Abstract
CHO-LY-B cells have been useful in studies of sphingolipid metabolism and function because they lack serine palmitoyltransferase (SPT) activity. Cloning and sequencing of the SPT1 transcript of LY-B cells identified the mutation as a guanine to adenine change at nucleotide 738, causing a G246R transformation. Western blots revealed low expression of the mutant SPT1 peptide, but activity was not detectable by mass spectrometric analysis of [13C]-palmitate incorporation into sphinganine, sphingosine, 1-deoxysphinganine, or 1-desoxymethylsphinganine. Treatment of LY-B cells with chemical chaperones (DMSO or glycerol) increased the amounts of mutant SPT1 as well as SPT2, but SPT activity was not restored. This study has established that G246R mutation in hamster SPT1 results in the loss of SPT activity.
Keywords: sphingolipid, sphingoid base, serine palmitoyltransferase, mutant enzyme
Introduction
Sphingolipids are a structurally diverse and important family of compounds [1] characterized by having a sphingoid base backbone [2]. Sphingoid base biosynthesis is initiated by serine palmitoyltransferase (SPT; EC2.3.1.50) [3], an activity that is essential for cells in culture unless the medium is supplemented with sphingolipids [4]. SPT is comprised of two [3], and possibly more [5,6], subunits and homozygous deletion of two subunits (Sptlc1 and Sptlc2, also known as LCB1/LCB2 or SPT1/SPT2) in mice is embryonic lethal [7]. Mutations in SPTLC1 are responsible for human hereditary sensory neuropathy type 1 (HSN1) [8,9].
LY-B cells lack SPT activity due to mutation of Sptlc1 [4], and have been useful tools for studies of de novo sphingolipid biosynthesis and the functions of sphingolipids [4]. This report describes the point mutation that results in the loss of SPT activity in LY-B cells, and notes that small amounts of SPT1 polypeptide can be detected by Western blotting, especially when the cells are treated with “chemical chaperones” [10] such as DMSO and glycerol, but without restoration of activity.
Experimental Procedures
Materials
Chinese hamster ovary (CHO) derived lines LY-B and LY-B/cLCB1 (which are stably transfected with the Chinese hamster LCB1 cDNA to restore SPT activity) were available from previous studies [4]. Ham’s F12 medium was from Gibco (Carlsbad, CA) and fetal bovine serum was obtained from Hyclone (Logan, UT). The sources for the antibodies were BD Biosciences (San Jose, CA) for the anti-LCB1 monoclonal antibody, Cayman (Ann Arbor, MI) for the anti-SPT2 monoclonal antibody, Sigma (St. Louis, MO) for the anti-beta actin and anti-HA antibodies, Ambion (Austin, TX) for the anti-GAPDH antibody, and David Uhlinger (Johnson & Johnson Pharmaceutical Research & Development, Raritan, NJ) for another anti-SPT1 antibody [11]. The enhanced chemifluorescence (ECF) Western blotting kit was purchased from GE healthcare (Piscataway, NJ). Mass spectrometry internal standards were obtained from Avanti Polar Lipids (Alabaster, AL) and [U-13C]- palmitic acid was purchased from Cambridge Isotope (Andover, MA). Fumonisin B1 (FB1) was obtained from Biomol (Plymouth Meeting, PA). All other reagents and solvents were of high quality.
Cell culture and treatments
LY-B and LY-B/cLCB1 cells were cultured in Ham’s F12 medium containing 10% fetal bovine serum, penicillin G (100 U/ml) and streptomycin sulfate (100 μg/ml) in 5% CO2 at 37 °C. During stable isotope labeling studies, the media was supplemented with 0.1 mM [U-13C]-palmitic acid complexed with equimolar fatty acid free BSA (Calbiochem, Gibbstown, NJ). After 12 or 24 h, depending on the experiment, the incorporation of stable isotope into sphingolipids was measured by liquid chromatography, electrospray ionization tandem mass spectrometry (LC ESI-MS/MS) as previously described [12] except as noted below.
To analyze whether LY-B and LY-B/cLCB1 cells produce both “typical” (i.e., sphinganine and sphingosine) and “atypical” sphingoid bases (1-deoxysphinganine and 1-desoxymethylsphinganine) [13,14], 25 μ M FB1 was added to the medium one hour before addition of 0.1 mM [U-13C]-palmitic acid-BSA complex to cause the sphingoid bases to accumulate [13].
For chemical chaperone treatment, the LY-B cells were grown to ~50% confluence and changed to media supplemented with different concentrations of DMSO (up to 3%) or glycerol (up to 1 M) for 24 h. To determine how this affected SPT amount, the SPT1 and SPT2 peptides were visualized by Western blotting as described below. GAPDH was used as the protein loading control because DMSO and glycerol influence actin expression [15-17]. To determine how SPT activity was affected, the cells were incubated for an 12 h in the same medium supplemented with 0.1 mM [U-13C]-palmitic acid-BSA complex then analyzed by LC ESI-MS/MS.
Cloning and sequencing of SPT1 from LY-B cells
Total RNA was extracted from LY-B cells by the Absolutely RNA Miniprep Kit (Qiagen, Valencia, CA) and reverse transcribed to cDNA using the cDNA Synthesis System Kit (TaKaRa, Mountain View, CA). PCR was performed to amplify the SPT1 transcript using primers (5’- atggcgatggcggcggagca and 3’- ctacagcagcacagcctgggca), which represent the start and stop codon for hamster SPT1 open reading frame. The amplified PCR transcript was ligated into pUC18 cloning vector and restriction digested with EcoRI to determine the clones with the correct inserts. 15 selected clones were sequenced (Nevada genomics center, Reno, NV) and compared to wildtype CHO SPT1 mRNA by multiple sequence alignment using T-coffee [18].
Western blotting
Cells were rinsed with PBS then scraped from the dishes using PBS containing Complete Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN), lysed by brief sonication, incubated for 30 min at 25°C with Benzonase Nuclease (Novagen, Madison, WI), then boiled briefly in Tris-HEPES-SDS (100 mM Tris, 100 mM HEPES, pH 8.0, 3 mM SDS) before loading onto a 12% SDS-PAGE (Pierce, Rockford, IL) minigel (40 μg of protein/well) and electrophoresed. The separated proteins were transferred onto a nitrocellulose membrane (Whatman, Florham park, NJ) and blocked with 5% milk-TBST (Tris buffered saline Tween-20) (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, 0.1% Tween-20) solution. The membrane was incubated with primary antibodies (anti-LCB1, diluted 1:2000; Anti-SPT1, diluted 1:3000; anti-SPT2, diluted 1:1000) overnight at 4° C and washed with TBST solution before being probed with a secondary anti–fluorescein antibody (ECF kit) for 1 h. The protein bands were visualized by incubating the membrane with the ECF substrate for 30 min and imaged using FLA-3000 imager (Fujifilm, Stamford, CT). Pixel density was measured using Multi Gauge (Fujifilm) image analysis software. Statistical significance was defined at a p-value of 0.05 using the Kruskal-Wallis rank sum test in R v2.61 (www.r-project.org).
LC ESI-MS/MS analysis of sphingolipids
The sphingolipids were analyzed in positive ionization mode by LC ESI-MS/MS [12] using ABI 3000 triple quadrupole and ABI 4000 quadrupole-linear ion trap mass spectrometers (Applied Biosystems, Foster City, CA). Since the major fragmentation products of ceramides (Cer) and ceramide monohexosides (HexCer) (e.g., ions with m/z 264.4 for sphingosine and m/z 266.4 for sphinganine) retain the carbon atoms originally derived from palmitoyl-CoA, the sphingolipids made de novo via incorporation of [U-13C]-palmitate into the sphingoid base backbones were determined by adding 16 mass units to the precursor-product pairs for species labeled in only the sphingoid base backbone, and by adding 32 mass units to the precursor ion and 16 to the product for compounds labeled in both the fatty acid and the sphingoid base. Since these product ions are of very low abundance from sphingomyelins (SM) [12], the incorporation of [U-13C]-palmitate into the sphingoid base backbones of SM was determined by treatment of an aliquot of the lipid extract with the phospholipase D from Streptomyces chromofuscus (Sigma) to produces ceramide 1-phosphates phosphates [19,20], which do provide definitive information about [13C] labeling of the sphingoid base backbone [12,21]. This treatment was conducted by incubating dried lipid extracts with 50 units of the enzyme suspended in 0.1 ml of 100 mM Tris HCl, pH 8.0, and 3 mM decylglucopyranoside (Sigma), at 37° C for 30 min. The reaction was dried by speedvac and analyzed by LC ESI-MS/MS.
Analysis of the biosynthesis of sphinganine (So), sphingosine (Sa), 1-deoxysphinganine (1-deoxy-Sa), and 1-desoxymethylsphinganine (1-desoxymethyl-Sa) [14,22] from cells incubated [U-13C]-palmitate and 25 μM FB1 was conducted by LC ESI-MS/MS in positive ionization mode using multiple reaction monitoring with Q1 and Q3 set to pass the following precursor and product ions: 316.5/298.3 ([13C]-So), 318.5/300.3 ([13C]-Sa), 302.4/284.4 ([13C]-1-deoxy-Sa), and 288.3/270.3 ([13C]-1-desoxymethyl-Sa). The LC conditions were the same as described previously [13,22], i.e. using a reverse phase LC column (Supelco 2.1 × 50 mm Discovery C18 column, Sigma, St. Louis, MO) and a binary solvent system at a flow rate of 0.6 mL/min; the binary system began with equilibration of the column with 60% mobile phase A (CH3OH/H2O/HCOOH, 58/41/1, v/v/v, with 5 mM ammonium formate) and 40% mobile phase B (CH3OH/HCOOH, 99/1, v/v, with 5 mM ammonium formate), sample injection and elution for 1.3 min in the same mobile phase, followed by a linear gradient over 2.8 min to 100% B (then a column wash with 100% B for 0.5 min followed by re-equilibration with the original A/B mixture before the next run).
Homology modelling of mutant SPT1
Homology modelling was performed by comparing SPT with two POAS enzymes [23], with one being the SPT from Sphingomonas paucimobilis (PDB ID: 2JG2) [24]. The crystal structures of PDB ID: 2JG2 and 1FC4 were selected as templates because they were identified to have 30% sequence identity with hamster SPT1 by a blast search of the PDB database. The sequences of the templates and mutant SPT1 protein were aligned using T-coffee [18], refined manually into a PIR format alignment file that was used to generate the homology model with Modeller v6.1 [25], which provides a consensus structure of the target protein without a bias towards a single template [26,27], and visualised by VMD v1.8.6 [28]. The 2JG2 template has 55% identity with hamster SPT1 for the amino acid residues located within a 5°A radius of the mutated amino acid residue, R246, of SPT1 from LY-B cells.
Results and Discussion
Analysis of the gene sequence for mutant SPT1 in LY-B cells
Sequencing of the mutated SPT1 from LY-B cells revealed a guanine to adenine mutation at nucleotide residue 738, which would result in the substitution of Arg for Gly246, which is conserved across numerous species (C. griseus, M. musculus, H. sapiens and S. cerviceae), as shown in Fig. 1A. To investigate the impact of G246R substitution on SPT1, a homology model was prepared and residues within 5 °A of G246R were compared to 2JG2 template by multiple sequence alignment, which revealed 55% sequence homology. The analysis revealed the presence of non-polar residues such as Val, Leu and Phe within 3-5 °A distance from the point mutation, while polar residues Glu and Ser were conserved between SPT1 and 2JG2, which may play a role in protein folding. The stearic hindrance resulting from G246R substitution in a conserved region of SPT1 may destabilize the protein structure.
Fig. 1.
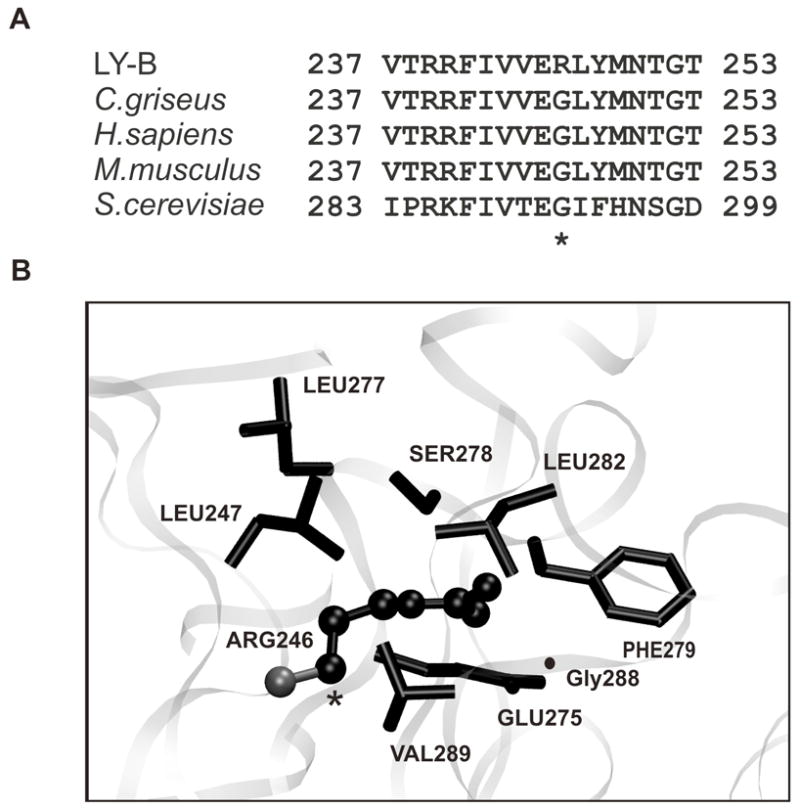
Sequence comparison and homology model of the SPT1 mutant protein of LY-B cells. (A) Protein sequences of SPT1 were compared from LY-B cells, CHO, human, mouse and S. cerviceae using T-coffee. The point mutation G246R in LY-B cells is indicated by an asterisk. (B) This homology model for mutant SPT1 from LY-B cells was prepared using Modeller v6.1. The illustrated figure shows the G246R mutation (indicated in a ball and stick representation) to be located in a pocket of hydrophobic residues (Phe279, Leu282, Val289, Leu247) was visualized by VMD.
Analysis of mutant SPT1 polypeptide in LY-B cells
Previous studies have not detected an SPT1 peptide in LY-B cells by Western blotting [4], however, a very faint band is discernable when larger amounts of protein (40 μg) are examined using two antibodies (anti-LCB1 and anti-SPT1) (Fig. 2A and B) but not using an antibody to another polypeptide epitope not found in the cells (HA) (Fig. 2C). Also shown is the immunoblotting of SPT1 in LY-B/cLCB1 cells, which have been stably transfected with the wild-type SPT1 gene (Fig. 2A and B).
Fig. 2.
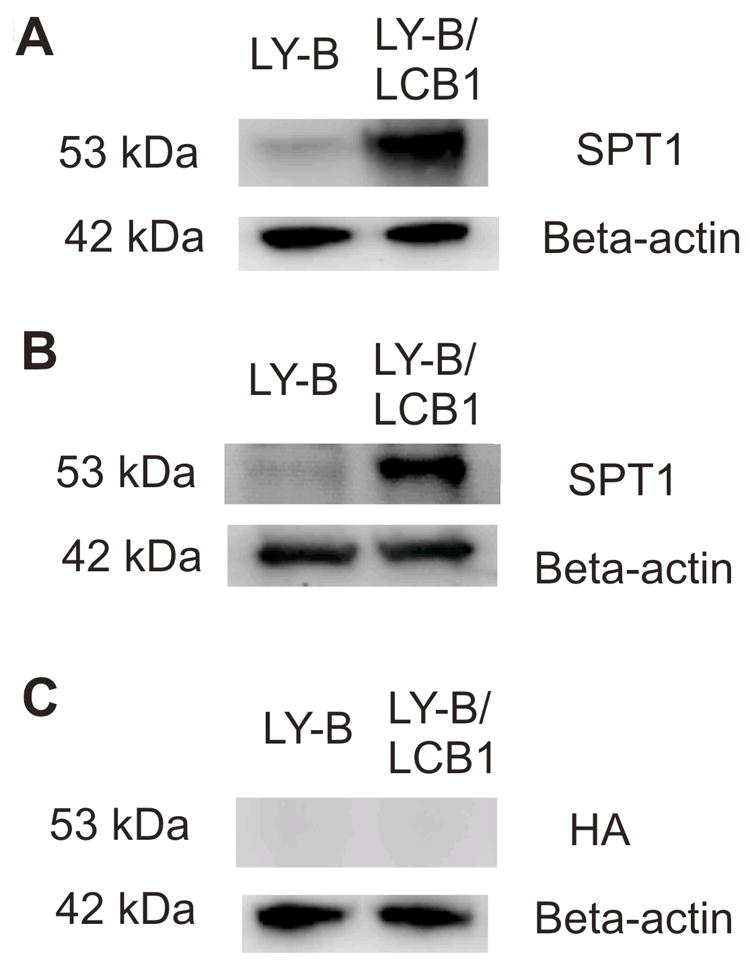
Western blots of SPT1 in LY-B and LY-B/cLCB1 cells. Whole cell lysates of LY-B and LY-B/cLCB1 cells were prepared and equal amounts (40 μg) of protein were separated by an SDS-PAGE gel. Replicate membranes were probed with (A) a commercial LCB1 antibody; (B) an independent anti-SPT1 polyclonal antibody, and (C) an anti-HA antibody as a negative control. The detection of β-actin with an anti-β-actin antibody was used as a protein loading control.
Analysis of mutant SPT activities in LY-B cells
To ascertain whether this small amount of mutant SPT1 afforded detectable de novo sphingolipid biosynthesis, cells were incubated with [13C]-palmitic acid and analyzed for incorporation of [13C] into the sphingoid base backbone (with free sphingoid bases in Fig. 3A and complex sphingolipids in Fig. 3B-D); LY-B/cLCB1 cells were used as a positive control because they have SPT activities comparable to wild-type CHO cells [4]. When all of the subspecies are summed, the amounts of backbone [13C]-labelled sphingolipids in the LY-B/cLCB1 cells at 6 h were 62 ± 0.3 pmol Cer/mg protein, 54 ± 0.8 pmol HexCer/mg protein and 300 ± 14 pmol SM/mg protein, for a total of ~416 pmol/mg protein; for LY-B cells, the amounts were below the limits of detection for Cer and HexCer (~ 0.1 pmol/mg protein) and very low for SM (5.5 ± 16 pmol SM/mg protein) which, based on the high noise in the signal (reflected in the high SD), might be due to other compounds with similar ionization characteristics. Thus, even counting the possibility that this represents SM, the maximum amount of labeled backbone sphingolipids in the LY-B cells would only ~1% that in LY-B/cLCB1 cells. This is in agreement with a previous study [4] which has shown that SPT activity in the microsomal fraction of LY-B cells was less than 5% of that in CHO-K1 cells.
Fig. 3.
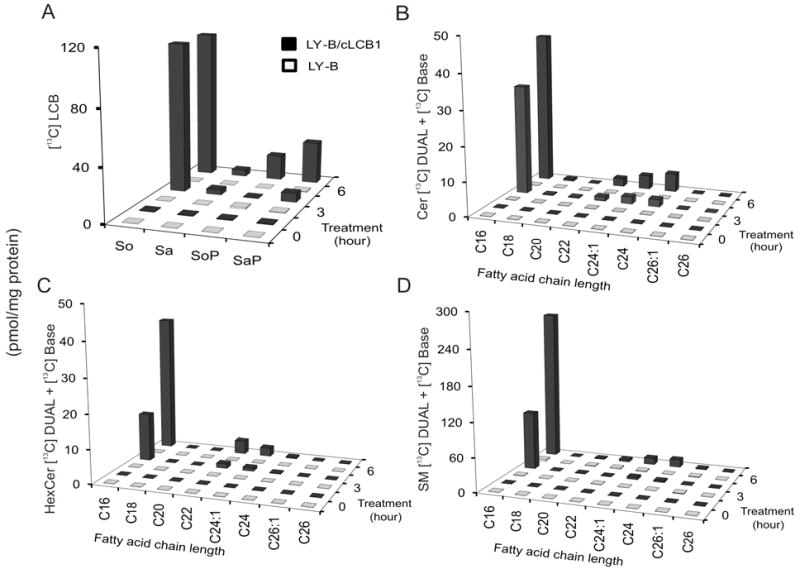
Comparison of de novo sphingolipid biosynthesis in LY-B and LY-B/cLCB1 cells. LY-B and LY-B/cLCB1 cells were incubated with 0.1 mM [13C]-palmitic acid for 0, 3 and 6 h and the sphingolipids labeled in the sphingoid base alone (BASE) plus the sphingoid base and fatty acid (DUAL) were measured by LC ESI-MS/MS. The black bars represent the sphingolipids in LY-B/cLCB1 cells while the white bars indicate the LY-B cells. (A) [13C]-labeled long-chain bases (LCB) sphingosine, So; sphinganine, Sa; sphingosine 1-phosphate, SoP; and sphinganine 1-phosphate, SaP. (B-D) provide the sum of the label in the BASE and DUAL subspecies of ceramide, Cer, monohexosylceramides, HexCer, and sphingomyelins, SM, based on variation in the chain length of the amide-linked fatty acid (shown by chain length:number of double bonds).
As has been shown before [4], although LY-B cells are defective in sphingolipid biosynthesis de novo, they contain appreciable amounts of sphingolipids if grown in medium that contains sphingolipids (e.g., from serum). In these studies, the pmol of unlabelled sphingolipids/mg protein for LY-B vs LY-B/cLCB1 cells (measured by LC ESI-MS/MS) were, respectively: 140 ± 18 vs 1073 ± 17 for Cer; 122 ± 7 vs 447 ± 87 for HexCer; and 3474 ± 452 vs 5143 ± 167 for SM (data not shown).
As another way to assess sphingoid base biosynthesis, the cells were incubated with FB1 to allow accumulation of the [13C]-labeled sphingoid bases which are otherwise rapidly N-acylated to complex sphingolipids, and thus less easily tracked. These results are shown in Fig. 4 as the relative ion yields for the precursor/product pairs as they elute from the reverse phase column. Production of [13C]-So (Fig. 4A), [13C]-Sa (Fig. 4C), [13C]-1-deoxy-Sa (Fig. 4B) and [13C]-1-desoxy-methyl-Sa (Fig 4D) were readily detected with the LY-B/cLCB1 cells, but the signal was not above background for the LY-B cells, which further confirms the conclusion from Fig. 3 that the LY-B cells do not synthesize detectable amounts of sphingoid bases de novo. The appearance of [13C]-sphingosine in LY-B/cLCB1 cells was somewhat surprising because desaturation of sphinganine is thought to occur only after N-acylation [29], but it is possible that this concentration of FB1 did not completely block [13C]-Cer biosynthesis and this [13C]-sphingosine arose from turnover.
Fig. 4.
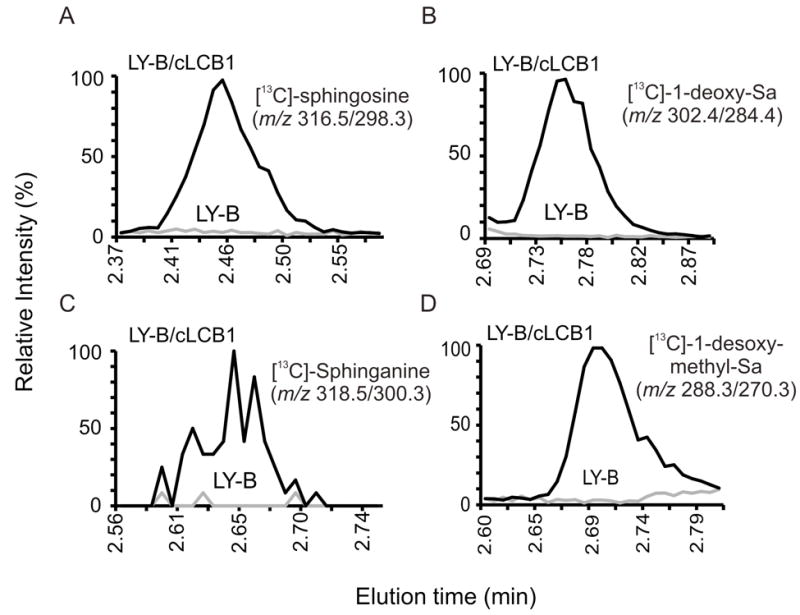
Relative amounts of typical and atypical [13C]-labelled sphingoid bases produced by LY-B and LY-B/cLCB1 cells upon co-treatment with FB1. Shown are the relative ion intensities of the shown precursor/product pairs in the LC ESI-MS/MS eluates. The LC retention times for these species matched that of standards for these compounds (not shown). The samples for these analyses were obtained by incubating LY-B and LY-B/cLCB1 cells with 0.1 mM [13C]-palmitic acid and 25 μM FB1 for 24 h; the grey line indicates LY-B cells and the black line represents LY-B/cLCB1 cells.
Partial stabilization of mutant SPT in LY-B cells
Some misfolded proteins can be stabilized by so-called chemical chaperones such as DMSO and glycerol [30-32]. When LY-B cells were treated for 24 h with DMSO or glycerol at concentrations where they have previously been effective [30,32], there were noticeable increases in both SPT1 and SPT2 by Western blotting (Fig. 5, upper), with the average increases in five experiments being between 2- and 3-fold (Fig. 5, lower). Lower concentrations had no effect, and higher were toxic (data not shown). In case these treatments also resulted in restoration of SPT activity, the incorporaton of [13C]-palmitate into sphingolipids was measured after LY-B cells had been treated with either 3% DMSO or 1 M glycerol for 24 h (the [13C]-palmitate was added to the same medium and the cells were incubated for 12 h before extraction for sphingolipid analysis by LC ESI-MS/MS). As noted earlier, the ion yields for the 13C-labelled species were very low, but by manually averaging the ion counts in the samples versus the background, we would estimate that the amounts of newly made sphingolipids (i.e., 13C-labelled in the sphingoid base backbone) were 2.0 ± 0.5 pmol/mg protein for the LY-B cells in medium alone, 1.6 ± 0.2 pmol/mg protein for the LY- cells incubated with 3% DMSO and 2.3 ± 1.2 pmol/mg protein for LY-B cells in 1 M glycerol. Therefore, there was no evidence that the small amounts of SPT1 detected upon treatment of the cells with these chemical chaperones restored SPT activity.
Fig. 5.
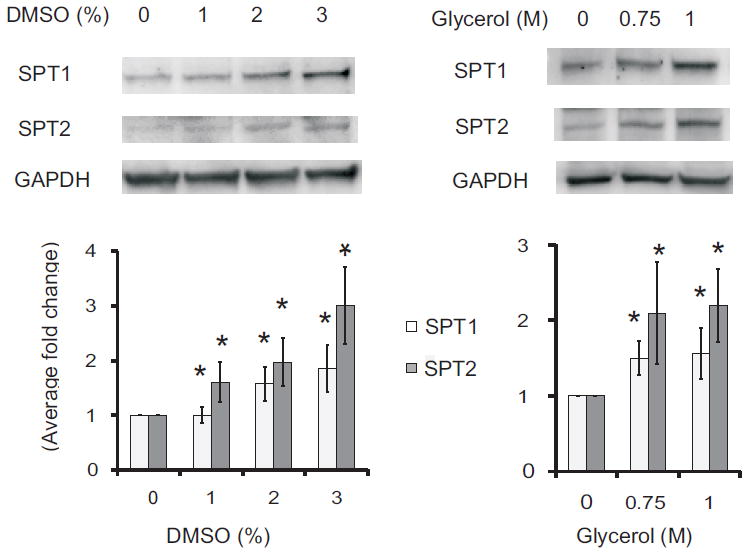
Elevation of the amounts of SPT1 and SPT2 in LY-B cells treated with chemical chaperones. LY-B cell were treated with DMSO (left panels) or glycerol (right panels) at the shown concentrations for 24 h then analyzed by Western blotting (upper panels), with GAPDH as the loading control. The lower panels are the relative differences in protein amounts as estimated from the pixel densities of the bands from five independent Western blots (bottom panels), with the standard error indicated by the bars. Ratios that were significantly different from the control (no treatment, 0) at P ≤ 0.05, as determined by the Kruskal Wallis rank sum test, are indicated by an asterisk.
Concluding comments
The finding that the sequence of the SPT1 transcript from LY-B cells has a G246R substitution is not surprising because these cells were produced using ethyl methanesulfonate [4], a mutagen that causes the random replacement of guanine(s) with adenine. Furthermore, the very low amounts of detectable mutant SPT1 polypeptide is consistent with the translated protein being unstable, which would be predicted upon localization of a positively charged Arg in a hydrophobic region of the enzyme, as suggested by the homology model [33]. It was worthwhile, nonetheless, to test for possible SPT activity given the recent discovery that SPT is able to produce not only sphinganine (and downstream sphingosine) but also the atypical sphingoid bases, 1-deoxysphinganine (from alanine) and 1-desoxymethylsphinganine (from glycine) [13,22]; furthermore, it has been suggested that these are elevated in the mutated SPTLC1 found in human sensory neuropathy type 1 (HSN1) [14]. However, the LY-B cells produced no detectable sphingoid bases of any of these types.
Therefore, for most studies, the presence of small amounts of mutant SPT1 polypeptide is not likely to interfere with the investigation. Its presence should be kept in mind, nonetheless, during investigations where the residual protein might become stabilized and complicate interpretation of studies of protein-protein interactions or other phenomena.
Acknowledgments
This work was supported by NIH grant GM076217, and MEXT grant of Japan.
Abbreviations
- 1-deoxy-Sa
1-deoxysphinganine and 1-desoxymethylsphinganine
- 1-desoxymethyl-Sa
1-desoxymethylsphinganine
- BSA
Bovine serum albumin
- CHO
Chinese hamster ovary
- Cer
Ceramide
- DMSO
Dimethyl sulfoxide
- FBS
Fetal bovine serum
- FB1
Fumonisin B1
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- HA
Human influenza hemagglutin
- HexCer
Hexosylceramide
- HSN1
Hereditary sensory neuropathy type 1
- LCB
Long chain base
- LCB1
Long chain base subunit 1
- LCB2
Long chain base subunit 2
- LC ESI-MS/MS
Liquid chromatography electrospray ionization mass spectrometry
- LY-B
Chinese hamster ovary cells strain with defective SPT
- LY-B/cLCB1
LY-B cells stably transfection with LCB1 cDNA
- PBS
Phosphate buffer saline
- PDB
Protein data bank
- Sa
Sphinganine
- SaP
Sphinganine 1-phosphate
- SM
Sphingomyelin
- So
Sphingosine
- SoP
Sphingosine 1-phosphate
- SPT
Serine palmitoyltransferase
- SPT1
Serine palmitoyltransferase subunit 1
- SPT2
Serine palmitoyltransferase subunit 2
- SPTLC1
serine palmitoyltransferase long chain base subunit 1
- SPTLC2
Serine palmitoyltransferase long chain base subunit 2
- TBST
Tris buffered saline Tween-20
References
- 1.Merrill AH, Jr, Wang MD, Park M, Sullards MC. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem Sci. 2007;32:457–68. doi: 10.1016/j.tibs.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, Merrill AH., Jr Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J Lipid Res. 2008;49:1621–39. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 4.Hanada K, Hara T, Fukasawa M, Yamaji A, Umeda M, Nishijima M. Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J Biol Chem. 1998;273:33787–94. doi: 10.1074/jbc.273.50.33787. [DOI] [PubMed] [Google Scholar]
- 5.Gable K, Slife H, Bacikova D, Monaghan E, Dunn TM. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J Biol Chem. 2000;275:7597–603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- 6.Hornemann T, Richard S, Rutti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem. 2006;281:37275–81. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 7.Hojjati MR, Li Z, Jiang XC. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim Biophys Acta. 2005;1737:44–51. doi: 10.1016/j.bbalip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Bejaoui K, Wu C, Scheffler MD, Haan G, Ashby P, Wu L, de Jong P, Brown RH., Jr SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat Genet. 2001;27:261–2. doi: 10.1038/85817. [DOI] [PubMed] [Google Scholar]
- 9.Dawkins JL, Hulme DJ, Brahmbhatt SB, Auer-Grumbach M, Nicholson GA. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat Genet. 2001;27:309–12. doi: 10.1038/85879. [DOI] [PubMed] [Google Scholar]
- 10.Tatzelt J, Zuo J, Voellmy R, Scott M, Hartl U, Prusiner SB, Welch WJ. Scrapie prions selectively modify the stress response in neuroblastoma cells. Proc Natl Acad Sci U S A. 1995;92:2944–8. doi: 10.1073/pnas.92.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batheja AD, Uhlinger DJ, Carton JM, Ho G, D’Andrea MR. Characterization of serine palmitoyltransferase in normal human tissues. J Histochem Cytochem. 2003;51:687–96. doi: 10.1177/002215540305100514. [DOI] [PubMed] [Google Scholar]
- 12.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–24. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Zitomer NC, et al. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J Biol Chem. 2009;284:4786–95. doi: 10.1074/jbc.M808798200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornemann T, Penno A, von Eckardstein A. The accumulation of two atypical sphingolipids cause hereditary sensory neuropathy type 1 (HSAN1) Chemistry and Physics of Lipids. 2008;154:62–62. [Google Scholar]
- 15.Iwatani M, Ikegami K, Kremenska Y, Hattori N, Tanaka S, Yagi S, Shiota K. Dimethyl sulfoxide has an impact on epigenetic profile in mouse embryoid body. Stem Cells. 2006;24:2549–56. doi: 10.1634/stemcells.2005-0427. [DOI] [PubMed] [Google Scholar]
- 16.Hegner B, Weber M, Dragun D, Schulze-Lohoff E. Differential regulation of smooth muscle markers in human bone marrow-derived mesenchymal stem cells. J Hypertens. 2005;23:1191–202. doi: 10.1097/01.hjh.0000170382.31085.5d. [DOI] [PubMed] [Google Scholar]
- 17.Wiebe JP, Kowalik A, Gallardi RL, Egeler O, Clubb BH. Glycerol disrupts tight junction-associated actin microfilaments, occludin, and microtubules in Sertoli cells. J Androl. 2000;21:625–35. [PubMed] [Google Scholar]
- 18.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 19.Malmqvist T, Mollby R. Enzymatic hydrolysis by bacterial phospolipases C and D of immobilized radioactive sphingomyelin and phosphatidylcholine. Acta Pathol Microbiol Scand [B] 1981;89:363–7. doi: 10.1111/j.1699-0463.1981.tb00201_89b.x. [DOI] [PubMed] [Google Scholar]
- 20.Brogden KA, Engen RL, Songer JG, Gallagher J. Changes in ovine erythrocyte morphology due to sphingomyelin degradation by Corynebacterium pseudotuberculosis phospholipase. D Microb Pathog. 1990;8:157–62. doi: 10.1016/0882-4010(90)90080-a. [DOI] [PubMed] [Google Scholar]
- 21.Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, Sullards MC, Merrill AH., Jr Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res. 2008 doi: 10.1194/jlr.D800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zitomer NC, et al. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxy-sphinganine: A novel category of bioactive 1-deoxy-sphingoid bases and 1-deoxy-dihydroceramides biosynthesized by mammalian cell lines and animals. J Biol Chem. 2008 doi: 10.1074/jbc.M808798200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gable K, Han G, Monaghan E, Bacikova D, Natarajan M, Williams R, Dunn TM. Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J Biol Chem. 2002;277:10194–200. doi: 10.1074/jbc.M107873200. [DOI] [PubMed] [Google Scholar]
- 24.Yard BA, et al. The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J Mol Biol. 2007;370:870–86. doi: 10.1016/j.jmb.2007.04.086. [DOI] [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 26.Venclovas C, Margelevicius M. Comparative modeling in CASP6 using consensus approach to template selection, sequence-structure alignment, and structure assessment. Proteins. 2005;61(Suppl 7):99–105. doi: 10.1002/prot.20725. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102:1029–34. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Rother J, van Echten G, Schwarzmann G, Sandhoff K. Biosynthesis of sphingolipids: dihydroceramide and not sphinganine is desaturated by cultured cells. Biochem Biophys Res Commun. 1992;189:14–20. doi: 10.1016/0006-291x(92)91518-u. [DOI] [PubMed] [Google Scholar]
- 30.Burrows JA, Willis LK, Perlmutter DH. Chemical chaperones mediate increased secretion of mutant alpha 1-antitrypsin (alpha 1-AT) Z: A potential pharmacological strategy for prevention of liver injury and emphysema in alpha 1-AT deficiency. Proc Natl Acad Sci U S A. 2000;97:1796–801. doi: 10.1073/pnas.97.4.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubota K, et al. Suppressive effects of 4-phenylbutyrate on the aggregation of Pael receptors and endoplasmic reticulum stress. J Neurochem. 2006;97:1259–68. doi: 10.1111/j.1471-4159.2006.03782.x. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi T, Uchida K, Uchida S, Sasaki S, Nihei H. Intracellular mislocalization of mutant podocin and correction by chemical chaperones. Histochem Cell Biol. 2003;119:257–64. doi: 10.1007/s00418-003-0511-x. [DOI] [PubMed] [Google Scholar]
- 33.Myers JK, Pace CN. Hydrogen bonding stabilizes globular proteins. Biophys J. 1996;71:2033–9. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


