Abstract
The tau protein is central to the etiology of several neurodegenerative diseases, including Alzheimer's disease, a subset of frontotemporal dementias, progressive supranuclear palsy and dementia following traumatic brain injury, yet the proteins it interacts with have not been studied using a systematic discovery approach. Here we employed mild in vivo crosslinking, isobaric labeling, and tandem mass spectrometry to characterize molecular interactions of human tau in a neuroblastoma cell model. The study revealed a robust association of tau with the ribonucleoproteome, including major protein complexes involved in RNA processing and translation, and documented binding of tau to several heat shock proteins, the proteasome and microtubule-associated proteins. Follow-up experiments determined the relative contribution of cellular RNA to the tau interactome and mapped interactions to N- or C-terminal tau domains. We further document that expression of P301L mutant tau disrupts interactions of the C-terminal half of tau with heat shock proteins and the proteasome. The data are consistent with a model whereby a higher propensity of P301L mutant tau to aggregate may reflect a perturbation of its chaperone-assisted stabilization and proteasome-dependent degradation. Finally, using a global proteomics approach, we show that heterologous expression of a tau construct that lacks the C-terminal domain, including the microtubule binding domain, does not cause a discernible shift of the proteome except for a significant direct correlation of steady-state levels of tau and cystatin B.
The tau protein is a member of the family of microtubule-associated proteins (MAPs)1 that in humans is coded by the MAPT gene on chromosome 17q21.31 (1). Initially, described as a factor that binds to and stabilizes microtubules (MTs) (2), interest in the tau protein grew when it was shown to represent the main constituent of intracellular protein aggregates, termed neurofibrillary tangles (NFTs), observed in Alzheimer's disease (3, 4). Similar tau aggregates have since been described in other, less common dementias, including progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), Pick's disease and dementia pugilistica, a form of dementia observed in athletes who had been exposed to repeated traumatic brain injury (5).
Despite its early recognition as a MT binding molecule, the physiological function of the tau protein is still being debated (6). At least, in part, this uncertainty is born from the observation that tau knockout mice are rather nonconspicuous in their phenotype (7, 8). Ongoing attempts to define additional roles for this protein have, over the years, generated several hypotheses, including that the tau protein modulates neurite outgrowth and axonogenesis (8, 9), bridges the microtubule and actin cytoskeletons (10), and acts as a scaffold for tethering the Src family tyrosine kinase Fyn to PSD-95/NMDA receptor complexes (11). The predominant expression of tau in neuronal axons suggests a role in brain function. Significantly, in all tauopathies, a group term used to describe dementias with pathological tau protein involvement, the tau protein is observed to detach from microtubules and to form aggregates. There also is compelling evidence from a body of work with transgenic models that the cellular toxicity observed in the aforementioned dementias relies on the presence of the tau protein (12). Consequently, it seems plausible that the cellular toxicity observed in AD and other dementias does not relate to a loss of function of the tau protein but represents a gain of toxic function the protein exhibits in its microtubule-detached form.
The tau molecule can be crudely subdivided into an amino-terminal projection domain (PD), a microtubule-binding domain (MTB), and a carboxy-terminal domain (C') (13). The protein has long been known to exhibit some remarkable biochemical characteristics, including an ability to withstand harsh acid and heat treatments that would cause a majority of other proteins to precipitate (2, 14). These characteristics have been attributed to tau being natively unfolded and possessing a highly dynamic character (15). The tau protein is also known to be a substrate for several post-translational modifications (PTMs), and the list of tau modifying enzymes that have been described is long. In particular, tau phosphorylation has been recognized to occur in vivo and in disease, and tau hyperphosphorylation at sites within the MTB domain and at nearby sites flanking the MTB has been shown to promote detachment of tau from microtubules (16). There further is broad agreement in the field that levels of several other tau PTMs are raised in tauopathies, including nitration (17), ubiquitination (18), sumoylation (19), and truncation (20, 21). Less agreement exists on the degree to which specific PTMs contribute to disease manifestation in individual tauopathies (22). Lacking also are insights into other physiological protein interactions the tau protein engages in and, surprisingly, to our knowledge, no systematic screen for tau binders has been reported. Thus, except for its well-established binding to microtubules (2), members of the Src family of protein kinases (23, 24), Hsp70 (25)/Hsp90 (26, 27), and reports on its interaction with F-actin (28), ApoE3 (29), a subset of peptidyl-prolyl cis-trans isomerases (30, 31), α-synuclein (32), PACSIN1 (33), and negatively charged polymers, including nucleic acids (34, 35), relatively little is known about other nonenzymatic interactions the protein engages in.
In an attempt to address this unmet need, we set out to define molecular interactors of the tau protein in the human neuroblastoma SH-SY5Y cell model. The study made use of advanced instrumentation and workflows that included comparative mass spectrometry based on isobaric tags. We observed that the tau interactome is dramatically enriched in cellular components involved in the regulation and execution of RNA-processing and translation. We document that the previously known ability of the tau protein to bind to nucleic acids is partially responsible but not sufficient by itself to explain this binding propensity of the tau protein. We narrowed down the binding preference of individual binders to N- and C-terminal domains within tau and document that several interactors, including 14-3-3 proteins, heat shock proteins, and the proteasome, exhibit a strong binding preference for the C terminus of tau. When comparing the interactomes of wild-type and mutant tau (P301L) linked to frontotemporal dementia, we observed that interactions with the aforementioned C-terminal tau binding partners are diminished for mutant tau. Despite the strong binding of tau to the ribonucleoproteome, its overexpression does not seem to affect the global steady-state levels of cellular proteins, and only the levels of cystatin B, a natural inhibitor of cysteine proteases, were modified and correlated directly with the levels of heterologously expressed tau.
EXPERIMENTAL PROTOCOL
Antibodies
The mouse monoclonal antibodies directed against GFP (cat. no. 632380, clone JL-8) and HSP90 (cat. no. 610418) were purchased from Clontech Laboratories, Inc. (Mountain View, CA) and BD Biosciences (Mississauga, ON, Canada), respectively. The rabbit polyclonal anti-tau antibody (K9JA) was from Dako Canada (cat. no. A0024, Burlington, ON). The secondary anti-mouse peroxidase-conjugated antibody (cat no. 170–6516) was from Bio-Rad (Mississauga, ON, Canada).
Cell Culture
Human SH-SY5Y neuroblastoma cells (cat. no. CRL-2266), African Green Monkey CV-1 kidney cells (cat. no. CCL-70) and human HEK-293 embryonic kidney cells (cat. no. CRL-1573) (American Type Culture Collection, Manassas, VA) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% v/v fetal bovine serum, antimycotic/antibiotic (Life Technologies, Burlington ON, Canada), and GlutaMAX-I supplement (Life Technologies). For interactome experiments, SH-SY5Y cells were passaged to 150 mm × 15 mm cell culture dishes (Sarstedt AG & Co, Nümbrecht, Germany). For global proteome analyses, HEK-293 cells were grown in 100 mm × 10 mm cell culture dishes (Sarstedt AG & Co). Mammalian expression plasmids were transiently transfected using Lipofectamine transfection reagent (Lipofectamine 2000, Life Technologies) according to the manufacturer's instructions.
Cloning of Tau Expression Constructs
The tau1-441-EGFP plasmid was a generous gift by Dr. George S. Bloom (36). Tau1-255-EGFP, tau256-441-EGFP, tau1-441(P301L)-ECFP, and EGFP were produced by deleting domains or mutating specific residues of the tau1-441-EGFP parent plasmid by conventional site-directed mutagenesis using HiFidelity polymerase (Qiagen, Valencia, CA).
Affinity Chromatography
SH-SY5Y cells were in vivo crosslinked with 1% formaldehyde as described before (37), then washed twice with ice-cold PBS, scraped from the cell culture dish, collected by mild centrifugation and gently lysed in Lysis buffer (150 mm Tris, HCl, pH 7.3, 50 μg/ml digitonin, 5 mm EGTA, 20 mm NaF, 1 mm vanadate, Complete protease inhibitor mixture (Roche Diagnostics Corporation, Indianapolis, IN), 1× PhosSTOP phosphatase inhibitor mixture (Roche), 10% glycerol). Lysates were first precleared by centrifuging at 3000 × g for 10 min at 4 °C, and then centrifuged at 30,000 rpm in an ultracentrifuge using a SW 55Ti rotor (Beckman Coulter, Mississauga ON, Canada) for 30 min at 4 °C. The protein concentration was determined by bicinchoninic acid assay (BCA assay; Fisher Scientific, Ottawa, ON), adjusted to 5 mg/ml and cell lysates added to GFP-trap beads (ChromoTek Inc., Planegg-Martinsried, Germany), which had been pre-equilibrated in Lysis buffer. Following overnight incubation at 4 °C with end-over-end tumbling, GFP-trap beads were sedimented by gravity before supernatants were removed and designated as “Unbound” fractions. Sedimented beads were washed three times using Lysis buffer without protease inhibitors and then subjected to a pre-elution wash with 10 mm HEPES, pH 7.4. Bound proteins were eluted with 500 μl of 0.2% TFA, 20% acetonitrile, pH 2 in water.
Sample Preparation for Global Proteome Analysis
Near-confluent layers of HEK-293 cells grown in six 100 mm dishes, were harvested 48 h following their transient transfection with tau1–255-EGFP or EGFP constructs. Cells were gently sedimented in ice-cold phosphate-buffered saline and subsequently subjected to hot lysis in preheated (90 °C) Lysis buffer comprised of 63 mm HEPES/NaOH, pH 8.0, 2% sodium dodecylsulfate in deionized water. Subsequently, nucleic acids and cellular debris were further destructed by three pulses of 1 min bead beating in a Mini-BeadBeater 8 (Biospec Products Inc., Bartlesville, OK) at “Homogenization level” in the presence of 0.5 mm glass beads. Cellular lysates were cleared by high speed centrifugation (1 h, 100,000 × g), protein levels adjusted by bicinchoninic acid colorimetric assay (Thermo Fisher Scientific, Nepean, ON) and volume equivalents containing 100 μg protein acetone precipitated.
Western blot and Immunofluorescence Analyses
Levels of total proteins in SH-SY5Y extracts were determined by BCA assay (Fisher Scientific) and equal amounts of proteins were separated at 180 V in 4–12% denaturing NuPAGE SDS-PAGE gels (Life Technologies), followed by transfer to 0.45 micron polyvinyl fluoride membranes. The Western blot membranes were blocked in skim milk and incubated with the respective primary antibodies overnight at 4 °C. Subsequently, Western blot membranes were incubated with peroxidase-conjugated anti-mouse secondary antibodies (1:10,000) and the ECL reagent (cat. no. RPN2106; GE Healthcare, Baie d'Urfe, QC). Signals were detected either with x-ray film or the Odyssey Fc Imager system (LI-COR Biosciences, Lincoln, NE).
Immunofluorescence Analyses
CV-1 cells were grown in four-well chamber slides (Lab-Tek II Chamber Slide System; Nunc, Roskilde, Denmark) and then transiently transfected with tau1–441-EGFP, tau1–255-EGFP, tau 256–441-EGFP, or EGFP as described before. Cell images were captured using a LSM 700 confocal microscope with an LCI stage-top incubator (Carl Zeiss Canada, Toronto, ON).
Quantitative Mass Spectrometry
For interactome analyses, six 150 mm × 15 mm SH-SY5Y cell culture plates were grown to near-confluency per sample (or control) and processed as described under “Affinity Chromatography.” For global proteome analyses, six 100 mm × 10 mm HEK-293 cell culture plates served as starting materials and were processed as outlined under “Samples Preparation for Global Proteome Analysis.” Identical procedures were applied to generate three biological replicates for each condition tested.
Workflows for protein reduction, alkylation, and trypsinization followed steps that have recently been described in detail (38). Following trypsinization, peptide mixtures were side-by-side conjugated to six-plex tandem mass tag (TMT) reagents (Thermo Fisher Scientific) or four-plex isobaric tags for relative and absolute quantification (iTRAQ) (39) (Applied Biosystems, Foster City, CA) using manufacturer instructions. Subsequently, equal volumes of the TMT (or iTRAQ) labeling mixtures were pooled into a master mix and subjected to C18 or SCX ZipTip clean-up using Bond Elut Omix 100 μl tips (Agilent Technologies Inc., Santa Clara, CA) followed by tandem mass spectrometry analysis on a Orbitrap Fusion Tribrid instrument (Thermo Fisher Scientific) using a previously described data-dependent MS3 workflow (38). Briefly, TMT-labeled (or iTRAQ-labeled) peptide mixtures were injected onto 25 cm Acclaim nanocapillary C18 columns using a split-free nLC 1100 nano-HPLC system (Thermo Fisher Scientific) and were separated by applying 4 h linear gradients of 3–50% acetonitrile in 0.1% formic acid at a flow rate of 300 nL/min. The eluent was directed toward the ion transfer tube of the Orbitrap Fusion Tribrid instrument by dynamic nanospray ionization. The acquisition method incorporated three separate scans. Following an orbitrap-based survey scan of the 400–2000 m/z range at 120,000 resolution, the 10 most intense ions carrying charges of two or more were isolated and fragmented by collision-induced dissociation (CID). Peptide fragments were scanned out from the linear ion trap to provide peptide sequence information. Finally, the 10 most abundant CID fragments were further fragmented by high energy collision-induced dissociation (HCD) to release TMT or iTRAQ signature ions, which were detected by the orbitrap at a resolution of 60,000. A dynamic exclusion window of 20 min prevented reanalysis of previously selected parent ions.
RNase Treatment
Cells were lysed using the aforementioned digitonin-containing Lysis buffer. Cellular lysates were precleared for 10 min at 4000 rpm in a refrigerated centrifuge (Napco 2028r, Thermo Fisher Scientific). Subsequently, supernatants were spun at 30,000 rpm in a swing-out rotor (SW 55Ti; Beckman Coulter, Pasadena, CA) to remove residual insoluble debris. 40 μg/ml RNase A and 100 U/ml RNase T1 (cat. no. EN0551; stock concentrations of 2 mg/ml RNase A, 5000 U/ml RNase T1) (Life Technologies) were added to cell lysates, which were incubated at room temperature for 1 h.
Postacquisition Data Analyses
The postacquisition data analyses of tau interactome and global proteome datasets was conducted against the human international protein index (IPI) database (Version 3.87), which was queried with Mascot (Version 2.4; Matrix Science Ltd, London, UK) and Sequest HT search engines within Proteome Discoverer (Version 1.4; Thermo Fisher Scientific). Spectra were filtered by the percolator algorithm using a maximum ΔCn of 0.05 for input data and a strict target false discovery rate (FDR) of 0.01 for the decoy database search (40). Protein identifications were cross-validated by the respective algorithm for determining the FDR within PEAKS Studio software (Version 6.0; Bioinformatics Solutions Inc., Waterloo, Ontario, Canada). A separate module within the latter software further served in assessing the reproducibility of the nano-HPLC runs. Searches were configured to accommodate a maximum of two missed tryptic cleavages and considered only peptides with a minimum length of six amino acids. Naturally occurring variable post-translational modifications (PTMs) considered were phosphorylations of serine, threonine, or tyrosine residues. Other variable PTMs considered were oxidation of methionines as well as deamidation of glutamines and asparagines. Static modifications considered were pyridylethyl-derivatised cysteines and TMT6plex-modified primary amines present on peptide N termini or lysine side chains. Precursor mass tolerance and fragment mass tolerance thresholds were set to 10 ppm and 0.6 Da, respectively. All protein identifications and relative quantitations were based on a minimum of three CID spectra and TMT signature ion distributions linked to them (note that the median count of quantified PSMs underlying individual protein quantitations was 14). Filtering of entries on the basis variances was avoided as we observed variances to be skewed by isolation interference (despite setting an isolation interference threshold of 30%) (41). Instead, cut-offs were based on the number of PSMs that contributed to protein group identifications and quantitations. A subset of quantitation data were exported from Proteome Discoverer to provide representative examples of TMT-based ratio assignments (supplemental Figs. S1 and S3). The Gene Ontology analyses as well as the identification of KEGG pathways and InterPro domains, which were enriched among proteins that co-isolated with the tau protein, were undertaken with an integrated suite of statistical algorithms embedded in ProteinCenter software (Version 3.12; Thermo Fisher Scientific). The analysis was based on 390 protein groups, for which a minimum of three reporter ion distributions were available, that exhibited average enrichment ratios >1.2. As the reference data set served in these calculations the human IPI database with 91,464 entries. When p values were corrected for multiple statistical testing using a previously published methodology (42) they are shown as FDR p values. Background colors in supplemental Tables S1, S3, and S4 were generated in Excel by defining two- or three-color rules based on enrichment ratios, i.e. high ratio: dark green; ratio = 1 (corresponding to no relative enrichment): white; low ratio: red. The color intensities correlated directly and proportionally with the relative deviation of enrichment levels from the ratio of 1.
All mass spectrometry datasets have been deposited to the ProteomeXchange Consortium (43) and can be identified for download by the PRIDE partner repository (44) identifier PXD002058.
RESULTS
Workflow of Tau Interactome Analysis
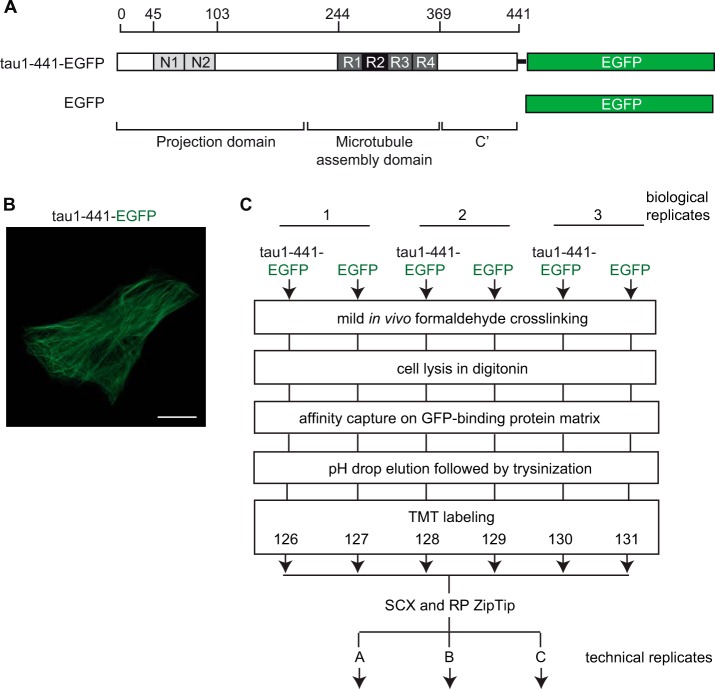
Any study that aims to identify molecular interactors of a given bait protein by affinity capture and mass spectrometry needs to consider the biological source material, the method of bait protein capture and a strategy for discriminating a relatively small number of specific interactors from an excess of nonspecific binders. Here we chose to express the tau protein carrying a C-terminal enhanced green fluorescent protein (EGFP) module in the human SH-SY5Y neuroblastoma cell line (Fig. 1A). The SH-SY5Y cell model represents one of the best understood human-derived paradigms for studying the cellular biology of neurodegenerative disease-linked proteins. This cell line further expresses the tau protein endogenously and, therefore, would be expected to have cellular components in place that interact with this protein under physiological conditions. The EGFP fusion was selected on the basis of prior results by others that documented that the C-terminal addition of this tag to the tau protein neither interferes with its binding to microtubules nor with the manifestation of tau-dependent toxicity in cells exposed to amyloid β (36). In addition to affording the ability to trace the subcellular localization of the fusion protein on account of the fluorescence signal it emanates when excited (Fig. 1B), EGFP-tagged proteins can readily be captured on matrices saturated with the GFP-binding protein (45) and offer the ability to generate informative negative control samples by heterologous expression of the EGFP-cassette alone.
Fig. 1.
Schematic presentation of bait proteins, expression analysis and workflow of tau interactome experiment. A, Tau1–441-EGFP fusion construct employed in this work. B, Live-cell fluorescence image of tau1–441-EGFP expression in CV-1 cells. Scale bar = 1 μm. C, Workflow for identifying tau interactors in human SH-SY5Y neuroblastoma cells.
Thus, cellular extracts were generated by digitonin extraction of SH-SY5Y cells that stably expressed human four-repeat tau with a C-terminal EGFP fusion (tau1–441-EGFP) or EGFP alone at levels comparable to endogenous tau levels observed in these cells (not shown). Following affinity capture, bait protein complexes were eluted by rapidly lowering the pH. Subsequently, proteins were fully denatured in concentrated urea, reduced, alkylated, trypsinized, and TMT-labeled. The use of six-plex TMT reagents afforded the concomitant analysis of three biological samples and three controls, which were generated side by side. It thereby facilitated inter-sample comparisons and eliminated a notorious confounder encountered when complex proteomics samples are analyzed consecutively and exhibit run-to-run variance. Here, the use of consecutive analyses was restricted to the generation of technical replicates (Fig. 1C).
The Tau Interactome is Enriched in Proteins with Roles in RNA-processing and Translation
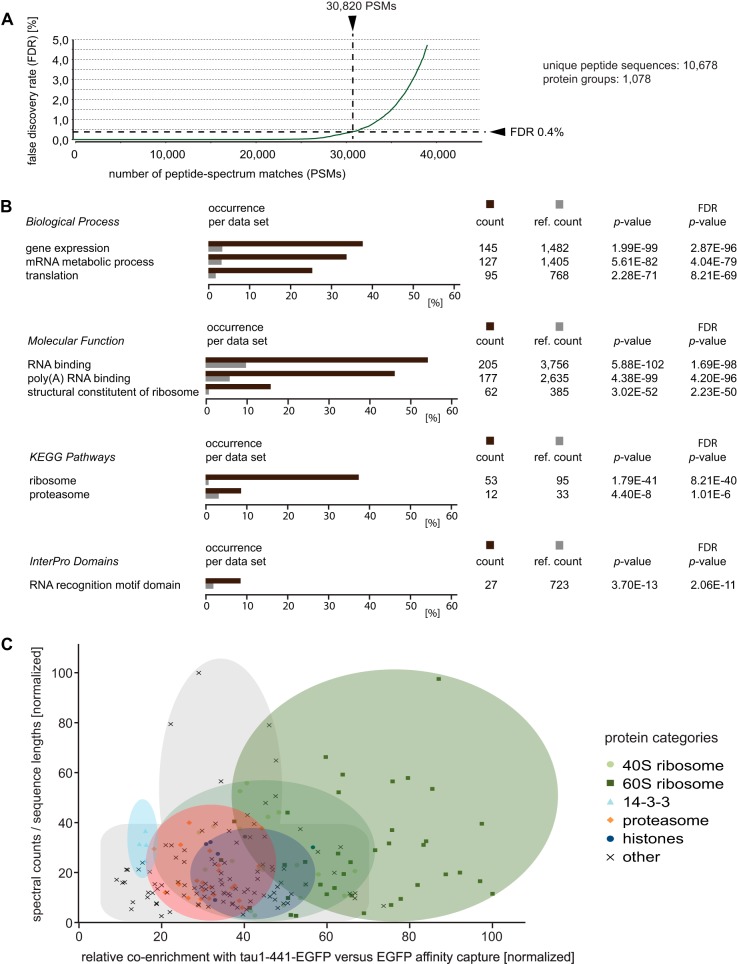
Interactome datasets obtained were searched against the nonredundant IPI human database (Version 3.87). When applying a stringent false-discovery rate (FDR) of 0.4% the combined tau1–441 interactome analyses resulted in the assignment of 10,678 unique peptide sequences and the detection of 1,078 protein groups, with quantitations minimally based on ten TMT reporter ion distributions (Fig. 2A, supplemental Table S1).
Fig. 2.
Benchmarks of tau1–441 interactome analysis and classification of candidate interactors. A, Chart depicting number of peptide-spectrum matches (PSM) versus false discovery rate (FDR). B, Subset of Gene Ontology (GO) classifiers most significantly overrepresented in tau1–441 interactome. Color code: brown, percentage of proteins with GO classifier categories that were overrepresented among tau interactors; gray, subset of proteins within reference data set assigned to a given “GO” classifier. C, Selective co-isolation of ribosomes with tau1–441. Chart depicting relative enrichment of candidate interactors in the tau1–441 interactome versus spectral counts (normalized by protein lengths) supporting their identifications.
Of these protein groups, 190 exhibited average enrichment ratios of 1.2 or greater (relative to the EGFP affinity capture control replicates) (see Table I for the top-listed hits). The cursory initial inspection of the candidate tau1–441 interactome revealed a surprising abundance of RNA binding proteins and proteins known to play a role in protein translation. To further explore in a systematic fashion if a particular biological process or molecular function was enriched among the subset of proteins that preferentially co-isolated with tau1–441, a comprehensive Gene Ontology (GO) search was conducted. This investigation revealed that proteins contributing to “mRNA metabolic processes,” “translation initiation,” and “translation elongation” were most significantly enriched among candidate tau1–441 interactors when using the entire human IPI database as a reference. Consistent with these observations, “RNA binding,” “poly(A) RNA binding,” and “structural constituent of ribosome” were the top-listed GO Molecular Functions. In addition to the KEGG pathway “Ribosome,” the tau1–441 interactome dataset was significantly enriched in components that had been mapped to the KEGG pathway “Proteasome.” Finally, when we interrogated the list of proteins to investigate if any known InterPro protein domain was enriched in the dataset, the “RNA recognition motif (RRM) domain” was returned as a subdomain that was present in 27 proteins (supplemental Table S2), a level of occurrence corresponding to a probability of p = 2.06E−11 (corrected for multiple testing) that this observation would be encountered by chance alone (Fig. 2B).
Table I. tau1–441-EGFP versus EGFP interactome dataset (sorted by enrichment level; truncated list - see also supplemental Table S1).
| Accession | Description | Coverage | # of Peptides |
tau1–441-EGFP/EGFP |
||||
|---|---|---|---|---|---|---|---|---|
| Unique | Total | 126/131 | 128/131 | 130/131 | Count | |||
| IPI00025499.3 | Tau-F of Microtubule-associated protein tau | 87.30% | 4 | 44 | 6.460 | 6.792 | 5.071 | 140 |
| IPI00412607.6 | 60S subunit L35 (one of >35 subunits from 60S ribosome) | 63.41% | 4 | 15 | 1.974 | 2.189 | 1.476 | 15 |
| IPI00941732.2 | Transcription elongation factor B polypeptide 3 | 38.72% | 3 | 34 | 1.571 | 1.942 | 1.509 | 10 |
| IPI00031651.1 | C7orf50 (probably poly(A) RNA binding) | 45.36% | 4 | 6 | 1.672 | 1.681 | 1.473 | 19 |
| IPI00060627.2 | Coiled-coil domain-containing protein 124 | 57.85% | 9 | 19 | 1.531 | 1.650 | 1.571 | 23 |
| IPI00024157.1 | Peptidyl-prolyl cis-trans isomerase FKBP3 | 60.27% | 10 | 21 | 1.566 | 1.623 | 1.472 | 29 |
| IPI00299000.5 | Proliferation-associated protein 2G4 | 77.16% | 21 | 28 | 1.565 | 1.613 | 1.431 | 71 |
| IPI00292071.6 | Secretogranin-3 | 64.74% | 15 | 26 | 1.388 | 1.706 | 1.426 | 32 |
| IPI00022317.3 | Leydig cell tumor 10 kDa protein homolog | 74.75% | 6 | 15 | 1.503 | 1.581 | 1.349 | 12 |
| IPI00655650.2 | 40S subunit S26 (one of >25 subunits from 40S ribosome) | 43.48% | 3 | 5 | 1.481 | 1.464 | 1.476 | 11 |
| IPI00021924.1 | Histone H1x | 49.77% | 9 | 17 | 1.374 | 1.615 | 1.425 | 24 |
| IPI00075248.11 | Calmodulin | 100.00% | 9 | 15 | 1.653 | 1.459 | 1.261 | 72 |
| IPI00100463.1 | CUE domain-containing protein 2 | 41.81% | 6 | 15 | 1.360 | 1.535 | 1.434 | 21 |
| IPI00329692.3 | Glycylpeptide N-tetradecanoyl transferase 1 | 36.90% | 9 | 25 | 1.314 | 1.615 | 1.354 | 32 |
| IPI00880007.2 | Microtubule-associated protein | 54.94% | 5 | 128 | 1.432 | 1.420 | 1.416 | 83 |
| IPI00879259.1 | RNA-binding protein EWS isoform 3 | 32.52% | 5 | 29 | 1.250 | 1.519 | 1.480 | 38 |
| IPI00470498.1 | Plasminogen activator inhibitor 1 RNA-binding protein | 65.39% | 3 | 32 | 1.370 | 1.500 | 1.343 | 84 |
| IPI00293434.2 | Signal recognition particle 14 kDa protein | 77.21% | 6 | 12 | 1.326 | 1.503 | 1.368 | 36 |
| IPI00328293.3 | Similar to Serine/arginine repetitive matrix protein 1 | 53.60% | 7 | 71 | 1.223 | 1.767 | 1.168 | 16 |
| IPI00654605.1 | MAP7 domain-containing protein 1 | 54.82% | 7 | 50 | 1.336 | 1.588 | 1.219 | 16 |
| IPI00220685.1 | Heterogeneous nuclear ribonucleoprotein D0 | 55.75% | 2 | 21 | 1.188 | 1.458 | 1.480 | 40 |
| IPI00792334.2 | Ataxin 2 | 50.22% | 9 | 48 | 1.216 | 1.564 | 1.329 | 14 |
| IPI00021728.3 | Eukaryotic translation initiation factor 2 subunit 2 | 71.17% | 10 | 22 | 1.253 | 1.420 | 1.418 | 36 |
| IPI00031812.3 | Nuclease-sensitive element-binding protein 1 | 64.20% | 11 | 25 | 1.182 | 1.484 | 1.401 | 155 |
| IPI00645208.3 | RNA-binding protein FUS isoform 3 | 41.38% | 5 | 25 | 1.151 | 1.539 | 1.365 | 43 |
| IPI00017596.3 | Microtubule-associated protein RP/EB family member 1 | 73.13% | 7 | 19 | 1.261 | 1.707 | 1.080 | 24 |
| IPI00033036.1 | Methionine aminopeptidase 2 | 51.67% | 9 | 22 | 1.384 | 1.371 | 1.292 | 17 |
| IPI00966877.1 | tRNA (cytosine-5-)-methyltransferase NSUN2 isoform 2 | 37.70% | 5 | 26 | 1.280 | 1.333 | 1.426 | 19 |
| IPI00550191.2 | Uncharacterized protein C9orf78 | 66.09% | 6 | 16 | 1.265 | 1.440 | 1.324 | 22 |
| IPI00643583.2 | Dynein light chain roadblock-type 1 | 70.83% | 4 | 8 | 1.458 | 1.421 | 1.140 | 16 |
| IPI00000001.2 | Double-stranded RNA-binding protein Staufen homolog 1 | 57.37% | 14 | 33 | 1.247 | 1.490 | 1.280 | 53 |
| IPI00479946.4 | STIP1 protein | 84.58% | 48 | 74 | 1.394 | 1.443 | 1.173 | 209 |
| IPI00218342.11 | C-1-tetrahydrofolate synthase | 65.39% | 11 | 60 | 1.316 | 1.324 | 1.346 | 109 |
| IPI00021570.1 | Endothelial differentiation-related factor 1 | 81.08% | 7 | 18 | 1.317 | 1.397 | 1.260 | 26 |
| IPI00297982.7 | Eukaryotic translation initiation factor 2 subunit 3 | 46.82% | 7 | 19 | 1.285 | 1.496 | 1.175 | 16 |
| IPI00873899.1 | ATP-binding cassette sub-family F member 1 | 54.79% | 4 | 43 | 1.468 | 1.182 | 1.295 | 47 |
| IPI00021167.3 | Double stranded RNA-dependent protein kinase activator | 58.47% | 9 | 19 | 1.271 | 1.425 | 1.248 | 24 |
| IPI00472094.2 | Microtubule-associated protein 2 | 50.03% | 23 | 87 | 1.361 | 1.539 | 1.039 | 66 |
| IPI00179890.2 | Ras GTPase-activating protein-binding protein 2 | 61.47% | 10 | 26 | 1.260 | 1.397 | 1.270 | 39 |
| IPI00013297.1 | 28 kDa heat- and acid-stable phosphoprotein | 71.82% | 12 | 22 | 1.303 | 1.420 | 1.199 | 41 |
| IPI00410618.4 | Uncharacterized protein KIAA1143 | 58.44% | 4 | 9 | 1.096 | 1.624 | 1.200 | 10 |
| IPI00012442.1 | Ras GTPase-activating protein-binding protein 1 | 70.17% | 14 | 31 | 1.268 | 1.326 | 1.320 | 74 |
| IPI00902947.1 | Similar to LIX1-like protein | 49.65% | 3 | 12 | 1.150 | 1.484 | 1.278 | 11 |
| IPI00550746.4 | Nuclear migration protein nudC | 70.39% | 14 | 29 | 1.481 | 1.150 | 1.275 | 39 |
| IPI00008524.1 | Polyadenylate-binding protein 1 | 82.86% | 21 | 50 | 1.188 | 1.401 | 1.290 | 118 |
| IPI00736859.1 | Heterogeneous nuclear ribonucleoprotein U-like 1 | 47.88% | 14 | 41 | 1.320 | 1.320 | 1.238 | 28 |
| IPI00030320.4 | ATP-dependent RNA helicase DDX6 | 65.84% | 13 | 26 | 1.207 | 1.533 | 1.136 | 63 |
| IPI00873326.3 | Ubiquitin fusion degradation protein 1 homolog | 51.60% | 5 | 18 | 1.243 | 1.558 | 1.075 | 18 |
| IPI00334713.1 | Heterogeneous nuclear ribonucleoprotein A/B | 53.68% | 8 | 22 | 1.146 | 1.291 | 1.434 | 33 |
| IPI00792186.4 | ATP-binding cassette sub-family F member 1 | 63.23% | 2 | 29 | 1.476 | 1.169 | 1.212 | 36 |
| IPI00006980.1 | UPF0568 protein C14orf166 | 86.07% | 15 | 23 | 1.342 | 1.408 | 1.094 | 42 |
| IPI00479306.1 | Proteasome subunit beta type-5 | 51.71% | 5 | 13 | 1.273 | 1.506 | 1.059 | 10 |
Tau Binds Preferentially to the Large 60S Subunit of the Ribosome
Several well-characterized protein machines, including the spliceosome and the ribosome, are known to contribute to the cellular processing and translation of RNA. To further explore the underlying causes for the presence of proteins with known roles in RNA-processing and translation in the tau1–441 interactome dataset, we next explored if specific subcomplexes of these cellular machines are preferentially enriched, or if the tau1–441 interactome is characterized by a broad and somewhat indiscriminate enrichment of RNA binding proteins. This analysis revealed several examples of tau1–441 exhibiting preferential interactions to a subset of constituents of RNA-containing complexes and can be exemplified by the interaction of tau1–441 with 60S and 40S subunits of the ribosome. More specifically, among more than 60 protein subunits of the ribosome that co-isolated with tau1–441, levels of enrichment were higher for 60S than for 40S subunits. This trend was particularly striking at the level of TMT signature ion distributions (supplemental Fig. S1) but could also be observed when levels of enrichment of individual subunits were plotted on the basis of spectral counts measured per protein (normalized by sequence lengths) (Fig. 3C). Taken together, this distribution of enrichment levels was consistent with the interpretation that ribosomal subunits did not co-isolate with tau1–441 individually but were sequestered by the bait protein in form of 60S and 40S subcomplexes, which exhibited different propensities for binding to tau.
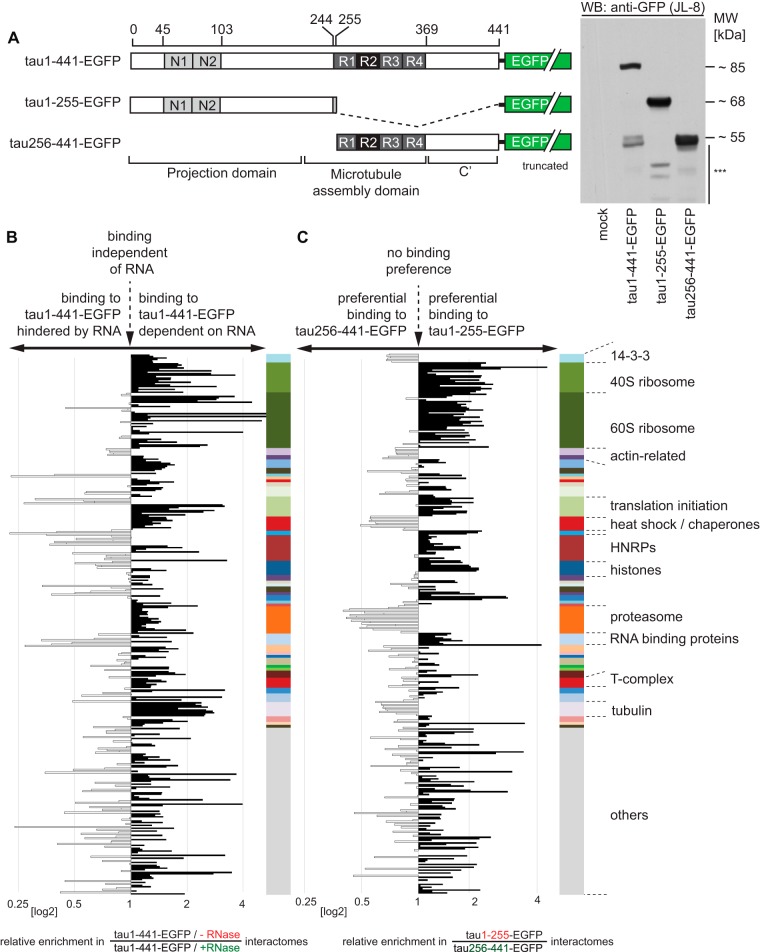
Fig. 3.
Characterization of role RNA played in composition of tau1–441 interactome, and identification of binders to tau1–255 and tau256–441. A, Schematic depiction of tau expression constructs employed in this second set of interactome experiments and confirmation of their cellular expression by Western blot analysis. The Western blot panel depicts levels of expression of the indicated tau-EGFP fusion proteins in SH-SY5Y cells. The asterisks denote low levels of proteolysis products of expressed proteins present in SH-SY5Y extracts. Note that because of the presence of the C-terminal EGFP fusion, the theoretical masses of the tau fusion constructs compute to 73 kDa (tau1–441-EGFP), 50 kDa (tau1–255-EGFP), and 46 kDa (tau256–441-EGFP). However, because of its largely disordered structure and post-translational modifications, the apparent molecular masses of tau constructs exceed the theoretical masses by 10–20 kDa. B, Graph depicting the dependence of the tau interactome on the presence or absence of RNA in the cellular extract. Binding of tau1–441 to RNA contributes to the indirect co-isolation of many but not all proteins present in the tau1–441 interactome. Note for example, that several subunits of the 60S ribosomal complex, HNRP and even RNA-binding proteins exhibit improved binding to tau1–441 in the absence of RNA. Please see Panel C and Supplemental Table S3 for the assignment of colors in the color bar to protein categories and detailed assignments of individual proteins to enrichment ratios. C, Graph depicting binding preference of individual tau interactors to N- or C-terminal tau expression constructs. Note the preferential binding of ribosomal subunits to tau1–255 and the predominant co-isolation of 14-3-3 proteins, the proteasome and several chaperones with tau256–441.
Tau-RNA Binding Accounts only Partially for Enrichment of Ribosomal Proteins
It had previously been reported that tau can associate with RNA (46) through electrostatic interactions that are largely independent of the nucleic acid sequence. To determine the extent to which the presence of RNA binding proteins, including ribosomal ribonucleoprotein complexes, in the tau1–441 interactome depends on tau-RNA interactions, a second set of tau1–441-EGFP interactome studies was conducted. To be able to directly compare the contribution of RNAs to the enrichment of individual proteins, SH-SY5Y lysates were either treated with a mix of RNAses A and T1 under conditions that broadly destructed RNAs (not shown), or subjected to mock digestion prior to the tau1–441-EGFP affinity capture step (supplemental Fig. S2). Not surprisingly, one outcome of this analysis was that it reproduced evidence in support of the aforementioned tau1–441 candidate interactors. Where applicable, candidate interactors were grouped on the basis of their known predominant function or association with protein complexes to facilitate data analysis (Fig. 3B). Consistent with the notion that the presence of RNA facilitated binding of many tau1–441 interactors, a majority of RNA binding proteins we had shortlisted as tau1–441 interactors (supplemental Table S1 and first data column of supplemental Table S3) were observed at lower enrichment levels when digitonin-solubilized cellular extracts were depleted of RNA prior to the affinity capture step (second data column in supplemental Table S3). In fact, several candidate interactors, including members of the eukaryotic translation initiation factor family, were dramatically reduced in RNase-treated samples, suggesting their binding to tau1–441 strictly depends on the presence of RNA in this experimental paradigm. Other tau1–441 candidate interactors, including all ribosomal 40S subunits emerged as partially dependent on RNA for their binding to tau1–441 in this analysis. Interestingly, the binding of 60S subunits to tau1–441 did not seem to correlate consistently with the presence or absence of RNA, suggesting that tau1–441 interacts with this subcomplex largely independent of RNA. Finally, a small subset of tau1–441 candidate interactors, including most heteronuclear ribonucleoproteins and a subset of RNA binding proteins (e.g. Fus and EWS), were observed at a higher enrichment level when tau1–441 capture occurred from RNA-depleted extracts (for an example, see supplemental Fig. S3C).
Binding to N- or C-terminal Tau1–441 Domains
To begin to map the sites within tau1–441 responsible for binding to the aforementioned candidate interactors, we next generated stable cell lines expressing tau1–255-EGFP or tau256–441-EGFP (Fig. 3A) and undertook another interactome investigation from digitonin-solubilized lysates using the truncated tau proteins as baits and the same experimental procedures for data acquisition and analysis described for the ± RNase study (supplemental Fig. S2). This experiment revealed a striking consistency in the N- or C-terminal tau binding preference of proteins that are either related in sequence or share an association with a given protein complex. Thus, whereas ribosomal subunits, eukaryotic translation initiation factors, heteronuclear ribonucleoproteins, histones, and RNA binding proteins were observed to bind to the tau1–255 domain, members of the 14-3-3 protein family, actin-related proteins, heat shock proteins, and subunits of the proteasome were observed to bind preferentially to tau256–441 (Fig. 3C).
P301L Mutant Impairs Interactions with Proteins That Bind to the C Terminus of Tau1–441
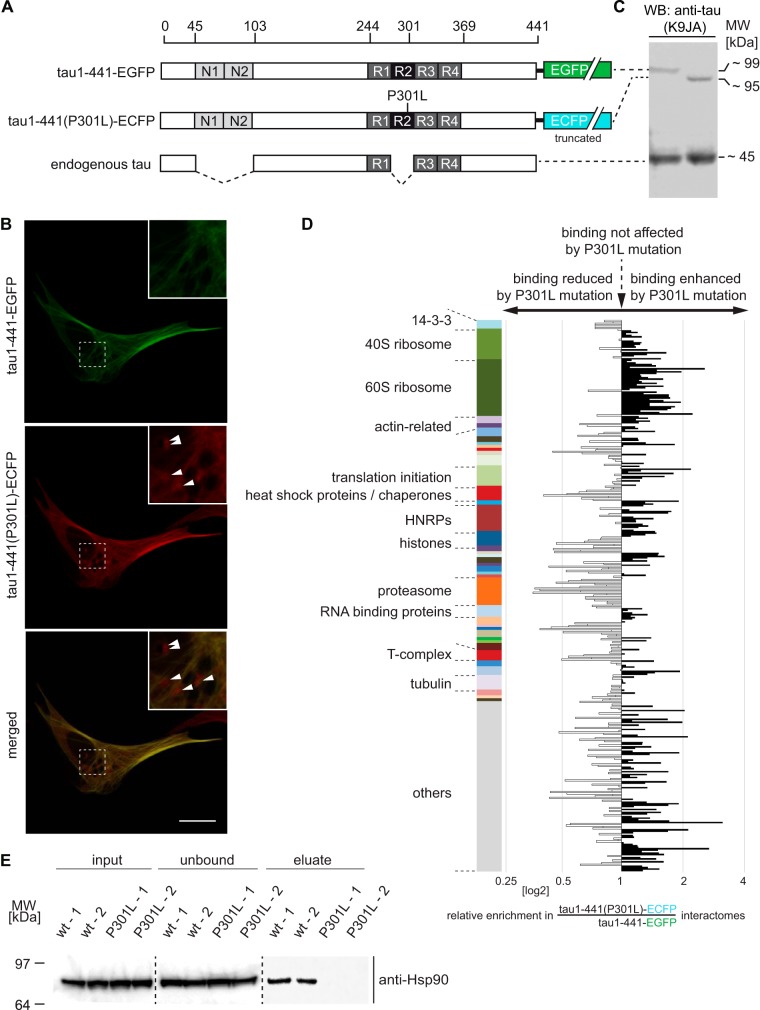
The human P301L mutation in tau is known to cause frontotemporal dementia with parkinsonism linked to chromosome 17 (FTLD17) and represents one of the most intensely studied tau mutations to date (47). To investigate if this mutation affects protein-protein interactions tau engages in, a tau1–441(P301L)-ECFP expression construct was derived from the tau1–441-EGFP parent plasmid by site-directed mutagenesis reactions that targeted both the P301 codon and critical residues for converting EGFP to ECFP (48) (Fig. 4A). The subsequent transient co-transfection of CV-1 cells with plasmids coding for wild-type and mutant tau fluorescent fusion proteins led to the expected predominant localization of both expression products to the microtubule network. Consistent with the notion that the cellular biology of the tau1–441(P301L) mutant differs from wild-type tau1–441, this coanalysis also revealed association of the P301L mutant—but not wild-type tau protein—with cytosolic punctae (Fig. 4B). To determine if these subtle differences in cellular distribution are reflected in a difference in protein–protein interactions, we next undertook a comparative interactome analysis of tau1–441-EGFP and mutant tau1–441(P301L)-ECFP using SH-SY5Y cells as hosts, which expressed the heterologous tau constructs at levels not exceeding endogenous tau levels (Fig. 4C). Because the GBP-based affinity capture works similarly well for EGFP and ECFP fusion proteins (ChromoTek, personal communication), this analysis made use of the previously established workflow (Fig. 1C). Data from this experiment revealed many similarities in the interactome of wild-type and P301L mutant tau but also illustrated differences. Perhaps counterintuitively, binding of the P301L mutant to heat shock proteins, and proteasomal proteins was relatively impaired when compared with the binding of these interactors to wild-type tau (supplemental Fig. S3A and S3B). Strikingly, a more in-depth side-by-side comparison of TMT enrichment ratios from this experiment and the aforementioned assignment of candidate interactions to tau domains uncovered a direct, albeit it imperfect, correlation between the preferential binding of candidate interactors to the C terminus of tau and the impairment of their binding in the presence of the P301L mutation. For instance, 14-3-3 proteins, heat shock proteins and proteasomal proteins bind preferentially to the tau C terminus and this interaction was particularly affected by the P301L mutation. An exception to this trend represented enhanced binding of the P301L mutant tau to ribosomal subunits, which were observed to bind preferentially to tau1–255 (Figs. 3C, Fig. 4E and 4D, supplemental Table S3).
Fig. 4.
Comparative interactome analysis of wild-type and P301L mutant tau. A, Schematic depicting full-length tau expression constructs used in this experiment as well as the principal tau isoform observed in undifferentiated SH-SY5Y cells (73). B, Representative live-cell images of CV-1 cells co-expressing tau1–441-EGFP and tau1–441(P301L)-ECFP. Note the presence of cytosolic punctae (indicated by white arrowheads) only in the ECFP fluorescence channel (pseudo colored in red for improved visibility); scale bar = 1 μm. C, Western blot analysis of total tau levels following transient transfection with K9J8 anti-tau antibody. As previously reported, P301L mutant tau migrated with lower apparent molecular mass than the corresponding wild-type tau construct (74). Note that the heterologous tau levels were comparable to endogenous tau levels in cells that were successfully transfected, because the transfection efficiency we typically achieve with SH-SY5Y cells is around 30% (as determined by the percentage of EGFP-positive cells). D, Graph depicting the relative co-enrichment of proteins together with wild-type or P301L mutant tau. Note the reduced binding of heat shock proteins and proteasomal subunits toward tau1–441(P301L) mutant tau (relative to tau1–441-EGFP). Color bar as in Fig. 3, Panels B and C. E, Validation of selective enrichment of Hsp90 with tau1–441 but not with tau1–441(P301L). Equal volumes of cellular extracts set aside before (input) or after (unbound) GBP-based tau affinity capture, or following elution (eluate) were analyzed by Western blotting and probed with anti-Hsp90 antibody.
Expression of Tau1–255 does not Shift the Global Proteome but Stabilizes Cystatin B Levels
It is increasingly understood that the interaction of some proteins (e.g. a subset of hnRNPs) with ribosomes can shift the preference of ribosomal translation (49). To begin to address if the interaction of tau with ribosomes might similarly influence ribosomal activity we sought to compare the steady-state protein levels of cells expressing tau1–255-EGFP or EGFP. The tau1–255-EGFP expression construct was selected for this experiment because, when it was employed as an affinity capture bait, it had led to a stronger co-enrichment of ribosomes than other tau constructs (possibly on account of its inability to bind to microtubules) (Fig. 3C and supplemental Table S3, data column 2). To eliminate the risk that endogenous tau levels might mask possible effects of tau expression on the steady-state proteome, this experiment was undertaken in human HEK-293 embryonic kidney cells, which are not known to express tau endogenously. Both constructs were transiently transfected in HEK-293 cells and the global proteome was measured following hot cell lysis in the presence of sodium dodecylsulfate by quantitative comparative mass spectrometry using the aforementioned workflow (Fig. 1C), except for the omission of the affinity capture step. The experiment provided quantitative information on more than 3,000 proteins, of which 1,680 were robustly quantified on the basis of >10 TMT reporter ion distributions (supplemental Table S4). Interestingly, in addition to tau itself (supplemental Fig. 4A), only the expression of one other protein, namely cystatin-B, correlated positively with the presence of tau1–255-EGFP in all three biological replicates (supplemental Fig. 4B). In addition to establishing that the binding of tau1–255 to ribosomes does not induce a major shift in ribosomal translation, the consistent detection of other proteins in the samples and controls at equal levels served as a powerful testament to the exquisite reproducibility of the workflow utilized in this study.
DISCUSSION
The work described in this report generated a first in-depth inventory of protein–protein interactions of the human tau protein using cutting-edge comparative mass spectrometry workflows (see supplemental Fig. S5 for a summary of analyses undertaken). A striking result of this study is the predominant association of tau with proteins that can be broadly characterized as belonging to the cellular ribonucleoproteome. Perhaps counterintuitively, this discovery does not seem to simply reflect an indiscriminate ability of tau to associate indirectly with these proteins on the basis of its capacity to interact with RNA. Instead, the binding of individual ribonucleoproteins may or may not: (1) depend on RNA; (2) involve the N- or C terminus of tau; and (3) be susceptible to mutation of tau at amino acid residue 301.
The tau protein had been the object of several mass spectrometry-based investigations that preceded this work. Most frequently, these studies served to characterize post-translational modifications within tau (50–53) and/or explored the merits of tau as a biomarker for neurodegenerative disease (54, 55). Conceptually more similar to the current study were prior investigations that identified proteins that copurify with microtubules (56), explored the composition of neurofibrillar tangles (57), or investigated how the presence or absence of methylene blue alters the tau-associated proteome (58). The latter study was based on an N-terminal V5 epitope-tagged tau construct and revealed 48 proteins, whose abundance levels underwent methylene blue-dependent changes in tau co-immunoprecipitates. A list of >500 total proteins that were identified in tau immunoprecipitates was appended in the Supplemental Information of this prior report. Regrettably, the data did not allow discerning if their presence in the sample was tau-dependent because the necessary negative control was not included. Thus, to our knowledge, the current report is the first that pursued the primary objective to generate an in-depth inventory of tau interactors. The study afforded unprecedented coverage of the tau interactome by combining a robust relative quantitation approach based on isobaric tagging with advanced peptide identification capabilities available on the Tribrid Orbitrap Fusion mass spectrometer. In contrast to 2D gel-based quantitation of protein abundances on the basis of signal intensities of protein spots, the TMT-based approach employed in this work can elucidate even small shifts in abundance levels (e.g. 20%). With this approach the statistical robustness of individual abundance level assignments increases with the number of peptides detected, as each peptide provides a separate quantitation data point.
Several of the previously proposed interactors, including Hsp70, Hsp90, the proteasome, and RNA were confirmed in this work. Others, including Src family kinases, were not observed, possibly because their enzyme-substrate relationship with tau translates into short-lived interactions that elude co-affinity capture workflows, a notorious shortcoming of this type of interactome investigation. In some instances, one or more homologs of a known tau interactor, e.g. peptidyl-prolyl cis-trans isomerases B and 3 (also known by the names cyclophilin B and FKBP3, respectively), were observed to co-isolate with the tau bait. Although one of the previously proposed FKBP family members to interact with tau (30, 31), namely FKBP5 (otherwise known as FKBP51), was also detected in this study, its level of tau co-isolation was lower than for FKBP3. More work is needed to determine if these differences reflect differences in the choice of biological source material or are caused by other factors.
With regard to the few tau protein–protein interactions for which the tau binding epitopes had been mapped, data presented in this report are in good agreement with prior knowledge. For instance, Hsp70 and Hsp90 were known to bind to relatively extended and partially overlapping tau binding epitopes, encompassing stretches of amino acids within the microtubule assembly domain and the proximal portion of the C-terminal domain (27). Our data corroborated that these two chaperones bind predominantly to the C-terminal half of tau and extended this knowledge by establishing that several other chaperones, including Hsp71, STIP1, and homologs of the DnaJ chaperone family, represent also candidates for binding to an epitope within tau256–441 (Fig. 3C, supplemental Table S3). For a majority of the candidate protein–protein interactions uncovered in this study, no prior binding information was available against which the assignment to N- versus C-terminal binding domains within tau can be compared. Interestingly however, our data document conspicuous agreement in the preference for binding to tau N- or C-terminal domains among protein homologs. This was, for example, observed for six members of the 14-3-3 protein family that are not known to coexist in a single protein complex but nevertheless exhibited similar levels of co-enrichment with tau. Moreover, tau binding of all six 14-3-3 proteins was only subtly affected by the destruction of RNA, preferentially occurred toward the C-terminal tau256–441 expression product and was partially impaired by the P301L tau mutation (supplemental Table S3).
In light of the relatively mild in vivo formaldehyde crosslinking and digitonin-based cell lysis procedures employed in this work, it was to be expected that many protein complexes would stay intact and, consequently, their subunits would exhibit similar enrichment trends in the various co-isolation experiments conducted. Indeed, subunits of the 60S ribosome complex were observed to consistently co-isolate with full-length tau, shared a preference for binding to the N terminus of tau, and exhibited a bias toward associating with the P301L mutant relative to wild-type tau. However, their distribution was not consistent in samples that had been subjected to RNase digestion, possibly, because the destruction of ribosomal RNAs had caused the 60S complex to disintegrate. Consistent with this interpretation, proteasomal subunits, which do not rely on RNA for maintaining complex integrity and are more likely to be stabilized by covalent crosslink bonds, did not seem to disintegrate upon RNase digestion and exhibited similar enrichment trends in all interactome samples.
A surprising observation in this study was the striking co-isolation of ribonucleoprotein complexes with tau. This preferential binding to ribonucleoprotein complexes was tau sequence specific as it did not occur when EGFP was used as the bait. More work is needed to evaluate the significance of this observation, in particular, in regards to possible mechanisms by which the tau protein may affect cellular toxicity in neurodegenerative diseases. Naturally, with SH-SY5Y cells lacking the morphology of neurons, one might expect that the tau interactions revealed in this study would be less informative about its physiological interactions within the axon than for understanding possible partners tau encounters when mislocalized to the somatodendritic compartment in AD or FTD (59). Two facets of these data seem worth highlighting at this time: (1) The observation that 27 RNA-recognition motif (RRM) domain containing proteins, including the FUS protein, co-isolated with tau (supplemental Table S2). The RRM family is notorious for its high proportion of members predicted to be able to acquire prion-like properties (60) and mutations in its members Fus and TDP43 cause neurodegenerative diseases whose pathologies overlap with tauopathies (61). (2) The robust co-isolation of ribosomes with tau. Because tau and ribosomes are both abundant in the cytosol of neurons, it would seem plausible that an interaction between these binding partners may also occur in vivo, raising the question if it has been described before and what its physiological significance might be. Intriguingly, there is a report on an echinoderm microtubule associated protein (EMAP) that exhibits sequence similarities within short sequence motifs to the Projection Domain of tau and MAP2 (62), and whose presence or absence correlated with the binding of ribosomes to microtubules in vitro (63). It could be argued that in light of the predominant association of tau with microtubules (Fig. 1C) the most compelling visual evidence for an interaction between tau and ribosomes might be found in tauopathies that are characterized by the detachment of tau from microtubules. Indeed, a survey of the pertinent literature revealed a few independent reports that predate this study by up to 25 years in which the respective authors described a co-localization of tau immunoreactivity with ribosomes based on several distinct anti-tau antibodies (Alz50, tau1, and tau2) (64, 65). As expected, some of these reports proposed that ribosomal co-localization of tau might be particularly pronounced in dementias characterized by the detachment of tau from microtubules (66–68). More recently, it was shown that tau isolated from Alzheimer's disease brains or from an FTD mouse overexpressing P301L mutant tau associated with the surface of rough endoplasmic reticulum membranes (69). Taken together, these observations lead us to propose that tau binding to ribosomes is not restricted to in vitro paradigms but does indeed occur also in vivo. The functional significance of this interaction is currently unclear. Overexpression of tau1–255, a construct that led to a stronger co-enrichment of ribosomes than other tau constructs tested in this study, did not alter the steady-state levels of more than 3,000 proteins, arguing against tau modulating access of a particular subset of mRNAs to the ribosome (49). Interestingly, a highly selective and direct correlation of tau and cystatin B was observed in this study. The possible relationship of tau and cystatin B levels warrants further investigation under separate cover, as it has been shown that genetic deletion of cystatin B improves several aspects of the AD-like neuropathology observed in the TCRND8 mouse model (70).
Prior to this study, it had been shown that Hsp70 and Hsp90 can act as a protective factor in tau aggregation in vitro (25) and that, relative to wild-type tau, the P301L mutant tau exhibited reduced binding to α-synuclein, a deficiency that could be rescued by addition of Hsp70 and Hsp90 (71). Comparative interactome analyses of wild-type and P301L tau presented in this work established that the P301L mutation partially impairs the ability of tau to bind to several of its interactors, including heat shock proteins, the proteasome complex and members of the 14-3-3 protein family. We further observed a concordance of proteins whose binding to tau is affected by this mutation with the subset of proteins that predominantly bind to the C-terminal half of tau, a relationship that had not been appreciated before. Two scenarios may seem most plausible to explain these data: (1) Considering that the P301L mutation maps to the second repeat in the microtubule assembly domain, these observations may reflect a reality whereby the mutated residue affects the local fold of tau, which in turn perturbs nearby protein binding epitopes in the C-terminal half of tau. (2) Alternatively, the mutation may cause the subset of cellular tau that is detached from microtubules to increase, thereby depriving it of its access to a microtubule-associated pool of proteins that normally bind to its C-terminal domain. Regardless of the causes for the binding impairment, our data are consistent with a model whereby the increased propensity of P301L mutant tau to form insoluble cytosolic aggregates (72) could be a consequence of its impaired interaction with proteins that are known to play critical roles in maintaining the delicate balance between protein stabilization and degradation.
CONCLUSION
As for any large-scale discovery project, this study has raised many more questions than it could answer. It is hoped that the extensive characterization of tau interactions presented in this work will serve the research community as a valuable resource that will stimulate existing research and precipitate novel ideas. Of the many possible avenues one might pursue, a few stand out: (1) Do interactions of tau with RRM-containing proteins play a role in the manifestation of overlapping frontotemporal dementia pathologies?; (2) What is the significance of the tau-ribosome interaction in health and disease?—does tau play a role in the cellular distribution of ribosomes along microtubule tracts?; (3) How do other chaperones, including STIP1, or the proteasome interact with tau, and do efforts to improve binding of chaperones to P301L mutant tau or to increase proteasomal turnover of tau have merit as disease intervention strategies?; and (4) What is the significance of the direct correlation of steady-state tau and cystatin B levels observed in this study?
Supplementary Material
Footnotes
Author contributions: C.G. and G.S. designed research; C.G., M.M., X.W., I.M., I.B.L., J.E.J., H.W., and G.S. performed research; G.S. contributed new reagents or analytic tools; C.G., M.M., X.W., I.M., I.B.L., H.W., and G.S. analyzed data; C.G., M.M., H.W., and G.S. wrote the paper.
* This work was supported by an operating grant from the “China-Canada Initiative on Alzheimer's Disease and Related Disorders,” a partnership initiative between the Canadian Institute of Health Research (CIHR), Canada, and the National Natural Science Foundation NNSF, China. The mass spectrometry component was enabled by a generous donation, kindly provided by the Irwin family, and a leading edge fund (LEF) infrastructure grant from the Canadian Foundation for Innovation (CFI). C.G.G. gratefully acknowledges support from a postdoctoral CIHR fellowship, M.M. contributed to this project supported by funding from an Ontario Trillium Scholarship, and G.S. received generous support from the W. Garfield Weston Foundation.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
1 The abbreviations used are:
- MAPs
- microtubule-associated proteins
- CBD
- corticobasal degeneration
- CID
- collision induced dissociation
- EGFP
- enhanced green fluorescent protein
- ESI
- electrospray ionization
- GO
- Gene Ontology
- HSP
- heat shock protein
- MTB
- microtubule binding domain
- MT
- microtubule
- MS/MS
- tandem mass spectrometry
- NFT
- neurofibrillary tangles
- PD
- projection domain
- PSM
- peptide-to-spectrum matches
- PSP
- progressive supranuclear palsy
- PTM
- posttranslational modifications
- TMT
- tandem mass tag.
REFERENCES
- 1. Neve R. L., Harris P., Kosik K. S., Kurnit D. M., and Donlon T. A. (1986) Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res. 387, 271–280 [DOI] [PubMed] [Google Scholar]
- 2. Weingarten M. D., Lockwood A. H., Hwo S. Y., and Kirschner M. W. (1975) A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. U.S.A. 72, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y. C., Zaidi M. S., and Wisniewski H. M. (1986) Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 261, 6084–6089 [PubMed] [Google Scholar]
- 4. Wolozin B. L., Pruchnicki A., Dickson D. W., and Davies P. (1986) A neuronal antigen in the brains of Alzheimer patients. Science 232, 648–650 [DOI] [PubMed] [Google Scholar]
- 5. Lee V. M., Goedert M., and Trojanowski J. Q. (2001) Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24, 1121–1159 [DOI] [PubMed] [Google Scholar]
- 6. Morris M., Maeda S., Vossel K., and Mucke L. (2011) The many faces of tau. Neuron 70, 410–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harada A., Oguchi K., Okabe S., Kuno J., Terada S., Ohshima T., Sato-Yoshitake R., Takei Y., Noda T., and Hirokawa N. (1994) Altered microtubule organization in small-calibe axons of mice lacking tau protein. Nature 369, 488–491 [DOI] [PubMed] [Google Scholar]
- 8. Dawson H. N., Ferreira A., Eyster M. V., Ghoshal N., Binder L. I., and Vitek M. P. (2001) Inhibition of neuronal maturation in primary hippocampal neurons from tau deficient mice. J. Cell Sci. 114, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 9. Brandt R., Léger J., and Lee G. (1995) Interaction of tau with the neural plasma membrane mediated by tau's amino-terminal projection domain. J. Cell Biol. 131, 1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farias G. A., Muñoz J. P., Garrido J., and Maccioni R. B. (2002) Tubulin, actin, and tau protein interactions and the study of their macromolecular assemblies. J. Cell. Biochem. 85, 315–324 [DOI] [PubMed] [Google Scholar]
- 11. Ittner L. M., Ke Y. D., Delerue F., Bi M., Gladbach A., van Eersel J., Wölfing H., Chieng B. C., Christie M. J., Napier I. A., Eckert A., Staufenbiel M., Hardeman E., and Götz J. (2010) Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell 142, 387–397 [DOI] [PubMed] [Google Scholar]
- 12. Ittner L. M., and Gotz J. (2011) Amyloid-beta and tau – a toxic pas de deux in Alzheimer's disease. Nat. Rev. Neurosci. 12, 65–72 [DOI] [PubMed] [Google Scholar]
- 13. Hirokawa N., Shiomura Y., and Okabe S. (1988) Tau proteins: the molecular structure and mode of binding on microtubules. J. Cell Biol. 107, 1449–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cleveland D. W., Hwo S. Y., and Kirschner M. W. (1977) Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J. Mol. Biol. 116, 227–247 [DOI] [PubMed] [Google Scholar]
- 15. Mukrasch M. D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2009) Structural polymorphism of 441-residue tau at single residue resolution. PLos Biol. 7, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., and Binder L. I. (1986) Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Horiguchi T., Uryu K., Giasson B. I., Ischiropoulos H., LightFoot R., Bellmann C., Richter-Landsberg C., Lee V. M., and Trojanowski J. Q. (2003) Nitration of tau protein is linked to neurodegeneration in tauopathies. Am. J. Pathol. 163, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morishima-Kawashima M., Hasegawa M., Takio K., Suzuki M., Titani K., and Ihara Y. (1993) Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron 10, 1151–1160 [DOI] [PubMed] [Google Scholar]
- 19. Dorval V., and Fraser P. E. (2006) Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J. Biol. Chem. 281, 9919–9924 [DOI] [PubMed] [Google Scholar]
- 20. Wang Y. P., Biernat J., Pickhardt M., Mandelkow E., and Mandelkow E. M. (2007) Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model. Proc. Natl. Acad. Sci. U.S.A. 104, 10252–10257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arnaud L. T., Myeku N., and Figueiredo-Pereira M. E. (2009) Proteasome-caspase-cathepsin sequence leading to tau pathology induced by prostaglandin J2 in neuronal cells. J. Neurochem. 110, 328–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcus J. N., and Schachter J. (2011) Targeting post-translational modifications on tau as a therapeutic strategy for Alzheimer's disease. J. Neurogenet. 25, 127–133 [DOI] [PubMed] [Google Scholar]
- 23. Lee G., Newman S. T., Gard D. L., Band H., and Panchamoorthy G. (1998) Tau interacts with src-family nonreceptor tyrosine kinases. J. Cell Sci. 111, 3167–3177 [DOI] [PubMed] [Google Scholar]
- 24. Reynolds C. H., Garwood C. J., Wray S., Price C., Kellie S., Perera T., Zvelebil M., Yang A., Sheppard P. W., Varndell I. M., Hanger D. P., and Anderton B. H. (2008) Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases. J. Biol. Chem. 283, 18177–18186 [DOI] [PubMed] [Google Scholar]
- 25. Patterson K. R., Ward S. M., Combs B., Voss K., Kanaan N. M., Morfini G., Brady S. T., Gamblin T. C., and Binder L. I. (2011) Heat shock protein 70 prevents both tau aggregation and the inhibitory effects of pre-existing tau aggregates on fast axonal transport. Biochemistry 50, 10300–10310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dou F., Netzer W. J., Tanemura K., Li F., Hartl F. U., Takashima A., Gouras G. K., Greengard P., and Xu H. (2003) Chaperones increase association of tau protein with microtubules. Proc. Natl. Acad. Sci. U.S.A. 100, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karagöz G. E., Duarte A. M., Akoury E., Ippel H., Biernat J., Morán Luengo T., Radli M., Didenko T., Nordhues B. A., Veprintsev D. B., Dickey C. A., Mandelkow E., Zweckstetter M., Boelens R., Madl T., and Rüdiger S. G. (2014) Hsp90-tau complex reveals molecular basis for specificity in chaperone action. Cell 156, 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fulga T. A., Elson-Schwab I., Khurana V., Steinhilb M. L., Spires T. L., Hyman B. T., and Feany M. B. (2007) Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol. 9, 139–148 [DOI] [PubMed] [Google Scholar]
- 29. Strittmatter W. J., Saunders A. M., Goedert M., Weisgraber K. H., Dong L. M., Jakes R., Huang D. Y., Pericak-Vance M., Schmechel D., and Roses A. D. (1994) Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 91, 11183–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chambraud B., Sardin E., Giustiniani J., Dounane O., Schumacher M., Goedert M., and Baulieu E. E. (2010) A role for FKBP52 in Tau protein function. Proc. Natl. Acad. Sci. U.S.A. 107, 2658–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blair L. J., Nordhues B. A., Hill S. E., Scaglione K. M., O'Leary J. C. 3rd, Fontaine S. N., Breydo L., Zhang B., Li P., Wang L., Cotman C., Paulson H. L., Muschol M., Uversky V. N., Klengel T., Binder E. B., Kayed R., Golde T. E., Berchtold N., and Dickey C. A. (2013) Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest. 123, 4158–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qureshi H. Y., and Paudel H. K. (2011) Parkinsonian neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and alpha-synuclein mutations promote Tau protein phosphorylation at Ser262 and destabilize microtubule cytoskeleton in vitro. J. Biol. Chem. 286, 5055–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y., Lv K., Li Z., Yu A. C., Chen J., and Teng J. (2012) PACSIN1, a Tau-interacting protein, regulates axonal elongation and branching by facilitating microtubule instability. J. Biol. Chem. 287, 39911–39924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sjöberg M. K., Shestakova E., Mansuroglu Z., Maccioni R. B., and Bonnefoy E. (2006) Tau protein binds to pericentromeric DNA: a putative role for nuclear tau in nucleolar organization. J. Cell Sci. 119, 2025–2034 [DOI] [PubMed] [Google Scholar]
- 35. Kampers T., Friedhoff P., Biernat J., Mandelkow E. M., and Mandelkow E. (1996) RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 399, 344–349 [DOI] [PubMed] [Google Scholar]
- 36. King M. E., Kan H. M., Baas P. W., Erisir A., Glabe C. G., and Bloom G. S. (2006) Tau-dependent microtubule disassembly initiated by prefibrillar beta-amyloid. J. Cell Biol. 175, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watts J. C., Huo H., Bai Y., Ehsani S., Jeon A. H., Won A.H., Shi T., Daude N., Lau A., Young R., Xu L., Carlson G. A., Williams D., Westaway D., and Schmitt-Ulms G. (2009) Interactome analyses identify ties of PrP and its mammalian paralogs to oligomannosidic N-glycans and endoplasmic reticulum-derived chaperones. PLoS Pathog. 5, e1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mehrabian M., Brethour D., MacIsaac S., Kim J. K., Gunawardana C. G., Wang H., and Schmitt-Ulms G. (2014) CRISPR-Cas9-based knockout of the prion protein and its effect on the proteome. PLoS One 9, e114594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zieske L. R. (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 40. Käll L., Canterbury J. D., Weston J., Noble W. S., and MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Meth. 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 41. Sandberg A., Branca R. M., Lehtiö J., and Forshed J. (2014) Quantitative accuracy in mass spectrometry based proteomics of complex samples: the impact of labeling and precursor interference. J. Proteomics 96, 133–144 [DOI] [PubMed] [Google Scholar]
- 42. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 57, 289–300 [Google Scholar]
- 43. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., and Leonhardt H. (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289 [DOI] [PubMed] [Google Scholar]
- 46. Wang X., Wang D., Zhao J., Qu M., Zhou X., He H., and He R. (2006) The proline-rich domain and the microtubule binding domain of protein tau acting as RNA binding domains. Protein Pept. Lett. 13, 679–685 [DOI] [PubMed] [Google Scholar]
- 47. Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., Hackett J., Adamson J., Lincoln S., Dickson D., Davies P., Petersen R. C., Stevens M., de Graaff E., Wauters E., van Baren J., Hillebrand M., Joosse M., Kwon J. M., Nowotny P., Che L. K., Norton J., Morris J. C., Reed L. A., Trojanowski J., Basun H., Lannfelt L., Neystat M., Fahn S., Dark F., Tannenberg T., Dodd P. R., Hayward N., Kwok J. B., Schofield P. R., Andreadis A., Snowden J., Craufurd D., Neary D., Owen F., Oostra B. A., Hardy J., Goate A., van Swieten J., Mann D., Lynch T., and Heutink P. (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 [DOI] [PubMed] [Google Scholar]
- 48. Shaner N. C., Patterson G. H., and Davidson M. W. (2007) Advances in fluorescent protein technology. J. Cell Sci. 120, 4247–4260 [DOI] [PubMed] [Google Scholar]
- 49. Chaudhury A., Hussey G. S., Ray P. S., Jin G., Fox P. L., and Howe P. H. (2010) TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanger D. P., Byers H. L., Wray S., Leung K. Y., Saxton M. J., Seereeram A., Reynolds C. H., Ward M. A., and Anderton B. H. (2007) Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J. Biol. Chem. 282, 23645–23654 [DOI] [PubMed] [Google Scholar]
- 51. Zhang X., Hernandez I., Rei D., Mair W., Laha J. K., Cornwell M. E., Cuny G. D., Tsai L. H., Steen J. A., and Kosik K. S. (2013) Diaminothiazoles modify Tau phosphorylation and improve the tauopathy in mouse models. J. Biol. Chem. 288, 22042–22056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saman S., Kim W., Raya M., Visnick Y., Miro S., Saman S., Jackson B., McKee A. C., Alvarez V. E., Lee N. C., and Hall G. F. (2012) Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 287, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Min S. W., Cho S. H., Zhou Y., Schroeder S., Haroutunian V., Seeley W. W., Huang E. J., Shen Y., Masliah E., Mukherjee C., Meyers D., Cole P. A., Ott M., and Gan L. (2010) Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Portelius E., Hansson S. F., Tran A. J., Zetterberg H., Grognet P., Vanmechelen E., Höglund K., Brinkmalm G., Westman-Brinkmalm A., Nordhoff E., Blennow K., and Gobom J. (2008) Characterization of tau in cerebrospinal fluid using mass spectrometry. J. Proteome Res. 7, 2114–2120 [DOI] [PubMed] [Google Scholar]
- 55. Schraen-Maschke S., Sergeant N., Dhaenens C. M., Bombois S., Deramecourt V., Caillet-Boudin M. L., Pasquier F., Maurage C. A., Sablonnière B., Vanmechelen E., and Buée L. (2008) Tau as a biomarker of neurodegenerative diseases. Biomark. Med. 2, 363–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kozielski F., Riaz T., DeBonis S., Koehler C. J., Kroening M., Panse I., Strozynski M., Donaldson I. M., and Thiede B. (2011) Proteome analysis of microtubule-associated proteins and their interacting partners from mammalian brain. Amino Acids 41, 363–385 [DOI] [PubMed] [Google Scholar]
- 57. Rudrabhatla P., Jaffe H., and Pant H. C. (2011) Direct evidence of phosphorylated neuronal intermediate filament proteins in neurofibrillary tangles (NFTs): phosphoproteomics of Alzheimer's NFTs. Faseb J. 25, 3896–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thompson A. D., Scaglione K. M., Prensner J., Gillies A. T., Chinnaiyan A., Paulson H. L., Jinwal U. K., Dickey C. A., and Gestwicki J. E. (2012) Analysis of the tau-associated proteome reveals that exchange of Hsp70 for Hsp90 is involved in tau degradation. ACS Chem. Biol. 7, 1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ballatore C., Lee V. M., and Trojanowski J. Q. (2007) Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 [DOI] [PubMed] [Google Scholar]
- 60. King O. D., Gitler A. D., and Shorter J. (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Leverenz J. B., Yu C. E., Montine T. J., Steinbart E., Bekris L. M., Zabetian C., Kwong L. K., Lee V. M., Schellenberg G. D., and Bird T. D. (2007) A novel progranulin mutation associated with variable clinical presentation and tau, TDP43, and alpha-synuclein pathology. Brain 130, 1360–1374 [DOI] [PubMed] [Google Scholar]
- 62. Li Q., Callaghan M., and Suprenant K. A. (1998) The 77-kDa echinoderm microtubule-associated protein (EMAP) shares epitopes with the mammalian brain MAPs, MAP-2, and tau. Biochem. Biophys. Res. Commun. 250, 502–505 [DOI] [PubMed] [Google Scholar]
- 63. Suprenant K. A., Dean K., McKee J., and Hake S. (1993) EMAP, an echinoderm microtubule-associated protein found in microtubule-ribosome complexes. J. Cell Sci. 104, 445–450 [DOI] [PubMed] [Google Scholar]
- 64. Nelson P. T., Marton L., and Saper C. B. (1993) Alz-50 immunohistochemistry in the normal sheep striatum: a light and electron microscope study. Brain Res. 600, 285–297 [DOI] [PubMed] [Google Scholar]
- 65. Papasozomenos S. C., and Binder L. I. (1987) Phosphorylation determines two distinct species of Tau in the central nervous system. Cell Motil. Cytoskeleton 8, 210–226 [DOI] [PubMed] [Google Scholar]
- 66. Papasozomenos S. C. (1989) Tau protein immunoreactivity in dementia of the Alzheimer type. I. Morphology, evolution, distribution, and pathogenetic implications. Lab. Invest. 60, 123–137 [PubMed] [Google Scholar]
- 67. Papasozomenos S. C. (1989) Tau protein immunoreactivity in dementia of the Alzheimer type: II. Electron microscopy and pathogenetic implications. Effects of fixation on the morphology of the Alzheimer's abnormal filaments. Lab. Invest. 60, 375–389 [PubMed] [Google Scholar]
- 68. Iseki E., Matsumura T., Marui W., Hino H., Odawara T., Sugiyama N., Suzuki K., Sawada H., Arai T., and Kosaka K. (2001) Familial frontotemporal dementia and parkinsonism with a novel N296H mutation in exon 10 of the tau gene and a widespread tau accumulation in the glial cells. Acta Neuropathol. 102, 285–292 [DOI] [PubMed] [Google Scholar]
- 69. Perreault S., Bousquet O., Lauzon M., Paiement J., and Leclerc N. (2009) Increased association between rough endoplasmic reticulum membranes and mitochondria in transgenic mice that express P301L tau. J. Neuropathol. Exp. Neurol. 68, 503–514 [DOI] [PubMed] [Google Scholar]
- 70. Yang D. S., Stavrides P., Mohan P. S., Kaushik S., Kumar A., Ohno M., Schmidt S. D., Wesson D., Bandyopadhyay U., Jiang Y., Pawlik M., Peterhoff C. M., Yang A. J., Wilson D. A., St George-Hyslop P., Westaway D., Mathews P. M., Levy E., Cuervo A. M., and Nixon R. A. (2011) Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain 134, 258–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Benussi L., Ghidoni R., Paterlini A., Nicosia F., Alberici A. C., Signorini S., Barbiero L., and Binetti G. (2005) Interaction between tau and alpha-synuclein proteins is impaired in the presence of P301L tau mutation. Exp. Cell Res. 308, 78–84 [DOI] [PubMed] [Google Scholar]
- 72. Lewis J., McGowan E., Rockwood J., Melrose H., Nacharaju P., Van Slegtenhorst M., Gwinn-Hardy K., Paul Murphy M., Baker M., Yu X., Duff K., Hardy J., Corral A., Lin W. L., Yen S. H., Dickson D. W., Davies P., and Hutton M. (2000) Neurofibrillary tangles, amyotrophy, and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 25, 402–405 [DOI] [PubMed] [Google Scholar]
- 73. Smith C. J., Anderton B. H., Davis D. R., and Gallo J. M. (1995) Tau isoform expression and phosphorylation state during differentiation of cultured neuronal cells. FEBS Lett. 375, 243–248 [DOI] [PubMed] [Google Scholar]
- 74. Terwel D., Lasrado R., Snauwaert J., Vandeweert E., Van Haesendonck C., Borghgraef P., and Van Leuven F. (2005) Changed conformation of mutant Tau-P301L underlies the moribund tauopathy, absent in progressive, nonlethal axonopathy of Tau-4R/2N transgenic mice. J. Biol. Chem. 280, 3963–3973 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.