Abstract
Mutations in the EGFR kinase domain are implicated in non-small cell lung cancer. Of particular interest is the drug-resistant double mutant (L858R/T790M, DM EGFR), which is not inhibited selectively by any approved kinase inhibitor. Here we apply bipartite tetracysteine display to demonstrate that DM and WT EGFR differ in structure outside the kinase domain. The structural difference is located within the cytoplasmic juxtamembrane segment (JM) that links the kinase domain with the extracellular and transmembrane regions and is essential for EGFR activation. We show further that third-generation DM EGFR-selective TKIs alter JM structure via allostery to restore the conformation found when WT EGFR is activated by the growth factors EGF and HB-EGF. This work suggests that the oncogenic activity of DM EGFR may extend beyond kinase activity per se to include kinase-independent activities. As JM structure may provide a biomarker for these kinase-independent functions, these insights could guide the development of allosteric, DM-selective inhibitors.
Mutations in the epidermal growth factor receptor (EGFR) kinase domain are implicated in 10 to 35% of non-small cell lung cancer cases.1 One common mutation (L858R) induces ligand-independent activation and oncogenic signaling.1 Patients whose tumors harbor L858R EGFR often respond to first-generation tyrosine kinase inhibitors (TKIs)2 but then regress, frequently due to a second kinase domain mutation (T790M) that lowers inhibitor potency.3 The kinase domains of wild-type (WT) EGFR and the drug-resistant, double mutant (DM) form are similar,4 making it difficult to develop molecules that effectively inhibit DM EGFR at concentrations at which WT EGFR is spared.5–9
Here we apply bipartite tetracysteine display10 to demonstrate that DM and WT EGFR differ in structure outside the kinase domain. The difference is located within the cytoplasmic juxtamembrane segment (JM) that links the kinase domain with the extracellular and transmembrane regions and is essential for EGFR activation.11 We also show that third-generation, DM EGFR-selective TKIs, as a group, alter JM structure via allostery to restore the conformation seen when WT EGFR is activated by the growth factors EGF and HB-EGF. As JM sequences are not highly conserved,12 these findings could lead to improved, DM-selective inhibitors.
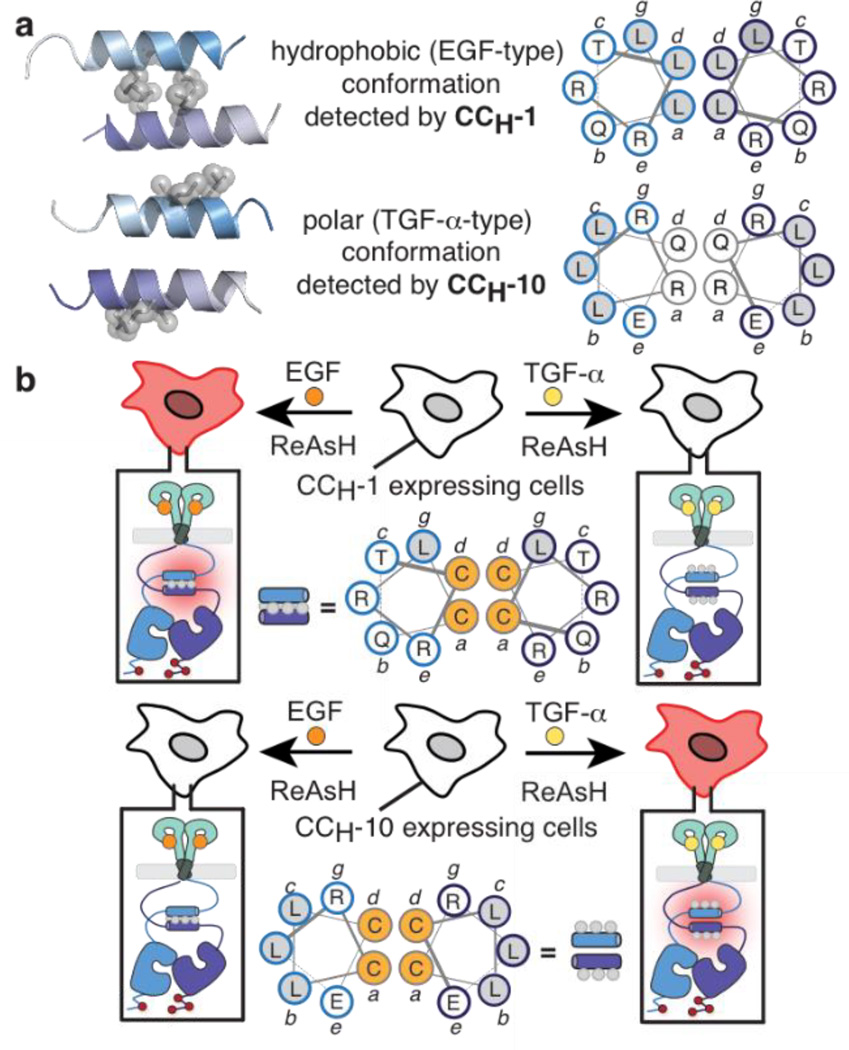
Previously, we applied bipartite tetracysteine display to characterize the conformation of the EGFR JM within intact receptors expressed on the cell surface.13,14 We discovered that the binding of epidermal growth factor (EGF) to the WT EGFR extracellular domain promotes formation of a distinct antiparallel coiled coil15 within the intracellular JM, whereas the binding of transforming growth factor-α (TGF-α) is communicated through the formation of a coiled coil – a rotational isomer - whose helical interface is ‘inside-out’ compared with the JM interface formed in the presence of EGF (Figure 1A).14 We also demonstrated that growth factors that activate EGFR fall into distinct categories in which coiled coil identity correlates with downstream signaling differences.14
Figure 1.
(A) Models of the EGF- and TGF-α-type coiled coils illustrating the relative Leu positions (gray balls). (B) Detection of the EGF-type coiled coil in cells expressing CCH-1 EGFR; detection of the TGF-α-type coiled coil in cells expressing CCH-10 EGFR.
These previous investigations were performed with a pair of Cys-Cys EGFR variants (CCH-1 and CCH-10) that report on formation of the EGF- and TGF-α-induced JM coiled coils, respectively (Figure 1B).13,14 When these coiled coils form within an EGFR dimer, the assembled Cys4 motif is poised to bind ReAsH and cause it to fluoresce. Expression of CCH-1 EGFR on the CHO-KI cell surface results in a significant increase in ReAsH fluorescence in the presence of EGF but not TGF-α, whereas expression of CCH-10 EGFR results in a significant increase in ReAsH fluorescence in the presence of TGF-α but not EGF (Figure 1B).13,14
To evaluate the state of the JM coiled coil in EGFR kinase domain mutants, we prepared three sets of CCH-1 and CCH-10 variants harboring substitutions associated with gefitinib/erlotinib sensitivity (L858R) or resistance (T790M and L858R/T790M) (Figure S1A). All Cys-Cys EGFR variants (CCX-1 and -10, where X = H (WT), 858 (L858R), 790 (T790M) or DM (L858R/T790M)) were constitutively active when expressed in CHO-K1 cells, as determined by the extent of auto-phosphorylation at Y1173 in the absence of added growth factor. The expression levels and activities of these CCX-1 and CCX-10 variants were comparable to variants lacking the cysteine residues required for ReAsH binding (Figure S1B).
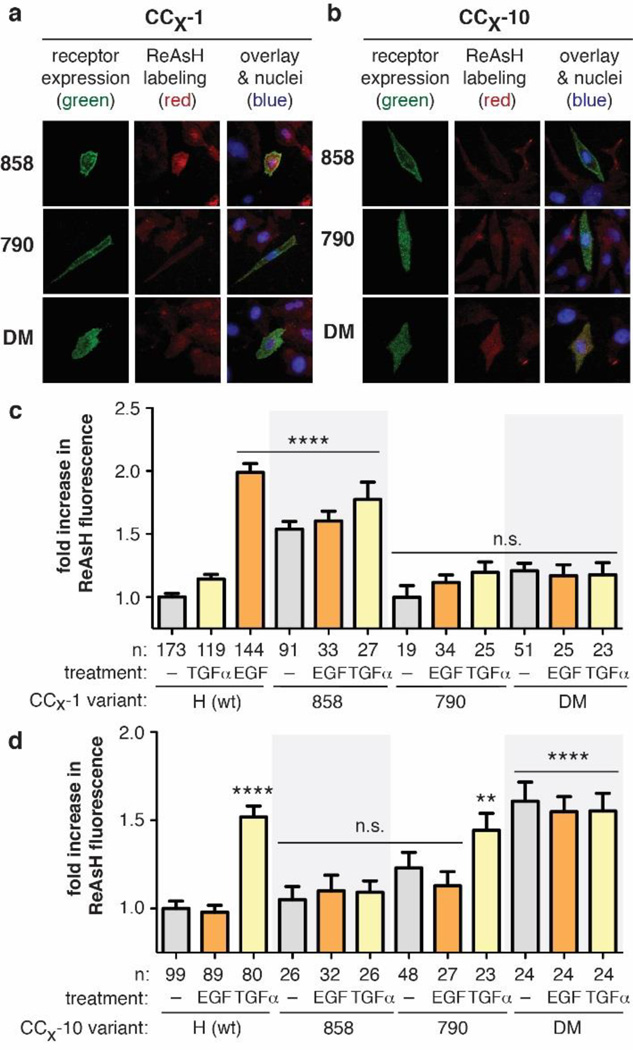
We first applied these CCX-1 and CCX-10 variants to evaluate the JM conformation in each EGFR mutant (L858R, T790M, and L858R/T790M) without added growth factor. Dynasore-treated16 CHO-K1 cells expressing each EGFR variant were treated with ReAsH and the level of EGFR-associated fluorescence was determined using total internal reflectance fluorescence microscopy (TIRF-M) (Figure 2A,B & S2). Among CCX-1 EGFR variants, only those cells expressing CC858-1 EGFR, harboring the L858R kinase domain mutation, displayed a significant increase (1.5-fold, p < 0.0001) in ReAsH-associated fluorescence in the absence of growth factor (Figure 2C). No ReAsH-associated increase in fluorescence over background was observed for cells expressing either CC790-1 or CCDM-1 EGFR, both of which contain the T790M mutation associated with drug resistance.
Figure 2.
(A,B) TIRF images of cells expressing FLAG-tagged CCX-1 or CCX-10 EGFR (green) and treated with ReAsH (red). (C,D) Fold increase in expression-corrected ReAsH fluorescence over background of cells expressing CCX-1 or CCX-10 ± EGF or TGF-α. Error bars, s.e.m. **** p < 0.0001, ** p < 0.01 from one way ANOVA with Bonferroni post-test.
We noticed that the ReAsH fluorescence associated with CC858-1 EGFR (1.5-fold) was lower than that observed for EGF-stimulated cells expressing CCH-1 EGFR (2.0-fold) (Figure 2C). To provide additional evidence for formation of the EGF-type JM coil in L858R EGFR, we generated a second set of EGFR variants whose Cys-Cys arrangement also reports on formation of the EGF-type coiled coil. These variants, derived from the previously studied CCH-2 (Figure S3A),13 carry Cys residues at the a and e positions of the JM heptad repeat as opposed to the a and d positions utilized in CCH-1. All CCX-2 variants harboring kinase domain substitutions were constitutively active at levels that were comparable to the analogous CCX-1 variants (Figure S3B). CHO-K1 cells expressing CC858-2 EGFR exhibited a 1.9-fold increase (p < 0.0001) in normalized ReAsH fluorescence in the absence of growth factor (Figure S3C,D). No ReAsH-associated increase in fluorescence over background was observed upon examination of cells expressing either CC790-2 or CCDM-2 EGFR. Taken with the results obtained with the CCX-1 variants, these results support the conclusion that constitutively active L858R EGFR adopts the EGF-type JM coil in the absence of growth factor stimulation.
We next employed the analogous CCX-10 variants to probe for the TGF-α-type coiled coil. Dynasore-treated CHO-K1 cells expressing each CCX-10 variant were treated with ReAsH and the level of EGFR-associated fluorescence detected using TIRF-M (Figure 2B). Only cells expressing CCDM-10 EGFR displayed a significant increase (1.6-fold, p < 0.0001) in ReAsH-associated fluorescence in the absence of growth factor (Figure 2D). No ReAsH-associated increase in fluorescence over background was observed when cells expressing either CC858-10 or CC790-10 EGFR were examined. Thus, while constitutively active L858R EGFR adopts only the EGF-type JM coil, the L858R/T790M double mutant adopts only the TGF-α-type coiled coil. The implication is that coiled coil structure reflects not only growth factor identity on the cell surface13,14 but also kinase mutational state. The structural changes induced in the kinase domain by T790M are transmitted allosterically through > 120 amino acids, beyond the kinase domain, into the adjacent JM.
Identical results were observed when cells expressing CC858-1, CC858-10, and CC858-2 were stimulated with EGF or TGF-α before ReAsH treatment (Figure 2 and S2B). Among variants that report on the EGF-type JM coil, only CC858-1 and CC858-2 EGFR displayed a significant increase in fluorescence (1.5 - 1.9-fold, p < 0.0001) and in a manner irrespective of growth factor treatment (Figure 2C and S3C,D). The observation that L858R EGFR retains the EGF-type JM structure even when stimulated with TGFα suggests that the structural effects of L858R substitution dominate over those related to growth factor identity. In a similar way, CCDM-10 EGFR displayed a significant increase (1.6-fold, p < 0.0001) in ReAsH fluorescence regardless of added ligand (Figure 2D and S2B), indicating that DM EGFR adopts the TGF-α-type coil even when stimulated with EGF.
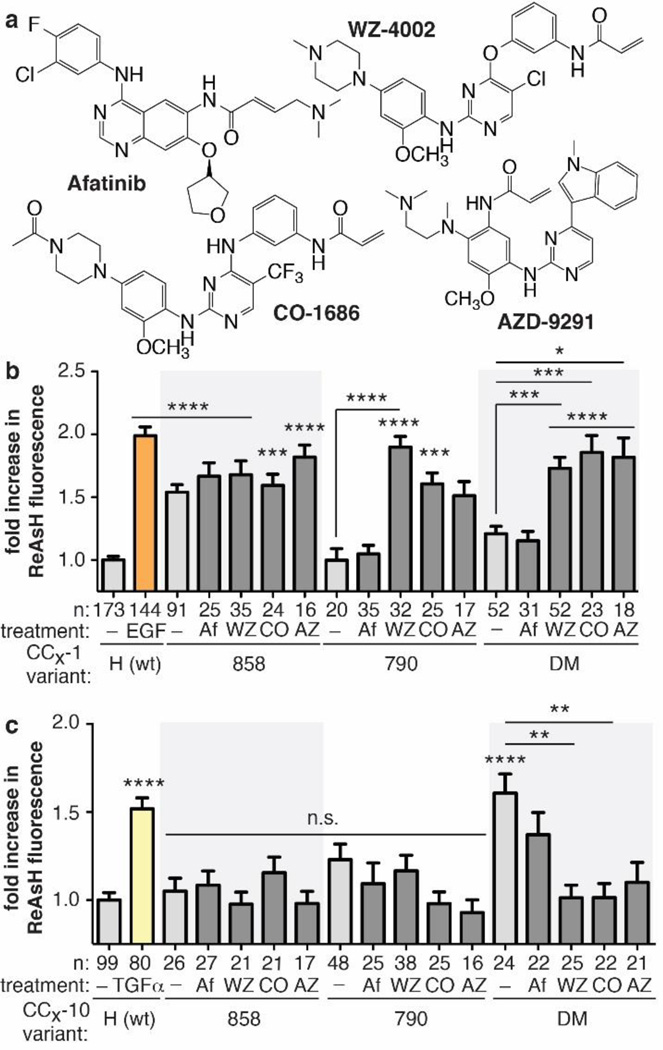
Several irreversible TKIs in pre-clinical or clinical development effectively inhibit DM EGFR (IC50 < 100 nM, Figure 3A).5–9 Afatinib,6 a recently approved second-generation TKI, inhibits WT, L858R and DM EGFR,6 while three third-generation molecules display measurable DM EGFR selectivity: WZ-4002,5 AZD-9291,8 and CO-1686.7 Comparison of the structure of the T790M EGFR kinase domain (residues 698–984) when bound to WZ40025 and afatinib17 reveals only small (albeit significant) differences in local inhibitor-receptor interactions; the structures are otherwise virtually superimposable (RMSD ~0.67 Å). Both structures–which lack the JM region as well as the TM and extracellular domain–contain the activation helix in the active, “DFG-in” conformation. Our observation that JM coiled coil structure is controlled by structural changes induced by a single side chain substitution in the kinase domain–replacement of Thr at 790 with Met–led us to ask whether they might also be affected by the presence of different tyrosine kinase inhibitors. In particular, we wondered whether DM EGFR selectivity would correlate with allosteric structural changes within the JM.
Figure 3.
(A) Structures of afatinib (Af)6, WZ4002 (WZ)5, CO-1686 (CO)7, and AZD-9291 (AZ)8. (B,C) Fold-increase in expression-corrected ReAsH fluorescence over background of cells expressing CCX-1 or CCX-10 variants in the presence or absence of TKI. Error bars, s.e.m. **** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05 from one way ANOVA with Bonferroni post-test.
To probe for long-range effects of inhibitor binding on JM coiled coil structure, we treated cells expressing CCDM-1 or CCDM-10 EGFR with 10 µM Afatinib, WZ4002, AZD-9291, or CO-1686 for 30 minutes, labeled the cells with 2 μM ReAsH, and quantified cell surface ReAsH fluorescence using TIRF-M (Figure 3B,C and Figure S4). Analogous experiments were performed with single mutant EGFR variants (CC858-1 and -10 and CC790-1 and -10); control experiments verified that the kinase activities of all CCX-1 and CCX-10 variants were abolished under these conditions (Figure S5).
As described above, in the absence of any inhibitor, DM EGFR contains only the TGF-α-type coiled coil: only CCDM-10 (and not CCDM-1 EGFR) showed evidence of ReAsH binding, in the presence or absence of growth factor (Figure 2C,D). Treatment with afatinib, under conditions where DM EGFR is fully inhibited (Figure S5), led to no significant change in the ReAsH-dependent fluorescence of CCDM-10 and CCDM-1: afatinib-treated cells expressing CCDM-1 showed no evidence of ReAsH binding and fluorescence, whereas afatinib-treated cells expressing CCDM-10 showed high ReAsH fluorescence. The ReAsH-induced fluorescence over background of cells expressing any CCX-1 or CCX-10 variant is almost identical in the presence or absence of afatinib (Figure 3B,C and S4). These results indicate that afatinib induces no detectable change in JM coiled coil structure: L858R EGFR retains the EGF-type structure and DM EGFR retains the TGF-α-like structure.
Different results were observed when cells expressing CCDM-1 or CCDM-10 EGFR were treated with the three DM-selective inhibitors tested–WZ-4002,5 CO-1686,7 and AZD-9291.8 Treatment with any of these three inhibitors, under conditions where DM EGFR is fully inhibited (Figure S5), resulted in significant changes in the ReAsH-dependent fluorescence of CCDM-10 and CCDM-1 EGFR: cells expressing CCDM-1–which detects formation of the hydrophobic, EGF-type coiled coil–showed high levels of ReAsH fluorescence, whereas treated cells expressing CCDM-10–which detects the polar, TGF-α-type coiled coil–did not (Figure 3B,C). These results indicate that WZ-4002, CO-1686, and AZD-9291, three TKIs that selectively inhibit DM EGFR in preference to WT EGFR,5,7,8 all induce a long-range change in structure that flips the JM coiled coil from the TGF-α-type conformation into the EGF-type structure. This flip corresponds to a 150° disrotatory rotation about each helix axis. Similar results were observed when CC790-1 and -10 EGFR were treated with the three DM EGFR-selective inhibitors. The binding of DM EGFR-selective, third-generation TKIs exerts an allosteric structural influence that preferentially stabilizes the EGF-type coiled coil in the sequence-distal juxtamembrane region.
This work provides increased resolution on how chemical information is encoded by the prototypic receptor tyrosine kinase EGFR. We describe that perturbations within the intracellular kinase domain due to mutation or TKI binding propagate over long distances to impact structure within the JM. A similar allosteric connection exists between the JM and the extracellular, growth factor-binding region.14 These findings identify the JM as a mediator of information flow initiated on both sides of the plasma membrane. More broadly, the finding that the EGFR JM region is both allosterically responsive to inhibitor binding and a determinant of downstream signaling14 emphasizes that EGFR is subject to a degree of conformational complexity currently unresolved by structures of its domains in isolation. Further, the results imply that the oncogenicity of DM EGFR may extend beyond kinase activity per se to include kinase-independent activities that are not sensitive to drugs like afatinib even when kinase activity is fully inhibited. As JM structure may provide a biomarker for these kinase-independent functions, these insights could guide the development of allosteric, DM-selective inhibitors.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the NIH (GM 83257, T32 GM07223 and GM08283) for research support and to an anonymous reviewer for insightful comments.
Footnotes
ASSOCIATED CONTENT
Methods, data and cell images. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. New Engl J Med. 2004;350:2129. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]; Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. Proc Natl Acad Sci USA. 2004;101:13306. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma SV, Bell DW, Settleman J, Haber DA. Nat Rev Cancer. 2007;7:169. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 3.Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, Mitsudomi T. Clin Cancer Res. 2006;12:5764. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]; Yun C-H, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong K-K, Meyerson M, Eck MJ. Proc Natl Acad Sci USA. 2008;105:2070. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewer MR, Yun C-H, Lai D, Lemmond MA, Eck MJ, Pao W. Proc Natl Acad Sci USA. 2013;110:E3595. doi: 10.1073/pnas.1220050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Janne PA. Nature. 2009;462:1070. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A, Himmelsbach F, Rettig WJ, Meyerson M, Solca F, Greulich H, Wong KK. Oncogene. 2008;27:4702. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter AO, Sjin RTT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, Sheets M, St Martin T, Karp R, van Kalken D, Chaturvedi P, Niu D, Nacht M, Petter RC, Westlin W, Lin K, Jaw-Tsai S, Raponi M, Van Dyke T, Etter J, Weaver Z, Pao W, Singh J, Simmons AD, Harding TC, Allen A. Cancer Disc. 2013;3:1404. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross DAE, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MRV, Ward RA, Mellor MJ, Hughes G, Rahi A, Jacobs VN, Red Brewer M, Ichihara E, Sun J, Jin H, Ballard P, Klinowska T, Richmond GHP, Cantarini M, Kim D-W, Ranson MR, Pao W. Cancer Disc. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K-O, Cha MY, Kim M, Song JY, Lee J-H, Kim YH, Lee Y-M, Suh KH, Son J. Cancer Res. 2014;74 LB. [Google Scholar]
- 10.Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Nat Chem Biol. 2007;3:779. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scheck RA, Schepartz A. Accts Chem Res. 2011;44:654. doi: 10.1021/ar2001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiel KW, Carpenter G. Proc Natl Acad Sci. 2007;104:19238. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard SR. Nat Rev Mol Cell Biol. 2004;5:464. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- 13.Scheck RA, Lowder MA, Appelbaum JS, Schepartz A. ACS Chem Bio. 2012;7:1367. doi: 10.1021/cb300216f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doerner AE, Scheck RA, Schepartz A. 2015 submitted. [Google Scholar]
- 15.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang XW, Kuriyan J. Cell. 2009;138:604. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Control experiments confirm that Dynasore itself has no effect on JM coiled coil formation. For details, see Figure S2
- 17.Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, Kraemer O, Himmelsbach F, Haaksma E, Adolf GR. J Pharmacol Exp Ther. 2012;343:342. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.