Abstract
DNA double-strand breaks (DSBs) disrupt the continuity of chromosomes and their repair by error-free mechanisms is essential to preserve genome integrity. Microhomology-mediated end joining (MMEJ) is an error-prone repair mechanism that involves alignment of microhomologous sequences internal to the broken ends before joining, and is associated with deletions and insertions that mark the original break site, as well as chromosome translocations. Whether MMEJ has a physiological role or is simply a back-up repair mechanism is a matter of debate. Here we review recent findings pertaining to the mechanism of MMEJ and discuss its role in normal and cancer cells.
Keywords: MMEJ, end joining, homologous recombination, DNA Polθ, chromosomal translocations, microhomology
Our cells are constantly exposed to extrinsic and intrinsic insults that cause several types of DNA lesions, including highly toxic breaks inflicted on both strands of the double helix. To counteract the harmful effects of these double stranded breaks (DSBs), cells evolved specialized mechanisms to sense and repair DNA damage. The repair of DSBs is required to preserve genetic material, but misrepair of DSBs can cause local sequence alteration or gross chromosomal rearrangements. The two main mechanisms to repair DSBs are classical non-homologous end joining (C-NHEJ) and homologous recombination (HR). HR is generally considered to be an error-free mechanism because repair is templated by the homologous sister chromosome in S or G2 phase cells. C-NHEJ involves the direct ligation of DNA ends and can occur with high fidelity or is associated with small alterations at the junctions. The core components of C-NHEJ are the Ku70/Ku80 heterodimer (hereafter referred to as Ku), which binds with high affinity to DNA ends and protects them from degradation, and DNA Ligase IV (Dnl4/Lig4), which catalyzes end ligation (Table 1) [1]. Early studies of C-NHEJ deficient cells identified alternative error-prone mechanisms of end joining, often referred to as alt-NHEJ. There is still some debate over whether alt-NHEJ is comprised of multiple overlapping mechanisms; however, it is evident that one form of alt-NHEJ, known as microhomology-mediated end joining (MMEJ), involves alignment of microhomologous sequences internal to the broken ends before joining and is associated with deletions flanking the original DSB. The mutagenic repair of DSBs by MMEJ likely contributes to the plasticity of genomes, but also has the potential to drive carcinogenesis and consequently has fueled interest in understanding the mechanism(s).
Table 1.
Eukaryotic end joining proteins
| Category | Function | S. cerevisiae protein | Mammalian protein | C-NHEJ | MMEJ | HR |
|---|---|---|---|---|---|---|
| DSB recognition | End binding, component of DNA- dependent protein kinase (DNA-PK) |
Yku70-Yku80 | Ku70-Ku80 | + | − | − |
| Poly-ADP ribose polymerase |
− | PARP1 | − | + | − | |
| End binding and tethering |
Mre11-Rad50-Xrs2 (MRX) |
Mre11-Rad50-Nbs1 (MRN) |
+ | +a | +a | |
| Nuclease | Endonuclease, 3’ exonuclease |
Mre11 | Mre11 | − | +a | +a |
| Promotes 5’ end resection |
Sae2 | CtIP | − | +a | +a | |
| Endonuclease, 5’ exonuclease |
− | Artemis | + | − | − | |
| DNA Polymerase |
Polδ scaffold | Pol32 (Polδ) | Pol3D (Polδ) | − | +/ND | +b/ND |
| PolX family | Pol4 (Polβ) | Polβ | + | − | − | |
| PolX family | − | Polμ | + | − | − | |
| PolX family | − | Polλ | + | − | − | |
| PolA family | − | Polθ | − | + | − | |
| Ligase | Lig4 catalytic subunit | Dnl4 | Lig4 | + | − | − |
| Lig4 structural scaffold | Lif1 | Xrcc4 | + | − | − | |
| Lig4 structural scaffold | Nej1/Lif2 | Xlf/Cernunnos | + | − | − | |
| Lig1 catalytic subunit | Cdc9 | Lig1 | − | +/−c | ND | |
| Lig3 catalytic subunit | − | Lig3 | − | + | − | |
| Lig3 structural scaffold | − | Xrcc1 | − | +/− | ND | |
| Kinase | DSB activated protein kinase recruited to DSBs by Ku |
− | DNA-PKcs | + | − | − |
indicates not essential in S. cerevisiae due to back-up activities but is required in mammalian cells
Pol32 is required for long tract gene conversion in yeast but not for recombination events that require short tracts of DNA synthesis
Presumed to be required for yeast MMEJ, but has not been directly tested.
ND: not determined
Evidence for NHEJ-independent joining mechanisms
Early studies in Saccharomyces cerevisiae provided the first in vivo evidence for a C-NHEJ-independent joining mechanism involving microhomologies (MH). The ends of restriction endonuclease-linearized plasmids transformed into yeast cells were found to be ligated with high fidelity by C-NHEJ (Box 1) [2]. The frequency of joining was decreased ~20-fold in Kudeficient mutants and the products recovered had sustained deletions with 3-16 bp MH at the junctions [2]. Concurrently, assays were developed to monitor repair of chromosomal site-specific DSBs in haploid cells [3, 4]. The frequency of end joining in NHEJ-defective cells was very low (<0.01%) and the junctions displayed perfect or interrupted MH of 5-18 bp, consistent with MMEJ being a backup repair mechanism [3, 4]. In contrast to many other eukaryotes studied, MMEJ in yeast is rarely associated with insertions at the junctions.
Box 1. Extrachromosomal and chromosomal MMEJ assays.
Plasmid-based assays
Restriction endonuclease-linearized plasmid DNA transformed into competent yeast cells can be end joined and maintained as an episome if it contains a selectable marker and a replication origin [2]. To specifically study MMEJ, oligonucleotides with blunt ends or ssDNA overhangs can be ligated to the linearized plasmid prior to transformation, or MH can be incorporated into the plasmid near the ends [34, 42]. Alternatively, a selectable marker and an adjacent replication origin can be co-amplified by PCR using primers that incorporate direct repeats to direct joining by MMEJ [35, 37]. Similar plasmid-based assays have been developed for mammalian cells [52, 94]. The advantage of the plasmid-based assays is the ease of manipulating the ends of the substrate to create different lengths of homology, mismatches within the MH and addition of heterologous nucleotides at the ends.
Chromosomal assays
The rare-cutting endonucleases, HO and I-SceI, have been used to create DSBs in a chromosomal context in yeast and mammalian cells [95, 96]. By inserting two sites in inverted orientation non-compatible ends are generated, preventing repair by error-free NHEJ and forcing error-prone repair [3, 28, 29, 97]. Newer technologies using zinc-finger nucleases or CRISPR/Cas9 to create site-specific chromosomal DSBs are also being applied to study end joining [77]. Naturally occurring MH close to the DSB can be used to align the ends, or direct repeats can be engineered to flank the endonuclease cut site to specifically monitor MMEJ repair [3, 4, 33, 35]. When continuously expressed in haploid yeast, the endonuclease cleaves both sister chromatids in G2 phase cells preventing repair by HR and forcing repair by end joining in order to survive. The frequency of repair is determined by cell survival in response to endonuclease induction (haploid yeast cells) or by using reporters to detect specific repair events. Chromosomal translocation in mammalian cells can be induced when endonuclease cut sites are inserted on non-homologous chromosomes. The resulting translocations are then monitored by nested PCR in a high throughput format [60]. CSR, which is initiated by AID-induced DSBs in the IgH switch regions, is a physiological process that can occur by MMEJ. Telomere uncapping as a result of shelterin inhibition [14] or following repeat loss [13] engages robust MMEJ activity resulting in telomere fusions. Excision of the P transposable element, which leaves 17-nt overhangs with several regions of 3-8 nucleotides MH, has also been used to study end joining in Drosophila melanogaster [47].
Early evidence for the engagement of MMEJ in mammalian cells emerged from studying programmed gene rearrangements by V(D)J and class switch recombination (CSR) in developing lymphocytes. Pro B cell lymphomas of p53 null mice lacking C-NHEJ factors sustained several chromosomal translocations in which recombination-activating gene (RAG)-induced breaks at the IgH locus were joined with random breaks in the vicinity of the c-Myc locus. A significant fraction of the oncogenic translocations displayed extensive MH at the junctions [5]. Later on it was noted that robust CSR occurred in B-cells deficient for a number of C-NHEJ factors, including Ku, Xrcc4, Lig4, DNA-PKcs, and Artemis [6]. Consistent with these joining events being driven by MMEJ, the frequency and length of MH at switch junctions was elevated. Unlike CSR, V(D)J recombination is greatly inhibited in the absence of Lig4 or Ku [7, 8]. This suggests that any role for MMEJ in V(D)J recombination is rather minimal, even when C-NHEJ is blocked. However, later studies highlighted an intriguing role for the RAG endonuclease in blocking MMEJ. Expression of a mutated form of RAG, that has lower affinity for DNA, activated MMEJ and facilitated V(D)J recombination, both in wild-type and in DNA-PK deficient cells [9]. This analysis provided the first indication that C-NHEJ and MMEJ could coexist in certain settings. Subsequent studies showed that MMEJ is used in human primary cells at substantial levels when both HR and NHEJ are active, further supporting the coexistence of MMEJ and NHEJ [10].
Early evidence for the activation of MMEJ at uncapped telomeres emerged from the analysis of telomerase-deficient mice and from the realization that telomere fusions persisted in the absence of Lig4 and DNA-PKcs [11]. These studies also hinted that MMEJ might play a role in aberrant repair of uncapped telomeres during the early stages of human cancers. Sequence analysis of telomere fusions in multiple human malignancies identified MH and deletions that extended into the adjacent non-telomere DNA [12, 13]. Insight into the mechanism by which mammalian telomeres suppress MMEJ came from genetic manipulation of the protective protein complex – termed shelterin – that binds to telomeric TTAGGG repeats. [14, 15]. Extensive analysis in mouse cells indicated that MMEJ is repressed in a highly redundant manner; its activity is fully unleashed only upon deleting the entire six-subunit shelterin complex in cells that also lack Ku [14].
Biochemical support for MMEJ came from fractionation of extracts from calf thymus or HeLa cells identifying fractions with distinct end-joining activities: one fraction was able to ligate linear DNA duplexes with no homology, or ends with small MH, while the other exclusively joined fragments via MH [16-18]. Subsequent studies showed that extracts prepared from NHEJ-deficient cells were proficient for MMEJ confirming that it is a biochemically distinct mechanism [19-21].
A mechanistic view of MMEJ
Much of our understanding of the mechanism of MMEJ derives from genetic analysis in model systems and from in vitro assays using fractionated extracts or purified proteins. The mechanism proposed involves 5’-3’ resection of the DNA ends, annealing of MHs, removal of heterologous flaps, gap filling DNA synthesis and ligation, each of which is discussed in detail below (Fig. 2).
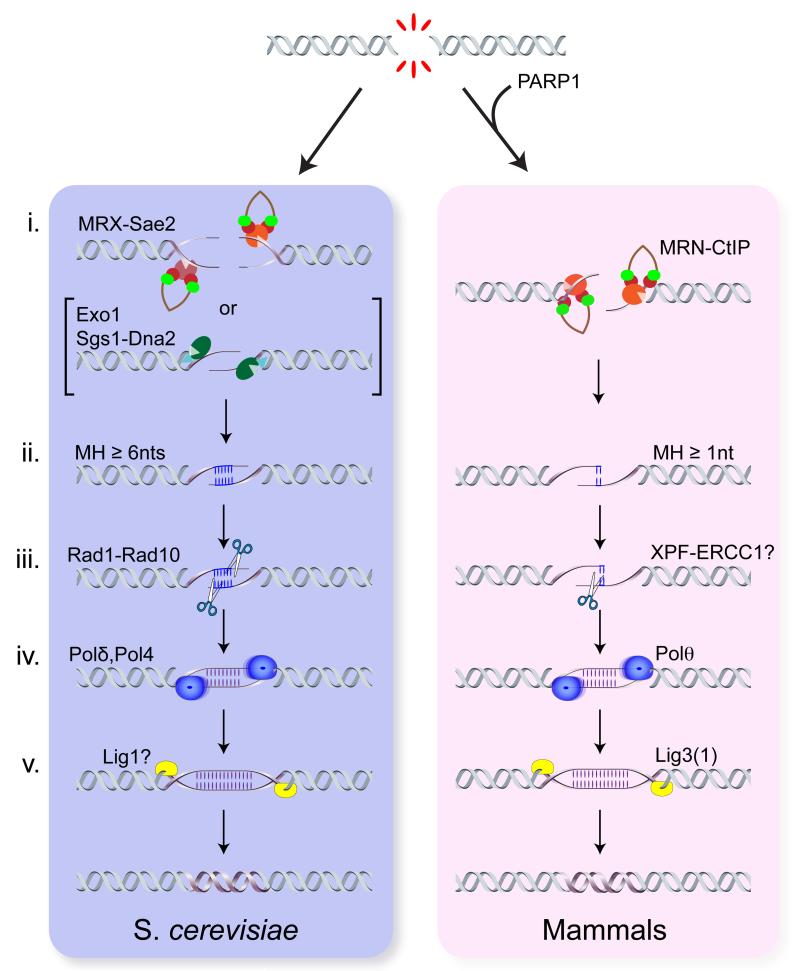
Figure 2. Mechanistic basis for MMEJ.
The illustration highlights the similarities and differences between budding yeast and mammals. The mechanism in both organisms involves i. end resection, ii. annealing of MHs, iii. flap removal, iv. fill-in synthesis, and v. ligation (see text for details). In S. cerevisiae, Exo1 or Sgs1-Dna2 can substitute for MRX and Sae2 to initiate end resection at endonuclease-induced DSBs.
End resection
Nucleolytic processing of the 5’-terminated strands at DSBs, referred to as end resection, is an essential step for HR and is required to expose MH internal to the DNA ends for MMEJ. Studies in S. cerevisiae have shown that the Mre11-Rad50-Xrs2 (MRX) complex and Sae2 initiate end resection by Mre11-catalyzed endonucleolytic cleavage of the 5’ strand internal to the DNA end [22, 23]. The resulting nick acts as entry site for bidirectional resection by the 3’-5’ exonuclease activity of Mre11 and the 5’-3’ exonuclease Exo1. The Dna2 nuclease, functioning in concert with the Sgs1/BLM helicase, acts redundantly with Exo1 to generate long tracts of ssDNA [24].
The initial cleavage by MRX and Sae2 is essential to remove covalent adducts, such as bound proteins and hairpin-capped ends. However, DSBs produced by endonucleases can be resected by Sgs1-Dna2 in the absence of the Mre11 nuclease or Sae2 after a delay of ~30 min. In Schizosaccharomyces pombe and vertebrate cells, there is a greater requirement for the MRN complex (Nbs1 is the functional ortholog of Xrs2) and Ctp1/CtIP (Sae2 ortholog) to initiate end resection and for HR, even at endonuclease-induced DSBs, than observed in S. cerevisiae [25-27].
Studies using mammalian cells deficient for the MRN complex, the Mre11 nuclease activity or CtIP confirmed that resection initiation is required for MMEJ [10, 28-32]. The decrease in MMEJ paralleled the defect in HR in agreement with end resection being an essential step for both processes [10, 33]. There are conflicting results on the role of resection initiation for MMEJ in yeast; some studies show no MMEJ defect in the absence of MRX, and others show a >5-fold decreased frequency of MMEJ [3, 34-37]. As noted previously, loss of MRX, Mre11 nuclease or Sae2 does not prevent resection in yeast, but does delay initiation due to inefficient recruitment of Exo1 and Dna2 to ends [24]. The delay to resection initiation could explain the reduced MMEJ observed in some studies. A significant increase in the frequency NHEJ has been reported for sae2 and mre11-H125N (nuclease defective) mutants, consistent with the delay in resection initiation, and consequently the proportion of MMEJ events among repaired products is low even though the actual MMEJ frequency is similar to wild-type cells [35, 36, 38]. Long-range resection by Exo1 and Sgs1/BLM-Dna2 is not required for MMEJ except when MHs are separated by >2 kb [10, 35, 39].
Annealing of microhomologies
One major difference between yeast and mammalian cells is the extent of MH required to drive MMEJ. C-NHEJ-independent junctions analyzed in mammalian cells and Caenorhabditis elegans can have as few as 1 nucleotide (nt) of homology [40, 41], whereas events attributed to MMEJ in yeast generally exhibit ≥6 nt of homology [36]. In a systematic study of MMEJ between direct repeats flanking a site-specific chromosomal DSB in budding yeast, MMEJ between 6 bp repeats was undetectable and there was a 10-fold increase in the frequency of repair for every base pair added between 12 and 17 [39]. By contrast, only 6-10 nt terminal repeats or overhangs are sufficient to drive MMEJ using transformation-based assays (Box 1) [37, 42]. Mismatches within the MH or low GC content also impede MMEJ indicating that MMEJ is driven by the thermostability of base pairing in yeast [37, 39, 42].
Replication protein A (RPA), the main eukaryotic ssDNA binding protein, removes secondary structure from ssDNA and prevents annealing of complementary oligonucleotides in vitro [43]. A large increase in the frequency of MMEJ repair (up to 350-fold higher) was reported for rfa1 mutants that encode proteins with reduced ssDNA binding affinity (RFA1/RPA1 encodes the largest subunit of the RPA heterotrimeric complex), consistent with an inhibitory role for RPA in preventing annealing between MH in yeast [35]. Rad52 anneals complementary ssDNA in vitro and is required for the single-strand annealing (SSA) mechanisms of end joining (Fig 1) [44]. Although the transformation-based assays identified a role for Rad52 in annealing MH of >8 nt [37, 42], Rad52 is not required for MMEJ of a chromosomal DSB flanked by repeats of <14 nt and is even inhibitory to MMEJ [3, 35, 39]. There appears to be a transition at around 14 nt where the inhibitory effect of RPA on strand annealing is counteracted by the stimulatory role of Rad52. Thus, Rad52-independence is one way to distinguish MMEJ from SSA [44].
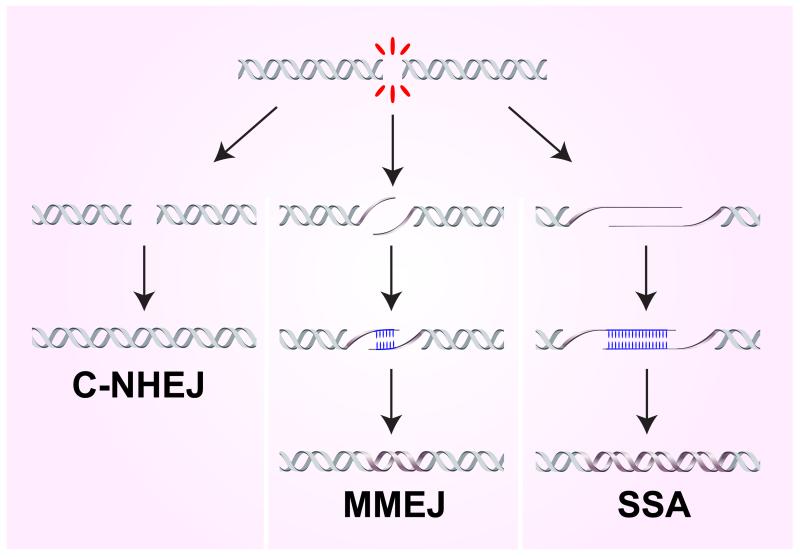
Figure 1. Mechanisms of end joining.
Classical non-homologous end-joining (C-NHEJ) involves no homology or only 1-4 nucleotides of homology at the junction; microhomology mediated end-joining (MMEJ) requires 1-16 nt of homology internal to the ends to align them for repair; and single-stranded annealing (SSA) involves annealing between more extensive homologies provided by direct repeats flanking the DSB. MMEJ and SSA are both highly mutagenic due to loss of one repeat and the intervening sequence.
The lower reliance on stable base pairing for MMEJ in mammalian cells indicates that dedicated proteins are involved in synapsis and extension of annealed MH. Poly ADP-ribose polymerase 1 (PARP1) is activated by DSBs and is able to compete with Ku for DNA end binding [19]. Extracts from PARP1−/− cells were shown to be deficient in synapsis and joining of linear DNA fragments by MMEJ, and purified PARP1 was able to tether DNA fragments, supporting a role for PARP1 in MMEJ [20]. Furthermore, PARP1 inhibition reduces MH at junctions formed during CSR, yields lower frequencies of IgH-Myc and restriction endonuclease-induced translocations, and reduces end joining in Ku deficient settings [5, 14, 45, 46].
Fill-in synthesis: the role of promiscuous polymerases
Once resected ends successfully anneal via complementary base pairing, the flanking single-stranded regions are then subject to fill-in synthesis. This is a key step that stabilizes paired-intermediates and commits a DSB to MMEJ. A distinguishing feature of MMEJ in metazoans is the presence of nucleotide insertions at break sites following repair. Insertions generate junctional diversity and contribute to the increased mutagenicity of the end-joining reaction. Insertions are often derived from sequences close to the breaks and in some cases are copied from other chromosomes. Additionally, a number of nucleotides can be added de novo as a result of non-templated extension of the 3’-termini of breaks. One can envision that when the ends of a break are incompatible, random nucleotide insertions could generate necessary MH and therefore promote base pairing of DNA ends.
The main DNA polymerases for MMEJ in S. cerevisiae are the replicative DNA polymerase, Polδ, and Pol4 (Polβ). The pol32 mutant, lacking a non-essential subunit of Polδ, exhibits the most severe MMEJ defect of the polymerase mutants tested [36]. Pol32 also associates with the Polζ translesion synthesis (TLS) polymerase, but deletion of REV3, encoding the catalytic subunit of Polζ, did not decrease MMEJ supporting a role for Polδ [39]. Pol4 is an X family DNA polymerase that participates in NHEJ events in which gaps must be filled to extend from 3’ overhangs, and has a minor role in MMEJ [36, 37].
Numerous genetic studies highlight a central role for the TLS polymerase theta (Polθ, encoded by PolQ) in stimulating MMEJ in higher organisms. Early reports suggested that Drosophila melanogaster Polθ promotes MMEJ at DSBs induced either during P-element transposition or by a sequence-specific endonuclease [47, 48]. In C. elegans, polq-1 is required for MMEJ in response to replication-fork collapse, especially at G-rich DNA [40, 49]. Recent reports linked mammalian Polθ to MMEJ activity at dysfunctional telomeres [50], during chromosomal translocation in mouse embryonic stem cells [50], and following endonuclease-mediated cleavage of reporter constructs (Box 1) [50-53]. No reduction in the frequency of CSR was found for PolQ−/− mice, in NHEJ proficient or deficient settings, and the length of MH at junctions was unaffected; however, Polθ was shown to be required for insertion of nucleotides during joining [52].
Polθ has a unique structure, as it is the first DNA polymerase to be identified that has an intrinsic SF2 helicase-like domain at its N terminus, which is separated from the C-terminal polymerase domain by a large central domain (Fig. 3) [54]. The function of the helicase-like domain is not fully understood. Drosophila Mus308 mutants with a substitution in a conserved residue of the helicase domain have reduced microhomolgy at MMEJ-mediated repair events [47], suggesting that the helicase domain of polθ is an important regulator of end joining. Biochemical analysis identified robust single-stranded DNA-dependent ATPase activity but failed to reveal any DNA unwinding activity [54]. It is possible that the helicase acts on a particular class of DNA substrate(s) that has not been identified thus far. Alternatively, the ATPase activity could behave similarly to the annealing helicase HARP/SMARCAL1 [55], displacing stably bound RPA from ssDNA. The loss of RPA is expected to stabilize annealed intermediates and promote MMEJ. With regards to the central domain, a Rad51-interaction motif was recently identified, and appears to act together with the ATPase domain to counteract Rad51 filament assembly and D-loop formation in vitro [51]. It will be important to address whether the observed anti-Rad51/anti-recombinase activity of Polθ is functionally linked to its role in MMEJ.
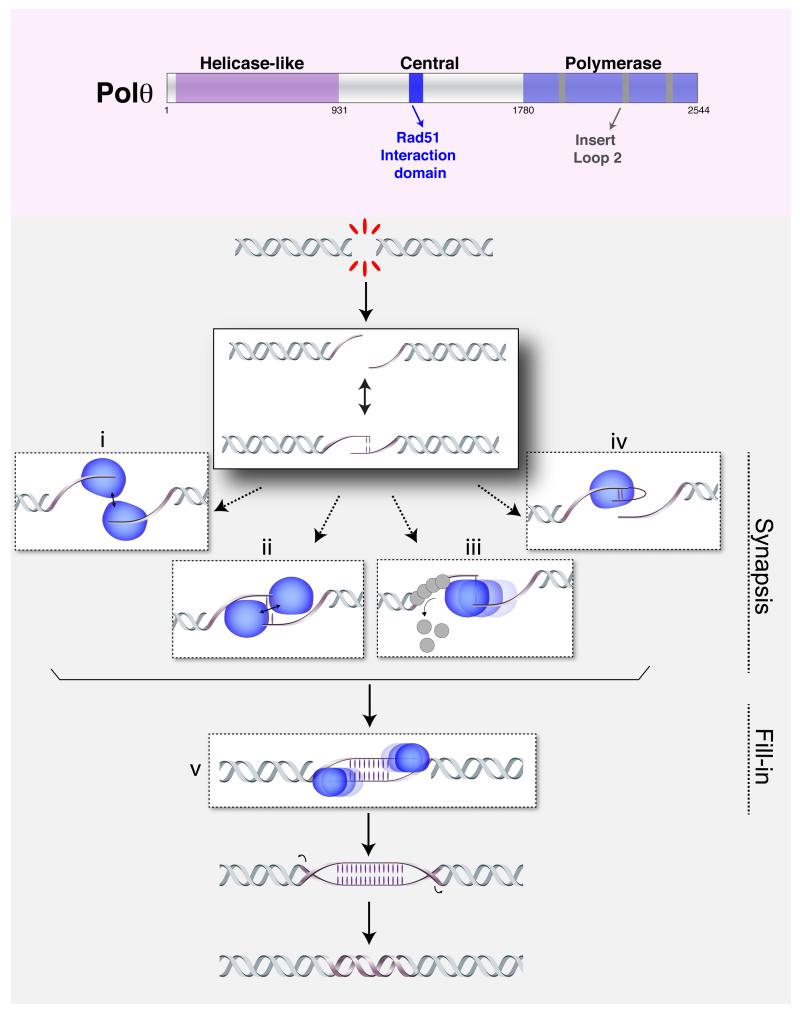
Figure 3. Model for Polθ activity.
Upper panel depicts the different domains of Polθ including the helicase-like domain at the N terminus, the polymerase domain at the C terminus, and the central domain that contains the Rad51 interaction motif. The polymerase domain contains an insert loop 2 that is necessary for DNA lesion bypass, ssDNA extension, and MMEJ. In the lower panel, we propose a model for the mechanism by Polθ (in blue) promotes MMEJ Nucleolytic processing of a DSB exposes MH, which could drive annealing of single stranded overhangs. Polθ potentially employ its different domains in order to promote formation and/or stabilization of the annealed intermediate. For example, its dimerization will tether DNA ends together (i) or even stabilize spontaneously annealed ends (ii). The ATPase domain could potentially displace ssDNA-binding proteins (iii). Lastly, the polymerase could extend resected ends, potentially in a snap-back mechanism, thereby generating additional MH (iv). In addition to promoting synapsis, Polθ activity is required for fill-in synthesis (v), a crucial step during MMEJ.
The C-terminal fragment of Polθ encodes a proofreading-deficient polymerase with two discernable activities. The promiscuous polymerase copies DNA in a template-dependent manner, but also extends mismatched termini and ssDNA [56]. Extension of ssDNA can be carried out using “snap-back” replication [53], or by copying from another template through cycles of slippage and re-priming [52]. The polymerase domain includes a conserved insertion loop domain (insert-loop 2) that is essential for both lesion bypass and extension of ssDNA. In addition, biochemical experiments indicated that insert-loop2 is essential for MMEJ, potentially promoting formation of Polθ dimers or multimers that then drive DNA synapsis during end-joining [53, 57]. Structural analysis confirmed that Polθ exists as a dimer, although the dimerization interface did not include insert-loop2 [58]. Dimerization of Polθ could stabilize annealed overhangs, even ones with as little as 1-2 complementary bases. This end-synapsis activity of vertebrate Polθ is reminiscent of archaeal NHEJ polymerase [59], and could explain the limited MH requirement for vertebrate MMEJ, especially when compared to yeasts, which lack a POLQ gene.
Removal of heterologous flaps
In the transformation-based yeast MMEJ assays the overhangs or repeats are at the ends of linear DNA fragments; therefore, no trimming is required for the 3’ ends to be extended by DNA synthesis. Addition of even a single heterologous nucleotide at each end of the linear transforming DNA reduced transformation efficiency by 50%, and 7 nt reduced MMEJ by >10-fold [37]. All of the chromosomal assays require heterologous tail removal and this could contribute to the low MMEJ frequency in yeast. Heterologous tails appear to be less inhibitory to MMEJ in mammalian cells where the efficiency of MMEJ at 8-9 bp repeats flanking an I-SceI-induced chromosomal DSB is >100-fold higher than observed at a similarly designed construct in yeast [10, 33, 35]. For MH internal to DNA ends, heterologous 3’ flaps form following annealing, and must be removed to permit DNA polymerases to extend from and stabilize the annealed MHs. In yeast this function is fulfilled by the Rad1-Rad10 endonuclease (XPF-ERCC1 in vertebrates) [3, 37]; however, murine Ercc1−/− cells exhibit only a minor MMEJ defect suggesting other nucleases can compensate [33].
Sealing the ends
MMEJ is completed by the final ligation step where DNA ends are covalently attached to each other. As MMEJ occurs in mammalian cells in the context of Lig4 deficiency, much effort has been geared towards understanding the role of Lig3 and DNA ligase I (Lig1) in end joining. Early biochemical experiments and plasmid-rejoining assays identified Lig3 as the major contributor to MMEJ [20, 21]. These results were later corroborated by in vivo results. For example, mouse cells lacking nuclear Lig3 displayed a reduced frequency of MMEJ-mediated chromosomal translocation [60]. The residual translocation events were devoid of MH and further reduced upon deletion of Lig1 [60]. These results indicate that in mouse cells, Lig3 supports MMEJ even when C-NHEJ is intact, and that Lig1 acts as a back-up enzyme. Lig3 is also the major ligase that drives the joining of dysfunctional telomeres in human cells experiencing telomere attrition [61] and in mouse cells following removal of shelterin [14]. The situation is different in B cells; knockdown of Lig3 in either wild type or Lig4-deficient setting did not lead to any reduction in CSR nor did it impact the extent of microhomology [62]. These data could be explained by possible redundancy between Lig3 and Lig1, although one cannot completely rule out that residual Lig3 is sufficient to support CSR. The role of Xrcc1, a Lig3 scaffold protein, in MMEJ is controversial [20, 62].
Intriguingly, Lig3 only plays a central role in MMEJ in higher organisms, since the gene is not found in yeast. MMEJ in yeast is independent of Lig4 [35, 37, 39] and it will be important to assess the requirement for Lig1 in mediating these events.
Decision time: regulating DNA repair pathway choice
MMEJ and HR both initiate by end resection raising the issue of how cells “choose” between different repair pathways. The choice between HR and C-NHEJ has been extensively studied and is linked to the cell cycle and end resection (Fig. 4). HR predominates in S-G2 phases when the sister template is available, whereas C-NHEJ operates in all stages of the cell cycle and is the preferred pathway in G1 when resection activity is low [63, 64]. Sae2/CtIP is activated by cyclin-dependent kinase (CDK) to ensure resection initiation is coordinated with DNA replication [65, 66]. Preventing the initiation of resection increases the efficiency of NHEJ repair [27, 35, 36, 67]; conversely, activation of end resection by elimination of Ku, 53BP1, Rif1, or Rev7 favors HR [24, 68-71].
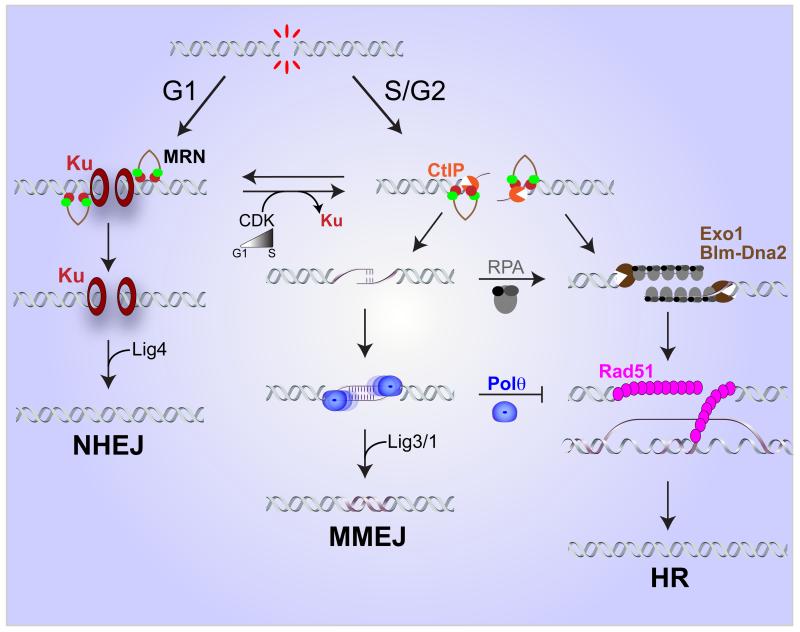
Figure 4. Repair pathway choice.
The choice between C-NHEJ, MMEJ and HR is regulated by multiple factors and influenced by the cell cycle. Low CDK activity in G1 enables Ku-dependent NHEJ activity. Increased CDK in S/G2 promotes limited resection by MRN-CtIP and results in a concomitant loss of Ku binding to DSBs. This enables both MMEJ and HR to act. Extensive resection by Exo1 or Blm-Dna2, binding of RPA and subsequent loading of Rad51 are required for HDR. By contrast, the activity of Polθ favors MMEJ, and inhibits HR. In S. cerevisiae RPA inhibits MMEJ by counteracting annealing between MH.
The choice between C-NHEJ and MMEJ seems to be largely influenced by Ku and the cell cycle. The higher affinity of Ku for ends compared with PARP1 could explain the predominance of C-NHEJ. One can envision that C-NHEJ rapidly ligates ends that are compatible, but ends that need additional processing and eventually undergo resection would escape Ku binding and become available for MMEJ or HR. HR and MMEJ both display maximal activity in S-phase and share the initial resection step mediated by Mre11 and Sae2/CtIP [10, 72]. How do cells keep MMEJ in check and preserve genomic stability? As noted above, resection occurs in two steps: initiation of resection by MRX/N and Sae2/CtIP, and extensive resection catalyzed by Exo1 or Sgs1/BLM-Dna2. Resection initiation in the absence of extensive resection creates a situation favoring MMEJ [10, 35]. RPA binds to the ssDNA formed by end resection and plays an important role in preventing MMEJ in yeast; moreover, by removing secondary DNA structure RPA facilitates extensive resection and Rad51 nucleoprotein filament formation favoring HR [35, 73]. Elimination of Rad51 results in an increased frequency of MMEJ in yeast suggesting HR removes substrates that can be channeled to MMEJ repair [35, 39]. In contrast, depletion of PolQ results in an increased frequency of HR, which could be due to MMEJ competing with HR for resection intermediates or due to disruption of Rad51 filaments by Polθ [50, 51]. Future studies aimed at fully understanding pathway choice control are essential and will provide new insights into the mechanistic basis of DNA repair.
MMEJ and cancer
Interest in the mechanisms and regulation of end joining has grown with the finding that tumor cells have multiple structural alterations to chromosomes. Translocations are particularly common and in some cases form pathological gene fusions that drive tumorigenesis [74]. DNA sequence analysis of tumor cells has shown rearrangements have either no homology or short MH at the junctions suggesting they originate by an end joining mechanism. The frequency of translocations induced by targeted DSBs on non-homologous chromosomes is increased in Lig4−/− and Ku70−/− mouse cells, and no alteration is observed in MH usage at the junctions, suggesting translocations arise by MMEJ [41]. Consistent with a role for MMEJ in formation of translocations, CtIP-depleted, PolQ−/−, Lig3−/− or PARP inhibitor treated cells show reduced frequencies of translocations and have a reduced amount of MH at the translocation junctions [32, 46, 60]. Studies of irradiation-induced translocations in NHEJ-deficient mouse embryonic fibroblasts also point to an important role for PARP1 and Lig3 in their formation [75]. However, endonuclease-induced translocations in human cells show lower MH usage than observed in mouse cells and the frequency is reduced in the absence of LIG4, suggesting that they are catalyzed by NHEJ and not MMEJ [76, 77].
Expression analyses suggest that certain cancers may be genetically predisposed towards MMEJ. For example, LIG3 and PARP1 display higher steady state levels in therapy-resistant breast cancer cell lines [78] and chronic myeloid leukemia [12, 79]. Genomic rearrangements consistent with MMEJ have also been noted in a number of hematological [12] and solid tumors, most notably being HR deficient cancers [80]. In addition, ovarian tumors with alterations in HR genes exhibit elevated levels of POLQ. Knockdown of POLQ [51], similar to PARP inhibition [81], is highly effective in killing HR-deficient cells, raising the possibility that cell proliferation in the absence of HR is dependent on MMEJ.
The physiological role of MMEJ
Ultimately, the physiological function of MMEJ remains unclear. The poor efficiency of MMEJ in budding yeast, even in Ku deficient settings, is consistent with it simply acting as a back-up pathway. Notably, Lig3, PARP1 and Polθ are absent from yeast, potentially accounting for the rare use of this mechanism, reliance on more extensive MH and lack of insertions at junctions. However, the frequency of MMEJ in metazoans is significantly higher than in yeast and the pathway can act even when HR and NHEJ are intact. The evolutionary conservation of this highly mutagenic mode of repair hints at an intrinsic function for MMEJ in higher organisms. Potentially, MMEJ activity could boost genetic variation and drive genome evolution. Circumstantial evidence in support of this notion can be derived from whole-genome sequencing. For example, massive paired-end mapping of Structural Variations (>3KB) in the human genome and subsequent analysis of break points revealed significant MH at junctions that were likely generated through an end-joining event [82]. In an independent study, comparative analysis between different subspecies of Caenorhabditis and Drosophila revealed elevated levels of MH at sequences flanking introns that were eliminated during metazoan evolution [83]. Intriguingly, a direct role for PolQ in driving genome evolution was recently reported in C. elegans. Sequencing of wild type and polq-1 mutant animals followed over ~50 generations implicated PolQ-mediated MMEJ in generating small indels frequently observed in wild isolates [84].
MMEJ also impacts transposon mobilization. Excision of DNA transposons leaves a DSB at the donor site, which--in the case of P-element in D. melanogaster [47] and Tc1 in germ cells of C. elegans [84]--is dependent on Polθ-mediated MMEJ for repair. Experiments also link MMEJ to an endonuclease-independent integration mode by which LINE1 retrotransposons invade mammalian genomes. This mechanism often yields truncated insertions with increased levels of microhomology, and is elevated in cells lacking core C-NHEJ factors. [85, 86].
Lastly, it is conceivable that MMEJ contributes to the stability of repetitive DNA. Around 45% of human genome is comprised of repetitive elements (terminal repeats, tandem repeats and transposable elements) that are at risk of producing toxic genomic rearrangements following DSB formation. Small deletions and insertions are potentially less harmful than repeat expansion and loss or translocation due to uncontrolled HR between repetitive elements.
Concluding remarks
During the past two decades, MMEJ activity has been reported in bacteria [87, 88], plants [89], yeasts [90], worms [91], flies [48], mice [92], and human cells [93]. While MMEJ was initially thought to act as a back-up pathway, later studies show that it is used even when HR and NHEJ are intact, and it seems to become essential in HR-defective backgrounds. Considerable progress has been made in the identification of key MMEJ factors and so far, all appear to have overlapping functions with other repair pathways, including HR (resection nucleases), interstrand crosslink repair (Polθ), base excision repair (PARP1, Lig3), thus complicating genetic studies to elucidate the physiological role of MMEJ. Disclosing the inherent function of this pathway in normal cells may require separation of function mutants or the identification of factors that are specific for MMEJ. A complete understanding of the molecular basis of MMEJ may provide novel avenues for cancer therapy, especially in HR-deficient tumors.
Outstanding Questions.
Is MMEJ comprised of several overlapping mechanisms? In yeast, MMEJ appears to be a back-up mechanism to resolve DSBs in the absence of higher fidelity HR and NHEJ, whereas MMEJ in metazoans relies on factors absent from yeast and requires less extensive MH suggesting there might be distinct mechanisms that share the end resection step.
Do PARP1, Lig3 and Polθ function together to promote MMEJ?
Is the inhibitory role of RPA on MMEJ restricted to yeast?
Is Polθ dimerization required for efficient MMEJ?
Is the elevated frequency of HR observed in Polθ-depleted cells due to Polθ inhibition of Rad51 or removal of the competing MMEJ pathways? Elevated HR is not observed in Lig3-depleted cells suggesting this is a unique function of Polθ.
What is the complete list of factors that are required for MMEJ?
How is the balance between MMEJ and HR regulated to maintain genome integrity?
Is MMEJ important for maintaining the stability of repetitive DNA?
TRENDS.
Microhomology-mediated end joining (MMEJ) is a mutagenic double-strand break (DSB) repair mechanism that uses 1-16 nt of homology flanking the initiating DSB to align the ends for repair.
MMEJ is associated with deletions and insertions that mark the original break site, as well as chromosome translocations.
RPA prevents MMEJ in by inhibiting annealing between microhomologies exposed by end resection
Recent studies implicate DNA polymerase θ (encoded by PolQ) in a subset of MMEJ events, particularly those associated with insertions at the break site.
DNA Polymerase θ is a promising therapeutic target to treat homologous recombination deficient tumors.
Acknowledgements
We thank S. K. Deng and W. K. Holloman for comments on the manuscript. We are grateful to Eliot Sela for the double helix illustration. Research in our laboratories cited here was supported by grants from the National Institutes of Health (R01 GM041784 and P01 CA174653 to L.S.S, DP2CA195767 and R01DK102562 to A.S.). A.S. is a Pew-Stewart Scholar.
Glossary
- Classical non-homologous end joining (C-NHEJ)
Ku and Lig4-dependent ligation of DSBs. Repair can be precise or associated with small insertions or deletions at the cut site.
- Homologous recombination (HR)
accurate repair involving Rad51-dependent invasion of a homologous donor duplex by the ends of the broken chromosome to prime DNA synthesis to restore sequence at the DSB. The donor duplex can be the sister chromatid after DNA synthesis, homologous chromosome in diploids or repeated sequence elsewhere in the genome.
- Microhomology-mediated end joining (MMEJ)
Ku and Lig4 independent joining of DSBs using microhomologies (1-16 nt) internal to the ends to align them for repair; associated with deletions at the break site.
- Single-strand annealing (SSA)
A mutagenic Rad51-independent repair mechanism that operates between long direct repeats flanking a DSB and results in loss of one of the repeats and intervening sequence. SSA shares some features with MMEJ, the main difference being the amount of homology used to align ends (>30 nt) and requirement for Rad52.
- Translesion synthesis (TLS) DNA polymerases
error-prone DNA polymerases that show reduced fidelity and processivity, and are able to incorporate nucleotides opposite lesions that block replicative DNA polymerases.
- Translocation
rearranged genetic material usually from two non-homologous chromosomes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiruvella KK, et al. Repair of double-strand breaks by end joining. Cold Spring Harbor perspectives in biology. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. The EMBO journal. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 3.Ma JL, et al. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Molecular and cellular biology. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert I, et al. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. The Journal of experimental medicine. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 7.Frank KM, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 8.Nussenzweig A, et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 9.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 10.Truong LN, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maser RS, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Molecular and cellular biology. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin TT, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 13.Letsolo BT, et al. Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements. Nucleic acids research. 2010;38:1841–1852. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rai R, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. The EMBO journal. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason RM, et al. The joining of non-complementary DNA double-strand breaks by mammalian extracts. Nucleic acids research. 1996;24:4946–4953. doi: 10.1093/nar/24.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlich B, et al. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. European journal of biochemistry / FEBS. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic acids research. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Audebert M, et al. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer research. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 22.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 23.Garcia V, et al. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 25.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langerak P, et al. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata A, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 29.Xie A, et al. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Y, et al. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee-Theilen M, et al. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennardo N, et al. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Paull TT. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA repair. 2005;4:1281–1294. doi: 10.1016/j.dnarep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Deng SK, et al. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decottignies A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics. 2007;176:1403–1415. doi: 10.1534/genetics.107.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuzaki K, et al. Cyclin-dependent kinase-dependent phosphorylation of Lif1 and Sae2 controls imprecise nonhomologous end joining accompanied by double-strand break resection. Genes to cells: devoted to molecular & cellular mechanisms. 2012;17:473–493. doi: 10.1111/j.1365-2443.2012.01602.x. [DOI] [PubMed] [Google Scholar]
- 39.Villarreal DD, et al. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012;8:e1003026. doi: 10.1371/journal.pgen.1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koole W, et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nature communications. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 41.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Molecular and cellular biology. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng SK, et al. Replication protein A prevents promiscuous annealing between short sequence homologies: Implications for genome integrity. BioEssays: news and reviews in molecular, cellular and developmental biology. 2015;37:305–313. doi: 10.1002/bies.201400161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Symington LS, et al. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansour WY, et al. The absence of Ku but not defects in classical non-homologous end-joining is required to trigger PARP1-dependent end-joining. DNA repair. 2013;12:1134–1142. doi: 10.1016/j.dnarep.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Wray J, et al. PARP1 is required for chromosomal translocations. Blood. 2013;121:4359–4365. doi: 10.1182/blood-2012-10-460527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan SH, et al. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic acids research. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roerink SF, et al. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome research. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mateos-Gomez PA, et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yousefzadeh MJ, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent T, et al. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seki M, et al. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic acids research. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hogg M, et al. Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic acids research. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hogg M, et al. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. Journal of molecular biology. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zahn KE, et al. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015;22:304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brissett NC, et al. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 60.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones RE, et al. Escape from telomere-driven crisis is DNA ligase III dependent. Cell reports. 2014;8:1063–1076. doi: 10.1016/j.celrep.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Boboila C, et al. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 63.Aylon Y, et al. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. The EMBO journal. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huertas P, et al. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008 doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. The Journal of biological chemistry. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shibata A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. The EMBO journal. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pierce AJ, et al. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clerici M, et al. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO reports. 2008;9:810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends in cell biology. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sale JE. REV7/MAD2L2: the multitasking maestro emerges as a barrier to recombination. The EMBO journal. 2015;34:1609–1611. doi: 10.15252/embj.201591697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu W, et al. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA repair. 2008;7:329–338. doi: 10.1016/j.dnarep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 73.Chen H, et al. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Molecular cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nature reviews. Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soni A, et al. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic acids research. 2014;42:6380–6392. doi: 10.1093/nar/gku298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brunet E, et al. Chromosomal translocations induced at specified loci in human stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Molecular cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tobin LA, et al. Targeting abnormal DNA repair in therapy-resistant breast cancers. Molecular cancer research: MCR. 2012;10:96–107. doi: 10.1158/1541-7786.MCR-11-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tobin LA, et al. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene. 2013;32:1784–1793. doi: 10.1038/onc.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 82.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Schendel R, Tijsterman M. Microhomology-mediated intron loss during metazoan evolution. Genome biology and evolution. 2013;5:1212–1219. doi: 10.1093/gbe/evt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Schendel R, et al. Polymerase Theta is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nature communications. 2015;6:7394. doi: 10.1038/ncomms8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zingler N, et al. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome research. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 87.Chayot R, et al. An end-joining repair mechanism in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aniukwu J, et al. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heacock M, et al. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. The EMBO journal. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic acids research. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pontier DB, Tijsterman M. A robust network of double-strand break repair pathways governs genome integrity during C. elegans development. Current biology: CB. 2009;19:1384–1388. doi: 10.1016/j.cub.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 92.Kabotyanski EB, et al. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic acids research. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic acids research. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Waters CA, et al. The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nature communications. 2014;5:4286. doi: 10.1038/ncomms5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rouet P, et al. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Molecular and cellular biology. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haber JE. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. BioEssays: news and reviews in molecular, cellular and developmental biology. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 97.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Molecular cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]