Abstract
Recent studies have revealed multiple roles of osteocytes in bone metabolism. However, detailed analyses of the embedded osteocytes in bone structure are still limited because of the high mineral content around these cells. In this study, we developed an innovative technique, the “FITC-Imaris technique”, which combines FITC ([2, 5]-Fluorescein isothiocyanate), confocal microscopy and Imaris software. With this method, we could not only visualize the 3-D morphology of embedded osteocytes, but more importantly, we were able to statistically quantitate the osteocyte structure in the cell surface, total cell volume, and dendrite numbers. Furthermore, we made a side-by-side comparison of the new method with the acid-etched SEM imaging technique, a common imaging method for studies of osteocyte morphology with a much smaller cell depth (< 3 μm). Finally, we used the FITC-Imaris technique to show both the morphological and statistical differences in the osteocyte structure between the Dmp1-null mice (the osteomalacia model) and their age-matched control littermates. We expect that this newly developed technique will become a powerful tool to disclose more roles that osteocytes play in bone health and diseases.
Keywords: Osteocyte, DMP1, FITC, Imaris, Osteocyte quantification
Introduction
Osteocytes account for more than 90% of the total number of cells in mineralized bone. However, most bone studies have focused on the surface cells, such as osteoblasts and osteoclasts, partly because these cells are more active in metabolism and are technically easier to access. However, recent studies have revealed more profound and important roles of osteocytes in addition to their role as mechanosensors. These roles include regulation of bone formation by secreting sclerostin proteins (1, 2), bone resorption through the production of a great amount of RANKL (3), control of phosphate homeostasis through FGF23 (4), and regulation of primary lymphoid organs and fat metabolism (5). Therefore, analyzing osteocytes in a more efficient and precise way is vital and necessary in bone biology studies.
One of the traditional methods of evaluating mineral-embedded bone cells is known as “acid-etched SEM”(4). Resin is packed into the non-mineralized structures during sample embedding; after acid treatment, the mineral is removed to expose the underlying resin-filled structures. Although a high-resolution, acid-etched SEM image can reveal the osteocyte-canalicular system, this technique has several limitations: 1) The optimal acid concentration and treatment time vary among samples, depending on the sample hardness and structure (thus, the process is time consuming). Oftentimes when the condition is optimized for mineralization defect samples like Dmp1-null bones, the mineral cannot be fully removed from the control samples under the same conditions; 2) The SEM technique can only be applied efficiently when the sample surfaces are polished so that all scratches are eliminated, although polishing removes many dendrites protruding from the osteocytes; and 3) Only the top layer of the osteocyte is uncovered, while cell processes deeply embedded in mineral remain hidden. Thus, a true 3-D analysis technique for osteocytes needs to be developed to enhance the understanding of the detailed osteocyte structure.
FITC is a small fluorescent molecule that penetrates all non-mineralized tissues. In bone studies, this molecule fills in the entire prospective osteocyte-canalicular system, surrounding blood vessels, and osteoid layers, as well as surface cells such as osteoblasts and osteoclasts. However, there is a lack of good quantitative methods for defining the cell surface, volume, dendrite number, and average length of the dendrite.
Dentin matrix protein 1 (DMP1) has been shown to be a key factor in osteocyte biology and bone development. Functional studies have revealed that DMP1 is essential for the maturation and function of osteocytes via both local and systemic mechanisms (4, 6, 7). Genetic research identified that DMP1 mutations in humans or its deletion in mice lead to autosomal recessive hypophosphatemic rickets, characterized by severe osteomalacia bone changes with significant reductions in bone mineralization (4). Although the osteocytes of Dmp1-null mice have been characterized by SEM, immunohistochemical staining and other methods, the morphologic changes in them still have not been quantified and analytically documented.
In this study, we developed an innovative technique, which we call the “FITC-Imaris technique”, because it combines FITC, confocal microscopy and Imaris software to statistically quantitate the osteocyte structure in the cell surface, total cell volume, and dendrite numbers. Our results revealed for the first time the statistical numbers of the detailed cell structures in healthy and Dmp1-null osteocytes. We predict that this study will have great impact on future studies of osteocyte roles in bone diseases such as osteoporosis.
Methods
Mouse, rat and human specimens
Eight-week-old Dmp1 null mice and Dmp1 heterozygote littermates were analyzed in this study. After they were sacrificed, the right side of the tibia was obtained for testing and evaluation. All animal protocols were approved by the Animal Welfare Committee at Texas A&M University Baylor College of Dentistry. In addition, the technique is further demonstrated through the use of human and rat bone.
FITC sample preparation and staining for confocal imaging
The muscle was removed from the mouse tibia and rinsed in PBS, followed by fixation in 70% ethanol for two days at room temperature with a one-time change of 70% ethanol. The bone samples then went through slow dehydration in 95% ethanol for one day and 100% ethanol for one day. The samples were stained with 1% FITC (Sigma, cat. no. F7250) in 100% ethanol overnight, followed by continuous dehydration with 100% ethanol for one more day, and acetone for 2 days; they also underwent the plastic embedding process described previously (8). Notably, during the FITC staining process, additional dehydration, and the plastic embedding process, the bones were kept away from light by wrapping them in aluminum foil. The human specimen obtained from femur cortical bone and the rat tibia were cut to cubic size by diamond saw for dehydration, which followed the same procedure as described above.
The embedded plastic blocks were sectioned using a water-cooled diamond-impregnated circular saw (Isomet, Buehler) into approximately 1–2-mm-thick slices. These slices were further sanded down to < 100 μm thickness using six grades (80, 200, 400, 600, 800, and 1200 grit) of sanding papers, and polished on a soft cloth rotating wheel with 1-μm alumina alpha micropolish II solutions (Buehler, no. 406323016). After polishing, the slides were immersed in a water-soluble mounting medium for confocal imaging and then covered with a plastic cover.
Confocal microscope imaging
The SP5 Leica confocal microscope located at Baylor College of Dentistry was used to gather stackable images. The FITC samples were subjected to 488-nm wave length excitation using an argon laser and emission at 520 nm. All the images were captured at light ranging from 500 to 540-μm wavelengths. Multiple stacked images were taken at 200 Hz and 1024x1024 dimensions using 63X glycerol objective lenses at 1.5 times magnification. For both the Dmp1 HET and the null mice, a thickness of 32 μm was obtained with a pixel size of 0.1542 μm × 0.1542 μm × 0.49 μm from the tibia cortical bone midshaft area. The cubic pixel size was revised from 0.1542 × 0.1542 × 0.1542 μm to 0.1542 × 0.1542 × 0.49 μm in order to reduce fluorescence bleaching from prolonged exposure of tissues to laser light. For human and rat specimen, a thickness of 35 μm was obtained for imaging under the same condition.
Image J, Autoquant and Imaris analyses
To obtain optical image quality, the stacked confocal images, which was initially saved as Tiff files, were converted to a multilayer Tiff file using Image J software. The images were then uploaded to Autoquant software for deconvolution. The dimensions for the pictures entered into Autoquant were set at 0.1542 × 0.1542 × 0.49 μm. Blind 3-D deconvolution was performed after selecting the magnification and the optical medium for confocal imaging. The Autoquant output file was then imported into Imaris for filament tracing and statistical quantification. The same threshold parameters were set for the osteocyte body (7.85 μm for the filament tracing start point) as well as the dendrite length (0.485 μm for the end point) for both the Dmp1 heterozygote (HET) and Dmp1-null specimens. The computed cell bodies were then manually corrected according to the FITC confocal images. After tracing, the filament surface area, volume, length, and terminal point number were all collected and selected for statistical analyses.
Data analyses
Osteocytes (20–25) from the middle of the tibia cortical bone region were chosen to be quantitatively analyzed. Statistical significance was determined by an independent-sample t-test using SPSS 12.0. A p value of < 0.05 was considered statistically significant.
Results
Comparison between the acid-etched SEM and FITC imaging techniques
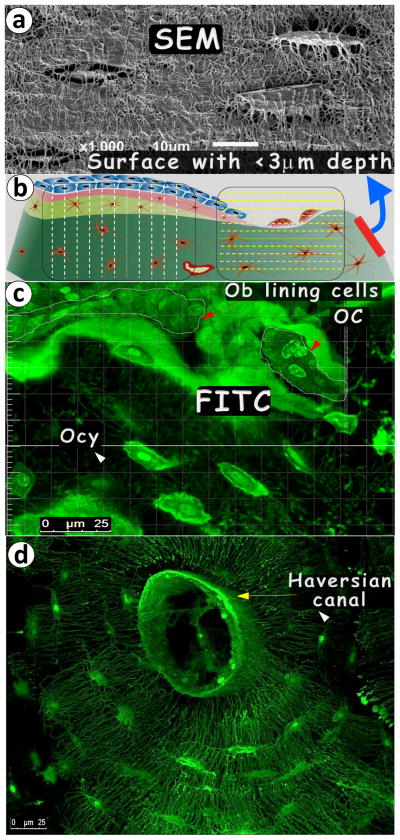
To better understand the advantage of the FITC-confocal imaging technique, we did a side-by-side comparison of both images. As shown in Fig 1-a, the maximum thickness of the osteocyte-lacuna image on the bone surface was less than 3 μm, which equals approximately half the thickness of the osteocyte. Therefore, this image could not be used for quantification. In contrast, an approximately 30-μm thickness of the stacked image could be obtained using the FITC-confocal technique with no distortion of the osteocyte structure using one photon laser beam (Fig 1b, which is a diagram showing the difference of the imaging area covered by both techniques). Fig 1-c gives an example of the rat tibia cortical bone (approximately 5 μm thick, note that only middle 10 slices of the series picture from confocal are stacked, to give a better view of cell structure), in which the osteoblast lining cells, osteoclasts, and osteocytes are well documented in the same frame. Fig 1-d displays an osteon (the basic bone unit in mammalian compact bone, approximately 35 μm in thickness) from a 30-year-old human male femur, which includes the Haversian canal; many osteocytes plus a large amount of dendrites.
Fig. 1. Pros and cons of acid-etched SEM and FITC techniques in imaging bone cells.
(a). Acid- etched, high-resolution SEM image displayed resin-casted osteocyte (Ocy)-canalicular system on the bone surface; (b). The diagram reveals the difference between the SEM and FITC imaging techniques with the former showing mineral- buried Ocy on the surface only and the latter technique imaging cells from the bone surface to inside the bone to a depth up to >50 mm with any region, angle or direction interested; (c). An example of an FITC image of the 6-mon-old rat tibia cortical bone revealing osteocyte (Ocy), osteoblast (Ob) lining cells, and osteoclast (OC); and (d). An example of a FITC-osteon image of the 30-year-old femur cortical bone showing the Ocy-dendritic system surrounding the Haversian canal.
Dmp1 null mice showed a significant increase in osteoid accumulation associated with a sharp reduction in dendrites and cell volume in osteocyte structures
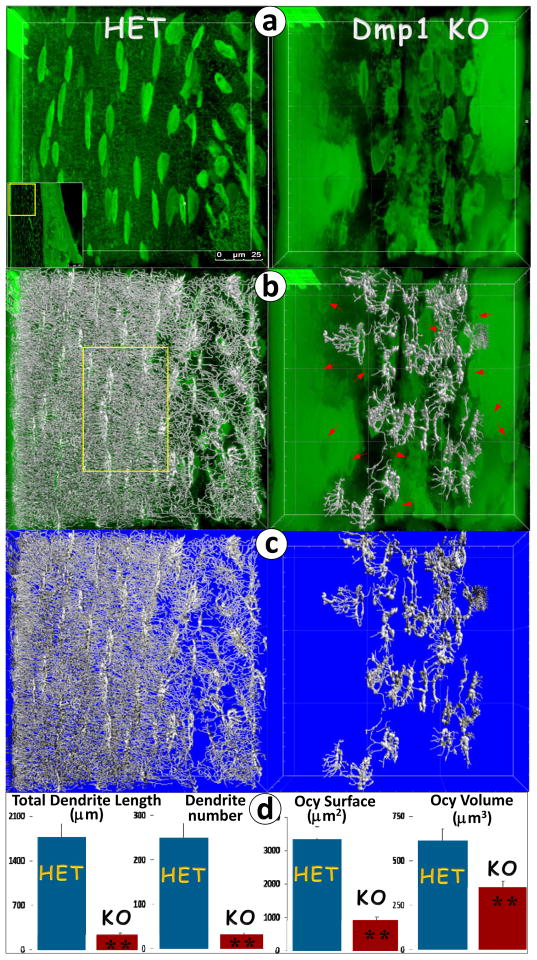
Imaris is Bitplane’s scientific software that was originally developed for neuronal visualization and interpretation of 3D microscopy datasets. We have adapted this software for analyzing osteocyte FITC images qualitatively and quantitatively. As an example, we took separate FITC images of the HET and Dmp1-null compact bones (Fig. 2a, step 1). Next, the osteocyte and its dendrites were traced using this software (Fig. 2b, step 2), and the original images were then removed (Fig. 2c, step 3). Finally, the data were processed for statistical comparison (Fig. 2d, step 4), which showed significant reductions in cellular surface, total volume, and total dendrite length and numbers, supporting the notion that DMP1 is essential for osteocyte maturation in the structure.
Fig. 2. Quantitation of osteocyte changes in the 8-wk-old Dmp1-null (KO) compared to the age-matched control (HET) mice using the FITC-Imaris technique.
(a). Confocal FITC image shows spindle-shaped osteocytes with numerous dendrites in a well-organized arrangement along the longitudinal axis in the HET (left) compared to the KO osteocytes, which are buried in poorly mineralized matrices (right) with much enlarged cell bodies that are less organized in structure and distribution, plus a few dendrites; (b). FITC confocal image reconstructed with the Imaris software revealed dense dendrites shown in artificial silver color (left, HET) compared to the KO tibia, in which there were 20 osteocytes with dendrites selected and reconstructed, and the rest of them were not (red arrows, right); (c). The original FITC confocal images were removed, with the Imaris-reconstructed images displayed only; and (d). The statistical analysis using Imaris software unveils a significant difference in total dendrite length, number, osteocyte surface area and total cell volume between the KO (n=20) and the HET control bone (n=25).
Discussion and Conclusion
In the present study, we developed a novel technique combining FITC confocal imaging and Imaris software, which not only enables the visualization of the true 3-D morphology of non-decalcified compact bone cells, but also statistically analyzes the morphologic differences among the healthy and the Dmp1-null osteocytes. This technique shares many similarities with the acid-etched SEM imaging technique, such as sample preparation and visualization of the 3-D osteocyte structure. The technique offers the following advantages: 1) much broader bone areas can be evaluated, including cells on the bone surface (osteoblasts and osteoclasts) and the interior structures, including both osteocytes and the osteon structure; 2) no extra physical and chemical treatments are required; thus, a natural cell distribution with essentially no tissue distortions can be seen; and 3) most importantly, the buried osteocytes and their dendrites can be precisely analyzed both qualitatively and quantitatively.
Acknowledgments
This study was supported by NIH grant DE018486.
Footnotes
Authors’ roles: Study design, manuscript writing and editing: RYS, PCD and JQF; Data collection and analyses: YSR, SXL and JY.
Competing interest
The authors have declared no competing interest.
References
- 1.Bellido T. Downregulation of SOST/sclerostin by PTH: a novel mechanism of hormonal control of bone formation mediated by osteocytes. Journal of musculoskeletal & neuronal interactions. 2006 Oct-Dec;6(4):358–9. Epub 2006/12/23. eng. [PubMed] [Google Scholar]
- 2.Galli C, Passeri G, Macaluso GM. Osteocytes and WNT: the mechanical control of bone formation. Journal of dental research. 2010 Apr;89(4):331–43. doi: 10.1177/0022034510363963. Epub 2010/03/05. eng. [DOI] [PubMed] [Google Scholar]
- 3.Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nature medicine. 2011 Oct;17(10):1231–4. doi: 10.1038/nm.2452. Epub 2011/09/13. eng. [DOI] [PubMed] [Google Scholar]
- 4.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nature genetics. 2006 Nov;38(11):1310–5. doi: 10.1038/ng1905. Epub 2006/10/13. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato M, Asada N, Kawano Y, Wakahashi K, Minagawa K, Kawano H, et al. Osteocytes regulate primary lymphoid organs and fat metabolism. Cell metabolism. 2013 Nov 5;18(5):749–58. doi: 10.1016/j.cmet.2013.09.014. Epub 2013/10/22. eng. [DOI] [PubMed] [Google Scholar]
- 6.Fen JQ, Zhang J, Dallas SL, Lu Y, Chen S, Tan X, et al. Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2002 Oct;17(10):1822–31. doi: 10.1359/jbmr.2002.17.10.1822. Epub 2002/10/09. eng. [DOI] [PubMed] [Google Scholar]
- 7.Ye L, Mishina Y, Chen D, Huang H, Dallas SL, Dallas MR, et al. Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. The Journal of biological chemistry. 2005 Feb 18;280(7):6197–203. doi: 10.1074/jbc.M412911200. Epub 2004/12/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erben RG. Embedding of bone samples in methylmethacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 1997 Feb;45(2):307–13. doi: 10.1177/002215549704500215. Epub 1997/02/01. eng. [DOI] [PubMed] [Google Scholar]