains a major challenge in current breast cancer management. Unveiling druggable protein targets that are critical for ER+ breast cancer cells to evade the inhibitory effects of endocrine therapy is an urgent and unmet need for developing novel therapeutic options to benefit patients with hormone refractory breast cancer. In this study, we employed SILAC labeling based LC-MS/MS proteomic approaches to comprehensively profile the phosphoproteome of MCF7 cells with tamoxifen resistance. We applied two phosphopeptide enrichment methods to identify and quantify alterations in serine, threonine and tyrosine phosphorylated peptides. This strategy allowed us to decipher signaling pathway changes at a much greater depth compared with other published studies (21, 22). With this comprehensive phosphoproteome profiling, we discovered for the first time, substantial and global elevation of protein phosphorylation in MCF7 tamoxifen resistant cells, strongly implicating that a number of kinases are activated in these cells. Our pathway analysis identified multiple crucial kinase-mediated signaling pathways to be hyperactive in tamoxifen resistant cells, including MAPK, ERBB, insulin and FAK signaling pathways. Our findings support previous reports and offer a more global and complete view of these activated pathways. For instance, in addition to BRAF and multiple MAP kinases, we also found 29 additional important signaling molecules in the MAPK signaling network to be regulated in tamoxifen resistant cells.
Previous studies have shown that the SRC and FAK signaling pathway were activated during the progression of hormone dependent breast cancer (33, 34). However, the mechanism of activation of SRC and FAK signaling has not been fully elucidated. In our study, both SRC and FAK1 were identified to be hyperphosphorylated in tamoxifen resistant cells. Moreover, several SRC and FAK substrates including SHC, Paxillin and BCAR1 and many downstream proteins were also found to be hyperphosphorylated in these cells. Most importantly, for the first time, we showed that FAK2 was transcriptionally overexpressed and hyperphosphorylated in tamoxifen resistant cells. Inhibition of FAK2 with small molecule inhibitor or specific siRNA significantly suppressed cell growth and tumor formation of resistant cells. We also demonstrated that specific siRNA targeting FAK2 substantially reduced the protein phosphorylation level of FAK1 and Paxillin, suggesting that FAK2 is a key kinase modulating the focal adhesion pathway. Our patient survival analysis using breast cancer expression database showed that high expression of FAK2 is associated with a significant decrease in the survival of ER+ patients on tamoxifen treatment, suggesting the clinical importance of FAK2 in this disease.
In summary, we show through our phosphoproteomic approaches that multiple kinase-mediated signaling pathways are activated in tamoxifen resistant cells. Our in vitro and in vivo functional studies demonstrated that the tyrosine kinase FAK2 plays a pivotal role in the development of resistance to tamoxifen and could potentially be a novel therapeutic target for this disease. However, given the complexity of signaling pathways and tumor heterogeneity, these discoveries require further testing and validation in larger cohorts of patients. Because our study was performed on one cell line, further investigation is needed to support the broader applicability of our findings. In particular, the exact mechanisms underlying FAK2 up-regulation and activation in tamoxifen resistant tumors and the efficacy of inhibition of FAK2 in pre-clinical and clinical settings need to be further investigated.
Supplementary Material
Acknowledgments
We thank the Department of Biotechnology of the Government of India for research support to the Institute of Bioinformatics, Bangalore, India. S. R. is a senior research fellow funded by University Grants Commission, Government of India. We thank the Majlis Amanah Rakyat (MARA) of Government of Malaysia for the research fellowship to M.S.Z. We thank the Mass Spectrometry and Proteomics Facility at Johns Hopkins University for their assistance.
Footnotes
Author contributions: X.W., M.S.Z., S.S., and A.P. designed research; X.W., M.S.Z., S.R., R.S.N., M.K., and S.S.M. performed research; S.S. contributed new reagents or analytic tools; X.W., M.S.Z., E.G., S.S., and A.P. analyzed data; X.W., M.S.Z., V.S., E.G., S.S., and A.P. wrote the paper.
* This study was supported by an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520 to A. P.), a grant from the National Cancer Institute (CA184165 to A.P.), a Career Catalyst Award (KG100459 to X.W.) from Susan G. Komen Foundation, a contract (HHSN268201000032C to A.P.) from the National Heart, Lung and Blood Institute; a DOD Era of Hope Scholar Award (BC051652 to A.P.), NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036 to A.P.), the Safeway Breast Cancer Research Foundation (X.W.) and an NIH Regional Oncology Research Center award (CA006973 to V.S.).
 This article contains supplemental Fig. S1 and Tables S1 to S3.
This article contains supplemental Fig. S1 and Tables S1 to S3.
Conflict of Interest Statement: All authors declare no conflict of interests.
Data and Materials Availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001812. PX reviewer account: Username: reviewer00970@ebi.ac.uk Password: wiEhx0MA.
1 The abbreviations used are:
- ER
- estrogen receptor
- SILAC
- stable isotope labeling by amino acids in cell culture
- phosphoPSM
- phosphopeptide-spectrum match
- TPCK
- L-1-tosylamido-2-phenylethyl chloromethyl ketone
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- IAP
- immunoaffinity purification
- E2
- estrogen
- FAK
- focal adhesion kinase
- TKI
- tyrosine kinase inhibitors
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1.Yamashita H. (2008) Current research topics in endocrine therapy for breast cancer. Int. J. Clin. Oncol. 13, 380–383 [DOI] [PubMed] [Google Scholar]
- 2.Ali S., and Coombes R. C. (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer 2, 101–112 [DOI] [PubMed] [Google Scholar]
- 3.Osborne C. K., Zhao H., and Fuqua S. A. (2000) Selective estrogen receptor modulators: structure, function, and clinical use. J. Clin. Oncol. 18, 3172–3186 [DOI] [PubMed] [Google Scholar]
- 4.Osborne C. K., and Schiff R. (2005) Estrogen-receptor biology: continuing progress and therapeutic implications. J. Clin. Oncol. 23, 1616–1622 [DOI] [PubMed] [Google Scholar]
- 5.Burstein H. J., Temin S., Anderson H., Buchholz T. A., Davidson N. E., Gelmon K. E., Giordano S. H., Hudis C. A., Rowden D., Solky A. J., Stearns V., Winer E. P., and Griggs J. J. (2014) Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: american society of clinical oncology clinical practice guideline focused update. J. Clin. Oncol. 32, 2255–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visvanathan K., Hurley P., Bantug E., Brown P., Col N. F., Cuzick J., Davidson N. E., Decensi A., Fabian C., Ford L., Garber J., Katapodi M., Kramer B., Morrow M., Parker B., Runowicz C., Vogel V. G. 3rd, Wade J. L., and Lippman S. M. (2013) Use of pharmacologic interventions for breast cancer risk reduction: American Society of Clinical Oncology clinical practice guideline. J. Clin. Oncol. 31, 2942–2962 [DOI] [PubMed] [Google Scholar]
- 7.Ring A., and Dowsett M. (2004) Mechanisms of tamoxifen resistance. Endocr. Relat. Cancer 11, 643–658 [DOI] [PubMed] [Google Scholar]
- 8. Early Breast Cancer Trialists' Collaborative Group, Davies C., Godwin J., Gray R., Clarke M., Cutter D., Darby S., McGale P., Pan H. C., Taylor C., Wang Y. C., Dowsett M., Ingle J., and Peto R. (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne C. K., Bardou V., Hopp T. A., Chamness G. C., Hilsenbeck S. G., Fuqua S. A., Wong J., Allred D. C., Clark G. M., and Schiff R. (2003) Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 95, 353–361 [DOI] [PubMed] [Google Scholar]
- 10.Osborne C. K., and Schiff R. (2003) Growth factor receptor cross-talk with estrogen receptor as a mechanism for tamoxifen resistance in breast cancer. Breast 12, 362–367 [DOI] [PubMed] [Google Scholar]
- 11.Knowlden J. M., Hutcheson I. R., Jones H. E., Madden T., Gee J. M., Harper M. E., Barrow D., Wakeling A. E., and Nicholson R. I. (2003) Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology 144, 1032–1044 [DOI] [PubMed] [Google Scholar]
- 12.McClelland R. A., Barrow D., Madden T. A., Dutkowski C. M., Pamment J., Knowlden J. M., Gee J. M., and Nicholson R. I. (2001) Enhanced epidermal growth factor receptor signaling in MCF7 breast cancer cells after long-term culture in the presence of the pure antiestrogen ICI 182,780 (Faslodex). Endocrinology 142, 2776–2788 [DOI] [PubMed] [Google Scholar]
- 13.Hawthorne V. S., Huang W. C., Neal C. L., Tseng L. M., Hung M. C., and Yu D. (2009) ErbB2-mediated Src and signal transducer and activator of transcription 3 activation leads to transcriptional up-regulation of p21Cip1 and chemoresistance in breast cancer cells. Mol. Cancer Res. 7, 592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua G., Zhu B., Rosa F., Deblon N., Adélaïde J., Kahn-Perlès B., Birnbaum D., and Imbert J. (2009) A negative feedback regulatory loop associates the tyrosine kinase receptor ERBB2 and the transcription factor GATA4 in breast cancer cells. Mol. Cancer Res. 7, 402–414 [DOI] [PubMed] [Google Scholar]
- 15.Shou J., Massarweh S., Osborne C. K., Wakeling A. E., Ali S., Weiss H., and Schiff R. (2004) Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J. Natl. Cancer Inst. 96, 926–935 [DOI] [PubMed] [Google Scholar]
- 16.Gee J. M., Robertson J. F., Ellis I. O., and Nicholson R. I. (2001) Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. Int. J. Cancer 95, 247–254 [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Tenorio G., Stål O., and Southeast Sweden Breast Cancer Group (2002) Activation of AKT/PKB in breast cancer predicts a worse outcome among endocrine treated patients. Br. J. Cancer 86, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkegaard T., Witton C. J., McGlynn L. M., Tovey S. M., Dunne B., Lyon A., and Bartlett J. M. (2005) AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J. Pathol. 207, 139–146 [DOI] [PubMed] [Google Scholar]
- 19.Leary A. F., Sirohi B., and Johnston S. R. (2007) Clinical trials update: endocrine and biological therapy combinations in the treatment of breast cancer. Breast Cancer Res. 9, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moy B., and Goss P. E. (2006) Estrogen receptor pathway: resistance to endocrine therapy and new therapeutic approaches. Clin. Cancer Res. 12, 4790–4793 [DOI] [PubMed] [Google Scholar]
- 21.Browne B. C., Hochgräfe F., Wu J., Millar E. K., Barraclough J., Stone A., McCloy R. A., Lee C. S., Roberts C., Ali N. A., Boulghourjian A., Schmich F., Linding R., Farrow L., Gee J. M., Nicholson R. I., O'Toole S. A., Sutherland R. L., Musgrove E. A., Butt A. J., and Daly R. J. (2013) Global characterization of signalling networks associated with tamoxifen resistance in breast cancer. FEBS J. 280, 5237–5257 [DOI] [PubMed] [Google Scholar]
- 22.Oyama M., Nagashima T., Suzuki T., Kozuka-Hata H., Yumoto N., Shiraishi Y., Ikeda K., Kuroki Y., Gotoh N., Ishida T., Inoue S., Kitano H., and Okada-Hatakeyama M. (2011) Integrated quantitative analysis of the phosphoproteome and transcriptome in tamoxifen-resistant breast cancer. J. Biol. Chem. 286, 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou C., Zhong Q., Rhodes L. V., Townley I., Bratton M. R., Zhang Q., Martin E. C., Elliott S., Collins-Burow B. M., Burow M. E., and Wang G. (2012) Proteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migration. Breast Cancer Res. 14, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengel S. M., Murray E., Langdon S., Hayward L., O'Donoghue J., Panchaud A., Hupp T., and Goodlett D. R. (2011) Data-independent proteomic screen identifies novel tamoxifen agonist that mediates drug resistance. J. Proteome Res. 10, 4567–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umar A., Kang H., Timmermans A. M., Look M. P., Meijer-van Gelder M. E., den Bakker M. A., Jaitly N., Martens J. W., Luider T. M., Foekens J. A., and Pasa-Tolić L. (2009) Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol. Cell. Proteomics 8, 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besada V., Diaz M., Becker M., Ramos Y., Castellanos-Serra L., and Fichtner I. (2006) Proteomics of xenografted human breast cancer indicates novel targets related to tamoxifen resistance. Proteomics 6, 1038–1048 [DOI] [PubMed] [Google Scholar]
- 27.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., and Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., and Jørgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 29.Spivak M., Weston J., Bottou L., Käll L., and Noble W. S. (2009) Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J. Proteome Res. 8, 3737–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X., Chen H., Parker B., Rubin E., Zhu T., Lee J. S., Argani P., and Sukumar S. (2006) HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 66, 9527–9534 [DOI] [PubMed] [Google Scholar]
- 31.Blume-Jensen P., and Hunter T. (2001) Oncogenic kinase signalling. Nature 411, 355–365 [DOI] [PubMed] [Google Scholar]
- 32.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 33.Hiscox S., Jordan N. J., Morgan L., Green T. P., and Nicholson R. I. (2007) Src kinase promotes adhesion-independent activation of FAK and enhances cellular migration in tamoxifen-resistant breast cancer cells. Clin. Exp. Metastasis 24, 157–167 [DOI] [PubMed] [Google Scholar]
- 34.Planas-Silva M. D., Bruggeman R. D., Grenko R. T., and Stanley Smith J. (2006) Role of c-Src and focal adhesion kinase in progression and metastasis of estrogen receptor-positive breast cancer. Biochem. Biophys. Res. Commun. 341, 73–81 [DOI] [PubMed] [Google Scholar]
- 35.Gutierrez M. C., Detre S., Johnston S., Mohsin S. K., Shou J., Allred D. C., Schiff R., Osborne C. K., and Dowsett M. (2005) Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J. Clin. Oncol. 23, 2469–2476 [DOI] [PubMed] [Google Scholar]
- 36.Donovan J. C., Milic A., and Slingerland J. M. (2001) Constitutive MEK/MAPK activation leads to p27(Kip1) deregulation and antiestrogen resistance in human breast cancer cells. J. Biol. Chem. 276, 40888–40895 [DOI] [PubMed] [Google Scholar]
- 37.Kurokawa H., Lenferink A. E., Simpson J. F., Pisacane P. I., Sliwkowski M. X., Forbes J. T., and Arteaga C. L. (2000) Inhibition of HER2/neu (erbB-2) and mitogen-activated protein kinases enhances tamoxifen action against HER2-overexpressing, tamoxifen-resistant breast cancer cells. Cancer Res. 60, 5887–5894 [PubMed] [Google Scholar]
- 38.Hutcheson I. R., Knowlden J. M., Madden T. A., Barrow D., Gee J. M., Wakeling A. E., and Nicholson R. I. (2003) Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway in tamoxifen-resistant MCF-7 cells. Breast Cancer Res. Treat. 81, 81–93 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Moerkens M., Ramaiahgari S., de Bont H., Price L., Meerman J., and van de Water B. (2011) Elevated insulin-like growth factor 1 receptor signaling induces antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling routes. Breast Cancer Res. 13, R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Flier S., Brinkman A., Look M. P., Kok E. M., Meijer-van Gelder M. E., Klijn J. G., Dorssers L. C., and Foekens J. A. (2000) Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J. Nat. Cancer Inst. 92, 120–127 [DOI] [PubMed] [Google Scholar]
- 41.Brinkman A., van der Flier S., Kok E. M., and Dorssers L. C. (2000) BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J. Nat. Cancer Inst. 92, 112–120 [DOI] [PubMed] [Google Scholar]
- 42.Barrett A., Pellet-Many C., Zachary I. C., Evans I. M., and Frankel P. (2013) p130Cas: a key signalling node in health and disease. Cell. Signalling 25, 766–777 [DOI] [PubMed] [Google Scholar]
- 43.Riggins R. B., Quilliam L. A., and Bouton A. H. (2003) Synergistic promotion of c-Src activation and cell migration by Cas and AND-34/BCAR3. J. Biol. Chem. 278, 28264–28273 [DOI] [PubMed] [Google Scholar]
- 44.Brábek J., Constancio S. S., Shin N. Y., Pozzi A., Weaver A. M., and Hanks S. K. (2004) CAS promotes invasiveness of Src-transformed cells. Oncogene 23, 7406–7415 [DOI] [PubMed] [Google Scholar]
- 45.Ruest P. J., Shin N. Y., Polte T. R., Zhang X., and Hanks S. K. (2001) Mechanisms of CAS substrate domain tyrosine phosphorylation by FAK and Src. Mol. Cell. Biol. 21, 7641–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chodniewicz D., and Klemke R. L. (2004) Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim. Biophys. Acta 1692, 63–76 [DOI] [PubMed] [Google Scholar]
- 47.Cowell L. N., Graham J. D., Bouton A. H., Clarke C. L., and O'Neill G. M. (2006) Tamoxifen treatment promotes phosphorylation of the adhesion molecules, p130Cas/BCAR1, FAK and Src, via an adhesion-dependent pathway. Oncogene 25, 7597–7607 [DOI] [PubMed] [Google Scholar]
- 48.Sun C. K., Ng K. T., Sun B. S., Ho J. W., Lee T. K., Ng I., Poon R. T., Lo C. M., Liu C. L., Man K., and Fan S. T. (2007) The significance of proline-rich tyrosine kinase2 (Pyk2) on hepatocellular carcinoma progression and recurrence. Br. J. Cancer 97, 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitra S. K., Hanson D. A., and Schlaepfer D. D. (2005) Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 50. (2013) Study Of PF-00562271, Including Patients With Pancreatic, Head And Neck, Prostatic Neoplasms. [Google Scholar]
- 51.Győrffy B., Surowiak P., Budczies J., and Lánczky A. (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one 8, e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.