Abstract
The coordinated and synchronized cardiac muscle contraction relies on an efficient gap junction-mediated intercellular communication (GJIC) between cardiomyocytes, which involves the rapid anisotropic impulse propagation through connexin (Cx)-containing channels, namely of Cx43, the most abundant Cx in the heart. Expectedly, disturbing mechanisms that affect channel activity, localization and turnover of Cx43 have been implicated in several cardiomyopathies, such as myocardial ischemia. Besides gap junction-mediated intercellular communication, Cx43 has been associated with channel-independent functions, including modulation of cell adhesion, differentiation, proliferation and gene transcription. It has been suggested that the role played by Cx43 is dictated by the nature of the proteins that interact with Cx43. Therefore, the characterization of the Cx43-interacting network and its dynamics is vital to understand not only the molecular mechanisms underlying pathological malfunction of gap junction-mediated intercellular communication, but also to unveil novel and unanticipated biological functions of Cx43. In the present report, we applied a quantitative SWATH-MS approach to characterize the Cx43 interactome in rat hearts subjected to ischemia and ischemia-reperfusion. Our results demonstrate that, in the heart, Cx43 interacts with proteins related with various biological processes such as metabolism, signaling and trafficking. The interaction of Cx43 with proteins involved in gene transcription strengthens the emerging concept that Cx43 has a role in gene expression regulation. Importantly, our data shows that the interactome of Cx43 (Connexome) is differentially modulated in diseased hearts. Overall, the characterization of Cx43-interacting network may contribute to the establishment of new therapeutic targets to modulate cardiac function in physiological and pathological conditions. Data are available via ProteomeXchange with identifier PXD002331.
Electrical conduction in the heart is mainly governed by the function of gap junctions (GJ)1, intercellular channels that are localized at the longitudinal termini of the cardiomyocytes, the intercalated discs (IDs) (1, 2). Here, GJ ensure the correct anisotropic impulse propagation, as well as the passage of second messengers and metabolites between neighbor cells, which is vital to ensure cardiac homeostasis and function. GJ channels are composed by two juxtaposed connexons, assembled by hexamers of transmembrane proteins called connexins (Cx) (3). Although several members of the Cx family are expressed in the heart, including Cx40 and Cx45, Cx43 is by far the most abundant isoform (1).
Besides its role upon GJ-mediated intercellular communication (GJIC), Cx43 has been associated with channel-independent functions. Indeed, mounting evidence suggests that Cx43 regulates other cellular mechanisms, including microtubule stability, cell cycle, differentiation and proliferation (4–6). In cardiomyocytes, Cx43 can also localize within mitochondrial membranes, where it has been implicated in enhanced ischemic preconditioning response. Accordingly, some authors reported that during stress conditions, as occurs in myocardial ischemia, the levels of mitochondrial Cx43 raise, which could contribute to keep the mitochondrial permeability transition pore (MPTP) in a closed state, delaying the release of apoptotic proteins and cytochrome c, thus reducing ischemia/reperfusion (I/R) injury (7, 8).
Several cardiomyopathies, including heart failure and myocardial ischemia, have been associated with defects on GJIC, as a consequence of GJ remodeling that includes channel closure, changes in Cx43 ubiquitination and phosphorylation profiles, and a redistribution of Cx43-containing channels from the IDs to the lateral membranes (9–11). Another causative factor for the GJIC impairment underlying heart disorders is the increased degradation of Cx43 (10). In any case, both the final fate and function of Cx43-containing channels depends upon the Cx43-interacting partners, that either through the direct interaction itself, or by mediating post-translational modifications, modulate the activity, levels and subcellular distribution of Cx43 (12). Therefore, increasing attention has been given to the Cx43-interactome, in order to understand how interacting partners contribute to regulate not only GJIC, both in physiological and pathological conditions, but also the role played by Cx43, namely its non-canonical functions (13).
Despite several Cx43-binding partners have been identified and associated with GJ-dependent and -independent functions, up until now, large-scale screenings intending to characterize the interactome of Cx43 are still scarce. To the best of our knowledge, only two proteomic analyses of Cx43 interacting partners have been performed, one in rat glial cell lines, and other in primary cultures of human chondrocytes (13, 14). Given the importance of Cx43 in the maintenance of cardiac function, the main objective of the present report was to unravel the Cx43-interaction network in the heart, and to establish the impact of ischemia and I/R upon these interactions. The results obtained in this study demonstrate that in the heart Cx43 mainly interacts with proteins related with metabolism, signaling and trafficking, and that this interactome can be differentially modulated in diseased hearts. Our results shed new light upon the understanding of Cx43 functions in the heart, both in health and disease, which ultimately may lead to the establishment of new therapeutic targets to modulate cardiac homeostasis.
EXPERIMENTAL PROCEDURES
Animal Models
Wistar rats were obtained from our local breeding colony (Faculty of Medicine of the University of Coimbra, Coimbra, Portugal). Animals were handled according to European Union guidelines for the use of experimental animals (86/609/EEC). Experiments were approved by the Ethics Committee of the Faculty of Medicine, University of Coimbra. For Langendorff-perfused heart experiments, 10-week-old Wistar rats (400 ± 25 g) were anesthetized with 85 mg/kg ketamine and 10 mg/kg xylazine and heparinized. Hearts were perfused on a Langendorff apparatus [perfusion pressure of 70 mmHg (1 mmHg = 0.133 kPa), constant flow rate of 15 ml/min], with modified Krebs-Henseleit (KH) buffer (118 mm NaCl, 25 mm NaHCO3, 4.7 mm KCl, 1.2 mm MgSO4, 1.2 mm KH2PO4, 10 mm Hepes, 1.25 mm CaCl2 and 10 mm glucose, pH 7.49), equilibrated with 95%O2/5%CO2 at 37 °C. Perfusion was stabilized for 10 min, followed by either 20 min-perfusion (control) or no-flow ischemia. Reperfusion (I/R) was induced by reestablishment of the initial flow rate for additional 60 min. After the experiments, hearts were either embedded in OCT (Tissue-Tek, Sakura, Alphen aan den Rijn, The Netherlands) for cryosectioning, or snap-frozen in liquid nitrogen for proteomic studies, before storage at −80 °C (9, 10).
Cell Culture
HL-1 cells (clone 6) were obtained from Dr Emmanuel Dupont (Imperial College London, London, UK), which established a culture of HL-1 subclones, from the HL-1 parental cell line (previously immortalized by Dr W. C. Claycomb (Louisiana State University Health Centre, New Orleans, LA)) (15, 16). HL-1 cells (clone 6) were cultured under an atmosphere of 5% CO2, at 37 °C in Claycomb medium supplemented with 10% fetal bovine serum (FBS), 2 mm l-glutamine and 100 μm norepinephrine, as previously described (10).
Immunopurification (IP) of Cx43 for the Interactomics Study
Lysates from whole hearts were prepared by homogenization (Potter-Elvehjem PTFE Tissue Grinder, Corning Life Sciences, Corning, New York) in lysis buffer (100 mm NaCl, 50 mm Tris-HCl, 5 mm EDTA, 1% Triton X-100, pH 7.4), supplemented with protease inhibitors. Homogenates were centrifuged at 1000 × g or 5 min, sonicated (3 pulses of 2″, 180 W) and further centrifuged at 10,000 × g, for 20 min. Determination of total protein was performed by the DC Protein Assay (BioRad, Hercules, CA), after which 25 mg of the supernatants were used for immunopurification (one heart/experimental condition, n = 3). Briefly, supernatants were incubated with 30 μg goat polyclonal antibodies directed against Cx43 (AB0016, Sicgen, Cantanhede, Portugal). Goat nonspecific polyclonal antibodies (anti-GFP; AB0020, Sicgen) were used for control IP. Incubations proceeded overnight, at 4 °C, followed by incubation with 120 μg protein G-Sepharose (GE Healhcare, Little Chalfont, UK) for 1.5 h at 4 °C. Protein G-Sepharose sediments were washed in lysis buffer, and immunopurified proteins were eluted in Laemmli buffer, and denatured at 95 °C, for 5 min.
Sample Preparation for MS Analysis
Denatured samples were alkylated with acrylamide and subjected to in-gel digestion following the short-GeLC approach (17) (supplemental Fig. S1). Briefly, samples were loaded into two wells of a “4–20% TGX Stain-Free Gel” (Bio-Rad), followed by partial electrophoretic separation (SDS-PAGE). Proteins were subsequently visualized with Colloidal Coomassie Blue staining (18). Gel lanes were sliced into seven bands of equal size, and further sliced into small pieces, for independent processing. Gel pieces were destained, dehydrated, and rehydrated with 25 μl of trypsin (0.01 μg/μl in 10 mm ammonium bicarbonate). Protein digestion was performed overnight at room temperature, and digested peptides were extracted from the gel, by sequential incubation with acetonitrile (ACN) solutions in 1% formic acid (FA) (30%, 50%, and 98% organic content). Peptides extracted from different bands were pooled together in two-peptide mixtures per sample, for subsequent liquid chromatography (LC)-MS/MS analysis. Peptide mixtures were dried and de-salted using OMIX tips with C18 stationary phase (Agilent Technologies, Santa Clara, CA).
To monitor samples loss during sample preparation samples were spiked with 1 μg of recombinant green fluorescent protein (GFP) before digestion. Additionally, peptides were resuspended in mobile phase (2% ACN in 0.1% FA) and spiked with iRTs peptides (Biognosys AG, Schlieren, Switzerland), for retention time adjustment.
SWATH-MS
SWATH Acquisition
Samples were analyzed on a Triple TOFTM 5600 System (ABSciex®, Framingham, MA) through information-dependent acquisition (IDA) followed by SWATH. Peptides were resolved by LC (nanoLC Ultra 2D, Eksigent®, Dublin, CA) on a ChromXPTM C18AR reverse phase column (300 μm ID × 15 cm length, 3 μm particles, 120 Å pore size, Eksigent®) at 5 μl/min, and eluted into the mass spectrometer with an ACN linear gradient in 0.1% FA (2% to 35% ACN, for 45 min), using an electrospray ionization source (DuoSprayTM Source, ABSciex) (17).
Pooled mixtures (one sixth of the two peptide mixtures of each biological replicate) were analyzed in IDA mode, to generate peptide fragmentation spectra for further protein identification/library creation. For IDA, the mass spectrometer was set to scanning full spectra (350–1250 m/z), for 250 ms, followed by up to 30 MS/MS scans (100–1500 m/z). Candidate ions with a charge state between +2 and +5, and counts per second above a minimum threshold of 70, were isolated for fragmentation. One MS/MS spectra was collected for 100 ms, before adding those precursor ions to the exclusion list for 15 s (mass spectrometer operated by Analyst® TF 1.6, ABSciex®). Rolling collision was used with a collision energy spread of 5. To improve sample coverage, an additional IDA experiment was done for each pool, using an exclusion list of the previously identified peptides.
For quantitative analysis, the peptide mixtures were combined into a single sample per biological replicate. The SWATH setup was essentially as described by Anjo et al. (17). The mass spectrometer was operated in a looped product ion mode, and specifically tuned to allow a quadrupole resolution of 25 m/z mass selection. Using an isolation width of 26 m/z (containing 1 m/z for the window overlap), a set of 30 overlapping windows was constructed, covering the precursor mass range of 350–1100 m/z. A 50 ms survey scan (350–1500 m/z) was acquired at the beginning of each cycle, and SWATH-MS/MS spectra were collected from 100–1500 m/z for 100 ms resulting in a cycle time of 3.1 s. Collision energy for each window was determined according to the calculation for a charge +2 ion-centered upon the window with a collision energy spread of 15.
Protein Identification/Library Generation
Peptide identification and library generation were performed with Protein Pilot software (v4.5, ABSciex®), using the following parameters: (1) search against a database composed by Rattus Norvegicus from SwissProt (release at February 2014, with 15 800 entries), GFP and iRT peptide sequences, and using (2) acrylamide alkylation as fixed modification; (3) trypsin digestion (with a miss cleavage factor of 0.75, ParagonTM Algorithm). An independent False Discovery Rate (FDR) analysis, using the target-decoy approach provided by Protein PilotTM, was used to assess the quality of identifications. Positive identifications were considered when identified proteins and peptides reached a 5% local FDR (19, 20). A specific library of precursor masses and fragment ions was created by combining all except the control IP files from the IDA experiments, and used for subsequent SWATH processing.
SWATH Data Processing
Data processing was performed using SWATHTM processing plug-in for PeakViewTM (v2.0.01, ABSciex®). Briefly, peptides were selected automatically from the library using the following criteria: (1) unique peptides for a specific targeted protein were ranked by intensity of the precursor ion from the IDA, estimated by Protein PilotTM; and (2) peptides with biological modifications and/or shared between different protein entries/isoforms were excluded. Up to 15 peptides were chosen per protein, and SWATHTM quantitation was attempted for all proteins considered as positive identifications. Peptides were confirmed by finding and scoring peak groups which are a set of fragment ions for the peptide.
Target fragment ions, up to five, were automatically selected and peak groups were scored following the criteria described in Lambert et al. (21). Peak group confidence threshold was determined based on a FDR analysis. Peptides within 1% FDR threshold (in at least two of the three biological replicates) were retained. Peak areas of the target fragment ions of those peptides were extracted across experiments, using an extracted-ion chromatogram (XIC) window of 3 min and 20 mDa XIC width. Retention time was adjusted to each sample with iRT peptides.
Protein levels were estimated by summing all peptide transitions for a given protein (adapted from (22), and normalized to GFP levels.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (23) via the PRIDE partner repository with the data set identifier PXD002331.
Clustering of Profiles and Comparative Analyses
Clustering analysis and complementary heat maps were done using GPRroX (version 1.1.15) (24). Clustering was performed using the unsupervised clustering fuzzy c-means algorithm implemented in the Mfuzz package (25), which is a soft clustering algorithm, noise-robust and well-fitted to the protein profile data. Clustering was presented as a complementary method to identify Cx43 interactors from the nonspecific ones (using the median-normalized protein levels), and to trace the different profiles of Cx43 interactions under the various experimental conditions (using interaction levels - proteins levels normalized to Cx43 levels -of the previously selected Cx43 interactors).
Gene Ontology (GO) enrichment analysis was performed, by importing UniProt GO classifications for biological processes (for each interactor), followed by enrichment analysis within GProX, using a Binomial statistical test with Benjamini-Hochberg adjustment and a cut-off of 0.05 p value. Kyoto encyclopedia of genes and genomes (KEGG) pathway and INTERPRO analyses were performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database v9.0 (http://www.string-db.org/).
Statistical analysis was performed in MarkerViewTM (version 1.2.1.1, ABSciex®). Statistical significance was considered for p values < 0.1 (26). Multiple t test was applied for comparison between experimental groups. Data normality was accessed by a combinatory analysis of histograms and Q-Q plots (27) (supplemental Fig. S2) obtained in InfernoRDN (version 1.1.5581.33355) (28). Data is presented as median ± median absolute deviation (MAD) of a set of three independent experiments.
Validation of Cx43-interacting Proteins
For HL-1 cells and tissue samples (5 μm cryosections), immunofluorescence staining was performed as previously described (9, 10). Incubation with primary antibodies was performed using antibodies against Cx43 (rabbit polyclonal (sc-9059, Santa Cruz Biotechnology, Dallas, TX) or mouse monoclonal (610062, BD Transduction Laboratories, San Jose, CA), as applicable), β-actin (A5441, Sigma-Aldrich, St. Louis, MO), Clathrin Heavy chain (610590, BD Transduction Laboratories), Cytochrome c oxidase subunit 1 (ab14705, Abcam), Mitofusin 1 (sc-50330, Santa Cruz Biotechnology) and Ryanodine receptor 2 (C3–33, Sigma-Aldrich). Incubation with Phalloidin (Sigma-Aldrich) was performed for 1 h at room temperature. Nuclei were stained with DAPI. Images were collected by confocal microscopy using a Zeiss LSM 710, and analyzed with Image J (National Institutes of Health, Washington, DC).
RESULTS
Identification of the Cardiac Cx43-Interaction Network
Remodeling of cardiomyocyte GJ at the onset of myocardial ischemia has been extensively reported (2, 29). Albeit its importance and all the efforts to elucidate the mechanisms associated with this GJ remodeling, the molecular players and pathways involved are still not completely characterized.
Therefore, we performed a quantitative proteomic analysis to investigate changes in the cardiac Cx43-interactome in the context of ischemia and I/R. For that, we used the Langendorff heart perfusion model, where rat hearts were either perfused for 20 min (controls, CT), subjected to no-flow ischemia (ISCH) for 20 min, or to 20 min of ischemia followed by 60 min of reperfusion (I/R), by restoration of the initial follow rate. Hence, immunopurification of endogenous Cx43 (Cx43 IP) from rat hearts, was combined with identification of Cx43-binding partners using the SWATH-MS approach. This experimental setup enabled, not only to identify the Cx43 interactors, but also to trace interaction profiles under the referred conditions. Although the common affinity purification (AP) coupled with MS leads to a comprehensive identification of the co-immunopurified proteins in a particular condition, this type of approach, focused in protein identification, fails to capture the dynamic nature of interactions (30, 31), which is particularly important in the evaluation of different physiological states. Therefore, a SWATH-MS strategy was followed, to achieve an accurate quantitative evaluation of the co-immunopurified proteins, allowing a high-confidence distinction between truly interactors and nonspecific proteins, and a precise measure of the changes in Cx43-interacting partners induced by ischemia and I/R.
In the present approach, we were able to initially identify 444 proteins (supplemental Table S1). From these 444 proteins, 299 (∼67% of the entire data set) were quantified (supplemental Table S2) and compared between the various experimental conditions. These 299 proteins were further evaluated by a series of complementary analyses to distinguish the truly Cx43 interactors from nonspecific binding to control IP (Ct -) (Figs. 1A–1B). Proteins were considered as putative Cx43 interactors if they met one of the following criteria: (1) a p value under 0.1, (2) a 50% increase when compared with control IP, or (3) 50% change among two Cx43 IPs (Fig. 1C). Statistical analysis was performed by combination of the t test results for each pair of conditions. According with this evaluation, 236 (out of the 299 quantified proteins) were considered as putative Cx43 interacting partners (supplemental Table S3).
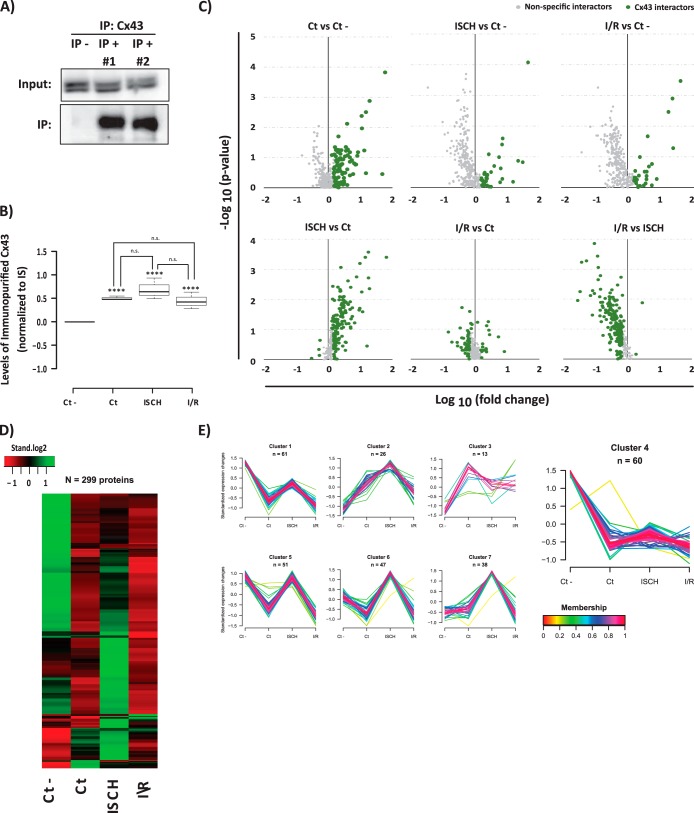
Fig. 1.
AP-SWATH approach for the study of the dynamic interactome of Cx43 in heart. A, Immunoblotting detection of the immunopurified Cx43 (Cx43 IP versus control IP). B, SWATH quantification of Cx43 IP in each condition. Data are presented as boxplots of the normalized values to internal standard (IS). Student t test was applied. ****ρ <0.001; “n.s.” (no statistical difference). C, Volcano plots showing log10 fold change plotted against -log10 p value for all the 299 quantified proteins in Cx43 IP samples versus samples generated from an irrelevant bait (GFP) (upper panel), and between the three Cx43 IPs (lower panel). Data points highlighted in green represent the proteins that met one of the following criteria: a p value < 0.1; a 50% increase (log10 fold change >1.5) when compared with control IP, or 50% change among two Cx43 IPs. Highlighted proteins correspond to the 236 putative Cx43 interacting partners. D, Heat map showing the levels of the copurified proteins among conditions. The row-clustered heat map represents the standardized median levels for all the 299 quantified proteins. E, Clustering of all the proteins copurified in the IPs. For the 299 proteins quantified the normalized levels were standardized and proteins were subjected to unsupervised clustering. An upper and lower ratio limit of log2 (2) and log2 (0.5) was used for inclusion into a cluster. “n” indicates the number of proteins within each cluster. Membership value represents how well the protein profile fit the average cluster profile. Highlighted Cluster 4 corresponds to the nonspecific Cx43 interactors.
The less stringent statistical evaluation of the data was further supported by a parallel heat map and clustering analysis of the immunopurified proteins (Fig. 1D–1E) that corroborate the dynamic profile of the majority of Cx43 interactions. Clustering analysis further shows that there is a high degree of membership among the proteins belonging to each cluster, supporting the inclusion of proteins with lower statistical evidence as putative Cx43 interactors. Finally, from the clustering analysis it was also possible to identify a cluster correspondent to the nonspecific interactors (cluster 4), which was composed by 60 proteins highly represented in the control IP, and with unvarying levels among the remaining experimental conditions. Importantly, this independent analysis corroborates the previous statistical analysis, where 63 proteins were eliminated.
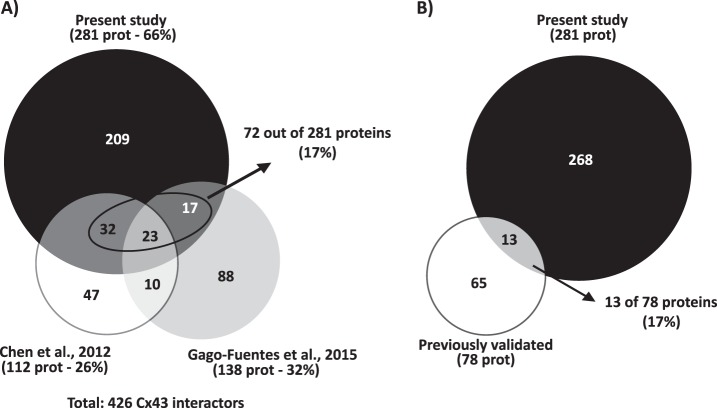
Up until now, only two large-scale studies aiming to address the Cx43-interactome were performed. In 2012, Chen et al. published the Cx43-interactome in a rat glial cell line (14). More recently, Gago-Fuentes et al. established the Cx43-interaction network in chondrocytes, in the context of osteoarthritis (13). Given that these previous works constituted identification-based experiments, for an appropriate comparison with our study, we also considered the proteins only identified in Cx43 IP, but not quantified in the SWATH experiment, as putative Cx43 interactors. The cross-comparison between the interactors identified among the three studies (Fig. 2A and detailed list on supplemental Table S4) reveals that our proteomic approach is by far the most comprehensive study of Cx43 interactome, representing 66% of the total number of Cx43 putative interactors identified. Although the large majority of the proteins correspond to new Cx43 interactors, 17% of the detected proteins are shared with Chen and Gago-Fuentes studies. For instance, GTP-binding nuclear protein Ran, peroxiredoxin-1, and several metabolism-related proteins, such as fructose-biphosphate aldolase A and isoform M2 of pyruvate kinase, are common to all proteomic studies (13). Finally, among the Cx43-interactors that we have uncovered, there are also proteins whose interaction with Cx43 was already well established and validated (Fig. 2B and Table I), including actin, tubulin, myosin motor proteins, clathrin, or vinculin. Altogether, these evidences support the reliability and high confidence of the new Cx43 interactors identified in the present study.
Fig. 2.
Meta-analysis of the Cx43-interactome data. A, Proportional Venn diagram comparing the proteins identified in the present study (black circle) with those identified in other two interactomic studies of Cx43: Chen et al., 2012 (white circle) and Gago-Fuentes et al., 2015 (gray circle). The contribution of each study to the characterization of Cx43 interactome was inferred from the calculation of the percentage of putative Cx43 interactors identified to the total. The percentage (17%) of proteins identified in the present study and already described in previous studies is highlighted. B, Proportional Venn diagram comparing the proteins identified in the present study (black circle) and Cx43-interacting proteins previously validated by classical biochemical and immunofluorescence techniques, either in cell lines or tissues from various origins (white circle). The percentage of well-validated Cx43 interactors identified in the present study is highlighted.
Table I. Cx43-interacting proteins previously identified and validated by classical biochemical and microscopy techniques (IP: immunoprecipitation; IV: in vitro assays (include cell-free assays, pull-down, etc.); co-loc: co-localization; (xxx): alternative name; # protein also identified in the present study).
| Interacting protein | Type of detection | References |
|---|---|---|
| Intercellular junctions and membrane-associated proteins | ||
| AGS8 | IV, co-loc | Sato et al., 2009 |
| Ankyrin G | IP | Sato et al., 2011 |
| Calmodulin | IV | Zhou et al., 2007 |
| Caveolin 1, 2 and 3 | IP, IV, co-loc | Schubert et al., 2002; Langlois et al., 2008; Liu et al., 2010 |
| Claudin 5 | IP, co-loc | Nagasawa et al., 2006 |
| Cx26 | IP | Olk et al., 2009 |
| Cx33 | IP, co-loc | Fiorini et al., 2004 |
| Cx40 | IP, IV, co-loc | He et al., 1999 |
| Drebrin | IV, co-loc | Butkevich et al., 2004 |
| F- and β-actin# | IP, co-loc, AFM | Yamane et al., 1999; Squecco et al., 2006; Wall et al., 2007 |
| Integrin α5/β1# | IP, co-loc | Burra et al., 2004 |
| Lin-7 | IV, co-loc | Singh et al., 2003 |
| N-cadherin | IP, co-loc | Lee and White, 2009 |
| Occludin | IP, co-loc | Kojima et al., 1999 |
| p120 | co-loc | Xu et al., 2001 |
| Plakophilin-2 | IP | Li et al., 2009 |
| Vinculin# | IP, IV, co-loc | Iacobas et al., 2007 |
| ZO-1 and -2 | IP, IV, co-loc | Giepmans and Moolenaar, 1998; Toyofuku et al., 1998; Singh et al., 2005 |
| α and β-tubulin# | IP, AB-array | Giepmans et al., 2001 |
| β-catenin | IP, co-loc | Ai et al., 2000; Lee and White, 2009 |
| Kinases and Phosphatases | ||
| c- and v-src | IP, co-loc | Toyofuku et al., 2001; Kanemitsu, et al., 1997 |
| CaMKII | IV, co-loc | Hund et al., 2008; Huang et al., 2011 |
| CASK (LIN2) | IP, IV, co-loc | Marquez-Rosado et al., 2011 |
| CDC2 (CDK1) | IV | Kanemitsu et al., 1998 |
| CIP85 | IP, co-loc | Lan et al., 2005 |
| CKI | IP, IV | Cooper and Lampe, 2002 |
| DMPK | IV, co-loc | Schiavon et al., 2002 |
| ERK 1/2 | IP | Li et al., 2005 |
| MAPK7/ERK5 | IV, IP | Cameron et al., 2003 |
| PKC | IP, IV, co-loc | Bowling et al., 2001; Niger et al., 2010 |
| PKG | IV | Kwak et al., 1995; Patel et al., 2006 |
| PP1/PP2A# | IV, co-loc | Ai et al., 2005 |
| Ionic channels and receptors | ||
| mAchR | IP, IV | Yue et al., 2004 |
| Nav 1.5 | IP, IV | Malhotra et al., 2004 |
| P2X7 | IP, co-loc, AB-array | Fortes et al., 2004; Iacobas et al., 2007 |
| RPTPμ | IP | Giepmans et al., 2001 |
| S100A1 | IP, IV, co-loc | Kiewitz et al., 2003 |
| β-arrestin | IP, IV, co-loc | Bivi et al., 2011 |
| Trafficking/turnover | ||
| 14–3-3# | IP, co-loc | Batra et al., 2013; Smyth et al., 2014 |
| AP2/Disabled2 | IP, co-loc | Fong et al., 2013 |
| Atg16L/Atg14/Atg9/Vps34 | IP, co-loc | Bejarano et al., 2014 |
| CIP75/Rpn1 | IP, IV, co-loc | Li et al., 2005; Su et al., 2014 |
| Clathrin# | IP, co-loc | Huang et al., 1996 |
| Consortin | IP, IV, co-loc | del Castillo, et al., 2010 |
| Dynamin | IP, co-loc | Gilleron et al., 2011 |
| Eps15 | IP, co-loc | Girao et al., 2009 |
| Hrs | co-loc | Leithe et al., 2009 |
| LC3 | IP, co-loc | Bejarano et al., 2012; Martins-Marques et al., 2015 |
| Myosin-VI# | co-loc | Piehl et al., 2007 |
| Nedd4 | IP, IV, co-loc | Leykauf et al., 2006; Catarino et al., 2011 |
| P62 | IP | Bejarano et al., 2012 |
| STAMBP | IP, co-loc | Ribeiro-Rodrigues et al., 2014 |
| TRIM21 | IV, IP | Chen et al., 2012 |
| Tsg101 | IP, co-loc | Auth et al., 2013 |
| Ubiquitin | IP, co-loc | Leithe et al., 2009; Girao et al., 2009 |
| Other proteins | ||
| Bax | IP | Sun et al., 2012 |
| BDIF-1 | IP, co-loc | Ito et al., 2011 |
| CIP150 | IV | Akiyama et al., 2005 |
| Cyclin E | IP, co-loc | Johnstone et al., 2012 |
| Hsc70# | IP, co-loc | Hatakeyama et al., 2013 |
| NOV (CCN3) | IP, IV, co-loc | Fu et al., 2004 |
| TOM20, Hsp90, ANT### | IP, co-loc | Rodriguez-Sinovas et al., 2006 |
Cx43-interactome is Differentially Affected in Pathological Conditions
Protein interactions with Cx43 strongly influence subcellular localization of junctional components and channel function, which can be dramatically altered in pathological conditions (32). Hence, we applied a SWATH strategy to characterize the cardiac Cx43-interacting network and its dynamics during heart ischemia and I/R. For that purpose, we started to calculate the interaction levels of the 236 putative Cx43-interacting partners, by normalizing to the levels of immunopurified Cx43, in each experimental condition. By performing this adjustment, a more accurate measurement of Cx43 interactions, in ischemia and I/R, was achieved. These interaction values were further subjected to an unsupervised clustering analysis. Strikingly, our results show that the 236 Cx43-interactors identified display a differential profile of interaction among the three experimental conditions (CT, ISCH and I/R; Fig. 3A–3B and supplemental Table S5).
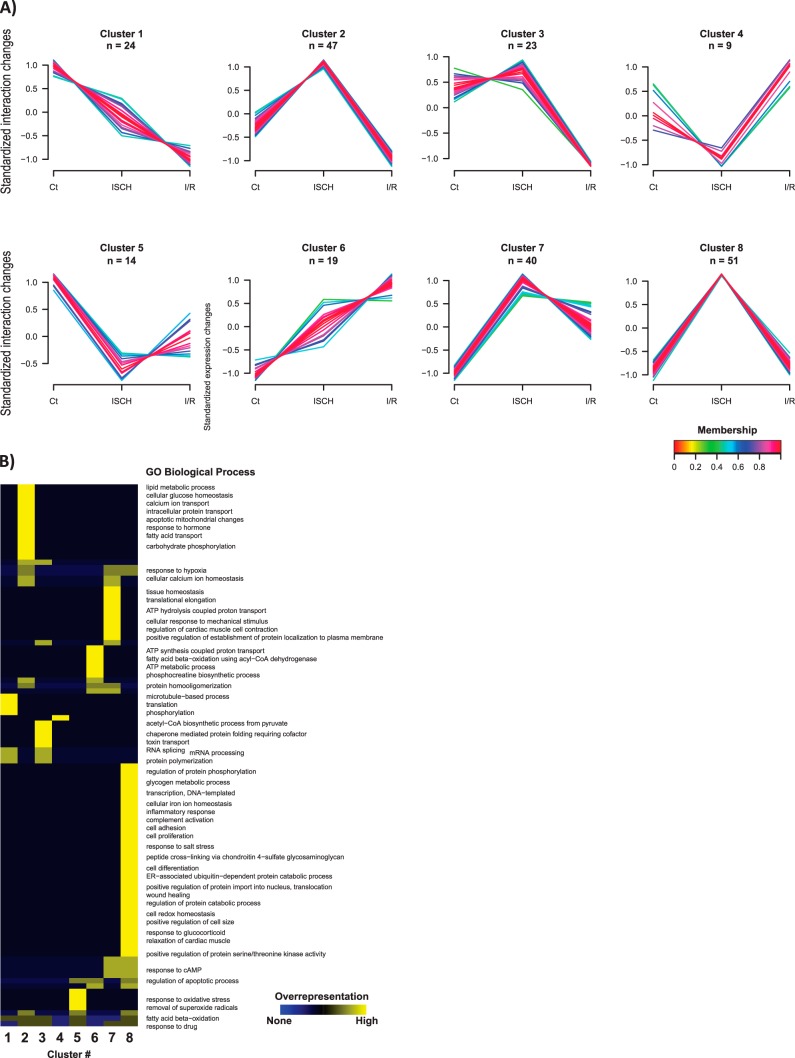
Fig. 3.
Cx43-interacting network in control, ischemic (ISCH) and I/R hearts. A, Dynamic profiles of Cx43 interactions among the experimental conditions determined by clustering analysis. Unsupervised clustering was performed for the standardized interaction levels - proteins levels normalized to Cx43 - of the 236 putative Cx43 interactors. An upper and lower ratio limit of log2 (2) and log2 (0.5) was used for inclusion into a cluster. “n” indicates the number of proteins within each cluster. Membership value represents how well the protein profile fit the average cluster profile. B, Representative overrepresented biological processes of each cluster. Each cluster from (A) was tested for overrepresented GO compared with unregulated proteins using a Binomial statistical test with Benjamini-Hochberg adjustment and a cut-off of 0.05 p value.
To highlight the most representative biological processes associated with each interaction profile, we performed a GO enrichment analysis for each cluster of interactors (Fig. 3B). Overall, our results show that there is an overrepresentation of Cx43-interactors related to lipid metabolism, calcium transport, intracellular protein transport (cluster 2), mRNA processing (cluster 3), ATP metabolic process (cluster 6), response to hypoxia (cluster 7 and 8), and regulation of protein phosphorylation (cluster 8). The most relevant and representative functional properties of the Cx43-interactors identified in our analysis were summarized in Table II. Despite the majority of the Cx43-interacting partners are involved on metabolic pathways, either upon energy production or RNA metabolism, more canonic groups of interactors, namely those related with intracellular trafficking and intercellular junctions, were also found.
Table II. Functional annotation of Cx43-interacting proteins. The 236 quantified proteins considered as putative Cx43 interacting partners were classified according to the most biological relevant and representative functional groups (based on the analysis of KEGG pathways).
| Group/complex | Protein name | |
|---|---|---|
| Metabolic pathways | Glycolysis/Gluconeogenesis | PFKM, ALDH2, ALDH9A1, ENO3, ALDOA, DLD, PDHA1, PDHB, GPI, PGK1, HK1, RGD1562690, HK2 |
| Pyruvate metabolism | MDH2, FH, MDH1, PDHA1, ALDH2, ALDH9A1, PDHB, PC, ACAT1, RGD1562690, DLD | |
| Fatty acid metabolism | ACADL, HADHA, ACADS, CPT1B, HADH, CPT2, HADHB, ACADM, ACSL1, ACAT1 | |
| Citrate cycle (TCA cycle) | IDH2, OGDH, IDH3A, MDH2, FH, PDHA1, MDH1, PDHB, PC, IDH1, SUCLG1, IDH3B, DLST, CS, DLD, SDHA | |
| Oxidative phosphorylation | ATP5C1, ND4, ATP5A1, CYTB, SDHA, ATP5D, ATP5G2, COX1, ATP5H, ATP5B, UQCRC2, UQCRC1, ND5, ND1 | |
| Valine, leucine and isoleucine degradation | HADHA, HSD17B10, PCCA, ALDH2, ALDH9A1, BCKDHA, ACAT1, ACADS, MCCC1, HADH, HADHB, ALDH6A1, MCCC2, DLD, ACADM | |
| Biosynthesis of amino acids | IDH2, PFKM, ENO3, IDH3B, ALDOA, GOT2, CS, IDH3A, PC, IDH1, PGK1, GOT1 | |
| Insulin secretion | ATP1B1, ATP1A1, RYR2, ATP1A2 | |
| PI3K-Akt signaling pathway | HSP90AA1, GYS1, HSP90B1, PPP2CA, YWHAE, ITGB1 | |
| AMPK signaling pathway | GYS1, CD36, PPP2CA, PFKM, CPT1B, EEF2 | |
| cGMP-PKG signaling pathway | VDAC2, MYH7, SLC25A5, PLN, VDAC3, ATP1B1, SLC8A1, SLC25A4, ATP1A1, ATP2A2, MYLK3, ATP1A2 | |
| Signaling pathways | Calcium signaling pathway | VDAC2, SLC25A5, PLN, VDAC3, SLC8A1, SLC25A4, ATP2A2, RYR2, MYLK3 |
| PPAR signaling pathway | FABP3, ACADL, CPT1B, CPT2, ACADM, CD36, ACSL1, FABP4 | |
| HIF-1 signaling pathway | ALDOA, PDHA1, PDHB, PGK1, RGD1562690, ENO3, TF, HK1, HK2 | |
| Insulin signaling pathway | GYS1, PYGB, HK1, PYGM, HK2 | |
| Adipocytokine signaling pathway | CPT1B, CD36, ACSL1 | |
| Ribosome | RPS3, UBA52, RPLP0, RPS6, RPS16 | |
| Spliceosome | HNRNPA1, HSPA8, HNRNPAK, HSPA1L | |
| RNA degradation | HSPA9, ENO3, HSPD1 | |
| RNA transport | KPNB1, EEF1A2 | |
| Intracellular trafficking and degradation | Endocytosis | EHD2, CLTC, HSPA8, EHD1, HSPA1L |
| Phagosome | TUBB5, CD36, TUBB4B, TUBA1B, TUBA4A, ITGB1, DYNC1H1 | |
| Lysosome | CLTC, AP1B1 | |
| Proteasome | PSMD2, PSMD1 | |
| Protein processing in endoplasmic reticulum | HSP90B1, HSPA8, DNAJC10, VCP, HSP90AA1, CRYAB, HSPA5, HSPA1L | |
| Intercellular junctions | Tight junction | MYH7, PPP2CA, MYH6, ACTN1, MYL2 |
| Adherens junction | ACTN1, VCL | |
| Focal adhesion | VCL, MYL2, ITGB1, ACTN1, MYLK3 | |
| Peroxisome | IDH2, SOD1, ECI2, CRAT, SOD2, PRDX1, ACSL1, IDH1, ECH1, PRDX5 | |
| Cardiac specific functions | Cardiac muscle contraction | MYH6, MYH7, MYL3, COX1, ATP1B1, CYTB, SLC8A1, UQCRC2, UQCRC1, ATP1A1, ATP2A2, MYL2, RYR2, ACTA2, ATP1A2 |
| Adrenergic signaling in cardiomyocytes | MYH7, MYL3, PPP2CA, PLN, ACTA2, MYH6, ATP1B1, SLC8A1, ATP1A1, ATP2A2, MYL2, RYR2, ATP1A2 | |
| Cardiac diseases | Hypertrophic cardiomyopathy (HCM) | MYBPC3, MYH6, MYH7, MYL3, SLC8A1, ITGB1, ATP2A2, MYL2, RYR2, ACTA2 |
| Dilated cardiomyopathy | MYBPC3, MYH6, MYH7, MYL3, SLC8A1, ITGB1, ATP2A2, PLN, MYL2, RYR2, ACTA2 | |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | SLC8A1, ITGB1, ACTN1, ATP2A2, RYR2 |
Additionally, InterPro (33) was used to identify specific protein families, domains or functional sites among Cx43 interactors. This search revealed that 12 of the 236 putative interactors contain a P-loop containing NTPase, five contain an Armadillo-type fold domain (found in β-catenins and importins, classical Cx43 interactors), five contain a nucleotide-binding alpha-beta plait domain, and five contain RNA recognition motifs, the latter commonly found in RNA binding proteins. Although other domains were also identified, these were less represented within the entire population of interactors.
Ischemia and I/R Modulate Cx43 Interactions
Subsequently, to explore the function of Cx43-associated proteins whose interaction profile is more affected in ischemia and I/R, the enriched GO terms were evaluated, within each experimental condition. First, we evaluated the Cx43-interactors that have a fold increase bellow 0.5 in ischemia (compared with control hearts), which revealed an enrichment of proteins involved in cellular amide metabolic processes. Moreover, when we established a threshold of fold increase > 1.25 in ischemia (ISCH-enriched Cx43-interacting partners; supplemental Table S6), we mainly identified proteins associated with membrane structures. In fact, this group of interactors contains a large number of transmembrane ionic channels, mitochondrial membrane proteins and proteins associated with membrane trafficking. Accordingly, our clustering analysis has also shown that protein interactions particularly increased under ischemia were grouped in clusters 2, 7, and 8, where these GO terms were particularly enriched (Fig. 3A–3B).
Analysis of KEGG pathways associated with the ISCH-enriched Cx43-interacting partners revealed that although the majority of these proteins are related with metabolism (56 interactors), proteins associated with cardiac muscle contraction (11 proteins) and adrenergic signaling in cardiomyocytes (10 proteins) could also be identified.
Concerning I/R, we did not find enrichments of any GO term in Cx43-interactors that have a fold increase bellow 0.5. On the other hand, I/R-enriched interactors, with a fold increase > 1.5 (supplemental Table S7), were found to be implicated with heart development, hypertrophy, anatomical structure and cardiac muscle morphogenesis. Moreover, our results show that, in I/R, there is an increased interaction with proteins associated with actin and cytoskeletal protein binding, and motor activity.
Changes in Cx43-Interactome during Reperfusion Following Ischemia
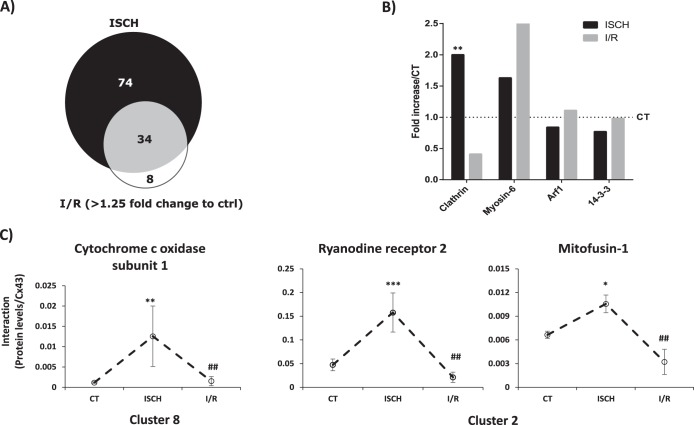
Up to this point, we have been analyzing the interactions that vary in ischemia or I/R in comparison with control. Given that in the course of ischemic heart disease, and in the sequence of treatment intending to improve blood flow, reperfusion following ischemia usually has irreversible harmful effects, we next focused on the differential interaction profile of metabolism-related proteins between ischemia and I/R. The results obtained in our study show that, of the 108 interactors whose interaction is particularly enriched during ischemia (supplemental Table S6), 42 of them are maintained above > 1.25-fold during I/R, meaning that the majority of the Cx43-interacting proteins identified in I/R arise from previously established interactions, during ischemia. Therefore, only a small fraction of interacting partners (8 proteins) are differentially enriched in I/R (Fig. 4A). Moreover, although the ISCH-enriched interactors are proteins involved in the carbon metabolism, including glycolysis/gluconeogenesis, fatty acid degradation, pyruvate metabolism, and amino acid biogenesis, the interactors that are enriched in I/R (relative to ischemia), are proteins mainly associated with oxidative phosphorylation.
Fig. 4.
Cx43 interactions Categorized by Functional Group. A. Proportional Venn diagram comparing the Cx43-interactors associated with metabolism found up-regulated in ISCH and I/R. (B). Graph depicts the quantification of the interaction between different traffic-related proteins and Cx43 in ISCH and I/R (represented as fold increase/CT). C, Individual profiles of the interaction of Cx43 with proteins involved in calcium signaling and metabolism validated in this studies (see Figs. 5 and 6). Data are presented as media±MAD of the interaction levels for three independent experiments. The respective cluster is indicated below the graphs. *ρ<0.1, ** ρ<0.05, *** ρ<0.01 (versus control condition). # statistical difference between ISCH and I/R conditions.
Ischemia Affects the Interaction of Cx43 with Regulators of its Phosphorylation and Intracellular Trafficking
GJIC is mainly dependent upon the number of active channels localized at the plasma membrane that, in turn, is determined by the mechanisms and players involved in the trafficking of Cx43 to and from the cell surface (34–37). In our proteomic study, we have identified some proteins previously described to be involved in Cx43 intracellular trafficking and degradation (36). For this reason, we explored in more detail the interaction profile of Cx43 with clathrin-mediated endocytosis (CME)-associated proteins during ischemia and I/R. The results obtained in this study show that interaction of Cx43 with both clathrin and myosin-6 are increased during ischemia (Fig. 4B), suggesting that heart ischemia is inducing CME-mediated internalization of Cx43, which likely precedes its degradation and/or lateralization.
Conversely, the interaction of Cx43 with ADP-ribosylation factor 1 (Arf1), a protein previously implicated in the delivery of newly synthesized Cx43 to the plasma membrane (Fig. 4B), presented a trend toward a decrease in ischemia, despite no statistical difference was found (38). Another group of proteins that have been shown to interact with Cx43 and modulate its anterograde transport and increased GJ assembly under stress conditions is the 14–3-3 family of proteins (39, 40). Our results indicate a reduced interaction between Cx43 and 14–3-3 epsilon in ischemia (Fig. 4B), which is consistent with a model where, by one hand, myocardial ischemia induces Cx43 internalization and, on the other hand, restrains its forward trafficking.
Oxygen and nutrient-deprivation, characteristics of ischemia, lead to an overall loss of kinases activity, which result in severe alterations on the phosphorylation profile of cardiac proteins (41, 42). Accordingly, our results show that interaction between serine/threonine-protein phosphatase 2A (PP2A) and Cx43 increases about 1.9-fold during ischemia. Indeed, previous studies have shown, in rabbit models of heart failure and in human samples, that colocalization of Cx43 with PP2A increases, which is accompanied by a down-regulation and dephosphorylation of Cx43 (41).
Cx43 Interacts with Proteins Associated with RNA Metabolism
Besides its role upon GJIC, Cx43 has been associated with non-junctional functions (13). For example, it has been shown that Cx43 can be localized in the nucleus, which can be explained by the existence of a putative nuclear targeting sequence in the carboxy-terminal of Cx43. Additionally, growing evidence suggests that the nuclear presence of Cx43 is associated with its role as gene expression regulator (43). Our report shows that Cx43 interacts with a wide variety of heterogeneous nuclear ribonucleoproteins (hnRNP), which can function upon transcription, splicing, mRNA trafficking or translational silencing. Importantly, ischemia and I/R negatively regulate the majority of these interactions, suggesting that the, interaction of Cx43 with these proteins might modulate the cell transcriptome usually associated with heart disease, namely by differentially affecting RNA splicing or transport (Table III).
Table III. Interaction of Cx43 with hnRNPs varies in ischemia (ISCH) and I/R. Quantification of interaction between different hnRNPs and Cx43 in ISCH and I/R (represented as fold increase/CT). The listed proteins met the criteria to be considered as Cx43 putative interactors (supplementary Table S3) and were quantified with at least two peptides (supplementary Table S2).
| Cx43-interacting protein | Fold increase/CT |
|
|---|---|---|
| ISCH | I/R | |
| hnRNP A3 | 0,78 | 0,59 |
| hnRNP A2/B1 | 0,84 | 0,73 |
| hnRNP D0 | 0,85 | 0,67 |
| hnRNP H | 0,86 | 0,74 |
| hnRNP H2 | 0,89 | 0,79 |
| hnRNP d-like | 0,9 | 0,94 |
| hnRNP F | 0,93 | 1,08 |
| hnRNP A1 | 0,99 | 0,67 |
| hnRNP Q | 1,32 | 0,79 |
| hnRNP K | 1,52 | 1,05 |
Validation of the Cardiac Cx43-interactome
To determine whether the Cx43-interacting proteins that we identified using our SWATH approach also associate with Cx43 in situ, we performed immunofluorescence confocal microscopy. Given the importance of calcium signaling for cardiac contractility, and the fact that the majority of Cx43-interactors found are associated with metabolism, we selected 3 proteins—ryanodine receptor 2 (RyR2), mitofusin 1 (Mfn1), and subunit 1 of the cytochrome c oxidase (COX1)—for validation.
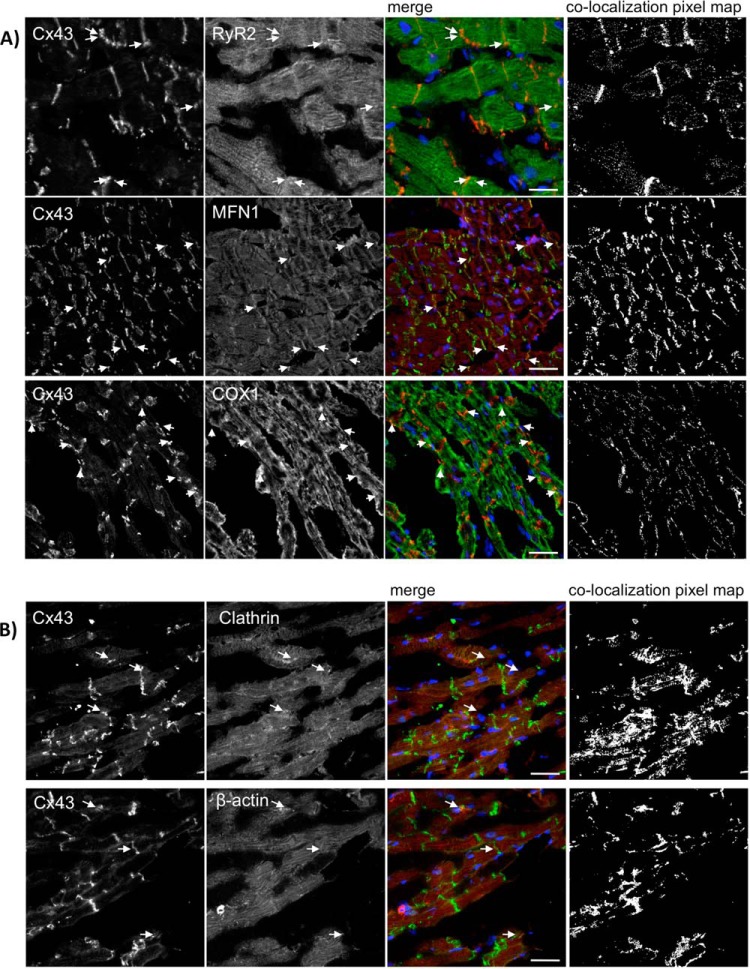
Our results show that Cx43 extensively colocalizes with the 3 proteins in the rat heart (Fig. 5), being the colocalization with Mfn1 particularly pronounced. As positive controls, we analyzed colocalization of Cx43 with clathrin heavy chain and β-actin, previously established to interact with Cx43 (Table I). Interestingly, clathrin, RyR2 and Mfn1 belong to the same cluster (cluster 2 - Fig. 2A), and presented similar interaction profiles among ischemia and I/R (Fig. 4C). Despite COX1 have been differently clustered, its interaction profile is very similar to the others (Fig. 4C). Furthermore, we confirmed the interaction of Cx43 with some of these proteins by co-immunoprecipitation assays in heart lysates (supplemental Fig. S5). Despite the low amount of protein that was co-immunoprecipitated with Cx43, the results obtained demonstrate that all the interactors tested, including clathrin, Mfn1, and COX1 can be precipitated with Cx43.
Fig. 5.
RyR2, Mfn1 and COX1 colocalize with Cx43 in the rat heart. Hearts from 10-week-old Wistar rats were maintained using a Langendorff apparatus for 10 min, followed by 20 min of perfusion (CT). Cryosections of control or ischemic hearts were immunostained using antibodies against (A) Cx43 (Sc9059) and RyR2 (upper panel), Cx43 (610062) and Mfn1 (middle panel), and Cx43 (Sc9059) and COX1 (lower panel). B, Cx43 (Sc9059) and Clathrin heavy chain (upper panel) or β-actin (lower panel). Nuclei were stained with DAPI. Scale bars, 25 μm. Arrows indicate colocalization spots. Colocalization pixel map is depicted on the right side.
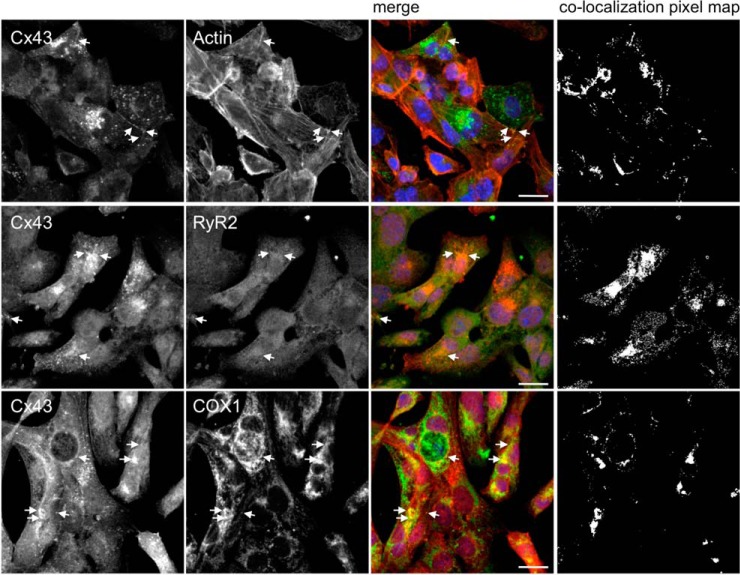
Besides cardiomyocytes, that are the functional unit of the heart, cardiac tissue is formed by other cell types, including endothelial cells and fibroblasts. In order to discard the contribution of other cell types to the results obtained in heart samples, we performed validation of some of these interactions in a cardiomyocyte cell line (HL-1 cells). Results of Fig. 6 show that Cx43 colocalizes with RyR2 and COX1 (actin was used as a positive control), corroborating the results obtained with rat hearts, and reinforcing our interactomic data.
Fig. 6.
RyR2 and COX1 colocalize with Cx43 in HL-1 cells. Immunostaining for Cx43 (Sc9059) and RyR2 (middle panel) and COX1 (lower panel), in HL-1 cells. Positive control was performed by colocalization analysis with F-actin, stained with Phalloidin (upper pannel, Cx43 staining with 610062). Nuclei were stained with DAPI. Scale bars, 20 μm. Arrows indicate colocalization spots. Colocalization pixel map is depicted on the right side.
The fluorescence intensity profile of Cx43 and the various proteins evaluated, both in heart samples and in HL-1 cells, has some similarities, at least in certain discrete areas, suggesting that the proteins colocalize in specific cellular compartments (supplemental Figs. S3–S4).
DISCUSSION
Given its importance for the heart function, the elucidation of the Cx43 interactome, both is resting and ischemic conditions, is of utmost importance to understand the mechanisms and players underlying the maintenance of intercellular communication. Although an interactomic analysis of Cx43 is not without precedent, the present work constitutes the first proteomic study carried out in heart samples. Moreover, this is by far the most exhaustive study of the Cx43 interactome, contributing with several new Cx43-binding partners. Furthermore, by applying the recent quantitative AP-SWATH strategy, it was possible to identify the Cx43 interactors and, more importantly, to precisely trace the interaction profiles under the referred conditions, which is crucial in the evaluation of different pathophysiological states. Strikingly, the results obtained in the present work, not only allowed the identification of new Cx43-interactors, but also demonstrate that the Cx43-interactome is a very dynamic entity, varying in ischemia and I/R. Besides predictable interactions with proteins involved in the regulation of cell adhesion, subcellular trafficking and signaling, we provide strong evidence that cardiac Cx43 interacts with proteins associated with metabolism and protein synthesis, thus implicating Cx43 in other biological processes.
It is well established that interaction of Cx43, either direct or indirect, with other membrane channels or transporters (Table I), might contribute to the concerted regulation of tissue homeostasis. For example, it has been shown that Cx43 interacts with the voltage-gated sodium channel Nav1.5 in the perinexus of cardiomyocytes (44). More recently, Lubkemeier et al. provided evidence that gap junctional Cx43 is required to ensure the arrival of Nav1.5 channels to the IDs, which is important for the maintenance of whole-cell sodium current density and, consequently, for the cardiac electrical coupling (45). In our study, despite we have not identified interaction with Nav1.5, several other ionic channels were present in the Cx43 interacting network, including voltage-dependent anion-selective channels and sodium/potassium-transporting ATPases (6). We also found interaction of Cx43 with the mitochondrial ATP-binding cassette (ABC) subfamily B member 7 (ABCB7). Interestingly, interaction of a member of the ABC subfamily, the ABCD3, with Cx32 has been uncovered in a recent proteomic study in liver samples (46). Previous studies also demonstrated the functional interplay between Cx43/Cx45 with other ABC transporters, namely with cystic fibrosis transmembrane conductance regulator (CFTR). Indeed, it was shown that GJIC can be regulated by CFTR through modulation of voltage sensitivity and gating of Cx channels (47). Although the molecular mechanisms involved are still unclear, it is known that CFTR/GJ crosstalk relies on a complex signaling network, involving c-src.
The interaction of Cx43 with proteins related to catabolism and energy production was reported in the Gago-Fuentes study (13). Although inhibition of GJ has been related with changes in the subcellular localization and up-regulation of glucose transporter 1 (GLUT-1) and type I hexokinase, the molecular mechanisms underlying such events remain unknown (48). Given that GJ enable the passage of glucose and ATP between neighbor cells, it is conceivable that altered metabolite trafficking controls the levels and/or localization of glycolytic enzymes, likely as part of a feedback loop. On the other hand, it is tempting to speculate that the mechanism whereby Cx43 participates in metabolic regulation relies on multiprotein complexes, formed with mitochondrial proteins, a dynamic that can be altered in pathological conditions affecting both glucose and oxidative metabolism, such as myocardial ischemia. In our study, we found a differential interaction profile of Cx43 with metabolism-associated proteins that likely reflects the metabolic shift undergone by heart cells during ischemia. The primary energy source of heart cells arises from fatty acid oxidation, with glycolysis representing only a small contribution for myocardial ATP production (49). However, as a consequence of oxygen deprivation during ischemia, glucose consumption increases, leading to increased lactate production, alanine accumulation and decreased levels of ATP, glycogen, glutamate, and aspartate (49). Accordingly, the ISCH-enriched Cx43 interactors are mainly associated with glycolysis and amino acid metabolism, whereas the Cx43 interactors specifically enriched in I/R, when O2 supply is re-established, include proteins primarily involved in oxidative metabolism.
Interaction of Cx43 with the regular mitochondrial protein import machinery had already been described in mitochondria isolated from pig hearts. Indeed, it was found that Cx43 is translocated to the inner mitochondrial membrane, a process dependent upon its interaction with translocase of the outer membrane 20 (Tom20), heat shock protein 90 (Hsp90), and adenine nucleotide transporter (ANT, also known as ADP/ATP translocase) (7). Our results show that, during ischemia, interaction of Cx43 with Hsp90α and Hsp90β increases about twofold, which is in accordance with previous studies demonstrating that Hsp90-mediated translocation of Cx43 to the mitochondria is enhanced in ischemia (7). Interaction with ADP/ATP translocase 2 also increases with ischemia, by more than 12-fold, whereas interaction with ADP/ATP translocase 1 did not present any variation (supplemental Table S5 and S6).
Interestingly, our data shows that Cx43 interacts with tripartite motif-containing protein 72 (TRIM72). Moreover, we demonstrate that interaction Cx43-TRIM72 increases 1.22-fold in ischemia, in comparison with control conditions (supplemental Table S6). In the heart, TRIM72 has been demonstrated to participate in membrane repair during ischemia. A recent study from Chung et al. has shown that the delivery of membrane repair machinery to damaged mitochondria during ischemia depends upon specialized microdomains, called ischemia-induced caveolin-3 enriched fraction (ICEF) signalosomes. The authors suggested that this process is important to restrict reactive oxygen species (ROS) production and, consequently, to reduce infarct size (50). It was also reported that both Cx43 and TRIM72 localizes at the ICEF, with ischemia and preconditioning enhancing such distribution (50). Having these results into account, it is plausible that Cx43, through interaction with TRIM72 within caveolin-enriched microdomains, plays a role in mediating membrane repair and cardioprotection.
The findings obtained in the present proteomic analysis demonstrate that Cx43 also interacts with proteins involved in signal transduction, namely G-protein-coupled receptors (GPCRs), which are essential for the integration and transduction of external stimuli, ultimately influencing cell function. Expectedly, GPCR malfunction has been associated with several cardiovascular pathologies. For instance, activator of G-protein signaling 8 (AGS8) is up-regulated in cardiomyocytes during ischemia and hypoxia, contributing to cell death under stress conditions. Also, it is known that AGS8 form complexes with Cx43, mediating hypoxia-induced Cx43 phosphorylation and GJ internalization, in a Gβγ-dependent manner (51). Our results show that Cx43 interacts with adenylate cyclase-stimulating Gα-protein (also known as Gnas, Gα-protein, isoform XLas), being this interaction increased in ischemia (supplemental Table S6), which suggests that, analogous with AGS8, ischemia-induced remodeling of GJ is regulated by Gnas. Moreover, during I/R, we observed a decreased interaction Gnas/Cx43.
Previous studies have demonstrated that Cx43 functions as a regulator of gene expression, being this role either dependent or independent of GJIC. Indeed, it was reported in various cell types that the absence of Cx43 leads to a differential cellular transcriptome, which ultimately determines phenotypical changes in cell morphology and cell adhesion suggesting that Cx43 has a direct role on gene transcription (6). On the other hand, studies carried out in osteosarcoma cell lines and in mouse bone marrow stromal cells (BMSCs), have shown that defects in GJIC may alter the subcellular localization and the recruitment of transcription factors to the promoter region of certain genes, impacting upon gene transcription (52, 53). In this case, it was hypothesized that differential GJ-mediated passage of second messengers modulates signaling pathways that will affect the binding affinity and/or activity of the transcription factor Sp1. In our study, we demonstrate that Cx43 interacts with proteins involved in the regulation of gene expression. Moreover, our results show that during ischemia and I/R the interaction profile of Cx43 with elongation factors and hnRNPs is altered, suggesting that Cx43 has an active role in modulating gene expression under stress conditions.
GJ degradation and/or lateralization have been associated with cardiac malfunction in the onset of myocardial ischemia. Our results show that the interaction profile of Cx43 with intracellular trafficking-related proteins varies during ischemia and I/R, corroborating these previous findings. In fact, we show that during ischemia, there is an increased interaction with the CME machinery, which is accompanied by a decreased interaction with proteins associated with forward trafficking of Cx43. It is likely that the changes in interactome profile described above will ultimately lead to a decrease in the levels of GJ present at the IDs. However, during reperfusion, it is plausible that normal trafficking of Cx43 is restored. In support of this hypothesis, during reperfusion, interaction of Cx43 with clathrin decreases, whereas the interaction with myosin-6, Arf1, and 14–3-3 presents a tendency to increase.
Although the heart samples used in our MS analysis contain different cardiac cell types, including, among others, cardiomyocytes, fibroblasts, endothelial cells and smooth muscle cells, given the high relative abundance of cardiomyocytes, it is conceivable that the interactome of Cx43 identified in heart samples mainly derives from this cell type
Overall, our work provides a comprehensive study of the cardiac Cx43-interactome, both in physiological and pathological conditions, namely in the context of ischemia and I/R. Our results strengthen the idea that, besides GJIC, Cx43 plays other biological roles. Indeed, we show that among the interactors of Cx43, there are proteins involved in regulation of protein homeostasis, including transcription, cell proliferation, and regulation of apoptosis. Overall, this study constitutes an important contribution for the elucidation of the Cx43-interactome, allowing not only a better understanding of the mechanisms and players involved in the regulation of intercellular communication, but also the identification of new putative roles of Cx43 that go beyond communication. Moreover, the identification of interactors differentially affected by ischemia and I/R paves the way toward the development of new therapeutic targets in heart disease.
Supplementary Material
Footnotes
Author contributions: T.M., S.I.A., B.M., and H.G. designed research; T.M. and S.I.A. performed research; B.M. and H.G. contributed new reagents or analytic tools; T.M., S.I.A., P.P., B.M., and H.G. analyzed data; T.M., S.I.A., P.P., B.M., and H.G. wrote the paper.
* This work was supported by Portuguese Foundation for Science and Technology (FCT) grants PTDC/SAU-ORG/119296/2010 and PEST-C/SAU/UI3282/2011-COMPETE, UID/NEU/04539/2013, PEST-C/SAU/LA0001/2013-2014; by QREN, the European Union (FEDER-Fundo Europeu de Desenvolvimento Regional), and by REDE/1506/REM/2005, from The National Mass Spectrometry Network (RNEM). TMM was supported by PD/BD/106043/2015 and S.A. by the Ph.D fellowship SFRH/BD/81495/2011.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S7.
1 The abbreviations used are:
- GJ
- Gap Junctions
- ABC
- ATP-binding cassette
- CAN
- Acetonitrile
- AGS8
- G-protein signaling 8
- ANT
- Adenine nucleotide transporter
- Arf1
- ADP-ribosylation factor 1
- BMSCs
- bone marrow stromal cells
- CFTR
- Cystic fibrosis transmembrane conductance regulator
- CME
- Clathrin-mediated endocytosis
- COX1
- subunit 1 of the cytochrome c oxidase
- Cx
- Connexins
- DIA
- Data independent acquisition
- FA
- Formic acid
- FDR
- False Discovery Rate
- GFP
- Green fluorescent protein
- GJIC
- GJ-mediated intercellular communication
- GLUT-1
- Glucose trasnporter 1
- Gnas
- adenylate cyclase-stimulating Galpha-protein
- GO
- Gene Ontology
- GPCRs
- G-protein coupled receptors
- hnRNPs
- Heterogenous nuclear ribonucleoproteins
- Hsp90
- Heat shock protein 90
- I/R
- Ischemia/reperfusion
- ICEF
- Ischemia-induced caveolin-3 enriched fraction
- IDs
- Intercalated discs
- IDA
- Information-dependent acquisition
- IP
- Immunopurification
- ISCH
- Ischemia
- KEGG
- Kyoto encyclopedia of genes and genomes
- LC
- Liquid chromatography
- LC-MS/MS
- Liquid chromatography coupled to tandem mass spectrometry
- m/z
- Mass to charge ratio
- MAD
- Median absolute deviation
- Mfn1
- Mitofusin 1
- MPTP
- Mitochondrial permeability transition pore
- MS/MS
- Scan of the fragmentation ions
- PP2A
- Serine/threonine protein phosphatase 2A
- Q–Q plots
- Quantile-Quantile Plot
- ROS
- Reactive oxygen species
- RyR2
- Ryanodine receptor 2
- SWATH
- Sequential Windowed data independent Acquisition of the Total High-resolution Mass Spectra
- Tom20
- Translocase of the outer membrane
- TRIM72
- Tripartite motif-containing protein 72
- XIC
- Extracted-ion chromatogram.
REFERENCES
- 1. Lo C. W. (2000) Role of gap junctions in cardiac conduction and development: insights from the connexin knockout mice. Circulation Res. 87, 346–348 [DOI] [PubMed] [Google Scholar]
- 2. Martins-Marques T., Catarino S., Marques C., Pereira P., and Girao H. (2015) To beat or not to beat: degradation of Cx43 imposes the heart rhythm. Biochem. Soc. Trans. 43, 476–481 [DOI] [PubMed] [Google Scholar]
- 3. Goodenough D. A., and Paul D. L. (2009) Gap junctions. Cold Spring Harbor Perspectives in Biology 1, a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olk S., Turchinovich A., Grzendowski M., Stuhler K., Meyer H. E., Zoidl G., and Dermietzel R. (2010) Proteomic analysis of astroglial connexin43 silencing uncovers a cytoskeletal platform involved in process formation and migration. Glia 58, 494–505 [DOI] [PubMed] [Google Scholar]
- 5. Kardami E., Dang X., Iacobas D. A., Nickel B. E., Jeyaraman M., Srisakuldee W., Makazan J., Tanguy S., and Spray D. C. (2007) The role of connexins in controlling cell growth and gene expression. Prog. Biophys. Mol. Biol. 94, 245–264 [DOI] [PubMed] [Google Scholar]
- 6. Herve J. C., Derangeon M., Sarrouilhe D., Giepmans B. N., and Bourmeyster N. (2012) Gap junctional channels are parts of multiprotein complexes. Biochim. Biophys. Acta 1818, 1844–1865 [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez-Sinovas A., Boengler K., Cabestrero A., Gres P., Morente M., Ruiz-Meana M., Konietzka I., Miro E., Totzeck A., Heusch G., Schulz R., and Garcia-Dorado D. (2006) Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circulation Res. 99, 93–101 [DOI] [PubMed] [Google Scholar]
- 8. Boengler K., Dodoni G., Rodriguez-Sinovas A., Cabestrero A., Ruiz-Meana M., Gres P., Konietzka I., Lopez-Iglesias C., Garcia-Dorado D., Di Lisa F., Heusch G., and Schulz R. (2005) Connexin 43 in cardiomyocyte mitochondria and its increase by ischemic preconditioning. Cardiovasc. Res. 67, 234–244 [DOI] [PubMed] [Google Scholar]
- 9. Martins-Marques T., Catarino S., Marques C., Matafome P., Ribeiro-Rodrigues T., Baptista R., Pereira P., and Girao H. (2015) Heart ischemia results in connexin43 ubiquitination localized at the intercalated discs. Biochimie 112, 196–201 [DOI] [PubMed] [Google Scholar]
- 10. Martins-Marques T., Catarino S., Zuzarte M., Marques C., Matafome P., Pereira P., and Girao H. (2015) Ischaemia-induced autophagy leads to degradation of gap junction protein connexin43 in cardiomyocytes. Biochem. J. 467, 231–245 [DOI] [PubMed] [Google Scholar]
- 11. Duffy H. S. (2012) The molecular mechanisms of gap junction remodeling. Heart Rhythm 9, 1331–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurtenbach S., and Zoidl G. (2014) Gap junction modulation and its implications for heart function. Front. Physiol. 5, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gago-Fuentes R., Fernandez-Puente P., Megias D., Carpintero-Fernandez P., Mateos J., Acea B., Fonseca E., Blanco F. J., and Mayan M. D. (2015) Proteomic analysis of connexin 43 reveals novel interactors related to osteoarthritis. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen V. C., Kristensen A. R., Foster L. J., and Naus C. C. (2012) Association of connexin43 with E3 ubiquitin ligase TRIM21 reveals a mechanism for gap junction phosphodegron control. J. Proteome Res. 11, 6134–6146 [DOI] [PubMed] [Google Scholar]
- 15. Claycomb W. C., Lanson N. A. Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., and Izzo N. J. Jr. (1998) HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dias P., Desplantez T., El-Harasis M. A., Chowdhury R. A., Ullrich N. D., Cabestrero de Diego A., Peters N. S., Severs N. J., MacLeod K. T., and Dupont E. (2014) Characterisation of connexin expression and electrophysiological properties in stable clones of the HL-1 myocyte cell line. PLoS ONE 9, e90266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anjo S. I., Santa C., and Manadas B. (2015) Short GeLC-SWATH: A fast and reliable quantitative approach for proteomic screenings. Proteomics 15, 757–762 [DOI] [PubMed] [Google Scholar]
- 18. Manadas B., Santos A. R., Szabadfi K., Gomes J. R., Garbis S. D., Fountoulakis M., and Duarte C. B. (2009) BDNF-induced changes in the expression of the translation machinery in hippocampal neurons: protein levels and dendritic mRNA. J. Proteome Res. 8, 4536–4552 [DOI] [PubMed] [Google Scholar]
- 19. Tang W. H., Shilov I. V., and Seymour S. L. (2008) Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 7, 3661–3667 [DOI] [PubMed] [Google Scholar]
- 20. Sennels L., Bukowski-Wills J. C., and Rappsilber J. (2009) Improved results in proteomics by use of local and peptide-class specific false discovery rates. BMC Bioinformatics 10, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambert J.-P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z.-Y., Zhong Q., Lindquist S., Vidal M., and Aebersold R. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nature Methods. 12:1239–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collins B. C., Gillet L. C., Rosenberger G., Röst H. L., Vichalkovski A., Gstaiger M., and Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14–3-3 system. Nature Methods 12:1246–53. [DOI] [PubMed] [Google Scholar]
- 23. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rigbolt K. T., Vanselow J. T., and Blagoev B. (2011) GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell Proteomics 10, O110 007450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar L., and M E. F. (2007) Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vadigepalli R., Chakravarthula P., Zak D. E., Schwaber J. S., and Gonye G. E. (2003) PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics 7, 235–252 [DOI] [PubMed] [Google Scholar]
- 27. Oveland E., Muth T., Rapp E., Martens L., Berven F. S., and Barsnes H. (2015) Viewing the proteome: how to visualize proteomics data? Proteomics 15, 1341–1355 [DOI] [PubMed] [Google Scholar]
- 28. Polpitiya A. D., Qian W. J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G. 2nd, Anderson G. A., and Smith R. D. (2008) DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24, 1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duffy H. S. (2011) The molecular mechanisms of gap junction remodeling. Heart Rhythm 9, 1331–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collins B. C., Gillet L. C., Rosenberger G., Rost H. L., Vichalkovski A., Gstaiger M., and Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 31. Lambert J. P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z. Y., Zhong Q., Lindquist S., Vidal M., Aebersold R., Pawson T., Bonner R., Tate S., and Gingras A. C. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agullo-Pascual E., Cerrone M., and Delmar M. (2014) Arrhythmogenic cardiomyopathy and Brugada syndrome: diseases of the connexome. FEBS Lett. 588, 1322–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mulder N., and Apweiler R. (2007) InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. 396, 59–70 [DOI] [PubMed] [Google Scholar]
- 34. Girao H., Catarino S., and Pereira P. (2009) Eps15 interacts with ubiquitinated Cx43 and mediates its internalization. Exp. Cell Res. 315, 3587–3597 [DOI] [PubMed] [Google Scholar]
- 35. Catarino S., Ramalho J. S., Marques C., Pereira P., and Girao H. (2011) Ubiquitin-mediated internalization of connexin43 is independent of the canonical endocytic tyrosine-sorting signal. Biochem. J. 437, 255–267 [DOI] [PubMed] [Google Scholar]
- 36. Fong J. T., Kells R. M., and Falk M. M. (2013) Two tyrosine-based sorting signals in the Cx43 C-terminus cooperate to mediate gap junction endocytosis. Mol. Biol. Cell 24, 2834–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ribeiro-Rodrigues T. M., Catarino S., Marques C., Ferreira J. V., Martins-Marques T., Pereira P., and Girao H. (2014) AMSH-mediated deubiquitination of Cx43 regulates internalization and degradation of gap junctions. FASEB J 28, 4629–4641 [DOI] [PubMed] [Google Scholar]
- 38. Majoul I. V., Onichtchouk D., Butkevich E., Wenzel D., Chailakhyan L. M., and Duden R. (2009) Limiting transport steps and novel interactions of Connexin-43 along the secretory pathway. Histochem. Cell Biol. 132, 263–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Batra N., Riquelme M. A., Burra S., and Jiang J. X. (2013) 14-3-3 theta facilitates plasma membrane delivery and function of mechanosensitive connexin 43 hemichannels. J. Cell Sci. 127, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smyth J. W., Zhang S. S., Sanchez J. M., Lamouille S., Vogan J. M., Hesketh G. G., Hong T., Tomaselli G. F., and Shaw R. M. (2014) A 14–3-3 mode-1 binding motif initiates gap junction internalization during acute cardiac ischemia. Traffic 15, 684–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ai X., and Pogwizd S. M. (2005) Connexin 43 downregulation and dephosphorylation in nonischemic heart failure is associated with enhanced colocalized protein phosphatase type 2A. Circ. Res. 96, 54–63 [DOI] [PubMed] [Google Scholar]
- 42. Lampe P. D., Cooper C. D., King T. J., and Burt J. M. (2006) Analysis of Connexin43 phosphorylated at S325, S328 and S330 in normoxic and ischemic heart. J. Cell Sci. 119, 3435–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dang X., Doble B. W., and Kardami E. (2003) The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell Biochem. 242, 35–38 [PubMed] [Google Scholar]
- 44. Rhett J. M., Ongstad E. L., Jourdan J., and Gourdie R. G. (2012) Cx43 associates with Na(v)1.5 in the cardiomyocyte perinexus. J. Membr. Biol. 245, 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lubkemeier I., Requardt R. P., Lin X., Sasse P., Andrie R., Schrickel J. W., Chkourko H., Bukauskas F. F., Kim J. S., Frank M., Malan D., Zhang J., Wirth A., Dobrowolski R., Mohler P. J., Offermanns S., Fleischmann B. K., Delmar M., and Willecke K. (2014) Deletion of the last five C-terminal amino acid residues of connexin43 leads to lethal ventricular arrhythmias in mice without affecting coupling via gap junction channels. Basic Res. Cardiol. 108, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fowler S. L., Akins M., Zhou H., Figeys D., and Bennett S. A. (2013) The liver connexin32 interactome is a novel plasma membrane-mitochondrial signaling nexus. J. Proteome Res. 12, 2597–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chanson M., Kotsias B. A., Peracchia C., and O'Grady S. M. (2007) Interactions of connexins with other membrane channels and transporters. Prog. Biophys. Mol. Biol. 94, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanchez-Alvarez R., Tabernero A., and Medina J. M. (2004) Endothelin-1 stimulates the translocation and upregulation of both glucose transporter and hexokinase in astrocytes: relationship with gap junctional communication. J. Neurochem. 89, 703–714 [DOI] [PubMed] [Google Scholar]
- 49. Rosano G. M., Fini M., Caminiti G., and Barbaro G. (2008) Cardiac metabolism in myocardial ischemia. Curr. Pharm. Des. 14, 2551–2562 [DOI] [PubMed] [Google Scholar]
- 50. Chung Y. W., Lagranha C., Chen Y., Sun J., Tong G., Hockman S. C., Ahmad F., Esfahani S. G., Bae D. H., Polidovitch N., Wu J., Rhee D. K., Lee B. S., Gucek M., Daniels M. P., Brantner C. A., Backx P. H., Murphy E., and Manganiello V. C. (2015) Targeted disruption of PDE3B, but not PDE3A, protects murine heart from ischemia/reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 112, E2253–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sato M., Jiao Q., Honda T., Kurotani R., Toyota E., Okumura S., Takeya T., Minamisawa S., Lanier S. M., and Ishikawa Y. (2009) Activator of G protein signaling 8 (AGS8) is required for hypoxia-induced apoptosis of cardiomyocytes: role of G betagamma and connexin 43 (CX43). J. Biol. Chem. 284, 31431–31440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schajnovitz A., Itkin T., D'Uva G., Kalinkovich A., Golan K., Ludin A., Cohen D., Shulman Z., Avigdor A., Nagler A., Kollet O., Seger R., and Lapidot T. (2011) CXCL12 secretion by bone marrow stromal cells is dependent on cell contact and mediated by connexin-43 and connexin-45 gap junctions. Nat. Immunol. 12, 391–398 [DOI] [PubMed] [Google Scholar]
- 53. Stains J. P., Lecanda F., Screen J., Towler D. A., and Civitelli R. (2003) Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J. Biol. Chem. 278, 24377–24387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.