Abstract
Prostasomes are exosomes derived from prostate epithelial cells through exocytosis by multivesicular bodies. Prostasomes have a bilayered membrane and readily interact with sperm. The membrane lipid composition is unusual with a high contribution of sphingomyelin at the expense of phosphatidylcholine and saturated and monounsaturated fatty acids are dominant. Lipid rafts are liquid-ordered domains that are more tightly packed than the surrounding nonraft phase of the bilayer. Lipid rafts are proposed to be highly dynamic, submicroscopic assemblies that float freely within the liquid disordered membrane bilayer and some proteins preferentially partition into the ordered raft domains. We asked the question whether lipid rafts do exist in prostasomes and, if so, which proteins might be associated with them. Prostasomes of density range 1.13–1.19g/ml were subjected to density gradient ultracentrifugation in sucrose fabricated by phosphate buffered saline (PBS) containing 1% Triton X-100 with capacity for banding at 1.10 g/ml, i.e. the classical density of lipid rafts. Prepared prostasomal lipid rafts (by gradient ultracentrifugation) were analyzed by mass spectrometry. The clearly visible band on top of 1.10g/ml sucrose in the Triton X-100 containing gradient was subjected to liquid chromatography-tandem MS and more than 370 lipid raft associated proteins were identified. Several of them were involved in intraluminal vesicle formation, e.g. tetraspanins, ESCRTs, and Ras-related proteins. This is the first comprehensive liquid chromatography-tandem MS profiling of proteins in lipid rafts derived from exosomes. Data are available via ProteomeXchange with identifier PXD002163.
Extracellular vesicles (EVs)1 are membrane surrounded structures that exist in all body fluids and all cells studied so far release EVs (1). They are heterogeneous, spherical organelles spanning between 30 to more than 1000 nm in diameter and include exosomes, microvesicles, and apoptotic bodies (2). There is increasing evidence supporting the important role of EVs in cell-to-cell communication by their delivery of proteins, lipids, and nucleic acids from one donor cell to many target cells. The generation of exosomes/prostasomes is a complicated process involving two invagination sessions of biological membranes. The first one comprises the plasma membrane contributing with endocytic vesicles in the formation of early endosomes that mature into late endosomes. The second one starts multiple inward buddings of the late endosomal membrane creating intraluminal vesicles (ILVs) therewith completing formation of multivesicular bodies (MVBs) or storage vesicles (3) thus retaining selected molecules from the maternal cell. Ceramide can induce such formation of small microdomains into larger domains (4). Ceramide is one of two cleavage products of sphingomyelin by sphingomyelinase, the other is phosphocholine (5) and prostasomes contain sphingomyelinase (6). The membrane of MVBs (storage vesicles) may fuse with the plasma membrane of the secretory cell and, in case of prostate epithelial cells, release the intraluminal vesicles as prostasomes to the extracellular space (7, 8). It is noteworthy that the bilayered membrane surrounding prostasomes (after the two sessions of invaginations) should be regarded as “right-side-out” with reference to the plasma membrane. This is illustrated by e.g. Mg2+ and Ca2+ -stimulated ATPase that is an ectoenzyme (9) that is also appearing at the outer surface of prostasomes (10). The corollary is that cell surface interactive molecules like enzymes and receptors may appear also on the membranes of exosomes/prostasomes.
The majority of prostasomes ranges in diameter-size from 30–200 nm, with a mean of 142 nm (11). The main purpose of prostasomes may be to transfer newly synthesized proteins from the prostate gland to sperm and thereby, among other things, render them protection in the female genital tract (12, 13). Prostasomal proteins may be transferred to sperm through different mechanisms, viz direct interaction with the sperm membrane (14), fusion at a lowered pH (15), and internalization (16). Prostasomes are immunosuppressive and regulate the complement system and they have proven antioxidant and antibacterial properties (17, 18). Prostasomes contain a surrounding lipid membrane bilayer that exhibits a high cholesterol/phospholipid ratio (19). The lipid composition of the membrane is unusual and among the phospholipids sphingomyelin is the dominant one, contrary to other cell membranes where phosphatidylcholine is most abundant. Prostasomes have a strong contribution of saturated and monounsaturated fatty acids (19, 20). These characteristics together with a high cholesterol/phospholipid ratio make the membrane of the prostasome very stable as demonstrated by electron spin resonance (19).
In the early 1970s the plasma membrane of the cell was described as a fluid mosaic by Singer and Nicholson (21), but as early as in 1953 Palade claimed that in the bilayered lipid membrane, proposed by Davson and Danielli (22), were areas of different composition, so called caveolae (23). These caveolae are invaginations of the plasma membrane (24). The first hypothesis of lipid rafts (specialized membrane domains enriched in glycosphingolipids, proteins and cholesterol) was brought up in 1988 by van Meer and Simons (25) and was subsequently elaborated in 1997 by Simons and Ikonen (26). Lipid rafts were defined as low density subdomains of the plasma membrane that are resistant to nonionic detergents at a low temperature (27, 28). Fatty acids present in lipid rafts are more saturated, compared with the membrane adjacent to the domains. It means that the fatty acids can be packed more densely and this may lead to phase separation. The abundance of intercalating cholesterol makes the rafts more rigid and less fluid than the rest of the plasma membrane (29). In other words, the membrane can undergo phase separation into co-existing liquid-disordered and liquid-ordered phases. The liquid-ordered phase (the lipid raft) becomes enriched in cholesterol and saturated fatty acids and is characterized by tight lipid packing and reduced molecular diffusion, as we noticed for prostasomes (19).
There are two different types of lipid rafts, planar and caveolae. The distinguishing factor is that the caveolae are formed by the protein caveolin whereas the planar rafts lack this protein (30). Instead they contain the protein flotillin (31). Researchers have found that selected proteins localize, and colocalize in lipid rafts (32). Lipid rafts are not anchored at a specific site in the plasma membrane, but float freely. This enables larger and more stable platform domains to aggregate (33). The formed aggregates are involved in many biological functions including endocytosis, cell communication, molecular trafficking, neurotransmission and they could be understood as organizing centers for signaling molecules and receptors (30, 31). When cells are depleted of cholesterol, e.g. by the agent methyl-β-cyclodextrin, formation of caveolae expression and also raft-dependent endocytosis are inhibited (34). This demonstrates the importance of these cholesterol-enriched domains to cell survival. Flotillins are also involved in endocytosis in a process controlled by the phosphorylation of tyrosine residues (35).
In this work we asked the question whether lipid rafts do exist in prostasomes and, if so, which proteins might be associated with them. Accordingly, we prepared lipid rafts from human prostasomes in order to characterize their protein content.
EXPERIMENTAL PROCEDURES
All experiments were approved by the Ethical Committee of Uppsala University.
Purification of Prostasomes
Seminal plasma was collected from the local Fertility Clinic (Uppsala University Hospital) following established routines (36) and collected samples were kept at −20 °C. Thawed seminal plasma was centrifuged for 10 min at 4 °C and 3000 × g and the supernatant was saved and centrifuged for 30 min at 4 °C and 10,000 × g to eliminate possible cell debris and larger vesicles and the new supernatant was subjected to ultracentrifugation for 2 h at 4 °C and 100,000 × g using Rotor 90 Ti (Beckman Coulter, Brea, CA) (7). The resulting pellet was resuspended in phosphate buffered saline (PBS), pH 7.6, and loaded onto an XK16/70 Superdex 200 gel column (GE Healthcare, Uppsala, Sweden) to separate prostasomes from amorphous material (37). Fractions were collected at a flow rate of 5 ml/h (fraction volumes of ∼1.3 ml) and absorbances at 260 nm (nucleic acid) and 280 nm (proteins) (indicating prostasome presence) were measured. Fractions with elevated absorbances were pooled and ultracentrifuged for 2 h at 4 °C and 100,000 × g. The pellet was resuspended in PBS and loaded onto a gradient of 1 m, 1.5 m, and 2 m sucrose and ultracentrifuged for 20 h at 4 °C and 85,000 × g using an SW28.1 rotor (Beckman Coulter). The main fraction at 1.5 m (density range 1.13–1.19g/ml) (38) was pelleted by ultracentrifugation for 2 h at 4 °C and 100,000 × g, resuspended in PBS and the concentration was estimated by a BCA protein assay kit (Merck, Darmstadt, Germany) and adjusted to 2 mg/ml. Purified prostasomes were kept at −70 °C.
Lipid Raft Extraction
Prostasomes (∼8 mg) with density range of 1.13–1.19g/ml (main fraction) were top loaded onto two sucrose density gradients of 5% (w/w), 30% (w/w) (corresponding to a density of 1.10g/ml), and 60% (w/w) (1.5 m sucrose, corresponding to 1.19g/ml). One of the two sucrose density gradients was prepared with PBS containing 1% Triton X-100 and the other one was prepared with PBS without Triton X-100 as a control. The 30% (w/w) sucrose (density of 1.10g/ml) is the classical floatation density of lipid rafts (39). Both sucrose gradients were ultracentrifuged for 28 h at 4 °C and 256,000 × g (SW40 Ti rotor, Beckman Coulter). Fractions at 30% (w/w) sucrose (lipid raft density) and 60% (w/w) (1.5 m) sucrose were collected in respective sucrose gradient. The collected fractions were separately ultracentrifuged for 2 h at 4 °C and 100,000 × g and pellets were resuspended in PBS. At least eight preparations of lipid rafts were obtained on different occasions and stored at −70 °C.
SDS-PAGE
Proteins of three independently prepared samples of prostasomal lipid rafts and one sample of control prostasomes (2.5 μg/well) were separated on a Novex® NuPAGE 4–12% Bis-Tris polyacrylamide gel (Invitrogen, Carlsbad, CA) at 200 V for 35 min, according to the manufacturer's instructions. SeeBlue® Prestained Standard (Invitrogen) served as indicator for molecular weights. Proteins were thereafter stained using a SilverQuest™ staining kit (Invitrogen). Analysis was performed on a Molecular Imager ChemiDoc™ XRS+ Imaging System using the software Image Lab 5.1 (Bio-Rad Laboratories, Hercules, CA).
Immunoblotting
Proteins from prostasomal lipid rafts and control prostasomes (2.5 μg/well) were separated by SDS-PAGE and transferred to a 0.2 μm Novex® nitrocellulose membrane (Invitrogen), at 25 V for 1 h. Unspecific binding sites were blocked by bovine serum albumin, BSA (1%) in PBS-Tween (0.1%).
Antibodies directed against membrane bound proteins (monoclonal antibodies: Flotillin-1, Caveolin-1 (BD Biosciences Pharmigen, San José, CA); Clathrin (Abcam, Cambridge, UK) and polyclonal antibodies: Flotillin-2, Caveolin-2 (Atlas Antibodies, Stockholm, Sweden)) were added and allowed to react for 1 h at 20 °C. A biotinylated secondary antibody (dilution 1:100,000; Goat anti-mouse IgG (H+L) (Zymed Laboratories Inc., San Fransisco, CA) and Goat anti-rabbit IgG (H+L) (Merck)) were added and allowed to react for 1 h at 20 °C. Streptavidin conjugated alkaline phosphatase (Invitrogen) and BCIP/NBT kit (Invitrogen) visualized the biotinylated antibodies. Analysis was performed on a Molecular Imager ChemiDoc™ XRS+ Imaging System using the software Image Lab 5.1 (Bio-Rad Laboratories).
LC-MS/MS of Prostasomal Lipid Raft Proteins
Prostasomal lipid rafts (20 μg) were denatured and reduced in SDS (1%), ammonium bicarbonate (0.1 m) and dithiothreitol (DTT) (10 mm) by heating for 5 min at 98 °C, and subsequent alkylation by iodoacetamide (20 mm) followed by incubation for 15–30 min at 20 °C in the dark. The sample was thereafter transferred to a spintube, 10 kDa cutoff (Pall Corporation, Port Washington, NY) (40). Urea (8 m) and ammonium bicarbonate (50 mm) were added four times (4 × 450 μl) and centrifuged for 15 min at 14,000 × g each time as described previously (40). Before the fifth centrifugation only 2 m urea and 50 mm ammonium bicarbonate were added and spun after which the solution was diluted to a final concentration of 1 m urea and 50 mm ammonium bicarbonate. The digestion was executed for 16 h at 20 °C by 0.5 μg trypsin, and stopped by acetic acid. UV-absorbance was measured in a NanoDrop 2000 (Thermo Fischer Scientific Inc., Waltham, MA) to establish protein concentration. A C18 filter absorbed the digest for 24 h (41) before the mass analysis was carried out on a Thermo Velos (Thermo Fischer Scientific Inc.). The column was 100 mm long with an internal diameter of 75 μm, packed with 3 μm C18-Aq particles. The elution was carried out by a constructed gradient from 5–65% during 60 min starting with two solutions; 0.1% formic acid and 100% acetonitrile with 0.1% formic acid, respectively. A maximum of 10 fragmentations was done for each MS-spectrum. Fragmentation was carried out by CID using standard settings. The list of peaks obtained was used to search the X! Tandem website.
Peptide Matches for Protein Identification
Raw data file was converted by ProteoWizard 3.0.3916. Derived data from ProteoWizard were transferred to Mascot version, 2.2.07. Proteins were identified by Uniprot Release 2013_06 (taxonomy filter: containing 20,258 sequences, Homo sapiens). Searches were by default. Trypsin was used as enzyme (cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine) and maximum missed cleavages was set at 1. Fixed modification was by carbamidomethyl and variable modification was oxidation. Mass tolerance for precursor ions was 10ppm and mass tolerance for fragments was 0.4Da, no known contaminants were excluded. The maximum significance threshold was set at 0.05 (indicating significance) and maximum number of hits was set at default. The used MudPIT (multidimensional protein identification technology) score was 1 and ion score cut-off was 0. False discovery rate was 6.54% and this was calculated by dividing decoy by peptide matches above identity threshold (more information can be found in supplemental Tables S1–S4). Conversion from Uniprot ID to Uniprot accession number was performed by Uniprot Retrieve/ID mapping (http://www.uniprot.org/uploadlists/).
In Mascot, an identity threshold and a homology threshold were calculated. The first depends on the expected chance of getting a false positive match and the second is an empirical measure of whether the match is an outlier. MudPIT score is the sum of the ion scores of all nonduplicate peptides with a filter against weak (or random) matches. The Select Summary takes the protein with the highest protein score as number one in the list. A peptide match, which is both red and bold, is the best match because the first time a match appears, it is shown in bold face and the top match is shown in red. A protein hit without any bold red match only contains peptides with better scoring matches assigned to higher scoring proteins (see supplemental Tables S1 and S2). Accordingly, we decided to set the threshold criteria for protein matches as at least one bold, red hit (supplemental Table S2). If the matches did not fulfill this criterion, they were deselected and not accounted for in the proteomic analysis.
RESULTS
Preparation of Prostasomal Lipid Rafts
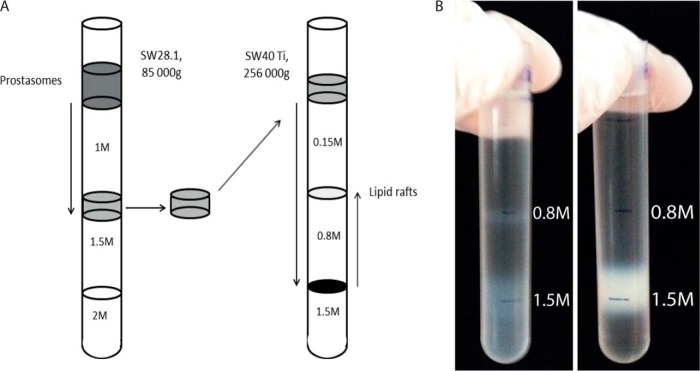
A schematic illustration of the prostasomal lipid raft extraction is presented in Fig. 1A. In Fig. 1B (left), in the Triton X-100 containing sucrose gradient, a distinct whitish band was outlined on top of 30% (w/w) sucrose (corresponding to lipid raft density). This band represented ∼13% (on a protein basis) of the originally loaded sample of prostasomes. Moreover, in the same tube, a residual fraction of possibly partially degraded prostasomes appeared from 1.5 m (60% (w/w)) and downwards representing higher densities in the sucrose gradient. On the contrary, control prostasomes subjected to an ordinary sucrose gradient centrifugation lacking Triton X-100 displayed the expected 1.5 m band ordinarily appearing at this density (Fig 1B, right).
Fig. 1.
Prostasomal lipid raft preparation by sucrose density gradient containing Triton X-100. A, Schematic illustration of prostasomal lipid raft preparation by density gradient ultracentrifugation. Purified prostasomes were top loaded on a sucrose gradient of 1 m, 1.5 m, and 2 m sucrose. The main fraction collected on 1.5 m (density range 1.13–1.19g/ml) was again top loaded on a 1% Triton X-100 sucrose gradient of 0.15 m, 0.8 m, and 1.5 m. Prostasomal lipid rafts floated on 0.8 m (density lower than 1.10g/ml). B, Photography of prostasomal lipid rafts floating on 0.8 m in a Triton X-100 containing sucrose gradient (left) are sharply outlined contrary to control (right) not containing Triton X-100 giving rise to only the expected 1.5 m main band.
Comparative SDS-PAGE Banding Patterns of Prostasomal Lipid Rafts and Control Prostasomes
The three independently prepared prostasomal lipid rafts (Fig. 2) displayed high reproducibility with distinct protein bands appearing especially in the molecular weight range of 38 kDa–188 kDa. When compared with control prostasomes (Fig. 2) the low molecular weight proteins (below 14kDa) were less expressed in prostasomal lipid raft domains. It should be mentioned that no protein bands were observed in SDS-PAGE of samples at the lipid raft floatation density position of the control gradient (sucrose without Triton X-100) (not shown).
Fig. 2.
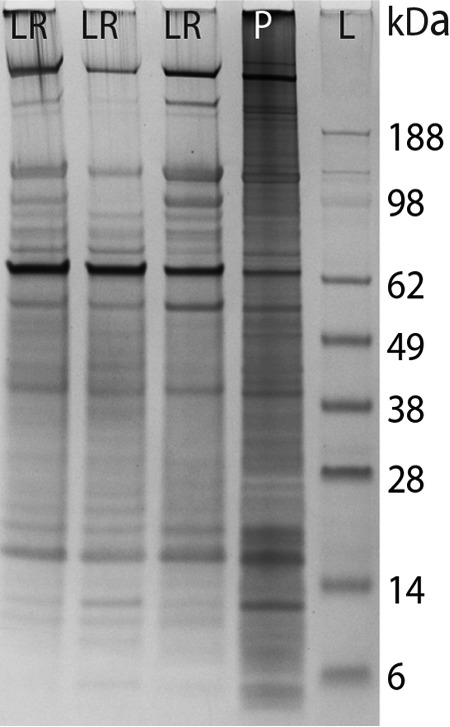
SDS-PAGE demonstration of three independent preparations of prostasomal lipid rafts. SDS-PAGE of prostasomal lipid rafts prepared on three different occasions (LR) in comparison with control prostasomes (P). SeeBlue® Prestained Standard (Invitrogen) indicated molecular weights (L).
Immunoblotting of Selected Proteins in Prostasomal Lipid Rafts and Control Prostasomes
All immunoblots were performed on DRM-fractions and control prostasomes as well as on the residual 1.5 m fraction in sucrose gradients containing Triton X-100. Flotillin-1, flotillin-2, and clathrin displayed bands contrary to caveolin-1 and caveolin-2 (Fig. 3). The bands representing flotillin-1 and flotillin-2 had a reduced molecular weight by 3–5 kDa in the DRM fraction compared with the bands in control prostasomes (Fig. 3). The residual 1.5 m fraction exhibited both bands (Fig. 3). There was also a difference in the appearance of clathrin immunoblots. Concerning clathrin, control prostasomes and residual 1.5 m fraction had a similar appearance with bands at 49 and 98 kDa whereas the DRM-fraction only presented a single band at 49 kDa (Fig. 3).
Fig. 3.
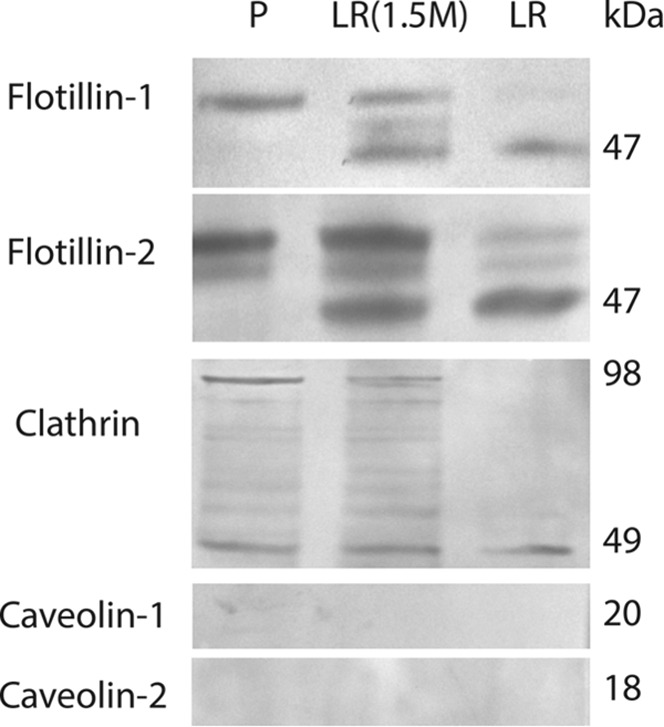
Detection of lipid raft specific proteins by immunoblot analysis. Flotillin-1, flotillin-2, and clathrin presented bands in all three samples tested (P- prostasomes, LR (1.5 m)- residual prostasomes in the Triton X-100 sucrose gradient, and LR- prostasomal lipid rafts). Caveolin-1 and caveolin-2 were not detected in any of the three samples.
Protein Characterization of Prostasomal Lipid Rafts in Comparison With Control Prostasomes
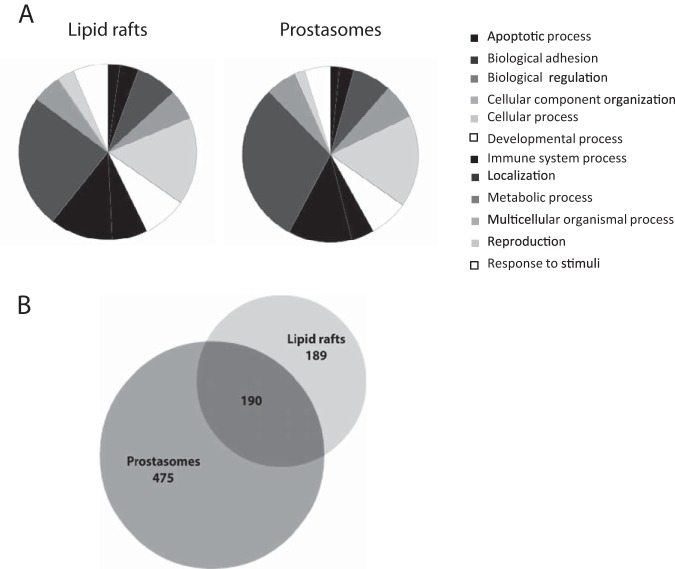
Identified proteins by mass spectrometry of prostasomal lipid rafts (supplemental Table S2) were compared against a list of proteins from prostasomes (42) in the database PANTHER (http://www.pantherdb.org/). A pie chart divided into different categories of biological processes was created for both lipid rafts and prostasomes (Fig 4A).
Fig. 4.
A proteomic comparison of prostasomal lipid rafts and prostasomes. Pie charts created in PANTHER, of biological processes for prostasomal lipid rafts and control prostasomes (A). The protein list of prostasomal lipid rafts was compared against a previously published protein list of prostasomes (42). A comparison between the prostasomal lipid raft protein list and the published prostasome protein list is visualized in a Venn diagram (B).
The Venn diagram (Fig 4B) depicts the partitioning of the different proteins found by mass spectrometry in prostasomal lipid rafts (supplemental Table S2) and prostasomes. Approximately half of the proteins in the prostasomal lipid raft list were found in the prostasome list (42). The diagram was created at the BioVenn website (http://www.cmbi.ru.nl/cdd/biovenn/).
DISCUSSION
The general mode of investigating detergent resistant membranes (DRMs) is homogenization of intact cells and their treatment with a non-ionizing detergent at 4 °C for 30 min followed by floatation of derived raft domains in a density gradient. A drawback with this method is that extracted DRMs may originate from any membrane in the cell (plasma membrane and organellar membranes). There may also appear molecular complexes that are not DRMs but still coincide at the same floatation density. Our approach was to examine the DRM content of prostasomes with a main origin from invaginated endosomal membranes therewith circumventing contamination from other cellular constituents. For that reason we used a sucrose gradient containing 1% Triton X-100 and identified proteins in DRM of prostasomes by LC-MS/MS. For comparison we used a previously published mass-spectrometry list of unmanipulated prostasomes as reference (control prostasomes) (42). Both flotillin-1 and 2 were identified (Table I) in the list of prostasomal DRM-proteins contrary to caveolin-1 and 2 that were not (also proven by immunoblotting). Clathrin was identified by immunoblotting of DRMs and, although not found in the current proteomic list, it was present in the list of control prostasomes (42). It is noteworthy that flotillin-1, flotillin-2 and clathrin presented bands with modestly reduced molecular weights in immunoblots of DRM compared with control prostasomes and the reason for that is not known.
Table I. Fifty-nine categorized proteins associated to prostasomal lipid rafts. “Pos” denotes the position of the protein in Supplementary Table I. Uniprot accession numbers were derived from the website (http://www.uniprot.org/).
| Tetraspanins | UniProt no. | Pos | CD Antigens | UniProt no. | Pos | Ras-related protein | UniProt no. | Pos |
|---|---|---|---|---|---|---|---|---|
| TSPAN6 | O43657 | 222 | CD 9 | P21926 | 117 | Rab-1A | P62820 | 113 |
| TSPAN7 | P41732 | 106 | CD 10 | P08473 | 3 | Rab-3B | P20337 | 12 |
| TSPAN8 | P19075 | 256 | CD 13 | P15144 | 1 | Rab-3D | O95716 | 101 |
| TSPAN9 | O75954 | 160 | CD 14 | P08571 | 226 | Rab-5A | P20339 | 207 |
| TSPAN24 (CD151) | P48509 | 358 | CD 26 | P27487 | 2 | Rab-5C | P51148 | 156 |
| TSPAN29 (CD9) | P21926 | 117 | CD 38 | P28907 | 108 | Rab-6A | P20340 | 263 |
| TSPAN30 (CD63) | P08962 | 91 | CD 47 | Q08722 | 167 | Rab-8A | P61006 | 120 |
| TSPAN31 | Q12999 | 128 | CD 55 | P08174 | 67 | Rab-8B | Q92930 | 137 |
| ESCRT-I | CD 58 | P19256 | 299 | Rab-14 | P61106 | 255 | ||
| Tsg101 | Q99816 | 80 | CD 59 | P13987 | 176 | Rab-27A | P51159 | 71 |
| Vps28 | Q9UK41 | 77 | CD 63 | P08962 | 91 | Rab-27B | O00194 | 126 |
| Vps37B | Q9H9H4 | 50 | CD 151 | P48509 | 358 | Rab-35 | Q15286 | 147 |
| Vps37C | A5D8V6 | 84 | CD 177 | Q8N6Q3 | 17 | Rab-39B | Q96DA2 | 262 |
| ESCRT-III | Adapter | Flotillins | ||||||
| CHMP4B | Q9H444 | 198 | cdc42 | P60953 | 44 | Flotillin-1 | O75955 | 118 |
| CHMP2A | O43633 | 187 | BAIAP2 | Q9UQB8 | 236 | Flotillin-2 | Q14254 | 261 |
| CHMP2B | Q9UQN3 | 333 | Ezrin | P15311 | 163 | Chaperons | ||
| Vps4 Complex (ESCRT) | Others | HSP70 | P08107 | 47 | ||||
| CHMP5 | Q9NZZ3 | 307 | Syntenin1 | O00560 | 321 | HSC71 | P11142 | 42 |
| MMP | ALIX | Q8WUM4 | 20 | HSP90 | P07900 | 54 | ||
| ADAM7 | Q9H2U9 | 135 | SNAP23 | O00161 | 25 | Clusterin | P10909 | 127 |
| ADAM9 | Q13443 | 13 | MAL2 | Q969L2 | 302 | |||
| Proteinkinase C | Q05655 | 82 | ||||||
| ZDHHC2 | Q9UIJ5 | 249 |
The three proteins with highest mascot scores in the list of prostasomal DRM-proteins were CD13, CD26, and CD10 (Table I, supplemental Table S1). These proteins have been used as genuine markers for prostasomes since they have a distinct and consistent (over time) appearance at around 100kDa in coomassie-stained SDS-PAGE gels of human prostasomes (36). CD26 (dipeptidyl peptidase 4) and CD10 (neprilysin) both associate with CD9, a tetraspanin that is a biomarker for exosomes in general. We produced monoclonal antibodies against purified seminal prostasomes and one third of the resulting hybridomas recognized an antigen that was identified as CD26/dipeptidylpeptidase 4, a serine protease with unique specificity (43). Ecto-adenosine deaminase (ecto-ADA) is known to be capable of association with the ecto-ADA binding protein CD26 during T-cell activation (44) and the interaction ecto-ADA/CD26 might be important for transducing adenosine receptor signals (45) and seminal plasma contains appreciable amounts of adenosine (46). It has been suggested that ecto-ADA may be the modulator between different cell types (47). We established the presence of adenosine receptors and ecto-ADA on stallion prostasomes together with CD26 and also, after transfer of CD26 from prostasomes to sperm cells, an interaction of CD26/ecto-ADA leading to fusion between prostasomes and sperm cells (48). Similarly, the well-anchored prostasomal protein, CD59 (Table I) was readily transferred to red blood cells deficient of CD59 giving these cells an almost complete protection against complement-induced hemolysis showing preserved functionality after transfer (49). Hence, exosomes/prostasomes seem to be essential intermediary links between cells.
We identified other tetraspanins including CD63 and CD151, also commonly used biomarkers for exosomes, although the tetraspanins CD81 and CD82 were not found in prostasomal DRMs while CD81 appeared in control prostasomes (42). Tetraspanins are a family of proteins with four transmembrane domains. Both the C-terminal and the N-terminal ends are on the cytoplasmic side of the plasma membrane and stretch less than 20 residues of most tetraspanins. Regions in the ends may interact with cytoskeletal adaptors (e.g. syntenin1, ezrin, BAIAP2 -all identified in the present study) and signaling proteins (e.g. protein kinase C, cdc42, G-proteins (Table I, supplemental Table S2)). The extracellular part of tetraspanins spans over two loops, the small and the large loop. Tetraspanins can interact with one another and they can also interact with a multitude of other transmembrane proteins including metalloproteinases and CD-antigens and a role in creating adhesion platforms has been proposed because of their ability to form complexes with certain integrins and immunoglobulins (50, 51). Because of this ability they have been suggested to play a role in metastasis of cancer cells (51). Moreover, the composition of the adhesion molecules in platforms may play a role in goal specific targeting. Accumulating evidence suggests a role of tetraspanins in EV cargo selection (50, 52). What is more, they may even be able to promote formation of MVBs independently of ceramide and a sorting complex called ESCRT (Endosomal Sorting Complex Required for Transport) (53, 54).
The ESCRT comprises four distinct complexes (ESCRT-0-III) that are involved in the MVB formation. ESCRT-0 together with flat clathrin coats initiate the ILV formation by creating a protein network that enlists ubiquitinated cargo proteins. ESCRT-I binds both ESCRT-0 and ubiquitinated cargo proteins suggesting an additional cargo sorting system. ESCRT-I binds ESCRT-II therewith continuing the action of invaginating the membrane and at the same time packaging cargo. ESCRT-III subunits assemble and help neck constriction and vesicle abscission. In a final step, the Vps4-Vta1 complex binds ESCRT-III and finishes the scission of the vesicle and then disassembles the ESCRT-III complex (55, 56). Interestingly, we identified almost all of the ESCRT-I complex proteins in the prostasomal lipid raft. Additionally, we found proteins belonging to the ESCRT-III and Vps4 complex, but no proteins from ESCRT-0 and ESCRT-II. Furthermore, we identified ALIX, a protein that interacts with TSG101 (a component of ESCRT-I) and CHMP4 (a component of ESCRT-III) and that is thought to participate in the abscission process (57). ALIX in complex with syntenin (Table I) and syndecan is believed to be a regulator of the biogenesis of exosomes (57).
Other identified proteins involved in the secretory-endocytic pathway are; Steap-4 (six transmembrane prostate protein 2), a metalloreductase that has the ability to reduce both Fe- and Cu-ions; MAL2, a component of the machinery of polarized transport; SNAP23, an important regulator of transport vesicle docking and fusion. SNAP23 is synthesized as a soluble protein and becomes membrane-associated via palmitoylation of its cysteine-rich domain (58). Palmitoylation is also important for the formation of tetraspanin complex. It is therefore noteworthy that we identified palmitoyltransferase (ZDHHC2) in prostasomal DRMs, which stimulates palmitoylation of tetraspanins CD9 and CD151 (59) being a candidate enzyme to regulate SNAP23 palmitoylation as well (58).
Ras related rab proteins are a family of proteins involved in intracellular vesicle trafficking and have therefore been of great interest for the formation of ILVs of MVBs. The rab GTPase activity supports processes including sorting of proteins, caveolin uncoating and motility of endocytic vesicles and later, tethering and fusion events of exocytotic vesicles (60). We identified several of these proteins in our prostasomal DRM-fraction (Table I) and among them especially rab-27A and B that apparently control important steps of the exosome secretion pathway (61). Moreover, the chaperons HSP70, 90 and heat shock cognate 71kDa protein were identified as well as clusterin, previously recognized as an autoantigen of prostasomes (62).
Supplementary Material
Acknowledgments
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (63) via the PRIDE partner repository with the data set identifier PXD002163 and 10.6019/PXD002163.
Footnotes
Author contributions: L.D., K.G.R., G.R., and A.L. designed research; L.D., K.G.R., B.E., G.R., and A.L. performed research; B.E. contributed new reagents or analytic tools; L.D., K.G.R., B.E., G.R., and A.L. analyzed data; L.D., K.G.R., G.R., and A.L. wrote the paper.
* This work is supported by the Lion's Cancer Foundation, Uppsala and Erik, Karin and Gösta Selanders fund, Uppsala. Percy Falk fund, Stockholm and ALF, the University Hospital of Uppsala.
 This article contains supplemental Tables S1 to S4.
This article contains supplemental Tables S1 to S4.
1 The abbreviations used are:
- EVs
- Extracellular vesicles
- MVBs
- Multivesicular bodies
- ILVs
- Intraluminal vesicles
- DRMs
- Detergent resistant membranes
- ESCRT
- Endosomal sorting complex required for transport
- SNAP-23
- Synaptosomal-associated protein 23
- ALIX
- Programmed cell death 6-interacting protein
- TSG101
- Tumor susceptibility gene 101 protein
- CHMP
- Charged multivesicular body protein
- MAL2
- Protein MAL2
- MudPIT
- Multidimensional protein identification technology
- HSP
- Heat shock protein.
REFERENCES
- 1. Aalberts M., Stout T. A., and Stoorvogel W. (2014) Prostasomes: extracellular vesicles from the prostate. Reproduction 147, R1–14 [DOI] [PubMed] [Google Scholar]
- 2. Raposo G., and Stoorvogel W. (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., and Simons M. (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 4. Gulbins E., and Kolesnick R. (2003) Raft ceramide in molecular medicine. Oncogene 22, 7070–7077 [DOI] [PubMed] [Google Scholar]
- 5. Clarke C. J., Snook C. F., Tani M., Matmati N., Marchesini N., and Hannun Y. A. (2006) The extended family of neutral sphingomyelinases. Biochemistry 45, 11247–11256 [DOI] [PubMed] [Google Scholar]
- 6. Poliakov A., Spilman M., Dokland T., Amling C. L., and Mobley J. A. (2009) Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate 69, 159–167 [DOI] [PubMed] [Google Scholar]
- 7. Ronquist G., and Brody I. (1985) The prostasome: its secretion and function in man. Biochim. Biophys. Acta 822, 203–218 [DOI] [PubMed] [Google Scholar]
- 8. Sahlén G. E., Egevad L., Ahlander A., Norlén B. J., Ronquist G., and Nilsson B. O. (2002) Ultrastructure of the secretion of prostasomes from benign and malignant epithelial cells in the prostate. Prostate 53, 192–199 [DOI] [PubMed] [Google Scholar]
- 9. Ronquist G., and Agren G. K. (1975) A Mg2+- and Ca2+-stimulated adenosine triphosphatase at the outer surface of Ehrlich ascites tumor cells. Cancer Res. 35, 1402–1406 [PubMed] [Google Scholar]
- 10. Ronquist G. (1987) Effect of modulators on prostasome membrane-bound ATPase in human seminal plasma. Eur. J. Clin. Investig. 17, 231–236 [DOI] [PubMed] [Google Scholar]
- 11. Ronquist G. (2012) Prostasomes are mediators of intercellular communication: from basic research to clinical implications. J. Intern. Med. 271, 400–413 [DOI] [PubMed] [Google Scholar]
- 12. Kelly R. W., Holland P., Skibinski G., Harrison C., McMillan L., Hargreave T., and James K. (1991) Extracellular organelles (prostasomes) are immunosuppressive components of human semen. Clin. Exp. Immunol. 86, 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rooney I. A., Atkinson J. P., Krul E. S., Schonfeld G., Polakoski K., Saffitz J. E., and Morgan B. P. (1993) Physiologic relevance of the membrane attack complex inhibitory protein CD59 in human seminal plasma: CD59 is present on extracellular organelles (prostasomes), binds cell membranes, and inhibits complement-mediated lysis. J. Exp. Med. 177, 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ronquist G., Nilsson B. O., and Hjertën S. (1990) Interaction between prostasomes and spermatozoa from human semen. Arch. Androl. 24, 147–157 [DOI] [PubMed] [Google Scholar]
- 15. Arienti G., Carlini E., and Palmerini C. A. (1997) Fusion of human sperm to prostasomes at acidic pH. J. Membr. Biol. 155, 89–94 [DOI] [PubMed] [Google Scholar]
- 16. Park K. H., Kim B. J., Kang J., Nam T. S., Lim J. M., Kim H. T., Park J. K., Kim Y. G., Chae S. W., and Kim U. H. (2011) Ca2+ signaling tools acquired from prostasomes are required for progesterone-induced sperm motility. Sci. Signal. 4, ra31. [DOI] [PubMed] [Google Scholar]
- 17. Saez F., Motta C., Boucher D., and Grizard G. (1998) Antioxidant capacity of prostasomes in human semen. Mol. Human Reprod. 4, 667–672 [DOI] [PubMed] [Google Scholar]
- 18. Carlsson L., Påhlson C., Bergquist M., Ronquist G., and Stridsberg M. (2000) Antibacterial activity of human prostasomes. Prostate 44, 279–286 [DOI] [PubMed] [Google Scholar]
- 19. Arvidson G., Ronquist G., Wikander G., and Ojteg A. C. (1989) Human prostasome membranes exhibit very high cholesterol/phospholipid ratios yielding high molecular ordering. Biochim. Biophys. Acta 984, 167–173 [DOI] [PubMed] [Google Scholar]
- 20. Arienti G., Carlini E., Polci A., Cosmi E. V., and Palmerini C. A. (1998) Fatty acid pattern of human prostasome lipid. Arch. Biochem. Biophys. 358, 391–395 [DOI] [PubMed] [Google Scholar]
- 21. Singer S. J., and Nicolson G. L. (1972) The fluid mosaic model of the structure of cell membranes. Science 175, 720–731 [DOI] [PubMed] [Google Scholar]
- 22. Davson H., and Danielli J. F. (1938) Studies on the permeability of erythrocytes: Factors in cation permeability. Biochem. J. 32, 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palade G. E. (1953) An electron microscope study of the mitochondrial structure. J. Histochem. Cytochem. 1, 188–211 [DOI] [PubMed] [Google Scholar]
- 24. Yamada E. (1955) The fine structure of the renal glomerulus of the mouse. J. Histochem. Cytochem. 3, 309. [DOI] [PubMed] [Google Scholar]
- 25. van Meer G., and Simons K. (1988) Lipid polarity and sorting in epithelial cells. J. Cell. Biochem. 36, 51–58 [DOI] [PubMed] [Google Scholar]
- 26. Simons K., and Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 27. Pike L. J. (2003) Lipid rafts: bringing order to chaos. J. Lipid Res. 44, 655–667 [DOI] [PubMed] [Google Scholar]
- 28. Brown D. A., and Rose J. K. (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544 [DOI] [PubMed] [Google Scholar]
- 29. Brown D. A., and London E. (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224 [DOI] [PubMed] [Google Scholar]
- 30. Head B. P., Patel H. H., Roth D. M., Murray F., Swaney J. S., Niesman I. R., Farquhar M. G., and Insel P. A. (2006) Microtubules and actin microfilaments regulate lipid raft/caveolae localization of adenylyl cyclase signaling components. J. Biol. Chem. 281, 26391–26399 [DOI] [PubMed] [Google Scholar]
- 31. Stuermer C. A. (2010) The reggie/flotillin connection to growth. Trends Cell Biol. 20, 6–13 [DOI] [PubMed] [Google Scholar]
- 32. Hancock J. F. (2006) Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 7, 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lingwood D., and Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 34. Damm E. M., Pelkmans L., Kartenbeck J., Mezzacasa A., Kurzchalia T., and Helenius A. (2005) Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 168, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riento K., Frick M., Schafer I., and Nichols B. J. (2009) Endocytosis of flotillin-1 and flotillin-2 is regulated by Fyn kinase. J. Cell Sci. 122, 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ronquist G. K., Larsson A., Ronquist G., Isaksson A., Hreinsson J., Carlsson L., and Stavreus-Evers A. (2011) Prostasomal DNA characterization and transfer into human sperm. Mol. Reprod. Dev. 78, 467–476 [DOI] [PubMed] [Google Scholar]
- 37. Stegmayr B., and Ronquist G. (1982) Promotive effect on human sperm progressive motility by prostasomes. Urol. Res. 10, 253–257 [DOI] [PubMed] [Google Scholar]
- 38. Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., and Geuze H. J. (1998) Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 [DOI] [PubMed] [Google Scholar]
- 39. McGuinn K. P., and Mahoney M. G. (2014) Lipid Rafts and Detergent-Resistant Membranes in Epithelial Keratinocytes. Methods. Mol. Biol. 1195, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Meth. 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 41. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 42. Ronquist K. G., Ek B., Morrell J., Stavreus-Evers A., Ström Holst B., Humblot P., Ronquist G., and Larsson A. (2013) Prostasomes from four different species are able to produce extracellular adenosine triphosphate (ATP). Biochim. Biophys. Acta 1830, 4604–4610 [DOI] [PubMed] [Google Scholar]
- 43. Schrimpf S. P., Hellman U., Carlsson L., Larsson A., Ronquist G., and Nilsson B. O. (1999) Identification of dipeptidyl peptidase IV as the antigen of a monoclonal anti-prostasome antibody. Prostate 38, 35–39 [DOI] [PubMed] [Google Scholar]
- 44. Kameoka J., Tanaka T., Nojima Y., Schlossman S. F., and Morimoto C. (1993) Direct association of adenosine deaminase with a T cell activation antigen, CD26. Science 261, 466–469 [DOI] [PubMed] [Google Scholar]
- 45. Ciruela F., Saura C., Canela E. I., Mallol J., Lluis C., and Franco R. (1996) Adenosine deaminase affects ligand-induced signalling by interacting with cell surface adenosine receptors. FEBS Lett. 380, 219–223 [DOI] [PubMed] [Google Scholar]
- 46. Fabiani R., and Ronquist G. (1995) Abundance of guanine, guanosine, inosine and adenosine in human seminal plasma. Int. J. Clin. Lab. Res. 25, 47–51 [DOI] [PubMed] [Google Scholar]
- 47. Franco R., Valenzuela A., Lluis C., and Blanco J. (1998) Enzymatic and extraenzymatic role of ecto-adenosine deaminase in lymphocytes. Immunol. Rev. 161, 27–42 [DOI] [PubMed] [Google Scholar]
- 48. Minelli A., Allegrucci C., Mezzasoma I., Ronquist G., Lluis C., and Franco R. (1999) CD26 and adenosine deaminase interaction: its role in the fusion between horse membrane vesicles and spermatozoa. Biol. Reprod. 61, 802–808 [DOI] [PubMed] [Google Scholar]
- 49. Babiker A. A., Ronquist G., Nilsson U. R., and Nilsson B. (2002) Transfer of prostasomal CD59 to CD59-deficient red blood cells results in protection against complement-mediated hemolysis. Am. J. Reprod. Immunol. 47, 183–192 [DOI] [PubMed] [Google Scholar]
- 50. Andreu Z., and Yanez-Mo M. (2014) Tetraspanins in extracellular vesicle formation and function. Frontiers Immunol. 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., and Rubinstein E. (2009) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 52. Perez-Hernandez D., Gutiérrez - Vázquez C., Jorge I., López -Martin S., Ursa A., Sánchez -Madrid F., Vázquez J., and Yáñez - Mó M. (2013) The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 288, 11649–11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buschow S. I., Nolte-'t Hoen E. N., van Niel G., Pols M. S., ten Broeke T., Lauwen M., Ossendorp F., Melief C. J., Raposo G., Wubbolts R., Wauben M. H., and Stoorvogel W. (2009) MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 10, 1528–1542 [DOI] [PubMed] [Google Scholar]
- 54. van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M. S., Rubinstein E., and Raposo G. (2011) The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Develop. Cell 21, 708–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henne W. M., Buchkovich N. J., and Emr S. D. (2011) The ESCRT pathway. Develop. Cell 21, 77–91 [DOI] [PubMed] [Google Scholar]
- 56. Babst M. (2011) MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opinion Cell Biol. 23, 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baietti M. F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., Zimmermann P., and David G. (2012) Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 [DOI] [PubMed] [Google Scholar]
- 58. Greaves J., Gorleku O. A., Salaun C., and Chamberlain L. H. (2010) Palmitoylation of the SNAP25 protein family: specificity and regulation by DHHC palmitoyl transferases. J. Biol. Chem. 285, 24629–24638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sharma C., Yang X. H., and Hemler M. E. (2008) DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol. Biol. Cell 19, 3415–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev.. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 61. Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M. C., Darchen F., Amigorena S., Moita L. F., and Thery C. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 [DOI] [PubMed] [Google Scholar]
- 62. Ronquist K. G., Carlsson L., Ronquist G., Semjonow A., Wülfing C., and Larsson A. (2008) Serum antibodies against prostasomal clusterin in prostate cancer patients. Scand. J. Clin. Lab. Invest. 68, 219–227 [DOI] [PubMed] [Google Scholar]
- 63. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.