Abstract
Bacteria assemble a wide range of adhesive proteins, termed adhesins, to mediate binding to receptors and colonization of surfaces. For pathogenic bacteria, adhesins are critical for early stages of infection, allowing the bacteria to initiate contact with host cells, colonize different tissues, and establish a foothold within the host. The adhesins expressed by a pathogen are also critical for bacterial-bacterial interactions and the formation of bacterial communities such as biofilms. The ability to adhere to host tissues is particularly important for bacteria that colonize sites such as the urinary tract, where the flow of urine functions to maintain sterility by washing away non-adherent pathogens. Adhesins vary from monomeric proteins that are directly anchored to the bacterial surface to polymeric, hairlike fibers that extend out from the cell surface. These latter fibers are termed pili or fimbriae, and were among the first identified virulence factors of uropathogenic Escherichia coli. Studies since then have identified a range of both pilus and non-pilus adhesins that contribute to bacterial colonization of the urinary tract, and have revealed molecular details of the structures, assembly pathways, and functions of these adhesive organelles. In this review, we describe the different types of adhesins expressed by both Gram-negative and Gram-positive uropathogens, what is known about their structures, how they are assembled on the bacterial surface, and the functions of specific adhesins in the pathogenesis of urinary tract infections.
INTRODUCTION
Bacteria assemble a variety of adhesive proteins (adhesins) on their surface to mediate binding to receptors and colonization of surfaces. For pathogenic bacteria, adhesins are critical for early stages of infection, allowing the bacteria to initiate contact with host cells, colonize different tissues, and establish a foothold within the host. Adhesins recognize specific receptors expressed by specific subsets of host cells. Therefore, the repertoire of adhesins expressed by a pathogen play a major role in dictating the tropism of the pathogen toward specific host tissues and organs. Moreover, binding of bacterial adhesins to host cell receptors influences subsequent events by triggering signaling pathways in both the host and bacterial cells. These signaling pathways may determine whether the bacteria remain extracellular or become internalized, and influence the intracellular trafficking of invaded bacteria and their ability to survive and replicate (1,2). The adhesins expressed by a pathogen are also critical for bacterial-bacterial interactions and the formation of bacterial communities such as biofilms. The ability to adhere to host tissues is particularly important for bacteria that colonize sites such as the urinary tract, where the flow of urine functions to maintain sterility by washing away non-adherent pathogens.
Adhesins vary from monomeric proteins that are directly anchored to the bacterial surface to polymeric, hairlike fibers that extend out from the cell surface. These latter fibers are termed pili or fimbriae, and were among the first identified virulence factors of uropathogenic Escherichia coli (UPEC) (3). Pili were first described in the late 1940’s and early 1950’s as bacterial surface structures distinct from flagella (4). Duguid and co-workers used the term fimbriae, Latin for thread or fiber, to describe surface appendages that allowed E. coli to bind to and agglutinate erythrocytes (5). Brinton later used the term pili, Latin for hair, to describe the non-flagellar surface structures expressed by E. coli (6). Ottow subsequently proposed that the term pili be reserved for the F or conjugative pili involved in bacterial mating, and that the term fimbriae should be used to describe surface fibers involved in adhesion (4). However, today the terms pili and fimbriae are generally used interchangeably. We will refer to these structures collectively as pili.
Various schemes have been proposed to classify the different types of pili (4,7–10). Although most of these classification schemes are no longer in common use, parts have entered the standard nomenclature. Pili were originally classified as mannose resistant (MR) or mannose sensitive (MS), based on their ability to agglutinate erythrocytes in the presence or absence of mannosides (11,12). This classification led to the term type 1 pili, which is still in current use, to refer to MS surface fibers. The MR pili were initially divided into the P and unknown (X) pili, with the unknown pili now defined to include the S, Dr, and additional pilus adhesins (3). Uropathogenic bacteria have been closely associated with the discovery and characterization of pili. The chromosomal gene clusters responsible for expression of both type 1 and P pili were first cloned from the J96 UPEC strain (13), and the genes coding for S pili were isolated from UPEC strain 536 (14). As discussed in detail in the following section, much of our current understanding of the structure, assembly, and functions of bacterial pili stems from studies of the type 1 and P pili originally isolated from UPEC.
Bacteria are now known to express a number of different types of pilus structures and other non-flagellar surface appendages (15). One such additional structure, termed curli, is expressed by UPEC and imparts unique characteristics to the bacteria that influence colonization within the urinary tract, including promoting biofilm formation (16). Curli are assembled by a completely different mechanism from pili such as the type 1 and P pili, and appear as aggregated masses on the bacterial surface rather than hairlike fibers. Pilus assembly is not restricted to Gram-negative bacteria. Pili were observed on the Gram-positive bacterium Corynebacterium renale in the 1960’s (17,18), but this observation was largely forgotten until studies dating from 2003 by Ton-That and Scheewind to characterize pilus biogenesis in Corynebacterium diphtheriae (19,20). A number of different Gram-positive bacteria are now known to assemble adhesive pili associated with virulence and this is an active area of research. The Gram-positive pili have unique structural features and assembly mechanisms compared to Gram-negative pili (21,22).
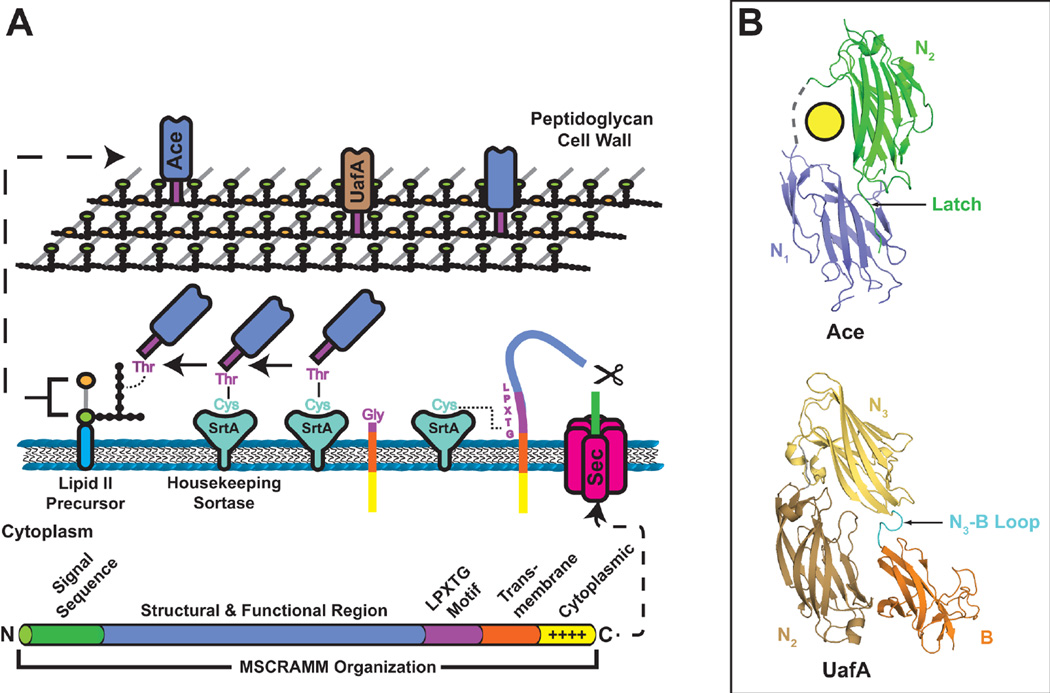
Pili and other extended surface fibers increase the functional reach of adhesins, enabling the bacteria to act at a distance. Pili place adhesins outside capsular or other protective surface structures, allowing contact with receptors while maintaining the protective integrity of the bacterial envelope. The ability to initiate contact at a distance also provides a means for pathogenic bacteria to avoid detection or uptake by host cells. Despite these advantages of pilus adhesins, bacteria also express a range of non-pilus adhesins, which are anchored directly on the bacterial surface. Non-pilus adhesins confer intimate binding to surfaces and are often associated with formation of bacterial colonies and biofilms. Gram-negative uropathogens display several adhesins important for pathogenesis on their outer membrane, with the majority of these adhesins assembled by the autotransporter (type V) secretion pathway. Gram-positive uropathogens also display adhesins on their surface important for colonizing the urinary tract. These Gram-positive adhesins typically are covalently linked to the peptidoglycan cell wall and are termed MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (22,23).
Table 1 lists adhesins that contribute to infection of the urinary tract by Gram-negative and Gram-positive uropathogens. In this chapter we will describe the different types of adhesins, what is known about their structures, and how they are assembled on the bacterial surface. We will also describe the functions of specific adhesins in the pathogenesis of urinary tract infections (UTIs). For the Gram-negative adhesins, we will focus our description on UPEC, which serves as a model system and for which extensive studies have been done.
Table 1.
Adhesins of uropathogenic bacteria
| Organism | Assembly pathway |
Adhesin | Associated UTI disease or function |
Receptor(s) | Reference(s) |
|---|---|---|---|---|---|
| GRAM-NEGATIVE BACTERIA | |||||
| Escherichia coli | |||||
| Chaperone/usher pathway | |||||
| P pili | Pyelonephritis | Digalactose (galabiose) | (165,365,366) | ||
| Type 1 pili | Cystitis | Mannosylated proteins, uroplakin, β1 and α3 integrin | (34,141,367) | ||
| S pili | Ascending UTI | α-sialic acid | (116,368,369) | ||
| F1C pili | Ascending UTI | galactosylceramide on bladder epithelium and globotriaosylceramide on kidney epithelium | (128,370) | ||
| Afa/Dr pili | Recurrent/chronic UTI, cystitis, pyelonephritis | Dra, DAF, type IV collagen, α5β1 integrin, CEACAM family proteins | (51,114,123) | ||
| Yad pili | Ascending UTI | Bladder epithelial cells | (43,118) | ||
| Ygi pili | Ascending UTI | Human embryonic kidney cells | (118) | ||
| F9 pili | Biofilm formation | ? | (120) | ||
| Auf pili | ? | ? | (119) | ||
| Type 3 pili | CAUTI | ? | (371) | ||
| Autotransporter | |||||
| Ag43 | UTI persistence, biofilm formation | Collagen, laminin | (263–265) | ||
| UpaB | ? | Fibronectin, fibrinogen, laminin | (270) | ||
| UpaC | Biofilm formation | ? | (270) | ||
| UpaH | Biofilm formation, bladder colonization | Collagen V, laminin, fibronectin | (217,272) | ||
| UpaG | Biofilm formation | Fibronectin, laminin | (232) | ||
| FdeC | Bladder and kidney colonization | ? | (273,274) | ||
| Outer membrane protein | |||||
| Iha | Fitness in urinary tract | ? | (278,279) | ||
| Type I secretion | |||||
| TosA | Fitness in urinary tract | Kidney epithelium | (284,287) | ||
| Extracellular nucleation/precipitation | |||||
| Curli | Biofilm formation, ? | Fibronectin, laminin, H-kininogen, fibrinogen, factor XII | (174,178,196) | ||
| Klebsiella pneumoniae | |||||
| Chaperone/usher pathway | |||||
| Type 1 pili | Ascending UTI, CAUTI | Mannosylated proteins | (372–374) | ||
| Type 3 (MR/K) pili | CAUTI | Type V collagen | (373,375,376) | ||
| Citrobacter freundii | |||||
| Chaperone/usher pathway | |||||
| Type 3 pili | CAUTI | ? | (377) | ||
| Proteus mirabilis | |||||
| Chaperone/usher pathway | |||||
| MR/P | Pyelonephritis, ascending UTI | Mannose-resistant | (378–380) | ||
| PMF (MR/K) | Ascending UTI | ? | (381,382) | ||
| UCA (NAF) | Colonization of urinary tract, complicated UTI | GalNAcβ(1–4)Gal | (383–385) | ||
| ATF | ? | ? | (386,387) | ||
| Autotransporter | |||||
| AipA | Bladder and kidney colonization | Collagen I, collagen IV, laminin | (220) | ||
| TaaP | Bladder colonization | Collagen I, collagen IV, laminin | (220) | ||
| GRAM-POSITIVE BACTERIA | |||||
| Staphylococcus saprophyticus | |||||
| Sortase-assembled MSCRAMM | |||||
| UafA | Ascending UTI | ? | (307,327) | ||
| UafB | Ascending UTI | Fibronectin, fibrinogen | (308) | ||
| SdrI | UTI persistence | Fibronectin | (296,309,329) | ||
| Enterococcus faecalis | |||||
| Sortase-assembled MSCRAMM | |||||
| Ace | CAUTI, ascending UTI | Collagen I and IV | (297,306,312) | ||
| Sortase-assembled pili | |||||
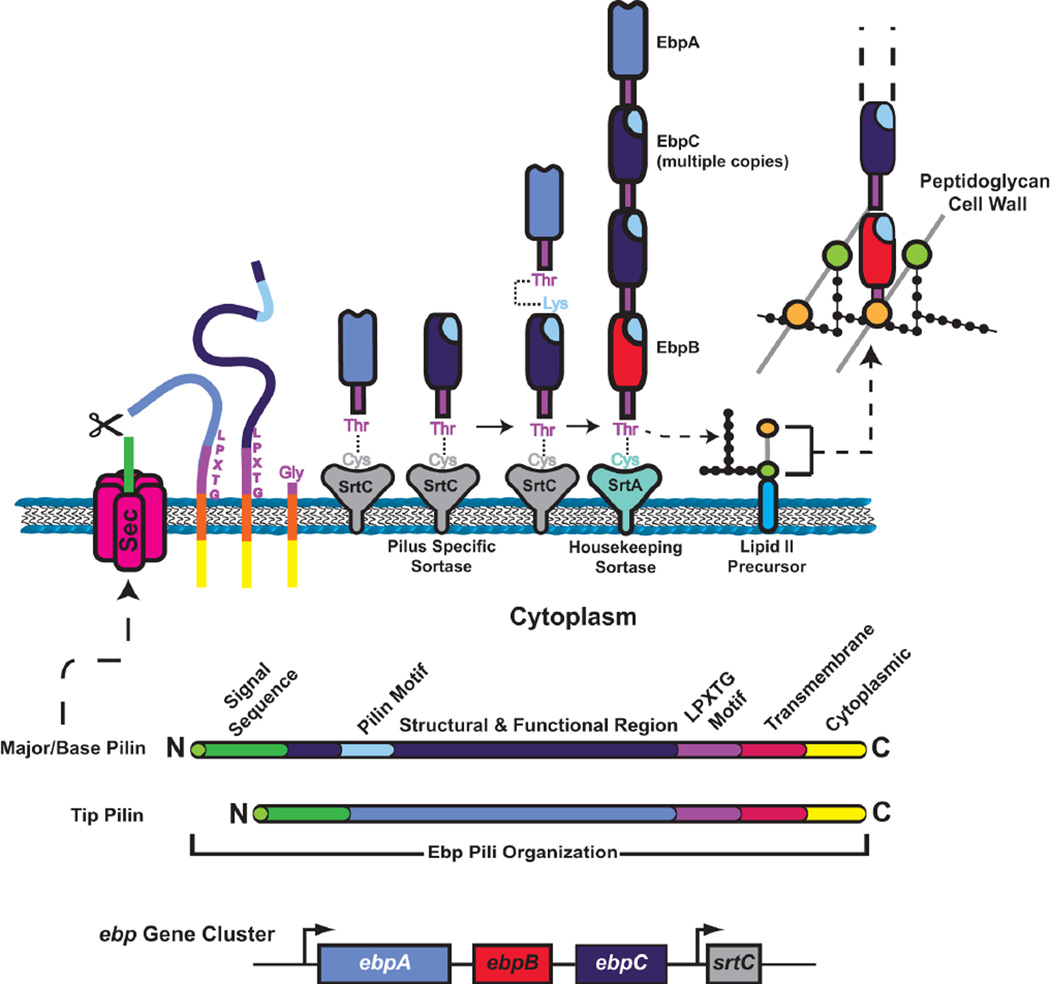
| Ebp | CAUTI, ascending UTI | ? | (340,345,352) | ||
| Unknown | |||||
| EfbA | Ascending UTI | Fibronectin | (331) | ||
| Esp | Urinary tract colonization and persistence, biofilm formation | ? | (332,333) | ||
| Enterococcus faecium | |||||
| Sortase-assembled pili | |||||
| Ebpfm | CAUTI, ascending UTI | ? | (339,347) | ||
ADHESINS EXPRESSED BY GRAM-NEGATIVE UROPATHOGENS
PILI ASSEMBLED BY THE CHAPERONE/USHER PATHWAY
A wide range of Gram-negative bacteria use the chaperone/usher (CU) pathway to assemble a superfamily of virulence-associated adhesive surface fibers (24–27). The CU pathway takes its name from the components of its secretion machinery, which consist of a dedicated periplasmic chaperone and an integral outer membrane protein termed the usher. The CU pathway builds a diverse array of peritrichous surface fibers, ranging from thin, flexible filaments to rigid, rod-like organelles. For uropathogenic bacteria, pili assembled by the CU pathway mediate adhesion to receptors in the urinary tract, initiating infection and promoting bacterial colonization. Pili are critical virulence factors of uropathogenic bacteria and have been the subject of intense study (Table 1). The CU pili expressed by uropathogenic bacteria are exquisitely adapted to colonization within the urinary tract, engineered to withstand and take advantage of forces encountered during colonization such as the flow of urine (28–31). In addition to binding to host molecules, CU pili are important for bacterial-bacterial interactions, biofilm formation, and adhesion to abiotic surfaces. Moreover, binding of bacteria to host cells via CU pili modulates host signaling pathways and promotes subsequent stages of pathogenesis such as invasion inside host cells (32–38).
Genes coding for CU pili are found on both the bacterial chromosome and on plasmids, and are clustered together with a similar organization: a 5’ regulatory region that is followed by a single downstream operon encoding the required pilus structural proteins and assembly components 7 (Fig. 1). CU gene clusters are often associated together with other virulence determinants in pathogenicity islands, which have characteristics indicating acquisition by horizontal gene transfer (39). A single bacterial genome often contains multiple CU pathways, which presumably provides the ability to adhere to a variety of different receptors and surfaces (40–42). A recent genomic analysis found that E. coli strains encode as many as 17 CU gene clusters and that UPEC strains encode from 9–12 intact CU gene clusters (43). Many of the CU gene clusters present in a bacterial genome are not expressed under laboratory growth conditions and their functions remain unknown (41). The expression of CU gene clusters is typically highly regulated, subject to phase variation, and responsive to environmental cues (44,45). Regulatory cross talk may occur among different CU gene clusters (46–48). This cross talk likely ensures that a given bacterium only expresses a single pilus at a given time, thus controlling adhesive specificity. Furthermore, expression of adhesive pili has been shown to be inversely correlated with the expression of flagella for motility (49).
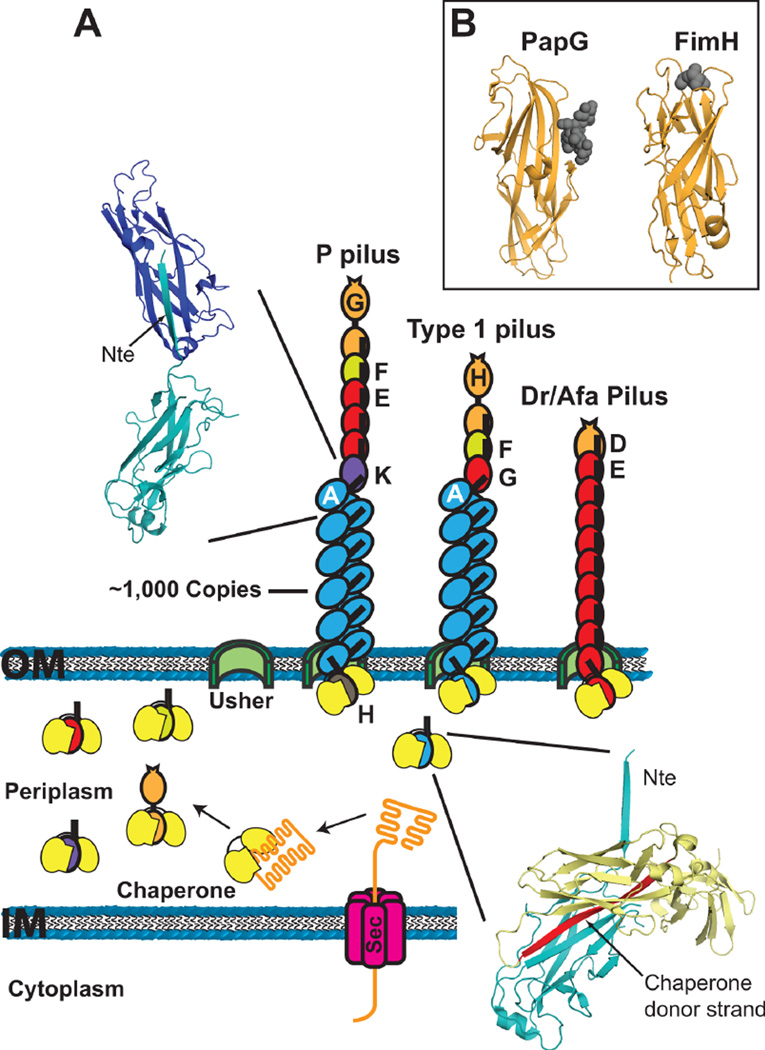
Figure 1.
Representative CU gene clusters and pili. Gene clusters coding for P (pap), type 1 (fim) and Dr/Afa pili are depicted, with the functions of the genes indicated. Electron micrographs are shown for (A) an E. coli bacterium expressing type 1 pili, (B) a P pilus fiber, and (C) a type 1 pilus fiber. Scale bars equal 700 nm (A), 100 nm (B), and 20 nm (C). The images in panels A–C are reprinted from references (138), (157), and (137), respectively, with permission of the publishers.
Much of what we know about the biogenesis and functions of CU pili comes from work on the prototypical type 1 and P pili expressed by UPEC, which bind to receptors in the bladder and kidney, respectively. We will focus on these pili as models, but will also discuss additional CU pili identified as important for pathogenesis in the urinary tract. Although we will limit this discussion to pili expressed by UPEC, CU pili have been identified as virulence factors in other uropathogens, particularly for Proteus mirabilis (Table 1) (50).
Structure of CU Pili
The pilus fiber
Pili assembled by the CU pathway range from 2–10 nm in diameter and generally 1–3 µm in length. The pili are linear fibers built from thousands of copies of non-covalently interacting subunit proteins, termed pilins. Some pili adopt a final helical quaternary structure, resulting in the formation of rigid, rod-like organelles. Alternatively, the pili may remain as linear, flexible fibers, which in some cases form amorphous or ‘afimbrial’ structures. Many pili assembled by the CU pathway are composite structures containing both a rigid, helical rod, which extends out from the bacterial surface, as well as a thin, flexible tip structure, which is located at the distal end of the rod and contains the adhesive activity. Type 1 and P pili expressed by UPEC are prototypical composite organelles with distinct rod and tip structures (Fig. 1). The Afa/Dr family of pili expressed by UPEC and other pathogenic E. coli are well-studied examples of thin, flexible fibers that often have an amorphous appearance by electron microscopy (51).
The structures of pilins and many aspects of pilus assembly by the CU pathway are understood in atomic detail (26,52–57). All pilins contain an immunoglobulin-like (Ig) fold termed the pilin domain (Fig. 2). Canonical Ig folds comprise seven β-strands arranged into two sheets as a β-sandwich (58). However, pilins lack the seventh, C-terminal β-strand (the G strand) and thus are unable to complete their own fold (52–55). This missing strand results in a deep groove on the surface of the subunit, exposing its hydrophobic core. To complete their folds, pilins rely on structural information provided by interaction with the periplasmic chaperone or with neighboring subunits in the pilus fiber.
Figure 2.
(A) Model for pilus biogenesis by the CU pathway. Pilus subunits enter the periplasm as unfolded polypeptides via the Sec system. Subunits fold upon forming binary complexes with the periplasmic chaperone (yellow). The crystal structure in the lower right depicts the chaperone-subunit donor strand exchange reaction (PapD-PapA; PDB ID: 2UY6), with the chaperone donor strand indicated in red. Pilus assembly takes place at the outer membrane usher, which catalyzes the exchange of chaperone-subunit for subunit-subunit interactions. Models for assembled P, type 1 and Afa/Dr pilus fibers are shown. The crystal structure in the upper left depicts the subunit-subunit donor strand exchange reaction that occurs in the pilus fiber (PapA-PapA; PDB ID: 2UY6), with the Nte donor strand indicated. (B) Crystal structures of the PapG (P pili; PDB ID: 1J8R) and FimH (type 1 pili; PDB ID: 1KLF) adhesin domains with bound globoside and mannose, respectively. The sugars are depicted as dark gray spheres.
Subunit-subunit interactions in the pilus fiber are mediated by a mechanism termed donor strand exchange (54,55). Pilins contain a conserved N-terminal extension (Nte) in addition to the pilin domain. In the pilus fiber, the Nte of one pilus subunit is ‘donated’ to the preceding subunit, completing the Ig fold of the preceding subunit (Fig. 2). Therefore, the pilus fiber consists of an array of Ig folds, with each subunit noncovalently bound to the preceding subunit by donor strand exchange. This arrangement provides great mechanical strength and stability to the pili, which is reflected by the property that subunit-subunit interactions in the pilus are resistant to dissociation by heat and denaturants (59,60). A high level of mechanical strength is essential for the pili to maintain adhesion in the face of shear forces encountered from the flow of urine. The helical pilus rod provides an additional mechanism to withstand hydrodynamic forces in the urinary tract; the helical rod is able to uncoil under stress to an extended linear fiber, thereby acting as a spring or shock absorber to prevent breakage of the pilus and extend the lifetime of pilus-receptor interactions (28,29,31,61).
The pilus adhesin
The receptor-binding activity of pili is conferred by the pilus adhesin. For composite pili such as type 1 and P pili, the adhesin is located in single copy at the distal end of the tip fiber (Fig. 2). Such pili have been termed monoadhesive pili (25). In contrast, for pili lacking a district tip structure, the main structural subunit that builds the pilus fiber may also contain receptor-binding sites along exposed surfaces and thus the entire pilus may function in adhesion (57,62–64). Such fibers are termed polyadhesive pili. Afa/Dr pili are polyadhesive fibers with a single major structural subunit/adhesin; however, these pili also have a separate subunit, termed the invasin, with distinct binding activity and which promotes uptake inside host cells (38,64,65). The invasin subunit is present in single copy at the distal end of the pilus fiber (Fig. 2).
Crystal structures have been solved for several adhesins from monoadhesive pili (52,66–71). In contrast to other pilus subunits, the adhesins are two domain proteins, containing an N-terminal receptor-binding or adhesin domain (in place of the Nte) and a C-terminal pilin domain. The pilin domain mediates incorporation of the adhesin into the pilus fiber and is an incomplete Ig-like fold as found for all CU pilins. Adhesin domains also have Ig-like folds, but the folds are complete (not lacking the terminal β-strand) and structurally distinct from the pilin domain. Despite their common architecture, adhesins vary greatly in sequence and employ distinct receptor binding mechanisms, reflecting their specific functions within the host (72). The FimH adhesin from type 1 pili folds as an elongated 11-stranded β-barrel with a jelly roll-like topology (52,67). The binding site for the mannose ligand is located at the tip of the adhesin domain and is formed by a deep, negatively-charged pocket surrounded by a hydrophobic ridge (Fig. 2B). In comparison, the adhesin domain of the P pilus adhesin PapG adopts a structure with two sub-regions; one region having a β-barrel fold similar to FimH and the other region having a unique, largely β-sheet structure that contains the binding site for the globoside receptor (66,68). In contrast to FimH, the receptor-binding site of PapG is located in a shallow groove along the side of the adhesin (Fig. 2B). For polyadhesive fibers such as Afa/Dr pili, both the main structural subunit and the tip-located invasin function as adhesins and both are single domain proteins with Ig-like pilin domains (57,62,65,73). However, the invasin subunit lacks an Nte donor strand, thus restricting its position to the tip of the fiber. For the AfaD and DraD major subunits, distinct receptor-binding sites for CD55/decay accelerating factor (DAF) and for members of the carcinoembryonic antigen family (CEACAM) have been located along opposite sides of the pilin domain (25,57,62). These binding sites would be repetitively presented along the length of the assembled fiber.
Pilus adhesins such as FimH exhibit the property of shear-enhanced binding, which enables tighter binding under conditions of shear stress such as encountered during bacterial colonization of the urinary tract (30). The application of shear stress causes FimH to switch from a low-affinity to a high-affinity binding state. This greater affinity presumably allows the bacteria to avoid being washed away by the flow of urine, and may also provide a mechanism for the bacteria to discriminate between surface-located and soluble receptors, as binding to the latter will not result in force generation on the pilus and FimH will stay in the low-affinity state. The shear-enhanced binding of FimH is mediated by a catch-bond mechanism that involves allosteric activation of the adhesin domain (74). When incorporated into the type 1 pilus tip fiber, the pilin domain of FimH interacts with the adhesin domain, causing structural alterations of the adhesin that weaken its mannose binding pocket. However, the application of force to the pilus fiber causes the FimH pilin and adhesin domains to separate, allowing the binding pocket to clamp tightly around its mannose ligand (75). Moreover, the physical properties of both the type 1 pilus tip fiber and the helical pilus rod appear to be designed to optimize the shear-enhanced behavior of FimH, and the flexibility of the pilus tip likely provides FimH maximum opportunity to find its target receptors (76,77).
Pilus Assembly by the Chaperone/Usher Pathway
Formation of chaperone-subunit complexes in the periplasm
Pilus subunits are synthesized with an N-terminal signal sequence that directs them to the Sec general secretory pathway for translocation to the periplasm (78). The signal sequence is cleaved in the periplasm, and the subunits form stable, binary complexes with the periplasmic chaperone (Fig. 2A). The chaperone enables proper folding of the pilus subunits, prevents premature subunit-subunit interactions, and maintains the subunits in an assembly-competent state (52–55). In the absence of the chaperone, pilus subunits misfold and form aggregates that are degraded by the DegP periplasmic protease (79,80).
The structure of the PapD chaperone and subsequent structures of chaperone-subunit complexes revealed the molecular basis for chaperone function in pilus biogenesis (52–54,81–83). As described above, pilins have an incomplete Ig fold, lacking the C-terminal G β-strand. The chaperone contains two Ig-like domains oriented in an L or boomerang shape. The binding site for subunits resides in the cleft between the two domains and extends out along the chaperone’s N-terminal domain (domain 1). The chaperone functions by a mechanism termed donor strand complementation, in which the chaperone inserts its G1 β-strand and a portion of its F1–G1 loop into the groove caused by the missing G strand of the subunit, completing the Ig fold of the pilin domain (Fig. 2A) (52,53,83,84). Conserved sequence differences in the F1–G1 loop region of chaperones defines two subfamilies of CU pathways: chaperones with a short F1–G1 loop belong to the FGS (F1–G1 short) subfamily and chaperones with a long F1–G1 loop belong to the FGL subfamily (64,85). Interestingly, these differences in the chaperones correlate with differences in the types of surface fibers assembled. FGL chaperones assemble only thin or amorphous pili comprising only one or two types of pilins (such as Afa/Dr pili), whereas FGS chaperones assemble both rod-like and thin pilus fibers that generally comprise multiple different pilins and may have composite architectures (such as type 1 and P pili).
The groove in the pilin domain caused by the missing β-strand contains a series of binding pockets, termed P1–5 (54). The G1 β-strand donated by the chaperone contains a conserved motif of alternating hydrophobic residues, and during donor strand exchange these residues insert into the P1–4 pockets of the subunit, forming a β-zipper interaction (55,86). In FGL chaperones, the longer G1 donor strand fills the P5 binding pocket as well, but this interaction is weaker than at the other pockets (86,87). The chaperone G1 β-strand is inserted parallel to the F strand of the subunit, forming a non-canonical Ig fold. This, together with the large size of the residues inserted by the chaperone maintains pilins in an open, “activated” state, which enables subsequent assembly into the pilus fiber (54,55,87). The groove of the pilin domain is also the site of subunit-subunit interactions, which are mediated by the donor strand exchange reaction as described above for the pilus fiber (54,55). Thus, donor strand complementation by the chaperone couples the folding of pilins with the simultaneous capping of their interactive surfaces, preventing premature fiber assembly in the periplasm. Recent studies have shown that chaperones also perform a quality control function during the initial binding of pilus subunits and that formation of chaperone-subunit complexes results in an allosteric change in the chaperone that permits binding to the outer membrane usher assembly platform (88,89).
Assembly of the pilus fiber at the outer membrane
Chaperone-subunit complexes must interact with the outer membrane usher for release of the chaperone, assembly of subunits into the pilus fiber, and secretion of the fiber to the cell surface through the usher channel (56,90). The usher acts as a pilus assembly catalyst, accelerating the rate of subunit incorporation into the pilus fiber (91). Subunit-subunit interactions form at the periplasmic face of the usher via the donor strand exchange mechanism (54,55). The donated subunit Nte contains a conserved motif of alternating hydrophobic residues, similar to the chaperone G1 β-strand (92,93). At the usher, the hydrophobic residues of the Nte from an incoming chaperone-subunit complex insert into the subunit groove of the preceding chaperone-subunit complex bound at the usher, displacing the donated G1 β-strand of the chaperone from the preceding subunit by a concerted strand displacement mechanism that initiates at the P5 pocket (54,55,86,94,95). In contrast to the donated chaperone β-strand, the Nte is inserted anti-parallel to the F strand of the preceding subunit and inserts smaller-sized residues into the subunit groove, thus completing the Ig fold of the pilin domain in a canonical fashion and allowing the subunit to adopt a highly stable final state (54,55,60,87). ATP is not available in the periplasm and pilus biogenesis at the outer membrane usher does not require input from other energy sources (96,97). The canonical Ig fold formed by donor strand exchange represents a more compact, lower energy state compared to the non-canonical Ig fold formed by donor strand complementation with the chaperone (54,55,87). This topological transition from the higher-energy chaperone-subunit complex to the lower-energy subunit-subunit interaction provides the driving force for fiber formation and secretion at the usher (98).
Pili are assembled in a top-down order, with the adhesin incorporated first, followed by the rest of the pilus tip and finally the rod. Each subunit specifically interacts with its appropriate neighbor subunit in the pilus, with the specificity of binding determined by the donor strand exchange reaction (99–101). In addition, the usher ensures ordered and complete pilus assembly by differentially recognizing chaperone-subunit complexes according to their final position in the pilus; i.e., chaperone-adhesin complexes have highest affinity for the usher, whereas chaperone-rod subunit complexes have low affinity (102–104). The usher channel is only wide enough to allow secretion of a linear fiber of folded pilus subunits (56,90). Therefore, the pilus rod is constrained to a linear fiber as it passes through the usher and only converts to its final helical form upon reaching the bacterial surface.
The pilus usher
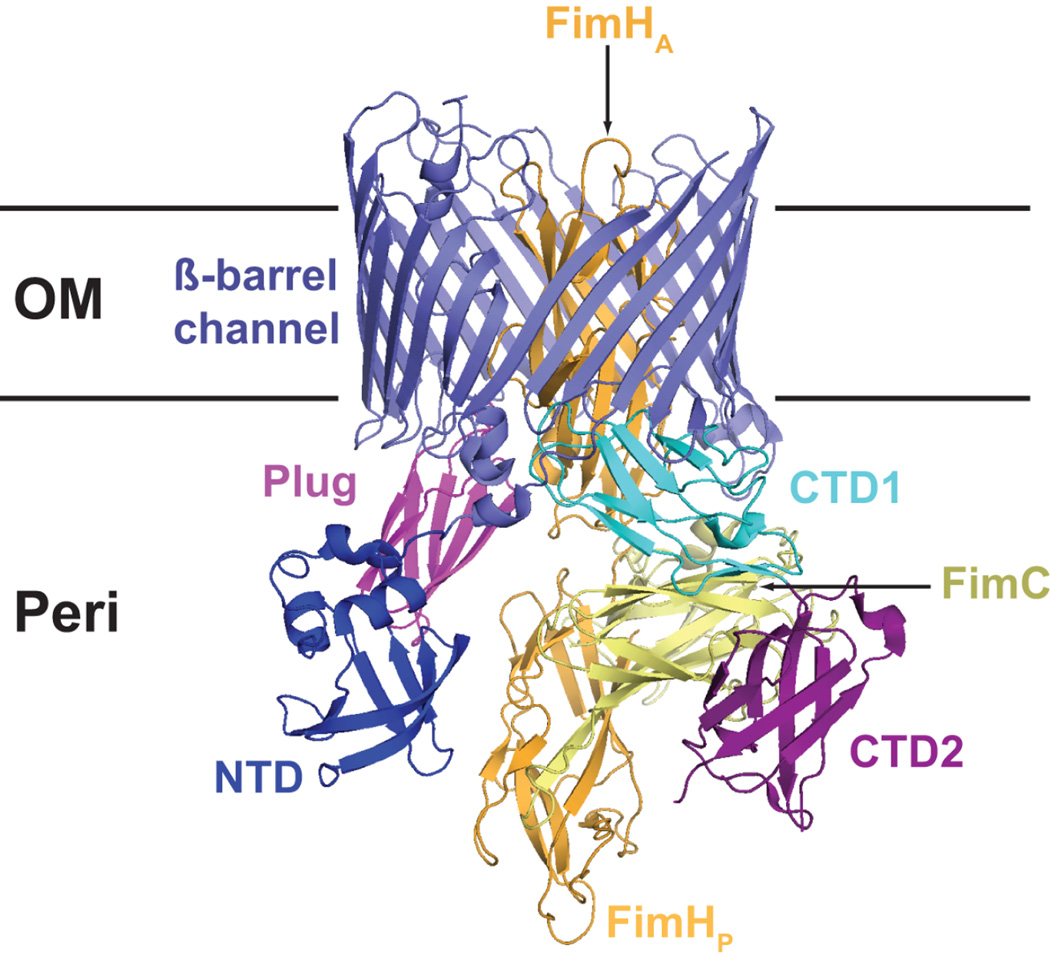
Ushers are large, integral outer membrane proteins containing five domains: a central transmembrane β-barrel domain that forms the secretion channel, a middle domain located within the β-barrel region that forms a channel gate (the plug domain), a periplasmic N-terminal domain (NTD), and two periplasmic C-terminal domains (CTD1 and CTD2) (Fig. 3) (56,90,105–109). The NTD provides the initial binding site for chaperone-subunit complexes and functions in the recruitment of periplasmic complexes to the usher (105,110). The CTDs provide a second binding site for chaperone-subunit complexes and anchor the growing pilus fiber (56). The usher is present as a dimeric complex in the OM, but only one channel is used for secretion of the pilus fiber and the function of the usher dimer remains to be determined, particularly since the usher monomer appears to be sufficient for pilus assembly (56,90,106,111,112).
Figure 3.
Crystal structure of the FimD-FimC-FimH type 1 pilus assembly intermediate (PDB ID: 3RFZ). The Usher NTD, plug, β-barrel channel, and CTD domains are indicated. The FimH adhesin domain (FimHA) is inserted inside the usher channel, and the FimH pilin domain (FimHP) and bound FimC chaperone are located at the usher CTDs.
The structure of the type 1 pilus usher FimD bound to the FimC-FimH chaperone-adhesin complex was recently solved, revealing the usher pilus assembly machine in action (Fig. 3) (56). The usher channel is formed by a 24-stranded β-barrel that is occluded by an internal plug domain (56,90). The binding of the FimH adhesin to FimD activates the usher for pilus biogenesis (91,103,113), resulting in displacement of the plug to the periplasm and insertion of the FimH adhesin domain inside the usher channel. The FimH pilin domain remains in complex with the FimC chaperone and bound to the usher CTDs (Fig. 3). CU pili extend by step-wise addition of new chaperone-subunit complexes to the base of the fiber. New chaperone-subunit complexes are recruited by binding to the usher NTD, which is unoccupied in the FimD-FimC-FimH structure (56,105,110). Modelling studies suggest that binding of a chaperone-subunit complex to the usher NTD would perfectly position the Nte of the newly recruited subunit to initiate donor strand exchange with the P5 pocket of the subunit bound at the usher CTDs, providing a molecular explanation for the catalytic activity of the usher in pilus assembly (56). Following donor strand exchange, the chaperone is displaced from the subunit bound at the CTDs and released into the periplasm. To reset the usher for another round of subunit incorporation, the newly incorporated chaperone-subunit complex must transfer from the NTD to the CTDs, concomitant with translocation of the pilus fiber through the usher channel toward the cell surface. Repeated iterations of this cycle would then result in assembly and secretion of a complete pilus fiber.
Functions of CU Pili Expressed by UPEC
A number of different CU pili have been identified that contribute to colonization of the urinary tract by UPEC (Table 1). In addition to type 1 and P pili, which are described in detail in the following paragraphs, CU systems with demonstrated or putative roles in UTIs include Afa/Dr, S, F1C, F9, type 3, Auf, Yad and Ygi pili (51,114–120). Further characterization is needed for many of these systems, which may have roles in direct adherence to host receptors or may facilitate bacterial-bacterial interactions and biofilm formation. The best characterized of these additional CU pili are the Afa/Dr family, which includes Dr, F1845, Afa, Nfa and Aaf pili. Afa/Dr pili are thin, polyadhesive fibers that are expressed by diffusely adhering strains of diarrheagenic E. coil (DAEC) in addition to being prominent virulence factors of UPEC (51,114,121). Afa/Dr adhesins bind to the Dra blood group antigen and have affinity for DAF, members of the CEACAM family, type IV collagen, and α5β1 integrin (122–124). In contrast to the Afa/Dr polyadhesins, S and F1C pili are structurally similar to type 1 and P pili. S pili bind to sialyl-galactoside moieties on extracellular matrix proteins such as fibronectin and laminin (125,126). S pili are expressed by clinical UPEC isolates and expression of S pili confers binding to bladder and kidney epithelial cells, indicating potential roles in ascending UTIs (115,127). F1C pili have affinity for globotriaosylceramide, which is present on the kidney epithelium, and for galactosylceramide, found in the bladder, kidney, and ureters (128).
Type 1 pili
Type 1 pili are expressed by most strains of E. coli and mediate binding to a variety of surfaces and host tissues in a mannose-sensitive manner. Type 1 pili are a major virulence factor of UPEC and antibodies to the type 1 pilus adhesin FimH provide protection against urinary tract infection by E. coli in both murine and primate models (129,130). However, a definitive requirement for type 1 pili in human UTIs has remained elusive (131), likely due to the large repertoire of adhesins expressed by uropathogenic strains. UPEC use type 1 pili to bind to α-d-mannosylated proteins present in the bladder, leading to bacterial colonization, bladder epithelial cell invasion, and the development of cystitis (33,132). In addition to urothelial cells, type 1 pili have been reported to bind to Tamm-Horsfall protein, surface glycoproteins of immune cells, extracellular matrix proteins, and abiotic surfaces (133–136).
Type 1 pili are encoded by the fim gene cluster (Fig. 1), which is present on the chromosome of pathogenic as well as non-pathogenic and laboratory strains of E. coli. Type 1 pili are built from 4 different types of pilins arranged into a rigid helical rod measuring 6.9 nm in diameter, and a short tip fiber measuring 2 nm in diameter and generally 10–19 nm in length (Fig. 1) (137,138). The type 1 pilus rod is built from greater than 1,000 copies of the FimA major pilin arranged into a right-handed helix (138). Type 1 pilus tips contain a single copy of the FimH adhesin at the distal end, followed by the FimG and FimF adaptor subunits, which are generally present in single copy (Fig. 2A) (75,137,138). The mannose binding site of the FimH adhesin is located in a deep pocket at the tip of the adhesin domain (Fig. 2B) (67). This places the receptor-binding site at the most distal end of the type 1 pilus organelle, which presumably facilitates access of the pilus to its receptor.
Studies using the murine urinary tract infection model have revealed many aspects of type 1 pilus function during UPEC pathogenesis. Upon entering the urinary tract, UPEC use their type 1 pili to bind to uroplakins, mannosylated proteins that coat the luminal surface of the bladder, allowing the bacteria to colonize the bladder and avoid being washed out by the flow of urine (33,139). Type 1 pili not only mediate binding of UPEC to the bladder surface, but also trigger host cell signaling pathways that lead to actin rearrangement in the urothelial cells and invasion of the bacteria inside the cells by a zipper-like mechanism (32,33,140). Additionally, bacterial uptake is facilitated by binding of type 1 pili to β1 and α3 integrins (34). Binding of E. coli to the urothelium leads to induction of innate host cell responses, such as upregulation of proinflammatory cytokines and cell death pathways (33,140,141). The FimH adhesin acts as a pathogen-associated molecular pattern that is recognized by Toll-like receptor 4 (TLR4), present on bladder epithelial cells as well as macrophages, and stimulates immune signaling pathways through a mechanism independent of LPS (36).
Following uptake inside bladder epithelial cells, UPEC are initially contained within vesicles, which may be routed for exocytosis in a TLR4- and cyclic AMP-dependent mechanism that may be used by the host cells to expel the invading bacteria (142). Bacteria that evade expulsion enter the cytoplasm where they rapidly replicate to form aggregates termed intracellular biofilm-like communities or pods (143,144). Bacteria within these intracellular communities are protected from host innate immune responses and shielded from antibiotics (145). Type 1 pili, which are known to contribute to the formation of extracellular biofilms (136), are also expressed by the intracellular bacteria and required for formation of the pods, separate from their function in host cell binding and invasion (35,146). Urothelial cells respond to UPEC invasion by undergoing programmed cell death and exfoliating into the bladder lumen, a host-defense mechanism to wash out the colonizing bacteria (33). However, UPEC counter this by fluxing out of the host cells and undergoing additional rounds of attachment to and invasion of neighboring cells, presumably mediated by type 1 pili as in the initial round of infection (144,147). During this process, the E. coli may gain access to the underlying bladder epithelium, leading to the formation of quiescent bacterial reservoirs from which recurrent infections can be seeded to begin the infection process anew (147,148). Thus, type 1 pili function at multiple different points during UPEC pathogenesis in the urinary tract and have both extracellular and intracellular roles.
P pili
P pili are expressed by UPEC and are strongly associated with the ability of the bacteria to colonize the kidney and cause pyelonephritis (66,149,150). P pili bind to Gal(α1–4)Gal moieties present in the globoseries of glycolipids found in kidney epithelial cells. The glycolipid receptor is also part of the P blood group antigen, thus allowing P pilus-mediated agglutination of human erythrocytes (151). P pili are encoded by the chromosomal pap (pyelonephritis-associated pili) gene cluster (Fig. 1), which is present on pathogenicity islands of UPEC strains, and also found in E. coli causing neonatal meningitis and avian pathogenic strains (152). Individual E. coli strains may carry more than one pap gene cluster, located in different pathogenicity islands (153,154). There are three predominant alleles of the P pilus adhesin PapG – class I, II and III – which have specificities for receptor isotypes that differ in carbohydrate residues distal from the Gal(α1–4)Gal core (155,156). Class II PapG is correlated with human kidney infections, whereas class III PapG is associated with colonization of the human bladder.
P pili are built from 6 different structural subunits that form a right-handed helical rod and distal tip fiber, similar to type 1 pili (Fig. 1). The P pilus tip fiber is longer and more flexible compared to type 1 pilus tips, measuring ~40 nm in length. The P pilus tip is composed mainly of PapE, which is present at approximately 5–10 copies per pilus. The PapG adhesin is present in single copy at the distal end of the tip and is joined to PapE via the PapF adaptor subunit (157,158) (Fig. 2A). Another adaptor subunit, PapK, links the tip fiber to the pilus rod (158). The helical P pilus rod measures 8.2 nm in diameter and is built from a linear homopolymer of over 1000 copies of the PapA major pilin (159). The P pilus rod is terminated by the PapH minor pilin, which also plays a role in anchoring the pilus fiber in the OM (160,161).
The glycolipid binding site on the PapG adhesin is formed by a shallow pocket on one side of the adhesin domain (Fig. 2B) (66,68). This is in contrast to the tip-located mannose-binding site of FimH on type 1 pili. P pili have a longer, more flexible tip fiber compared to type 1 pili. The flexibility of the P pilus tip and side-on orientation of the PapG binding site likely function in tandem to facilitate docking of the adhesin onto the globoside moiety of the glycolipid receptor, which is oriented parallel to the membrane surface (66).
Expression of P pili promotes ascending urinary tract infection and facilitates colonization of the kidneys by E. coli (150,162). Consistent with a role in pathogenesis, vaccination with P pili was shown to provide protection against pyelonephritis in both murine and primate models (163,164). However, studies using P pilus mutants have had variable results in establishing an essential requirement for the pili in kidney infections, likely due to the many different adhesins expressed by UPEC strains (165). As for type 1 pili, P pilus-mediated adhesion of UPEC to the urothelium stimulates cytokine production and resultant inflammatory responses in the urinary tract, which likely exacerbates kidney damage during acute pyelonephritis (37,166,167). Binding of P pili to its glycolipid receptor in kidney epithelial cells causes release of the second messenger ceramide, which forms the membrane anchor portion of the receptor. Ceramide is as an agonist for TLR4, and thus provides a potential link between bacterial adhesion and induction of innate immune pathways (168). PapG-mediated binding also activates signal transduction pathways within the bacteria (169). These pathways result in upregulation of iron acquisition systems and may prepare UPEC for colonization of the urinary tract.
CURLI
Curli fibers, also called thin aggregative fimbriae, are produced by Gram-negative enteric bacteria such as E. coli and Salmonella and form part of a complex extracellular matrix that contributes to adhesion, biofilm formation, host colonization, and invasion (16,170–172). The expression of curli imparts special properties to biofilm structures, allowing attachment to normally resistant surfaces such as Teflon and stainless steel (170). Curli were first characterized by Normark and colleagues as novel bacterial surface structures that conferred binding to fibronectin (173). Curli bind to range of host molecules in addition to fibronectin, including laminin, human contact phase proteins, and MHC class I (174–176). Most bacteria optimally express curli at temperatures of 30°C or lower, consistent with a central role in biofilm formation and colonization of environmental surfaces. However, many clinical E. coli strains, including UPEC isolates, express curli at host temperature (37°C) (177,178). In addition to conferring adhesive and aggregative properties to bacteria, curli expression is sensed by the host and modulates host immune responses (177,179).
Curli share many properties with eukaryotic amyloid fibers. Amyloid fibers are typically associated with human neurodegenerative illnesses such as Alzheimer’s, Parkinson’s, and prion-mediated diseases (180,181). In contrast to these diseases, which are thought to be due to uncontrolled protein folding, curli belong to a growing class of fibers termed ‘functional amyloid’, whose expression is controlled and directed for the benefit of the expressing cell (182,183). Curli assemble as thin, tangled fibers that are extraordinarily stable and impart important physiological properties to bacteria, some of which play significant roles during host-pathogen interactions. The pathway for assembly of curli on the bacterial surface is distinct from the CU pathway and other pilus assembly systems, and instead utilizes an extracellular nucleation-precipitation mechanism, in which curli subunit proteins are first secreted to the cell surface before being incorporated into the growing fiber.
Curli Structure
Curli form densely aggregated masses on the bacterial surface (Fig. 4B). Individual curli fibers measure 3–4 nm in diameter and are of varying lengths (173,184,185). Similar to eukaryotic amyloid, curli fibers are non-branching, rich in β-sheet structure, and highly resistant to the action of proteases and denaturants (182,184,186). Curli and other amyloid fibers also share the property of binding to specific dyes such as Congo red and thioflavin T (187).
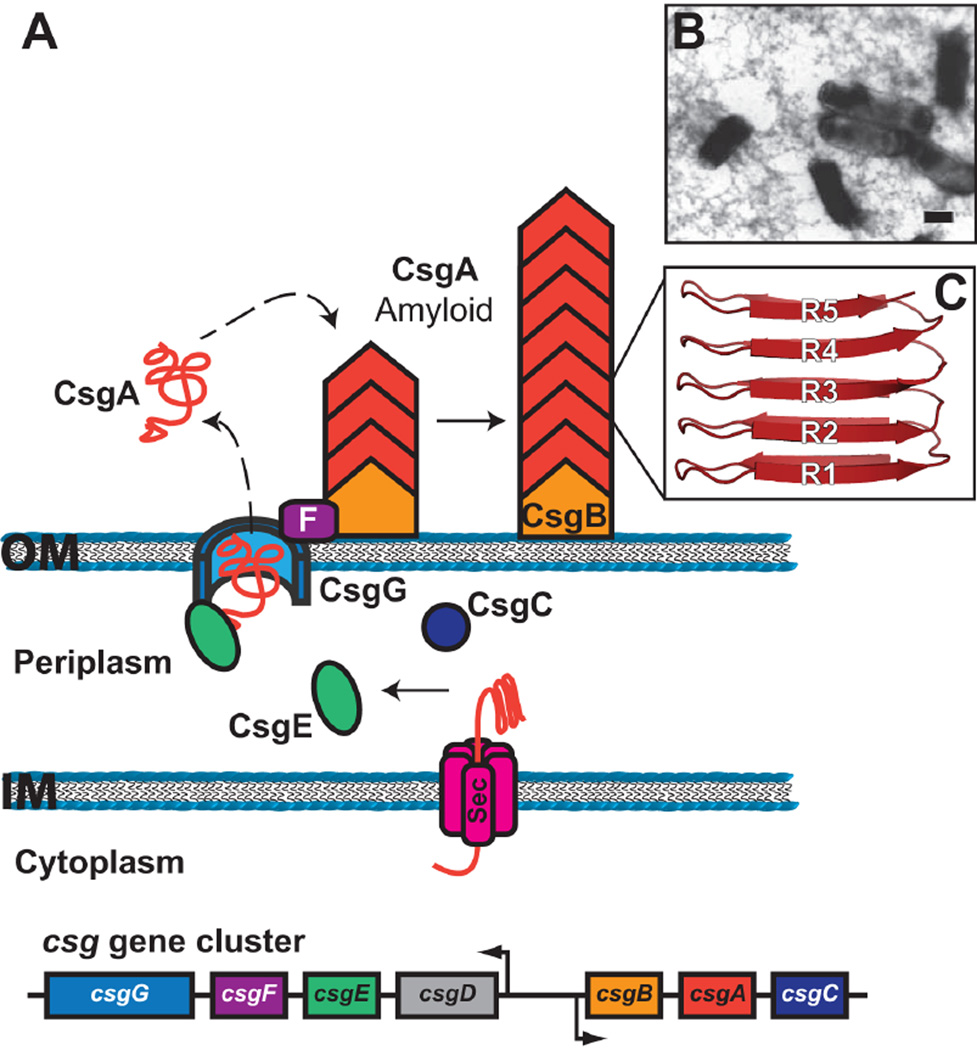
Figure 4.
(A) Model for curli biogenesis by the extracellular nucleation/precipitation pathway. The csg gene cluster coding for curli biogenesis is shown at the bottom. The curli subunit proteins enter the periplasm via the Sec system and are secreted to the bacterial surface via the CsgG outer membrane channel. CsgE may act as a chaperone for the curli subunits in the periplasm, whereas CsgF assists assembly of CsgB on the cell surface. Polymerization of CsgA occurs on the cell surface and is nucleated by interaction with CsgB. (B) Electron micrograph of E. coli expressing curli. Scale bar equals 1 µm; reprinted from reference (205) with permission of the publisher. (C) Structure of a CsgA subunit, with the R1–R5 repeats indicated.
Curli fibers expressed by E. coli are composed of repeating copies of the major subunit protein, CsgA (Fig. 4A). Each CsgA molecule contains five conserved repeating units (R1 through R5), which are predicted to form two parallel, stacked β-sheets containing five β-strands each (Fig. 4C) (16,188). Curli fibers also contain a minor, nucleating subunit; in E. coli this is CsgB. CsgB shares 30% sequence identity with CsgA, both proteins are of identical predicted size, and both are built from similar repeat motifs (189). The R1 and R5 repeat units, which flank the N and C termini of CsgA, mediate intersubunit contacts and are important for CsgB-mediated nucleation of the CsgA fiber as well as for CsgA-CsgA polymerization (190). In contrast, the R2-to-R4 internal repeats govern the kinetics of fiber polymerization, slowing the rate of polymerization so as to limit toxicity during curli production (190). Structural analysis indicates that the individual CsgA subunits in the curli fiber stack on top of each other, forming an extended β-helix-like structure (185). Therefore, the final fiber consists of an expanse of β-sheets oriented parallel to the fiber axis, but with the individual β-strands oriented perpendicular to the fiber axis. Such a cross β-strand structure is a hallmark of amyloid fibers (181,185).
The exact binding sites present on curli fibers and the mechanism by which curli bind to a wide range of receptors is unknown. A study examining synthetic peptides corresponding to overlapping regions of the CsgA sequence identified N- and C-terminal regions of 24 and 26 residues, respectively, that recapitulated binding to several different human proteins (176). In addition, CsgB may have a direct role in adhesion separate from its role in nucleating polymerization of CsgA. This is suggested by studies in S. enterica, in which a deletion of csgB, but not csgA, decreased adherence to alfalfa sprouts (191).
Curli Assembly on the Bacterial Surface
The genes required for curli biogenesis in E. coli are encoded by the divergently transcribed csgBAC and csgDEFG operons (Fig. 4A) (192). The csgBAC operon encodes the major structural subunit CsgA and the nucleator protein CsgB (189,192). CsgC is a periplasmic protein with structural similarity to oxido-reductases (193). The function of CsgC is not understood, but it may be important for proper function of the CsgG outer membrane channel (193). Expression of the csgBAC operon is dependent on the positive regulator CsgD, which is part of the csgDEFG operon (192,194).
Extracellular nucleation-precipitation
Assembly of curli on the bacterial surface occurs by an extracellular nucleation-precipitation pathway (16). In the absence of the CsgB nucleator protein, curli are not assembled; instead, CsgA is released from the bacteria in an unpolymerized, soluble form (184,189). This released CsgA can be assembled into curli fibers on recipient cells expressing only CsgB (189). This process, termed interbacterial complementation, demonstrates that curli assembly may take place entirely on the bacterial surface, and that assembly likely involves a conformational change in CsgA that is triggered by CsgB. Thus, CsgA is secreted outside the bacteria as a soluble, unstructured monomer, which is then nucleated into a fiber on the cell surface by interaction with CsgB or with the structurally altered CsgA in the growing curli fiber (Fig. 4A) (182,195).
Curli assembly machinery
The secretion and polymerization of CsgA is dependent on the CsgE, CsgF and CsgG proteins, the functions of which are not fully understood (192). CsgG is an outer membrane lipoprotein that is thought to form the channel for secretion of CsgA and CsgB to the cell surface (196). In the absence of CsgG, curli fibers are not assembled and the CsgA and CsgB subunits become unstable (184). Consistent with a channel protein, CsgG forms oligomeric, ring-shaped complexes and overexpression of CsgG correlates with increased pore-formation in the outer membrane (196). Structural analysis of CsgG predicts that it belongs to the recently characterized class of transporters that assemble in the outer membrane as α-helical rather than β-barrel channels (193). The CsgG-mediated secretion of CsgA is dependent on the N-terminal 22 amino acids of the mature CsgA protein. These residues are not predicted to be an integral part of the curli fiber, suggesting that they act as a secretion signal (196,197).
CsgE is a periplasmic protein and its expression is important for stability of the CsgA and CsgB subunits (184). Consistent with a role in proper folding of the curli subunits, csgE mutant bacteria do not act as donors or acceptors for interbacterial complementation and the few curli fibers produced by csgE mutants are morphologically distinct from curli expressed by wild-type cells (184). CsgE physically interacts with CsgG at the outer membrane, and CsgE may chaperone periplasmic CsgA subunits to the CsgG secretion channel by interacting with the N-terminal CsgA signal sequence (198). CsgE may also function to prevent premature fiber assembly in the periplasm (198). The CsgF protein also interacts with CsgG at the outer membrane; however, CsgF localizes to the cell surface rather than the periplasm (199). csgF mutants have a distinct phenotype, producing reduced levels of curli fibers and secreting soluble, unpolymerized CsgA (184,199). Similar to csgB mutants, csgF mutants act as donors but not acceptors for interbacterial complementation. In agreement with this behavior, CsgF influences the folding of CsgB and localization of CsgB to the bacterial surface, suggesting that CsgF functions as an extracellular chaperone for CsgB (199).
Functions of Curli in UPEC
Curli are multifunctional surface fibers, conferring adhesion to specific host molecules, promoting bacterial community behaviors such as aggregation and biofilm formation, and modulating interactions with the host immune system (16,170–172,178,200). Curli bind to the extracellular matrix proteins fibronectin and laminin (173,174). Curli also bind to human contact phase proteins including H-kininogen, fibrinogen, and factor XII (175,176). By binding to the contact phase proteins, curliated bacteria slow clotting, which could facilitate bacterial dissemination throughout the host (177,201). In addition, curli interact directly with molecules of the immune system. MHC class I molecules, which present antigens to T cells, bind to curli and curliated bacteria adhere better to tissue culture cells that over-produce MHC class I (202).
UPEC and other E. coli isolates produce curli at the host temperature of 37°C, including UPEC strains freshly isolated from the urine of infected patients (178,201). This supports a functional role for curli in colonization of the urinary tract. Curli expression enhances adhesion to urothelial cells in cell culture, and the ability to express curli correlates with increased colonization of the urinary tract during early stages of the murine infection model (178,203,204). Curli-mediated binding to host molecules may also facilitate uptake inside host cells (16,172,173). Expression of curli genes promoted invasion of human epithelial cells by a non-pathogenic K12 strain of E. coli, and invasion was inhibited by addition of peptides that blocked curli formation (172,205).
In addition to binding to specific host molecules, a major functional role of curli in UPEC is likely promoting bacterial aggregation and biofilm formation. Indeed, curli were shown to contribute to biofilm formation by UPEC distinct from the action of type 1 pili (203). Finally, curli expression by UPEC appears to be an important modulator of host immune responses during infection. Curli fibers are recognized by host cells as a PAMP (pathogen-associated molecular pattern) (200). Curli recognition is mediated by TLR2, resulting in the activation of pro-inflammatory molecules such as IL-6, IL-8, and TNF-α (177,200). A recent study demonstrated multiple functions for curli during infection of the murine urinary tract: facilitating colonization, protecting the bacteria from the action of host anti-microbial peptides, and provoking an increased pro-inflammatory response (178). Taken together, these results demonstrate that curli play important and varied roles during both initial colonization and subsequent stages of the infectious process.
AUTOTRANSPORTERS AND OTHER NON-PILUS ADHESINS
In addition to assembling adhesins in the form of extended pili or curli fibers, Gram-negative uropathogens also display adhesins directly on their cell surface. The majority of these non-pilus adhesins are assembled on the outer membrane by the autotransporter (type V) secretion pathway (206,207). Autotransporters are a widespread family of secreted proteins with activities ranging from proteases and toxins to adhesins and invasins. The term autotransporter was first used by Meyer and colleagues to describe the IgA1 protease of Neisseria meningitidis, and refers to the idea that a single polypeptide encodes both functional and secretion activities (208). The range of autotransporter functions is reflected by additional well-studied autotransporters such as the NalP protease of N. meningitidis, the VacA cytotoxin of Helicobacter pylori, and the Pertactin and AIDA-I adhesins of Bordetella pertussis and E. coli, respectively (209–213). Autotransporters are characterized by the presence of a conserved C-terminal translocator or β-domain that inserts into the outer membrane and directs the secretion of an N-terminal passenger or α-domain, which carries the functional activity, to the cell surface (206,207). Following secretion, the passenger domain may remain tethered to the outer membrane by the translocator domain or may undergo proteolytic cleavage to be released into the extracellular environment (Fig. 5) (207,209). In some cases, such as with AIDA-I and related autotransporters, the passenger domain remains associated with the cell surface even after proteolytic cleavage, through noncovalent interactions with the translocator domain (214).
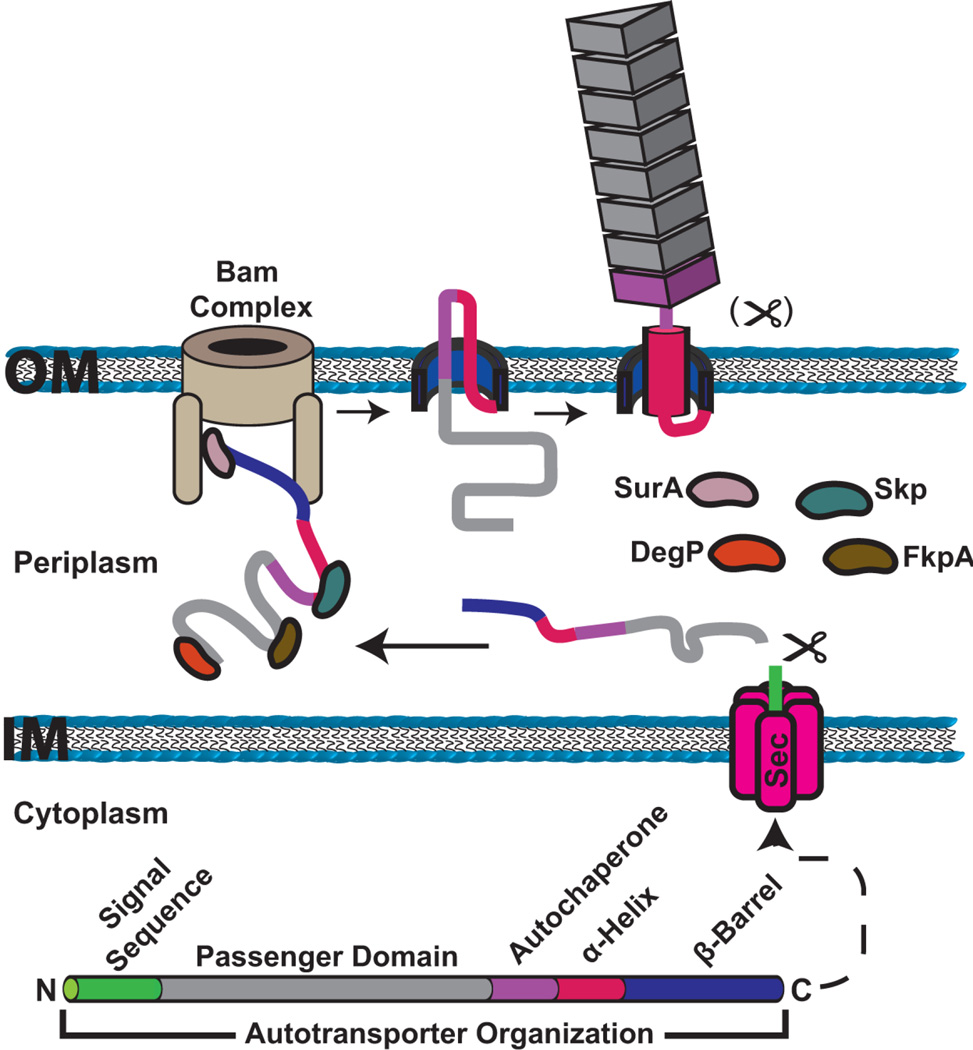
Figure 5.
Model for autotransporter secretion and assembly on the bacterial surface. The domain organization of an autotransporter protein is shown at the bottom. Autotransporter polypeptides have an N-terminal signal sequence for translocation to the periplasm via the Sec system. The protein is maintained in an extended, largely unfolded state during transit across the periplasm, assisted by periplasmic folding factors (SurA, Skp, DegP and FkpA). The C-terminal translocator domain inserts into the outer membrane as a β-barrel channel, with the assistance of the Bam complex. The Bam complex may also assist in secretion of the passenger domain to the cell surface. In the hairpin model of secretion, the C-terminal region of the passenger domain forms a hairpin structure in the translocator channel, exposing part of the passenger to the cell surface. Folding initiates at the autochaperone region, which then nucleates folding and secretion of the rest of the passenger domain. Following secretion, the linker region adopts an α-helical structure to plug the translocator domain channel. The passenger domain may remain linked to the translocator domain or may be proteolytically cleaved.
The contributions of autotransporters to bacterial pathogenesis in the urinary tract are still being defined, and their identification has largely proceeded from genomics studies and efforts to characterize UPEC-specific virulence factors (Table 1). One reason for the relative paucity of information on surface-located adhesins compared to pili is that the expression of pili or other extended surface structures may obscure or sterically hinder the functions of proteins present at the bacterial surface (215,216). Thus, autotransporter adhesins are likely to be important under conditions in which pili expression is turned off. Eleven autotransporters have been identified in the genome of UPEC strain CFT073, a prototypical pyelonephritis isolate (153,217,218). Seven of these belong to the AIDA-I family of autotransporter adhesins, and at least four of the UPEC autotransporters function in adhesion to host cells and contribute to fitness in the urinary tract, as discussed below. At least one of the other UPEC autotransporters, Sat, is not an adhesin but is an important protease and toxin of UPEC (219). In addition to UPEC, autotransporter adhesins that contribute to colonization of the urinary tract have also been identified in P. mirabilis (220).
This section will provide an overview of the structure and assembly of autotransporters, and will describe the functions of autotransporter adhesins that have been characterized in UPEC. We will also describe two additional non-pilus adhesins expressed by UPEC that are assembled by distinct mechanisms.
Autotransporter Structure
The translocator domain
Autotransporters contain a C-terminal translocator domain and N-terminal passenger domain. The translocator domain is the most conserved feature of autotransporters, whereas passenger domains exhibit a high level of sequence variation (221). Translocator domains belonging to the classical (type Va) autotransporter family are typically 250–300 residues in length and insert into the outer membrane to form a β-barrel channel. Crystal structures for several translocator domains have been solved, revealing a typical outer membrane β-barrel structure comprising 12 antiparallel transmembrane β-strands, enclosing a channel of ~10–13 Å diameter (Fig. 6) (210,222,223). An α-helical linker region important for secretion of the passenger domain precedes the β-barrel, and the helix and barrel together have been termed the translocation unit (224–226). In the autotransporter structures, the α-helical linker occupies the lumen of the β-barrel channel (Fig. 6) (210,222,223). The N terminus of the α-helix is oriented toward the bacterial surface, suggesting that translocation of the passenger domain would occur through the β-barrel channel. However, as discussed below, there is debate about the exact mechanism of passenger domain secretion.
Figure 6.
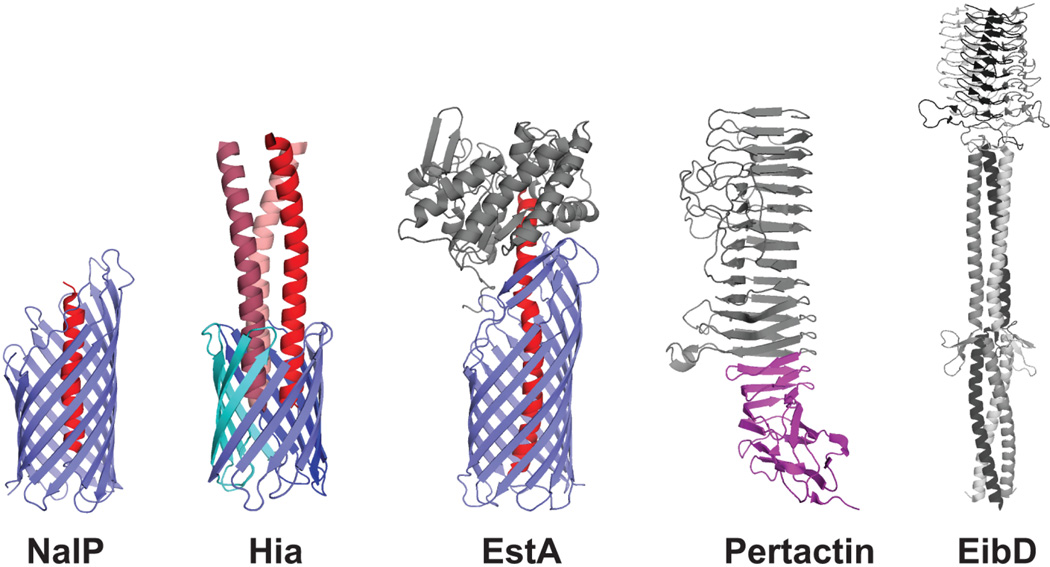
Crystal structures of representative autotransporter proteins. Translocator domains from the monomeric NalP and trimeric Hia autotransporters are shown (PDB IDs: 1UYN and 2GR7, respectively), with the β-barrel channels in blue and the α-helical linker regions in red. Passenger domains from the monomeric Pertactin and trimeric EibD autotransporters are shown (PDB IDs: 1DAB and 2XQH, respectively), with the approximate location of the Pertactin autochaperone region indicated in purple. The complete structure of the EstA autotransporter is shown (PDB ID: 3KVN), with the translocator domain in blue, the α-helical linker in red, and the globular passenger domain in gray.
Some autotransporters have smaller translocation domains, consisting of only ~70–80 amino acids. These proteins trimerize to form a single 12-stranded β-barrel, with each monomer contributing 4 strands (Fig. 6) (227–229). The Hia and YadA adhesins of H. influenzae and Y. pestis are prototypical members of this subfamily (type Vc), termed trimeric autotransporter adhesins (206,230,231). The β-barrel formed by the Hia translocator domain has a central pore of 18 Å diameter, which is sufficient to accommodate passage of the three α-helical linker segments that connect to the extracellular passenger domains (Fig. 6) (229). Of the 11 autotransporters identified in UPEC, only UpaG belongs to the trimeric subfamily (232).
The passenger domain
The N-terminal passenger domains of autotransporters are structurally and functionally diverse, but share important conserved features. Most passenger domains are large in size and contain repetitive sequence motifs that assemble into repetitive structural elements (206,207). The sequence motifs typically form β-sheets that are arranged into an extended β-helix, with each rung of the helix comprising three β-strands in a triangular arrangement (Fig. 6) (212,233–235). In different autotransporters, this β-helix core structure may be modified with interspersed extended loops, globular domains, or other elements, which contain specific functions such as receptor binding sites or protease activity. However, not all passenger domains have a β-helix architecture. For example, the complete structure of the EstA autotransporter from Pseudomonas aeruginosa revealed a typical 12-stranded β-barrel translocator domain, but a globular passenger domain primarily composed of α-helices and loop sequences (Fig. 6) (222).
Passenger domains from the trimeric autotransporter subfamily exhibit a distinct architecture compared to the classical autotransporters, but also assemble into extended structures built from repetitive sequence elements. Members of this family typically function as adhesins and the passenger domains remain attached to the outer membrane translocator rather than undergoing proteolysis. The passenger domains trimerize, matching the trimeric structure of the translocator domain, and form extended rod-shaped structures. A common architecture is shared by the trimeric passenger domains, comprising a globular N-terminal head region with extensive β-sheet structure, followed by an extended coiled-coil stalk region that connects to the translocator anchor domain in the outer membrane (Fig. 6) (230,231,236–238). Some trimeric autotransporters have more complex architectures, with modular arrangements of interspersed head, neck, and stalk regions (236,237). The head region typically contains the receptor binding site (Fig. 6), which may be present in each monomer and thus displayed in triplicate around the surface of the trimer, such as for the Hia adhesin (230). The stalk region may also have binding activity for host molecules or mediate bacterial-bacterial interactions (238,239).
The Autotransporter Secretion Pathway
Transit across the periplasm and insertion into the outer membrane
Nascent autotransporter polypeptides are synthesized with an N-terminal signal peptide that is cleaved following translocation of the protein from the cytoplasm to the periplasm via the Sec general secretory pathway (78). The passenger domain must be kept in a largely unfolded state to remain competent for secretion. Resident periplasmic chaperones and folding factors (DegP, FkpA, SurA, and Skp) interact with the extended autotransporter polypeptide in the periplasm (Fig. 5) to maintain the passenger domain in a secretion-competent state and prevent non-productive interactions, as well as assist in proper folding and insertion of the translocator domain in the outer membrane (206,207,240,241). Some autotransporters have an extended N-terminal Sec signal sequence that slows the rate of translocation across the cytoplasmic membrane through the Sec system (207,242,243). This slowing of translocation may facilitate proper transit of the autotransporter polypeptide across the periplasm.
Secretion of the N-terminal passenger domain to the cell surface requires the C-terminal translocator domain, which inserts into the outer membrane to form a β-barrel channel. Recent studies have defined a set of proteins in the bacterial outer membrane, termed the β-barrel assembly machine (Bam) complex, which is responsible for proper insertion of most β-barrel proteins into the outer membrane (244). In keeping with this, correct assembly of the translocator domain in the outer membrane and its proper functioning in autotransporter secretion requires the Bam complex (245–247).
Secretion of the passenger domain to the cell surface
The exact mechanism by which the passenger domain is secreted from the periplasm to the bacterial surface remains a topic of active investigation. Two main models have been proposed for this process: a ‘classical’ hairpin model and a newer model that invokes a central role for the Bam complex (206,207).
In the hairpin model, secretion of the passenger domain occurs through the lumen of the β-barrel channel formed by the translocator domain. Secretion of the passenger domain initiates when a C-terminal region of the passenger domain, likely including the α-helical linker region, forms a temporary hairpin structure within the pore (Fig. 5). Formation of the hairpin exposes part of the C terminus of the passenger domain to the cell surface, where it may begin folding. Folding of the passenger domain would then proceed vectorially from the C to the N terminus, progressively pulling the polypeptide through the channel (248,249). The hairpin model requires the presence of two strands of the passenger domain polypeptide within the lumen of the translocator domain pore. Given the narrow dimensions of the pore, these strands would need to be in a largely unfolded and extended conformation, which is consistent with studies showing a general lack of tolerance for structured elements in passenger domains (241,250,251).
Autotransporter secretion across outer membrane does not require the input of energy from the cytoplasmic membrane, and folding of the passenger domain at the cell surface likely provides the energy to drive secretion through the translocator channel (248,249). Folding at the cell surface could also act as a ratchet to prevent diffusion of the passenger back into the periplasm. Most passenger domains contain a conserved junction sequence adjacent to the α-helical linker, termed the autochaperone domain. This region, which is critical for the folding of β-helical passenger domains, would be exposed to the cell surface upon formation of the hairpin loop and may act as an intramolecular chaperone to nucleate folding of the rest of the passenger domain on the cell surface (Figs. 5 and 6) (252–254). Progressive folding of rest of the passenger domain would then occur through a self-templating mechanism, driven by the repeated β-helix structure (255). Once secretion of the passenger domain is complete, the linker region would assume its final α-helical conformation to plug the pore, as observed in the autotransporter crystal structures (Figs. 5 and 6) (210,222,223).
The classical hairpin model proposes that the β-barrel channel formed by the translocator domain is sufficient for passenger secretion and that no other accessory factors are required (which is the basis for the term autotransporter). However, conflicts with this model, particularly in studies showing tolerance for secretion of some folded elements and the presence of post-translationally modified passenger domains (256–258), have lead to revised models that invoke a central role for the Bam complex (210,245,259). In these models, the β-barrel translocator domain serves to target the autotransporter polypeptide to the Bam complex. Interaction with the Bam complex then allows folding and insertion of the translocator domain in the outer membrane, coupled with secretion of the passenger domain to the cell surface. Rather occurring through the translocator domain channel, secretion of the passenger domain would occur, at least in part, through the BamA channel or through some interface between the Bam complex and the translocator domain (207,210). As the BamA channel is unlikely to be able to gate laterally to allow release of the passenger domain, secretion of the passenger domain would need to occur in concert with insertion of the translocator β-barrel into the outer membrane, and is likely to involve some aspects of the hairpin model (206,207,245,259,260). Thus, the Bam complex may facilitate coupled formation of the hairpin structure and insertion of the translocator domain into the outer membrane (Fig. 5), and possibly also assist in secretion of structured regions. This would then establish an initiating point from which secretion of the remainder of the passenger domain would proceed through the translocator channel as proposed in the classical hairpin model.
Functions of Autotransporter Adhesins in UPEC
Ag43
Antigen 43 (Ag43) is an autotransporter adhesin encoded by the flu gene (also termed agn43). Ag43 functions in adhesion to host cells and self-associates to promote bacterial aggregation (autoaggregation), leading to flocculation in static liquid cultures and biofilm formation on surfaces (261–264). Ag43 is present in approximately 80% of UPEC strains, and many strains encode more than one copy (265). UPEC strain CFT073 expresses two Ag43 variants, Ag43a and Ag43b. The Ag43a variant appears to be the functionally relevant form in UPEC, promoting high levels of aggregation, biofilm formation, and colonization of the urinary tract (265). Expression of Ag43 is phase variable and opposite from expression of type 1 pili; expression of the longer pilus fibers on the bacterial surface sterically blocks adhesion mediated by the shorter Ag43 molecules (216,266).
Ag43 belongs to the AIDA family of autotransporters (267). Similar to AIDA-I, the passenger domain of Ag43 is proteolytically processed following transport to the cell surface, but remains associated with the translocator domain via non-covalent interactions. Also similar to AIDA-I, the Ag43 passenger domain is glycosylated in some E. coli strains, including UPEC isolates (258). The importance of glycosylation for Ag43 function remains to be determined, as different studies have found variable effects of glycosylation on autoaggregation, biofilm formation, and adhesion to host cells (258,263). Ag43 promotes adhesion to various cell lines, including human kidney cells, and binds to the extracellular matrix components collagen and laminin (263). The Ag43 passenger domain contains multiple repeats of ~19 residues each and folds with an extended, L-shaped β-helical structure (268). The region of Ag43 responsible for autoaggregation is located in the first N-terminal third of the mature passenger domain (261). Recent structural analysis of the Ag43 passenger domain suggests that self-association is mediated by a “Velcro-like” mechanism (268).
Several lines of evidence point to a role for Ag43 in UTIs. Anderson and colleagues found that Ag43 is expressed during bladder infection by UPEC strain UTI89 (143). Specifically, Ag43 was present on the surface of bacteria engaged in formation of intracellular biofilm-like communities following invasion of bladder epithelial cells. Consistent with this observation, the Ag43a variant was found to promote long-term persistence of UPEC strain CFT073 in the murine UTI model; a fluA but not fluB mutant of CFT073 is present at lower numbers in the bladder compared to the parental strain at day 5 post infection (265). In addition, expression of Ag43 may promote formation of linked bacterial chains that are formed by asymptomatic bacteriuria E. coli isolates when grown in human urine (269). The autoaggregation properties of Ag43 are likely to enhance bacterial colonization of the urinary tract as well as formation of intracellular and extracellular biofilms.
UpaB
UpaB is an autotransporter adhesin identified in UPEC strain CFT073, and is widely distributed among both uropathogenic and non-uropathogenic E. coli strains (270). UpaB belongs to the AIDA-I family of autotransporters and contains a predicted pertactin-like passenger domain (270). UpaB confers binding to extracellular matrix proteins, including fibronectin, fibrinogen, and laminin. A upaB deletion mutant of CFT073 is outcompeted by the wild-type strain for colonization of the bladder, and the mutant strain is specifically defective for an early stage of bladder colonization (270). However, a direct role for UpaB in adhesion to the urinary tract has not been demonstrated. UPEC encode an additional autotransporter related to UpaB, termed UpaC; however, UpaC is not expressed by CFT073 and a UpaC mutant had no phenotype in the murine UTI model (270).
UpaG
UpaG is a trimeric autotransporter adhesin prevalent among extraintestinal pathogenic E. coli (ExPEC) strains belonging to the B2 and D phylogenetic groups, including UPEC strain CFT073 (232). The structure of UpaG has been reconstructed from crystal structures of fragments of the homologous Salmonella enterica protein SadA (237). UpaG assembles as an extended coiled-coil fiber, ~115 nm in length, containing four YadA-like head repeats and adaptor neck regions typical of trimeric autotransporters (237). Expression of UpaG in CFT073 promotes adhesion to the T24 human bladder epithelial cell line, with specificity for fibronectin and laminin, and promotes autoaggregation and biofilm formation (232). UpaG was identified as a potential protective antigen of ExPEC, suggesting that it is expressed during infection (271). However, native expression of UpaG was not detected in CFT073 grown under in vitro conditions, and no role was found for UpaG in colonization of either the bladder or kidneys using the murine infection model (232).
UpaH
The UpaH autotransporter adhesin is expressed by the CFT073 UPEC strain, where it provides a competitive advantage for colonization of the bladder and contributes to biofilm formation (217). UpaH is a large-sized (~280 kDa) member of the AIDA-I family of autotransporters (272). UpaH binds to the extracellular matrix proteins collagen V, fibronectin, and laminin (272). However, a direct role for UpaH in adhesion to the urinary tract has not been demonstrated. The upaH gene is present in the chromosomes of many UPEC isolates, as well as in non-uropathogenic E. coli strains (217). Bioinformatics analysis predicts a typical 12-stranded β- barrel translocator domain and a large passenger domain with 50 imperfect sequence repeats predicted to encode an extended β-helix structure (217). Sequence variation is present in the UpaH passenger domain from different E. coli isolates (272). These variations were found to impact function in biofilm formation but not binding to extracellular matrix proteins.
FdeC
FdeC was identified in a screen for ExPEC vaccine antigens that provided protection in a murine sepsis model (273). The fdeC gene is widely distributed among ExPEC as well as intestinal E. coli strains (274). FdeC shares a low level of sequence homology with the invasin and intimin proteins of Yersinia pseudotuberculosis and enteropathogenic E. coli, respectively, which function in adhesion to host cells (275,276). Similar to these proteins, FdeC is anchored in the outer membrane via a presumed N-terminal β-barrel domain, with the extracellular portion of the protein forming an elongated structure comprising 9 repeated Ig-like domains (274). A model was recently proposed that proteins such as intimin and invasin form a new subfamily of autotransporters (type Ve) (206,277). In this model, the proteins are secreted in an analogous mechanism to autotransporters, but with a reverse topology; i.e., the outer membrane translocator domain is located at the N terminus instead of the C terminus as for typical autotransporters. In contrast to intimin and invasin, no obvious lectin domain is present in FdeC (274). Recombinant FdeC binds to human urothelial cell lines, as well as other types of epithelial cells, with specificity for collagen (274). FdeC is expressed during interactions with host cells and during infection of the urinary tract, and an fdeC mutant of UPEC strain 536 was defective for colonization of the bladder and kidneys during co-infection with the wild-type strain (274).
Other Outer Membrane-Associated Adhesins of UPEC
At least two additional non-pilus adhesins that contribute to pathogenesis in the urinary tract have been identified in UPEC. These adhesins are assembled on the bacterial outer membrane by mechanisms that are distinct from the autotransporter pathway. The Iha adhesin is secreted by the type I secretion pathway and TosA, a multifunctional siderophore receptor and adhesin, is an integral outer membrane protein.
Iha
The IrgA homologue adhesin (Iha) protein, encoded by the iha gene, is prevalent among UPEC strains and has the novel phenotype of functioning in both adhesion and iron uptake (278,279). Iha was originally identified in the diarrheagenic E. coli strain O157:H7 as an adhesin with homology to the Vibrio cholerae iron-regulated virulence factor IrgA (280). Iha is an outer membrane protein with homology to β-barrel siderophore receptor proteins such as FepA (281). Iha is a virulence factor of UPEC, as demonstrated by reduced fitness of a CFT073 iha deletion mutant for colonization of both bladder and kidneys in the murine UTI model (279). Similarly, an iha mutant of the UPEC clonal group A outbreak strain UCB34 (282) was attenuated for infection of the urinary tract in competition with the parental wild-type strain (278). There is also evidence supporting a role for iha in the pathogenesis of UTIs caused by Proteus mirabilis (283). Iha is expressed in vivo during infection of the murine urinary tract (278). Expression of recombinant Iha promoted adhesion to human epithelial cells, including the T24 bladder cell line, whereas overexpression of other siderophore receptors did not promote adhesion (278,279). In addition to its function as an adhesin, Iha functions in iron uptake as an iron-regulated catecholate siderophore receptor (278). Given the likely topology of Iha as in integral outer membrane β-barrel protein, adhesive activity is presumably conferred by surface-exposed loops of the protein. Whether the adhesin function, iron uptake, or both are important for pathogenesis remains to be determined.
TosA
TosA belongs to the repeats-in-toxin (RTX) family of secreted bacterial proteins that have diverse functions, including acting as toxins, proteases, and adhesins (284). RTX proteins share the characteristic features of repetitive glycine- and aspartate-rich sequences, located in the C-terminal region of the protein, and use of the type I secretion system for export out of bacteria. Type I secretion systems function in the secretion of a variety of toxins and other virulence factors directly from the cytoplasm to the extracellular milieu in a single energized step (285,286). The type I system comprises three components: an outer membrane channel-forming protein (TolC in E. coli), a periplasmic adaptor or membrane fusion protein, and an inner-membrane pump that typically belongs to the ATP-binding cassette family. In contrast to most RTX proteins, TosA remains associated with the bacterial surface following secretion, rather than being released into the external environment (287). The tosA gene is present in a pathogenicity island in UPEC strains, particularly those of the B2 phylogenetic group, in an operon together with genes encoding a type I secretion system (287,288). tosA is expressed in vivo during infection of the urinary tract (287) and a CFT073 ΔtosA mutant was defective for colonization of both the bladder and kidneys in the mouse UTI model (289). Evidence suggests that tosA also enhances fitness during disseminated infections (287). Expression of TosA promotes adherence to both murine and human kidney epithelial cells, but does not appear to be important for colonization of the lower urinary tract (287).
ADHESINS EXPRESSED BY GRAM-POSITIVE UROPATHOGENS
While Gram-negative bacteria are responsible for the majority of UTIs, Gram-positive bacteria are also significant uropathogens. Staphylococcus saprophyticus is the second leading cause of community acquired UTIs in sexually active women, accounting for ~15% of all incidences (290). Entercoccus faecalis, a normal member of the gut flora, is also a causative agent of UTIs, particularly in nosocomial infections (291). The adhesive organelles of Gram-positive bacteria differ significantly from those of Gram-negative bacteria in structure and assembly mechanism, but fall into the same two general classes of pilus or surface-located adhesins. Gram-positive pili, like their Gram-negative counterparts, consist of multiple pilin subunits linked together to form an extended, hairlike fiber (21,22,292). The majority of non-pilus adhesins associated with UTIs are multi-domain proteins known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), which are anchored directly to the bacterial cell wall (22,23).
MSCRAMMs
The extracellular matrix is a complex protein network that serves as the major scaffolding component of eukaryotic cells and tissues, and mediates numerous essential cellular processes including morphogenesis and differentiation (293,294). Gram-positive bacteria utilize components of the extracellular matrix such as collagen, fibronectin, and laminin as binding ligands to promote adherence, colonization, and biofilm formation (295–297). To achieve this, Gram-positive bacteria express the MSCRAMM family of surface-associated adhesins. MSCRAMMs are multi-domain proteins that are covalently linked to the peptidoglycan cell wall, exposing both conserved and non-conserved regions to the extracellular milieu. Different MSCRAMM domains confer specific functionality, including ligand binding, cell wall anchoring, and structural integrity (22). Crystal structures have been solved for domains from several different MSCRAMMs, including SdrG from Staphylococcus epidermidis, Ace from E. faecalis, and Cna and ClfA from Staphylococcus aureus, providing a structural context for understanding the assembly and functions of MSCRAMMs (298–305). MSCRAMMs that have been associated with uropathogenic bacteria are Ace, expressed by E. feacalis, and UafA, UafB and SdrI, expressed by S. saprophyticus (306–309).
Structure of MSCRAMMs
All MSCRAMMs share a common domain organization. The N terminus contains a signal peptide that directs the proteins for translocation across the cytoplasmic membrane by the Sec pathway. The N-terminal signal peptide is followed by a multi-domain central region, where the major functional and structural diversity resides (22). The C terminus contains a conserved cell wall sorting signal (CWSS), which consists of the amino acid sequence LPXTG (X represents any amino acid), followed by a hydrophobic transmembrane domain, followed by a positively charged cytoplasmic tail (Fig. 6A). This CWSS is required for the covalent linkage of MSCRAMMs to the cell wall (310).
The MSCRAMM functional region is typically divided into A and B regions. The N-terminal A region is responsible for ligand binding and specificity; the C-terminal B region can have both binding and structural properties. Variations on this organization occur, including the presence of an R structural region instead of or in addition to a B region (22). The C-terminal structural domains function to project the binding domain away from the bacterial surface. Similar to the CU pilins of Gram-negative bacteria, MSCRAMMs, as well as Gram-positive pilins, have exploited the Ig fold as a common building block (58). Two Ig variants present in both MSCRAMMs and Gram-positive pilins are the DEv-IgG and IgG-rev folds (Figs. 7B and 8) (22). These folds were first observed in the A and B regions of the S. aureus Cna and ClfA MSCRAMMs (305,311). A typical IgG constant domain contains two β-sheets (sheets 1 and 2) of four and three β-strands, arranged into a barrel configuration. DEv-IgG folds contain the same overall structure, but with the addition of at least two β-strands between strands D and E of sheet 1. The IgG-rev fold has a typical two sheet, seven-stranded barrel, but the strands are arranged in a reverse order compared to typical Ig folds.
Figure 7.
(A) Model for MSCRAMM secretion and incorporation into the cell wall. The domain organization of a typical MSCRAMM is shown at the bottom. MSCRAMMs have an N-terminal Sec signal sequence for translocation across the cytoplasmic membrane. The protein remains anchored in the cytoplasmic membrane by the CWSS. The positively charged C terminus remains in the cytoplasm, orienting the LPXTG motif to the extracellular side of the membrane. The SrtA sortase cleaves between the Thr and Gly of the MSCRAMM LPXTG motif, forming a covalent thioacyl intermediate. The MSCRAMM is then transferred to a lipid II peptidoglycan precursor and finally integrated into the cell wall at an amino acid cross-bridge. (B) Crystal structures of the Ace and UafA MSCRAMMs (PDB IDs: 2Z1P and 3IRP, respectively). The upper structure shows the N1 and N2 subdomains of Ace in blue and green, respectively; the yellow circle represents bound collagen. Both domains have DEv-Ig folds. The C terminus of the N2 subdomain inserts into the N1 subdomain, forming a latch. The lower structure depicts the N2, N3 and B subdomains of UafA. The N2 and N3 subdomains adopt DEv-Ig folds and the B subdomain adopts a variant of the IgG-rev fold. The loop connecting the N3 and B domains (cyan) is thought to insert into the N2 subdomain upon ligand binding to form a latch.
Figure 8.
Crystal structures of representative tip and major pilins. The RrgA tip pilin of S. pneumoniae (PDB ID: 2WW8) is shown on the left, with the fold adopted by each of the four subdomains indicated. The VWA domain is depicted in green, with the residues forming the MIDAS motif and bound magnesium ion shown in purple. The residues involved in intramolecular isopeptide bond formation are shown in red. The Spy0128 major pilin of S. pyogenes (PDB ID: 3B2M) is depicted on the right in purple and the two subdomains are labeled as for RrgA. The lysine side chain of Syp0128 thought to be involved in intermolecular isopeptide bond formation is shown in cyan.
Ace and the collagen hug model
The E. faecalis Ace (adhesin to collagen of E. faecalis) protein was the first MSCRAMM to be associated with UTIs (295,297,306,312). Much of the structural organization of Ace was delineated based on its homology to Cna (295,302,303). The functional portion of Ace is made up of A and B regions. The A region is divided into two DEv-IgG subdomains, N1 (146 amino acids) and N2 (135 amino acids). These domains are essential for binding extracellular matrix proteins such as laminin and collagen I and IV (297). Crystal structures have been solved for the Ace N1 and N2 subdomains (Fig. 7B) (300,301). Both subdomains are composed of ten β-strands forming two β-sheets in a sandwich like configuration. A missing G strand in sheet 1 of the N1 subdomain is complemented by a C-terminal extension of the N2 subdomain, forming an interface between the two subdomains (Fig. 7B). This interface, in addition to a collagen-binding site on the N2 subdomain and a linker region connecting the two subdomains, forms a deep, tunnel-like collagen binding trench. Together, the N1 and N2 subdomains form a dynamic cooperative structure utilizing a “collagen hug” model of ligand binding (301).
Structural and functional studies of both Cna and Ace have led to a mechanistic understanding of ligand binding by the A region. The structures of the N1 and N2 subdomains of the A region show a closed form of the molecule, in which the N1 and N2 subdomains and the inter-domain linker interact with each other, creating a tunnel to accommodate and secure collagen (Fig. 7B) (302). This closed form of the molecule is unable to initiate binding to collagen, and evidence indicates that the N1 and N2 subdomains exist in equilibrium between open and closed conformations (301). In the open conformation, a shallow groove in the N2 subdomain binds the repeating glycine-proline-hydroxylproline (GPO)n, triple helical peptide of collagen with low affinity. Following binding, the C-terminal extension of the N2 subdomain orients and inserts into a trench in the N1 subdomain, complementing a missing N1 β-strand and allowing the N1 and N2 subdomains to come into close proximity of each other, shrinking the hole and “hugging” the collagen molecule in place (301,302). The N2 C-terminal extension also acts as a latch, securing the complex. Truncations of the latch cause a decreased affinity for collagen, likely due to the N1 and N2 subdomains insufficiently securing the collagen in place (301).
The B region of Ace has not been resolved, but a high degree of sequence homology between this region and the B region of Cna provides insight into its structural organization and function. The Ace B region comprises five repeating domains, B1–5 (301). The crystal structure of the Cna B domain indicates that it functions as a structural element rather than in ligand binding (303). The Cna B1 domain is divided into two IgG-rev fold subdomains, D1 and D2. Domains B2–5 share the same structure, and the repeating B domains are arranged in an accordion-like fashion, which acts as a stalk supporting the A region and extending it distally from the surface of the cell (303). C terminal to the B region is the characteristic CWSS, which allows anchoring to the peptidoglycan cell wall by the housekeeping sortase enzyme.
UafA and the dock, lock and latch model
Another MSCRAMM shown to impact bacterial adherence in the urinary tract is UafA (uro-adherence factor A), expressed by S. saprophyticus (307). Information obtained from MSCRAMMs such as the S. aureus Cna and ClfA, as well as the recent crystal structure of the functional region of UafA, have provided insight into the structural organization and function of UafA (Fig. 7B) (313). UafA contains an N-terminal Sec signal sequence, a 72 kDa A region, a 13 kDa B region, a 148 kDa R region, and a C-terminal CWSS. Unlike Ace however, the UafA A region comprises three subdomains, N1–3, and the B region contains a single, non-repeating domain. The additional R region is rich in serine and glutamine residues and of low complexity (313).
The A region of UafA is required for ligand binding and shares the same variant DEv-IgG folds in its N2 and N3 subdomains as seen in Ace, Cna, and ClfA (305,313). The single B domain does not act as a stalk to extend the A region from the surface, but instead is a required component in ligand binding (313). A short loop links the B domain to the N3 subdomain, and in the three-dimensional structure the B domain also resides adjacent to the N2 subdomain (Fig. 7B). The low-complexity R region is thought to form the UafA stalk, supporting both the upstream A and B regions (313).
A “dock, lock and latch” model has been proposed for UafA, based on similarities with ClfA and the S. epidermidis fibrinogen-binding protein SdrG (298,299,304). This model is mechanistically related to that of the collagen hug model of Ace and Cna, but has distinct features. The N3 subdomain is composed of two β-sheets with a total of nine β-strands in the configuration of A, B, D, E and C, D1, D2, F and G (313). The N2 subdomain contains two similar β-sheets, but sheet 2 lacks a D β-strand and in its place exists a D loop, which does not appear to hydrogen bond with the adjacent E strand. The dock, lock and latch model proposes that in the apo form of the complex, the ligand is first captured by binding between the N2 and N3 subdomains. This docking event then triggers a conformational change to engage the loop connecting the N3 subdomain and the B domain (Fig. 7B), causing the loop to insert into the pocket between the D and E β-strands of the N2 domain, completing the missing β-strand and forming the “latch” (298).
The ligand for UafA has not yet been defined. UafA functions in hemagglutination and this activity is dependent on the N2 and N3 subdomains, as well as the B domain (313). Hemmagglutination studies with proteinase K-treated erythrocytes suggest that the ligand is not a protein, but may instead be a carbohydrate or lipid molecule (313). In addition, the B domain may also have secondary ligand binding properties of its own, independent of the N2 and N3 subdomains (313).
Assembly of MSCRAMMs
The majority of surface exposed proteins in Gram-positive bacteria utilize a highly conserved assembly mechanism that facilitates translocation of these proteins across the cytoplasmic membrane and their covalent linkage to the peptidoglycan cell wall. As with most Gram-negative adhesins, MSCRAMMs are synthesized with an N-terminal signal sequence that directs the proteins for translocation across the cytoplasmic membrane via the Sec general secretory pathway. The signal sequence is then cleaved following translocation by the signal peptidase (22,314). Since Gram-positive bacteria lack an outer membrane, proteins translocated across the cytoplasmic membrane will be lost to the extracellular milieu unless they are anchored to the peptidoglycan cell wall (315). Proteins destined to be anchored to the cell wall, including MSCRAMMs, contain the highly conserved CWSS, comprising the LPXTG motif, a stretch of hydrophobic residues, and a positively charged C-terminal tail. Mutations to this motif abrogate cell wall anchoring (316). Following passage across the cytoplasmic membrane via the Sec pathway, the hydrophobic region of the CWSS forms a transmembrane domain, with the positively charged tail orienting the domain such that the C terminus remains in the cytoplasm and the LPXTG motif is exposed on the extracellular surface of the membrane (Fig. 6A). Anchoring of the secreted protein to the call wall is then achieved by processing of the LPXTG motif by membrane associated proteins known as sortases (317,318).
Sortases are cysteine transpeptidases responsible for sorting covalently linked cell wall proteins to the bacterial surface, including many virulence factors (319). While different classes of sortases exist, most Gram-positive bacteria express a general housekeeping sortase required for displaying a broad spectrum of proteins with distinct functions (320). Sortase A (SrtA) is the best characterized of these housekeeping sortases (318). SrtA is responsible for processing a newly secreted protein’s CWSS by catalyzing a transpeptidation reaction between the LPXTG motif and an amino acid cross-bridge of the peptidoglycan (Fig. 7A). SrtA cleaves the carbonyl carbon between the threonine and glycine residues of the LPXTG motif via nucleophilic attack by the conserved active site cysteine of the sortase (318). This cleavage facilitates the creation of a thioacyl bond between the SrtA cysteine and the threonine of the surface protein, resulting in the two proteins being covalently linked together (Fig. 7A). SrtA then transfers the covalently linked protein to lipid II, a membrane bound peptidoglycan precursor. An amino group of lipid II nucleophilically cleaves the sortase-surface protein thioacyl linkage, forming an isopeptide bond with the surface protein threonine and creating a lipid II-surface protein complex (Fig. 7A). The lipid II-surface protein complex is then modified by transpeptidases and transglycosylases during peptidoglycan synthesis. The lipid moiety is processed and the resulting protein and peptidoglycan fragment is integrated into to the cell wall, covalently anchoring the protein to the peptidoglycan amino acid cross-bridge and exposing it to the surface (Fig. 7A) (321,322).
Functions of MSCRAMMs in Uropathogenic Bacteria
The initial stage of adherence and colonization is absolutely necessary for pathogenic Gram-positive bacteria to establish successful infection (323). MSCRAMMs mediate this process by facilitating the recognition and binding of surface exposed host ligands to promote colonization of specific tissues. In the context of UTIs, Gram-positive uropathogens express and assemble specialized MSCRAMM molecules with a distinct tropism for urogenital epithelia. This tropism is dependent both on the ligand expressed by the host tissue, in most cases a component of the extracellular matrix, as well as the affinity for that ligand by the bacterial adhesin. However, compared to well-characterized Gram-negative adhesins such as type 1 and P pili, much less is known about the mechanisms of MCRAMMs during bacterial infection of the urinary tract.
Ace
E. faecalis, a major contributor of endocarditis and infections of the blood, wounds and abdomen, is also associated with high incidences of nosocomial catheter associated urinary tract infections (CAUTIs) (324,325). The Ace A domain is required for recognition and binding of extracellular matrix molecules. In vitro mutational analysis as well as competitive inhibition studies of the A domain show that the protein has binding affinity for laminin and types I and IV collagen, each of which is a major component of the extracellular matrix (297). Ace has been identified as an important virulence determinant of E. faecalis in UTIs, using the murine infection model. Lebreton and colleagues showed that significantly higher doses of an E. faecalis ace deletion mutant are required to establish infection in mice when compared to wild-type E. faecalis (306). Furthermore, organ burden analysis revealed that an ace deletion mutant is attenuated in its ability to colonize renal tissue (306,312). While current research implicates collagen as the major Ace binding ligand, further work needs to be done to determine the role of Ace and its binding partners in the establishment of UTIs.
UafA
S. saprophyticus holds significant prevalence as a causative agent of uncomplicated UTIs (326). Comparative genomic analysis between S. aureus, S. epidermidis, and S. saprophyticus revealed that S. saprophyticus contains the unique MSCRAMM UafA (307). Expression of uafA by S. saprophyticus promotes adherence to human bladder carcinoma cells, while deletion of the uafA gene causes decreased adherence (307). Furthermore, strains of S. saprophyticus that express UafA, in addition to other surface proteins such as SdrI and Ssp, are internalized into human bladder carcinoma cells (327). Although the precise ligand or tissue tropism of UafA remains unknown, preliminary data suggest that the ligand may be a carbohydrate or lipid molecule rather than a protein (313).
UafB
UafB is a recently discovered, plasmid-encoded MSCRAMM of S. saprophyticus that affects bacterial adherence to uroepithelial cell lines (308). In addition to a predicted N-terminal Sec signal sequence and C-terminal CWSS, the majority of the 2279 residue UafB protein comprises three serine-rich tandem repeats (repeats 1–3) and a single non-repeating region. The non-repeating region lies between repeats 1 and 2, and is a putative binding domain. The third repeat region is the longest of the three and is located just upstream of the CWSS. S. saprophyticus UafB is predicted to be glycosylated on the surface of S. saprophyticus (308). Although less prevalent than UafA among S. saprophyticus isolates, strains expressing UafB exhibit increased adhesion to human bladder carcinoma cells when compared to a uafB mutant (308). Analysis of the putative non-repeat binding domain indicates that UafB binds both fibronectin and fibrinogen, but not collagen types I, II or IV, laminin, or vitronectin (308). This information may suggest that UafB has a tropism for bladder epithelium since human bladder carcinoma cells abundantly express fibronectin (328). However, a role for UafB during infection remains to be established, as both wild-type and uafB knockout strains equally colonize mouse bladders in a murine ascending UTI model (308).
SdrI
SdrI is a recently discovered MSCRAMM in S. saprophyticus classified by its serine-aspartate repeat (SD or Sdr) region, which is indicative of the Sdr family of MSCRAMMs. Sequence analysis of SdrI shows that it contains a C-terminal CWSS, an N-terminal A region, a B region containing two repeat domains, and the SD region (329). Initial binding experiments showed that when SdrI was deleted, S. saprophyticus exhibited a significant deficiency in adhesion to collagen compared to wild-type bacteria (329). Further work indicated that SdrI also has affinity for fibronectin, which is dependent on its A domain (296). This is unique since SdrI does not contain any known fibronectin binding motifs. In vivo data using a murine UTI model revealed that sdrI mutant S. saprophyticus bacteria do not have defects in initial colonization of the bladder or kidneys when compared to wild-type bacteria; however, the mutant strain is cleared faster from these organs (309). This suggests a role for SdrI in the persistence of S. saprophyticus in the urinary tract, rather than in initial adhesion or colonization.
Additional surface-located adhesins
A unique class of Gram-positive surface exposed adhesins has been discovered that lack both a Sec secretion signal as well as a cell wall localization LPXTG motif (330). These adhesins are not classified as MSCRAMMs or pili. While little has been elucidated as to their structure, one such adhesin, the enterococal fibronectin-binding protein A (EfbA), expressed by E. faecalis, has been implicated in mouse models of ascending urinary tract infections. An efbA deletion strain was significantly attenuated in its ability to bind immobilized fibronectin in vitro as well as mouse kidneys and bladders in vivo (331).
An additional protein, Esp, has been implicated in colonization and persistence of E. faecalis in the urinary tract (332). Esp has not been classified as an MSCRAMM, but it contains an N-terminal Sec signal sequence, a central region with a non-repeat domain followed by a number of repeating sequences, and a CWSS-like region at the C terminus with a variation of the LPXTG motif. The N-terminal region of Esp promotes biofilm formation in vitro, but may do this indirectly (333).
GRAM-POSITIVE PILI
Although first described in 1968, the assembly of pili by Gram-positive bacteria has only been widely recognized and characterized since the work of Ton-That and Scheewind beginning in 2003 (17–20). Multi-subunit, peptidoglycan-linked adhesive pili have now been described on the surface of a number of Gram-positive bacteria (21,22). Like MSCRAMMs, pili play important roles in binding to and recognizing extracellular matrix molecules, colonization of host tissues, and biofilm formation. Gram-positive pili are expressed from gene clusters encoding one or more minor pilins, a major pilin, and pilus assembly machinery in the form of pilus-specific sortases. Gram-positive pili expressed by C. diphtheria and Streptococcal pathogens have been well studied and serve as prototypes (334–338). Although similar to Gram-negative pili in their general ultrastructure (hairlike polymeric fibers) and functions (adhesion to and colonization of surfaces), Gram-positive pili have unique features, including the presence of intramolecular and intermolecular covalent bonds that stabilize the fibers against factors encountered in the extracellular environment. Currently, E. faecalis and E. faecium are the only Gram-positive bacteria known to express pili associated with UTIs. These pili were first identified as biofilm determinants during endocarditis infections and are named Ebp for endocarditis and biofilm associated pilus (339,340).
Structure of Gram-Positive Pili
The pilus fiber
Gram-positive pili form thin, extended fibers one protein subunit wide (~3 nm diameter) and up to several micrometers in length. The pilus fiber is built from repeating copies of covalently linked major pilus subunits, with minor pilins covalently incorporated at different points in the fiber. Minor pilins that function to recognize and bind target host ligands are known as tip pilins, while pillins that anchor the entire pilus fiber to the cell wall are referred to as base pilins. The major pilin forms the pilus shaft, which functions to extend the adhesive tip pilin distally from the bacterial surface.
A number of crystal structures have been solved for Gram-positive pilins, including from C. diphtheria, Streptoccocal species, and Bacillus cereus (Fig. 8) (22,334–338,341). Pilins share a common domain organization with MSCRAMMs, comprising an N-terminal Sec secretion signal, a central structural region, and a C-terminal CWSS (Fig. 9). The central structural region typically contains multiple N subdomains that adopt DEv-IgG or IgG-rev folds (Fig. 8) (335,342). An important characteristic of Gram-positive pilins is their ability to form covalent, intramolecular isopeptide bonds between lysine and asparagine residues of the DEv-IgG and IgG-rev folds (Fig. 8) (343). These isopeptide bonds promote resistance to proteases and increase pilin stuctural stability (334). DEv-IgG folds typically posses a D-type isopeptide bond, in which a lysine residue in the A β-strand of sheet 1 forms a bond with an asparagine on the antiparallel F β-strand of sheet 2 (Fig. 8). A C β-strand aspartic acid residue on sheet 2 catalyzes this reaction. IgG-rev folds exhibit an E-type bond where a lysine on the A β-strand of sheet 1 is covalently linked to the asparagine of the parallel G β-strand, also of sheet 1 (Fig. 8). A glutamine residue on the sheet 2 E β-strand catalyzes this reaction (22).
Figure 9.
Model for Gram-positive pilus polymerization and incorporation into the cell wall. The domain organization of typical major and minor pilins is shown at the bottom, along with the ebp gene cluster coding for Ebp pili of E. faecalis. The steps of secretion across the cytoplasmic membrane and covalent linkage to a sortase are the same as for MSCRAMMs, except the pilins are processed by the SrtC pilus-specific sortase. Pilus subunits are polymerized by formation of intermolecular isopeptide bonds between the Lys of a pilin motif of one subunit and the Thr of the LPXTG motif of a preceding subunit in the fiber. Linkage to the cell wall occurs when a growing pilus fiber is transferred to a base pilin bound to the SrtA housekeeping sortase. Integration of the pilus into the cell wall follows the mechanism as described for MSCRAMMs.
In addition to their LPXTG motif, major pilins also have a conserved pilin motif, typically of the sequence VYPK, housing an essential lysine residue (22). An intermolecular isopeptide bond is formed between the lysine of the pilin motif and the threonine of the LPXTG motif of a neighboring subunit in the pilus fiber, catalyzed by a pilus-specific sortase (Fig. 9). Tip pilins have an LPXTG motif but typically lack a pilin motif, and therefore can only be incorporated at the beginning of the pilus fiber. Base pilins, which contain both LPXTG and pilin motifs, are linked to the peptidoglycan cell wall via their LPXTG motif by the housekeeping sortase, thus anchoring the pilus fiber to the bacterial surface (Fig. 9) (336). In some instances, base pilins can also be incorporated along the length of the pilus fiber (19,344). Some pilins, particularly major pilins, may also possess a third motif called an E-box, which contains an invariant glutamic acid residue. Little work has been done to elucidate the role of the E-box in pilus assembly, but it may facilitate integration of minor pilins throughout the pilus fiber (19).
Ebp pili
The Ebp operon, found in both E. faecalis and E. faecium, consists of four genes, ebpA, ebpB, ebpC, and srtC on what is termed a pilus island on the bacterial chromosome (Fig. 9) (340). Little is known about the structure of Ebp pili, but studies have shown that EbpA and EbpB are minor pilins, with EbpA serving as the tip pilin and EbpB as the base pilin (340). EbpC is the major structural pilin. SrtC is a class C pilus specific sortase required for polymerization of the pilus fiber (340,345,346). Sequence analysis of these pilins suggests that they share structural characteristics with known MSCRAMMs, having potential Dev-IgG and IgG-rev domains (347). EbpA and EbpC also possess predicted E-box motifs (340,347).
Sequence and mutational analysis of EbpA predicts that the tip pilin contains a von Willebrand factor A (VWA) domain with a metal ion-dependent adhesion site (MIDAS) (Fig. 8), which facilitates adhesion to host molecules (345,348). VWA domains are found in proteins from different kingdoms and contain a β-sheet surrounded by a number of α-helices. Many VWA domain proteins also contain a MIDAS motif (348). Functional analysis of the EbpA MIDAS motif indicates that it is essential for EbpA function (348,349). While the ligand specificity of EbpA remains unknown, other VWA domain- and MIDAS motif-containing proteins bind extracellular matrix proteins such as collagen (350).
Assembly of Gram-Positive Pili
The assembly mechanisms of Gram-positive pili in bacteria such as C. diphtheriae have been well studied and share many commonalities with the anchoring of MSCRAMMs to the cell wall (19,20,292). Additional pilus specific features such as the pilin motif and class C sortases are required for biogenesis of the multi-subunit pilus fiber. The initial stages of pilus assembly are essentially the same as for all LPXTG cell wall anchored proteins. Like MSCRAMMs, Gram-positive pilins contain a cleavable N-terminal signal sequence for targeted translocation across the cytoplasmic membrane via the general Sec pathway. Following translocation, the transmembrane domain and the positively charged cytoplasmic tail of the CWSS retains and orients the pilin as so to make the LPXTG motif accessible for cleavage by a sortase (Fig. 9). Gram-positive pilus gene clusters typically encode a class C pilus specific sortase (srtC), which is essential for pilus polymerization (340). Differences between the pilus specific and housekeeping (SrtA) sortases determine how the LPXTG motif of individual pilins are processed, either for covalent linkage to the lipid II peptidoglycan precursor via SrtA, or for the formation of structural pilin-pilin isopeptide bonds via SrtC.
For both major and minor pilins, SrtC cleaves between the threonine and glycine residues of the pilin LPXTG motif, forming an acyl-enzyme intermediates between the active site cysteine of the sortase and the threonine of the pilin (Fig. 9). Major pilins contain a pilin motif (VYPK) in addition to the LPXTG motif, whereas tip pilins typically only contain the LPXTG motif. To incorporate the tip pilin into the pilus fiber, SrtC catalyzes formation of an intramolecular isopeptide bond between the lysine of the pilin motif of the major pilin with the threonine of the LPXTG motif of the tip pilin (Fig. 9). This reaction releases the tip pilin from its SrtC molecule, forming a tip pilin-major pilin-SrtC complex (351). This process is then repeated to add additional major pilins to the complex, elongating the pilus structure and forming the pilus shaft to project the tip pilin distally from the bacterial surface. Pilus elongation is terminated via incorporation of a minor base pilin into the pilus structure by the same mechanism (21,292). Following elongation, the pilus fiber must be securely anchored to the peptidoglycan cell wall. This is achieved by the SrtA housekeeping sortase, which catalyzes isopeptide bond formation between the LPXTG motif of the base pilin and a lipid II amino group (Fig. 9) (345). As for MSCRAMMs, the lipid II-pilus fiber complex is then processed by transpeptidases and transglycosylases, covalently securing the pilus to the peptidoglycan cross-bridge.
Function of Ebp Pili in UTIs
The Ebp pili expressed by E. faecalis and E. faecium, two major causative agents of hospital acquired CAUTI, are the only Gram-positive pili that have been associated with UTIs (345,352). The connection between Ebp pili-mediated biofilm formation and infection was first investigated in a rat endovascular model (340,353). Data from this work showed that strains with deletions of each individual pilin, or a deletion of srtC alone, had significant defects in biofilm formation in vivo. Additionally, a deletion strain of just the tip pilin ebpA was significantly attenuated for colonization of aortic vegetations and kidneys, when compared to wild-type bacteria, suggesting a role for Ebp pili in UTIs as well as endocarditis (340). Later studies confirmed the role of Ebp pili in colonization of the urogenital tract, using ebpA and ebpC deletion strains to show a colonization defect in both the kidneys and bladders of mice (346,352). Similar results were observed in experiments using ebp deletion mutants in E. faecium (339).
Experiments have also been done to identify the role of Ebp pili in CAUTI. Using a murine CAUTI model, an E. faecalis strain lacking the ebpABC pilus genes displayed significantly reduced adherence compared to the wild-type strain to bladders and silicone implants placed in the bladder to mimic catheters (345). Interestingly, bacteria lacking only the major pilin EpbC colonized mouse bladders and implants similar to wild-type bacteria, suggesting that expression of just the minor pilins on the bacterial surface is sufficient to mediate colonization. Although the ligand(s) for Ebp pili remain unknown, mutations introduced into the MIDAS motif of the EbpA tip pilin drastically inhibited bacterial adhesion in the CAUTI model, indicating a role for this motif in EbpA function and pilus-mediated adhesion (345).
CONCLUDING REMARKS
The diverse array of adhesins expressed by uropathogenic bacteria reflects the importance of adhesion to colonization. Pathogens invading into the urinary tract must have strong adherence properties to overcome the washing action of the flow of urine. In addition, adhesins expressed by bacteria must be able to withstand mechanical forces exerted by urine flow, to avoid being sheared off once they have bound to their receptors on the urothelial surface. These factors underlie the prominence of pilus adhesins such as type 1 and P pili, which are adapted to function in the urinary tract, among the virulence factors of uropathogens. Uropathogenic bacteria encode multiple different types of adhesins, providing specificity for different niches within the urinary tract, as well as redundancy in function to ensure maintenance of adhesion under varying conditions. Bacterial colonization is also enhanced by formation of bacterial-bacterial interactions and biofilm structures, explaining the association of adhesins such as curli and Ag43 with uropathogens.
Adhesion is crucial at early stages of infection, and thus represents an attractive target for therapeutic intervention. Advances in understanding the structure and assembly of bacterial adhesins will be critical for the development of effective vaccines and antimicrobial agents that target adhesion. Pilus-based vaccines have shown promise in preventing UTIs, although none has reached clinical use. Studies using purified P pili or recombinant PapG adhesin purified in complex with its PapD chaperone demonstrated protection against pyelonephritis in primates (163,354). Similarly, vaccination with the type 1 pilus adhesin FimH, purified in complex with its FimC chaperone, provided protection in primates against challenge with a UPEC cystitis isolate (129). Interestingly, a recent study found that rather than blocking type 1 pilus-mediated adhesion, the host antibody response may actually enhance binding of FimH to its ligands by stabilizing the adhesin’s high-affinity binding state (355). An improved understanding of pilus adhesion mechanisms under conditions in the urinary tract may allow tailoring of the antigen used for vaccination to provoke a more effective immune response. In addition to pili, surface-located adhesins such as the MSCRAMMs expressed by Gram-positive bacteria also represent viable targets for vaccine development (356,357).
An alternative approach to vaccination is to disrupt bacterial adhesion to host cells through the use of small molecule competitive inhibitors of adhesin-receptor interactions. Examples include the use of galabiose- and mannose-based inhibitors to interfere with adhesion mediated by P and type 1 pili, respectively, with the goal of preventing UPEC colonization of the urinary tract (358–361). Another alternative approach is to develop small molecule inhibitors that disrupt the machinery used for adhesin biogenesis, thereby preventing assembly of the adhesins on the bacterial surface. Once such class of small molecules developed by Almqvist and colleagues, termed pilicides, interferes with the CU assembly pathway and blocks expression of both P and type 1 pili by E. coli (362). Pilicides target the periplasmic chaperone and appear to disrupt pilus assembly by interfering with binding of chaperone-subunit complexes to the outer membrane usher. Modified pilicides were recently developed that also had activity against curli (203). Treatment of UPEC with one of these inhibitors blocked assembly of both curli and type 1 pili, reduced biofilm formation, and attenuated bacterial colonization in the murine urinary tract infection model (203). These compounds highlight the potential for a new class of anti-infective agents that target virulence factor secretion systems and the assembly of virulence-associated surface structures, rather than disrupting essential biological processes as for conventional antibiotics (363,364). Such anti-virulence molecules should place less pressure on the bacteria and thus may be less prone to the development of resistance mechanisms. This strategy may also allow the selective targeting of pathogenic bacteria, avoiding detrimental side effects of broad-spectrum antibiotics on the normal bacterial flora.
ACKNOWLEDGEMENTS
We thank Matthew Chapman (University of Michigan, Ann Arbor) for providing structural coordinates for CsgA. This work was supported by award number R01GM62987 from the National Institute of General Medical Sciences. P.C. is supported by award number T32AI007539 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottow JCG. Ecology, physiology and genetics of fimbriae and pili. Annu. Rev. Microbiol. 1975;29:79–108. doi: 10.1146/annurev.mi.29.100175.000455. [DOI] [PubMed] [Google Scholar]
- 5.Duguid JP, Smith IW, Dempster G, Edmunds PN. Non-flagellar filamentous appendages ("fimbriae") and hemagglutinating activity in bacterium coli. J. Pathol. Bacteriol. 1955;70:335–348. doi: 10.1002/path.1700700210. [DOI] [PubMed] [Google Scholar]
- 6.Brinton CC. Non-flagellar appendages of bacteria. Nature. 1959;183:782–786. doi: 10.1038/183782a0. [DOI] [PubMed] [Google Scholar]
- 7.Duguid JP, Anderson ES, Campbell I. Fimbriae and adhesive properties in Salmonellae. J. Pathol. Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- 8.Orskov I, Orskov F. Serologic classification of fimbriae. Curr. Top. Microbiol. Immunol. 1990;151:71–90. [PubMed] [Google Scholar]
- 9.Low D, Braaten B, van der Woude M. Fimbriae. In: Neidhardt FC, editor. Escherichia Coli and Salmonella; Cellular and Molecular Biology. 2nd ed. Washington DC: ASM Press; 1996. pp. 146–157. [Google Scholar]
- 10.Thanassi DG, Nuccio S-P, Shu Kin So S, Bäumler AJ. Fimbriae: Classification and Biochemistry. In: Bôck A, Curtiss R III, Kaper JB, Neidhardt FC, Nyström T, Rudd KE, Squires CL, editors. EcoSal–Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 2007. [Online] http://www.ecosal.org. [DOI] [PubMed] [Google Scholar]
- 11.Duguid JP, Clegg S, Wilson MI. The fimbrial and non-fimbrial haemagglutinins of Escherichia coli. J. Med. Microbiol. 1979;12:213–227. doi: 10.1099/00222615-12-2-213. [DOI] [PubMed] [Google Scholar]
- 12.Old DC. Inhibition of the interaction between fimbrial hemagglutinatinins and erythrocytes by D-mannose and other carbohydrates. J. Gen. Microbiol. 1972;71:149–157. doi: 10.1099/00221287-71-1-149. [DOI] [PubMed] [Google Scholar]
- 13.Hull RA, Gill RE, Hsu P, Minshaw BH, Falkow S. Construction and expression of recombinant plasmids encoding type 1 and D-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker J, Schmidt G, Hughes C, Knapp S, Marget M, Goebel W. Cloning and characterization of genes involved in production of mannose-resistant neuraminidase-susceptible (X) fimbriae from a uropathogenic 06:K15:H31 Escherichia coli strain. Infect. Immun. 1985;47:434–440. doi: 10.1128/iai.47.2.434-440.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fronzes R, Remaut H, Waksman G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008;27:2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnhart M, Chapman M. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagawa R, Otsuki K. Some properties of the pili of Corynebacterium renale. J. Bacteriol. 1970;101:1063–1069. doi: 10.1128/jb.101.3.1063-1069.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagawa R, Otsuki K, Tokui T. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn. J. Vet. Res. 1968;16:31–37. [PubMed] [Google Scholar]
- 19.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 20.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 21.Danne C, Dramsi S. Pili of gram-positive bacteria: roles in host colonization. Res. Microbiol. 2012;163:645–658. doi: 10.1016/j.resmic.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Vengadesan K, Narayana SV. Structural biology of Gram-positive bacterial adhesins. Protein. Sci. 2011;20:759–772. doi: 10.1002/pro.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009;17:423–430. doi: 10.1016/j.tim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zav'yalov V, Zavialov A, Zav'yalova G, Korpela T. Adhesive organelles of Gram-negative pathogens assembled with the classical chaperone/usher machinery: structure and function from a clinical standpoint. FEMS Microbiol. Rev. 2010;34:317–378. doi: 10.1111/j.1574-6976.2009.00201.x. [DOI] [PubMed] [Google Scholar]
- 26.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol. Rev. 2012;36:1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller E, Garcia T, Hultgren S, Oberhauser AF. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fallman E, Schedin S, Jass J, Uhlin BE, Axner O. The unfolding of the P pili quaternary structure by stretching is reversible, not plastic. EMBO Rep. 2005;6:52–56. doi: 10.1038/sj.embor.7400310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 31.Castelain M, Ehlers S, Klinth J, Lindberg S, Andersson M, Uhlin BE, Axner O. Fast uncoiling kinetics of F1C pili expressed by uropathogenic Escherichia coli are revealed on a single pilus level using force-measuring optical tweezers. Eur. Biophys. J. 2011;40:305–316. doi: 10.1007/s00249-010-0648-1. [DOI] [PubMed] [Google Scholar]
- 32.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 34.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 2007;3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 36.Mossman KL, Mian MF, Lauzon NM, Gyles CL, Lichty B, Mackenzie R, Gill N, Ashkar AA. Cutting edge: FimH adhesin of type 1 fimbriae is a novel TLR4 ligand. J. Immunol. 2008;181:6702–6706. doi: 10.4049/jimmunol.181.10.6702. [DOI] [PubMed] [Google Scholar]
- 37.Bergsten G, Samuelsson M, Wullt B, Leijonhufvud I, Fischer H, Svanborg C. PapG-dependent adherence breaks mucosal inertia and triggers the innate host response. J. Infect. Dis. 2004;189:1734–1742. doi: 10.1086/383278. [DOI] [PubMed] [Google Scholar]
- 38.Plancon L, Du Merle L, Le Friec S, Gounon P, Jouve M, Guignot J, Servin A, Le Bouguenec C. Recognition of the cellular beta1-chain integrin by the bacterial AfaD invasin is implicated in the internalization of afa-expressing pathogenic Escherichia coli strains. Cell Microbiol. 2003;5:681–693. doi: 10.1046/j.1462-5822.2003.00308.x. [DOI] [PubMed] [Google Scholar]
- 39.Oelschlaeger TA, Dobrindt U, Hacker J. Pathogenicity islands of uropathogenic E. coli and the evolution of virulence. Int. J. Antimicrob. Agents. 2002;19:517–521. doi: 10.1016/s0924-8579(02)00092-4. [DOI] [PubMed] [Google Scholar]
- 40.van der Velden AW, Baumler AJ, Tsolis RM, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korea CG, Badouraly R, Prevost MC, Ghigo JM, Beloin C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 2010;12:1957–1977. doi: 10.1111/j.1462-2920.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- 42.Hatkoff M, Runco LM, Pujol C, Jayatilaka I, Furie MB, Bliska JB, Thanassi DG. Roles of chaperone/usher pathways of Yersinia pestis in a murine model of plague and adhesion to host cells. Infect. Immun. 2012;80:3490–3500. doi: 10.1128/IAI.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wurpel DJ, Beatson SA, Totsika M, Petty NK, Schembri MA. Chaperone-Usher Fimbriae of Escherichia coli. PLoS ONE. 2013;8:e52835. doi: 10.1371/journal.pone.0052835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blomfield IC. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 2001;45:1–49. doi: 10.1016/s0065-2911(01)45001-6. [DOI] [PubMed] [Google Scholar]
- 45.van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 46.Xia Y, Gally D, Forsman-Semb K, Uhlin BE. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB operon. EMBO J. 2000;19:1450–1457. doi: 10.1093/emboj/19.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Totsika M, Beatson SA, Holden N, Gally DL. Regulatory interplay between pap operons in uropathogenic Escherichia coli. Mol. Microbiol. 2008;67:996–1011. doi: 10.1111/j.1365-2958.2007.06098.x. [DOI] [PubMed] [Google Scholar]
- 48.Snyder JA, Haugen BJ, Lockatell CV, Maroncle N, Hagan EC, Johnson DE, Welch RA, Mobley HL. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 2005;73:7588–7596. doi: 10.1128/IAI.73.11.7588-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane MC, Simms AN, Mobley HL. complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J. Bacteriol. 2007;189:5523–5533. doi: 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armbruster CE, Mobley HL. Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat. Rev. Microbiol. 2012;10:743–754. doi: 10.1038/nrmicro2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 2005;18:264–292. doi: 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 53.Sauer FG, Fütterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 54.Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 55.Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 56.Phan G, Remaut H, Wang T, Allan WJ, Pirker KF, Lebedev A, Henderson NS, Geibel S, Volkan E, Yan J, Kunze MBA, Pinkner JS, Ford B, Kay CWM, Li H, Hultgren S, Thanassi DG, Waksman G. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson KL, Billington J, Pettigrew D, Cota E, Simpson P, Roversi P, Chen HA, Urvil P, du Merle L, Barlow PN, Medof ME, Smith RA, Nowicki B, Le Bouguenec C, Lea SM, Matthews S. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol. Cell. 2004;15:647–657. doi: 10.1016/j.molcel.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 59.Henderson NS, Ng TW, Talukder I, Thanassi DG. Function of the usher N-terminus in catalysing pilus assembly. Mol. Microbiol. 2011;79:954–967. doi: 10.1111/j.1365-2958.2010.07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puorger C, Eidam O, Capitani G, Erilov D, Grutter MG, Glockshuber R. Infinite kinetic stability against dissociation of supramolecular protein complexes through donor strand complementation. Structure. 2008;16:631–642. doi: 10.1016/j.str.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 61.Bullitt E, Makowski L. Structural polymorphism of bacterial adhesion pili. Nature. 1995;373:164–167. doi: 10.1038/373164a0. [DOI] [PubMed] [Google Scholar]
- 62.Pettigrew D, Anderson KL, Billington J, Cota E, Simpson P, Urvil P, Rabuzin F, Roversi P, Nowicki B, du Merle L, Le Bouguenec C, Matthews S, Lea SM. High resolution studies of the Afa/Dr adhesin DraE and its interaction with chloramphenicol. J. Biol. Chem. 2004;279:46851–46857. doi: 10.1074/jbc.M409284200. [DOI] [PubMed] [Google Scholar]
- 63.Korotkova N, Le Trong I, Samudrala R, Korotkov K, Van Loy CP, Bui AL, Moseley SL, Stenkamp RE. Crystal structure and mutational analysis of the DaaE adhesin of Escherichia coli. J. Biol. Chem. 2006;281:22367–22377. doi: 10.1074/jbc.M604646200. [DOI] [PubMed] [Google Scholar]
- 64.Zavialov A, Zav'yalova G, Korpela T, Zav'yalov V. FGL chaperone-assembled fimbrial polyadhesins: anti-immune armament of Gram-negative bacterial pathogens. FEMS Microbiol. Rev. 2007;31:478–514. doi: 10.1111/j.1574-6976.2007.00075.x. [DOI] [PubMed] [Google Scholar]
- 65.Cota E, Jones C, Simpson P, Altroff H, Anderson KL, du Merle L, Guignot J, Servin A, Le Bouguenec C, Mardon H, Matthews S. The solution structure of the invasive tip complex from Afa/Dr fibrils. Mol. Microbiol. 2006;62:356–366. doi: 10.1111/j.1365-2958.2006.05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dodson KW, Pinkner JS, Rose T, Magnusson G, Hultgren SJ, Waksman G. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 67.Hung CS, Bouckaert J, Hung D, Pinkner J, Widberg C, DeFusco A, Auguste CG, Strouse R, Langermann S, Waksman G, Hultgren SJ. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 2002;44:903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- 68.Sung MA, Fleming K, Chen HA, Matthews S. The solution structure of PapGII from uropathogenic Escherichia coli and its recognition of glycolipid receptors. EMBO Rep. 2001;2:621–627. doi: 10.1093/embo-reports/kve133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buts L, Bouckaert J, De Genst E, Loris R, Oscarson S, Lahmann M, Messens J, Brosens E, Wyns L, De Greve H. The fimbrial adhesin F17-G of enterotoxigenic Escherichia coli has an immunoglobulin-like lectin domain that binds N-acetylglucosamine. Mol. Microbiol. 2003;49:705–715. doi: 10.1046/j.1365-2958.2003.03600.x. [DOI] [PubMed] [Google Scholar]
- 70.Merckel MC, Tanskanen J, Edelman S, Westerlund-Wikstrom B, Korhonen TK, Goldman A. The structural basis of receptor-binding by Escherichia coli associated with diarrhea and septicemia. J. Mol. Biol. 2003;331:897–905. doi: 10.1016/s0022-2836(03)00841-6. [DOI] [PubMed] [Google Scholar]
- 71.Li YF, Poole S, Rasulova F, McVeigh AL, Savarino SJ, Xia D. A Receptor-binding Site as Revealed by the Crystal Structure of CfaE, the Colonization Factor Antigen I Fimbrial Adhesin of Enterotoxigenic Escherichia coli. J. Biol. Chem. 2007;282:23970–23980. doi: 10.1074/jbc.M700921200. [DOI] [PubMed] [Google Scholar]
- 72.Westerlund-Wikstrom B, Korhonen TK. Molecular structure of adhesin domains in Escherichia coli fimbriae. Int. J. Med. Microbiol. 2005;295:479–486. doi: 10.1016/j.ijmm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 73.Jedrzejczak R, Dauter Z, Dauter M, Piatek R, Zalewska B, Mroz M, Bury K, Nowicki B, Kur J. Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped beta-tails. Acta Crystallogr. D Biol. Crystallogr. 2006;62:157–164. doi: 10.1107/S0907444905036747. [DOI] [PubMed] [Google Scholar]
- 74.Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas WE. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J. Biol. Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, Rodriguez V, Interlandi G, Klevit R, Vogel V, Stenkamp RE, Sokurenko EV, Thomas WE. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Forero M, Yakovenko O, Sokurenko EV, Thomas WE, Vogel V. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 2006;4:e298. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aprikian P, Interlandi G, Kidd BA, Le Trong I, Tchesnokova V, Yakovenko O, Whitfield MJ, Bullitt E, Stenkamp RE, Thomas WE, Sokurenko EV. The bacterial fimbrial tip acts as a mechanical force sensor. PLoS Biol. 2011;9:e1000617. doi: 10.1371/journal.pbio.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lycklama ANJA, Driessen AJ. The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones CH, Dexter P, Evans AK, Liu C, Hultgren SJ, Hruby DE. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 2002;184:5762–5771. doi: 10.1128/JB.184.20.5762-5771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slonim LN, Pinkner JS, Branden CI, Hultgren SJ. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 1992;11:4747–4756. doi: 10.1002/j.1460-2075.1992.tb05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holmgren A, Brändén C. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature. 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 82.Kuehn MJ, Ogg DJ, Kihlberg J, Slonim LN, Flemmer K, Bergfors T, Hultgren SJ. Structural basis of pilus subunit recognition by the PapD chaperone. Science. 1993;262:1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 83.Zavialov AV, Kersley J, Korpela T, Zav'yalov VP, MacIntyre S, Knight SD. Donor strand complementation mechanism in the biogenesis of non-pilus systems. Mol. Microbiol. 2002;45:983–995. doi: 10.1046/j.1365-2958.2002.03066.x. [DOI] [PubMed] [Google Scholar]
- 84.Barnhart MM, Pinkner JS, Soto GE, Sauer FG, Langermann S, Waksman G, Frieden C, Hultgren SJ. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hung DL, Knight SD, Woods RM, Pinkner JS, Hultgren SJ. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 1996;15:3792–3805. [PMC free article] [PubMed] [Google Scholar]
- 86.Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu XD, Fooks LJ, Moslehi-Mohebi E, Tischenko VM, Askarieh G, Knight SD, Macintyre S, Zavialov AV. Large is fast, small is tight: determinants of speed and affinity in subunit capture by a periplasmic chaperone. J. Mol. Biol. 2012;417:294–308. doi: 10.1016/j.jmb.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 88.Crespo MD, Puorger C, Scharer MA, Eidam O, Grutter MG, Capitani G, Glockshuber R. Quality control of disulfide bond formation in pilus subunits by the chaperone FimC. Nat. Chem. Biol. 2012;8:707–713. doi: 10.1038/nchembio.1019. [DOI] [PubMed] [Google Scholar]
- 89.Di Yu X, Dubnovitsky A, Pudney AF, Macintyre S, Knight SD, Zavialov AV. Allosteric mechanism controls traffic in the chaperone/usher pathway. Structure. 2012;20:1861–1871. doi: 10.1016/j.str.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 90.Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, Hultgren SJ, Thanassi DG, Waksman G, Li H. Fiber Formation across the Bacterial Outer Membrane by the Chaperone/Usher Pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of Pilus Assembly Reveals a Bacterial Outer Membrane Catalyst. Science. 2008;320:376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- 92.Soto GE, Dodson KW, Ogg D, Liu C, Heuser J, Knight S, Kihlberg J, Jones CH, Hultgren SJ. Periplasmic chaperone recognition motif of subunits mediates quaternary interactions in the pilus. EMBO J. 1998;17:6155–6167. doi: 10.1093/emboj/17.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sauer FG, Remaut H, Hultgren SJ, Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 94.Vetsch M, Erilov D, Moliere N, Nishiyama M, Ignatov O, Glockshuber R. Mechanism of fibre assembly through the chaperone-usher pathway. EMBO Rep. 2006;7:734–738. doi: 10.1038/sj.embor.7400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu J, Kape JB. Cloning and characterization of the eae gene of enterohaemorrhaic Escherichia coli O157:H7. Mol. Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 96.Jacob-Dubuisson F, Striker R, Hultgren SJ. Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 1994;269:12447–12455. [PubMed] [Google Scholar]
- 97.Thanassi DG, Stathopoulos C, Karkal A, Li H. Protein secretion in the absence of ATP: the autotransporter, two-partner secretion, and chaperone/usher pathways of Gram-negative bacteria. Mol. Membr. Biol. 2005;22:63–72. doi: 10.1080/09687860500063290. [DOI] [PubMed] [Google Scholar]
- 98.Zavialov AV, Tischenko VM, Fooks LJ, Brandsdal BO, Aqvist J, Zav'yalov VP, Macintyre S, Knight SD. Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem J. 2005;389:685–694. doi: 10.1042/BJ20050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee YM, Dodson KW, Hultgren SJ. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J. Bacteriol. 2007;189:5276–5283. doi: 10.1128/JB.01648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rose RJ, Verger D, Daviter T, Remaut H, Paci E, Waksman G, Ashcroft AE, Radford SE. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12873–12878. doi: 10.1073/pnas.0802177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nishiyama M, Glockshuber R. The outer membrane usher guarantees the formation of functional pili by selectively catalyzing donor-strand exchange between subunits that are adjacent in the mature pilus. J. Mol. Biol. 2010;396:1–8. doi: 10.1016/j.jmb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 102.Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ. Outer membrane PapC usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Q, Ng TW, Dodson KW, So SS, Bayle KM, Pinkner JS, Scarlata S, Hultgren SJ, Thanassi DG. The differential affinity of the usher for chaperone-subunit complexes is required for assembly of complete pili. Mol. Microbiol. 2010;76:159–172. doi: 10.1111/j.1365-2958.2010.07089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nishiyama M, Horst R, Eidam O, Herrmann T, Ignatov O, Vetsch M, Bettendorff P, Jelesarov I, Grutter MG, Wuthrich K, Glockshuber R, Capitani G. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shu Kin So S, Thanassi DG. Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol. Microbiol. 2006;60:364–375. doi: 10.1111/j.1365-2958.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- 107.Mapingire OS, Henderson NS, Duret G, Thanassi DG, Delcour AH. Modulating effects of the plug, helix and N-and C-terminal domains on channel properties of the PapC usher. J. Biol. Chem. 2009;284:36324–36333. doi: 10.1074/jbc.M109.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ford B, Rego AT, Ragan TJ, Pinkner J, Dodson K, Driscoll PC, Hultgren S, Waksman G. Structural homology between the C-terminal domain of the PapC usher and its plug. J. Bacteriol. 2010;192:1824–1831. doi: 10.1128/JB.01677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang Y, Smith BS, Chen LX, Baxter RH, Deisenhofer J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7403–7407. doi: 10.1073/pnas.0902789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ng TW, Akman L, Osisami M, Thanassi DG. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 2004;186:5321–5331. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H, Qian L, Chen Z, Thahbot D, Liu G, Liu T, Thanassi DG. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 2004;344:1397–1407. doi: 10.1016/j.jmb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 112.Allen WJ, Phan G, Hultgren SJ, Waksman G. Dissection of Pilus Tip Assembly by the FimD Usher Monomer. J. Mol. Biol. 2013;425:958–967. doi: 10.1016/j.jmb.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Munera D, Hultgren S, Fernandez LA. Recognition of the N-terminal lectin domain of FimH adhesin by the usher FimD is required for type 1 pilus biogenesis. Mol. Microbiol. 2007;64:333–346. doi: 10.1111/j.1365-2958.2007.05657.x. [DOI] [PubMed] [Google Scholar]
- 114.Nowicki B, Selvarangan R, Nowicki S. Family of Escherichia coli Dr adhesins: decay-accelerating factor receptor recognition and invasiveness. J. Infect. Dis. 2001;183(Suppl 1):S24–S27. doi: 10.1086/318846. [DOI] [PubMed] [Google Scholar]
- 115.Marre R, Kreft B, Hacker J. Genetically engineered S and F1C fimbriae differ in their contribution to adherence of Escherichia coli to cultured renal tubular cells. Infect. Immun. 1990;58:3434–3437. doi: 10.1128/iai.58.10.3434-3437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schmoll T, Morschhauser J, Ott M, Ludwig B, van Die I, Hacker J. Complete genetic organization and functional aspects of the Escherichia coli S fimbrial adhesion determinant: nucleotide sequence of the genes sfa B, C, D, E, F. Microb. Pathog. 1990;9:331–343. doi: 10.1016/0882-4010(90)90067-z. [DOI] [PubMed] [Google Scholar]
- 117.Riegman N, Kusters R, H VV, Bergmans H, Van Bergen En Henegouwen P, Hacker J, Van Die I. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster andidentification of minor subunits. J. Bacteriol. 1990;172:1114–1120. doi: 10.1128/jb.172.2.1114-1120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HL. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect. Immun. 2011;79:4753–4763. doi: 10.1128/IAI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Buckles EL, Bahrani-Mougeot FK, Molina A, Lockatell CV, Johnson DE, Drachenberg CB, Burland V, Blattner FR, Donnenberg MS. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 2004;72:3890–3901. doi: 10.1128/IAI.72.7.3890-3901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ulett GC, Mabbett AN, Fung KC, Webb RI, Schembri MA. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology. 2007;153:2321–2331. doi: 10.1099/mic.0.2006/004648-0. [DOI] [PubMed] [Google Scholar]
- 121.Labigne-Roussel A, Falkow S. Distribution and degree of heterogeneity of the afimbrial-adhesin-encoding operon (afa) among uropathogenic Escherichia coli isolates. Infect. Immun. 1988;56:640–648. doi: 10.1128/iai.56.3.640-648.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Loy CP, Sokurenko EV, Moseley SL. The major structural subunits of Dr and F1845 fimbriae are adhesins. Infect. Immun. 2002;70:1694–1702. doi: 10.1128/IAI.70.4.1694-1702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Berger CN, Billker O, Meyer TF, Servin AL, Kansau I. Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC) Mol. Microbiol. 2004;52:963–983. doi: 10.1111/j.1365-2958.2004.04033.x. [DOI] [PubMed] [Google Scholar]
- 124.Carnoy C, Moseley SL. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol. Microbiol. 1997;23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 125.Schmoll T, Hoschutzky H, Morschhauser J, Lottspeich F, Jann K, Hacker J. Analysis of genes coding for the sialic acid-binding adhesin and two other minor fimbrial subunits of the S-fimbrial adhesin determinant of Escherichia coli. Mol. Microbiol. 1989;3:1735–1744. doi: 10.1111/j.1365-2958.1989.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 126.Virkola R, Parkkinen J, Hacker J, Korhonen TK. Sialyloligosaccharide chains of laminin as an extracellular matrix target for S fimbriae of Escherichia coli. Infect. Immun. 1993;61:4480–4484. doi: 10.1128/iai.61.10.4480-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Korhonen TK, Parkkinen J, Hacker J, Finne J, Pere A, Rhen M, Holthofer H. Binding of Escherichia coli S fimbriae to human kidney epithelium. Infect. Immun. 1986;54:322–327. doi: 10.1128/iai.54.2.322-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Backhed F, Alsen B, Roche N, Angstrom J, von Euler A, Breimer ME, Westerlund-Wikstrom B, Teneberg S, Richter-Dahlfors A. Identification of target tissue glycosphingolipid receptors for uropathogenic, F1C-fimbriated Escherichia coli and its role in mucosal inflammation. J. Biol. Chem. 2002;277:18198–18205. doi: 10.1074/jbc.M111640200. [DOI] [PubMed] [Google Scholar]
- 129.Langermann S, Mollby R, Burlein JE, Palaszynski SR, Auguste CG, DeFusco A, Strouse R, Schenerman MA, Hultgren SJ, Pinkner JS, Winberg J, Guldevall L, Soderhall M, Ishikawa K, Normark S, Koenig S. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 2000;181:774–778. doi: 10.1086/315258. [DOI] [PubMed] [Google Scholar]
- 130.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner JS, Burlein J, Barren P, Koenig S, Leath S, Jones CH, Hultgren SJ. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 131.Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Schembri MA, Hultgren SJ. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol. Rev. 2012;36:616–648. doi: 10.1111/j.1574-6976.2012.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abraham SN, Sun D, Dale JB, Beachey EH. Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae . Nature. 1988;336:682–684. doi: 10.1038/336682a0. [DOI] [PubMed] [Google Scholar]
- 133.Pak J, Pu Y, Zhang ZT, Hasty DL, Wu XR. Tamm-Horsfall protein binds to type 1 fimbriated Escherichia coli and prevents E. coli from binding to uroplakin Ia and Ib receptors. J. Biol. Chem. 2001;276:9924–9930. doi: 10.1074/jbc.M008610200. [DOI] [PubMed] [Google Scholar]
- 134.Baorto DM, Gao Z, Malaviya R, Dustin ML, van der Merwe A, Lublin DM, Abraham SN. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature. 1997;389:636–639. doi: 10.1038/39376. [DOI] [PubMed] [Google Scholar]
- 135.Kukkonen M, Raunio T, Virkola R, Lahteenmaki K, Makela PH, Klemm P, Clegg S, Korhonen TK. Basement membrane carbohydrate as a target for bacterial adhesion: binding of type 1 fimbriae of Salmonella enterica and Escherichia coli to laminin. Mol. Microbiol. 1993;7:229–227. doi: 10.1111/j.1365-2958.1993.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 136.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 137.Jones CH, Pinkner JS, Roth R, Heuser J, Nicholoes AV, Abraham SN, Hultgren SJ. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae . Proc. Natl. Acad. Sci. U. S. A. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Haner M, Taschner N, Burkhard P, Aebi U, Muller SA. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- 139.Zhou G, Mo WJ, Sebbel P, Min G, Neubert TA, Glockshuber R, Wu XR, Sun TT, Kong XP. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J. Cell Sci. 2001;114:4095–4103. doi: 10.1242/jcs.114.22.4095. [DOI] [PubMed] [Google Scholar]
- 140.Thumbikat P, Berry RE, Zhou G, Billips BK, Yaggie RE, Zaichuk T, Sun TT, Schaeffer AJ, Klumpp DJ. Bacteria-induced uroplakin signaling mediates bladder response to infection. PLoS Pathog. 2009;5:e1000415. doi: 10.1371/journal.ppat.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Connell H, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Song J, Bishop BL, Li G, Grady R, Stapleton A, Abraham SN. TLR4-mediated expulsion of bacteria from infected bladder epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14966–14971. doi: 10.1073/pnas.0900527106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 144.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blango MG, Mulvey MA. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob. Agents Chemother. 2010;54:1855–1863. doi: 10.1128/AAC.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen SL, Hung CS, Pinkner JS, Walker JN, Cusumano CK, Li Z, Bouckaert J, Gordon JI, Hultgren SJ. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22439–22444. doi: 10.1073/pnas.0902179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 2001;69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mysorekar IU, Hultgren SJ. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14170–14175. doi: 10.1073/pnas.0602136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bock K, Breimer ME, Brignole A, Hansson GC, Karlsson K-A, Larson G, Leffler H, Samuelsson BE, Strömberg N, Svanborg-Edén C, Thurin J. Specificity of binding of a strain of uropathogenic Escherichia coli to Gala(1–4)Gal-containing glycosphingolipids. J. Biol. Chem. 1985;260:8545–8551. [PubMed] [Google Scholar]
- 150.Roberts JA, Marklund B-I, Ilver D, Haslam D, Kaack MB, Baskin G, Louis M, Mollby R, Winberg J, Normark S. The Gal(alpha1–4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kallenius G, Mollby R, Svenson SB, Windberg J, Lundblud A, Svenson S, Cedergen B. The Pk antigen as receptor for the haemagglutinin of pyelonephritogenic Escherichia coli . FEMS Microbiol. Lett. 1980;7:297–302. [Google Scholar]
- 152.Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao EM, Laturnus C, Diehl I, Glodde S, Homeier T, Bohnke U, Steinruck H, Philipp HC, Wieler LH. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 2007;297:163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 153.Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli . Proc. Natl. Acad. Sci. U. S. A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lund B, Marklund BI, Stromberg N, Lindberg F, Karlsson KA, Normark S. Uropathogenic Escherichia coli Can Express Serologically Identical Pili of Different Receptor-Binding Specificities. Mol. Microbiol. 1988;2:255–263. doi: 10.1111/j.1365-2958.1988.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 155.Stromberg N, Marklund BI, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson KA, Normark S. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galalpha(1–4)Gal-containing isoreceptors. EMBO J. 1990;9:2001–2010. doi: 10.1002/j.1460-2075.1990.tb08328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Johnson JR, Russo TA, Brown JJ, Stapleton A. papG alleles of Escherichia coli strains causing first-episode or recurrent acute cystitis in adult women. J. Infect. Dis. 1998;177:97–101. doi: 10.1086/513824. [DOI] [PubMed] [Google Scholar]
- 157.Kuehn MJ, Heuser J, Normark S, Hultgren SJ. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature. 1992;356:252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- 158.Jacob-Dubuisson F, Heuser J, Dodson K, Normark S, Hultgren SJ. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 1993;12:837–847. doi: 10.1002/j.1460-2075.1993.tb05724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mu XQ, Bullitt E. Structure and assembly of P-pili: a protruding hinge region used for assembly of a bacterial adhesion filament. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9861–9866. doi: 10.1073/pnas.0509620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Baga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- 161.Verger D, Miller E, Remaut H, Waksman G, Hultgren S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli . EMBO Rep. 2006;7:1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hagberg L, Hull R, Hull S, Falkow S, Freter R, Svanborg Eden C. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect. Immun. 1983;40:265–272. doi: 10.1128/iai.40.1.265-272.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Roberts JA, Hardaway K, Kaack B, Fussell EN, Baskin G. Prevention of pyelonephritis by immunization with P-fimbriae. J. Urol. 1984;131:602–607. doi: 10.1016/s0022-5347(17)50513-3. [DOI] [PubMed] [Google Scholar]
- 164.O'Hanley P, Lark D, Falkow S, Schoolnik G. Molecular basis of Escherichia coli colonizatin of the upper urinary tract in BALB/c mice: Gal-Gal pili immunization prevents Escherichia coli pyelonephritis. J. Clin. Invest. 1985;83:2102–2108. doi: 10.1172/JCI111707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lane MC, Mobley HL. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 2007;72:19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 166.Hedlund M, Svensson M, Nilsson Å, Duan R-D, Svanborg C. Role of the ceramide-signaling pathway in cytokine responses to P-fimbriated Escherichia coli . J. Exp. Med. 1996;183:1037–1044. doi: 10.1084/jem.183.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hedlund M, Wachtler C, Johansson E, Hang L, Somerville JE, Darveau RP, Svanborg C. P fimbriae-dependent, lipopolysaccharide-independent activation of epithelial cytokine responses. Mol. Microbiol. 1999;33:693–703. doi: 10.1046/j.1365-2958.1999.01513.x. [DOI] [PubMed] [Google Scholar]
- 168.Fischer H, Ellstrom P, Ekstrom K, Gustafsson L, Gustafsson M, Svanborg C. Ceramide as a TLR4 agonist; a putative signalling intermediate between sphingolipid receptors for microbial ligands and TLR4. Cell. Microbiol. 2007;9:1239–1251. doi: 10.1111/j.1462-5822.2006.00867.x. [DOI] [PubMed] [Google Scholar]
- 169.Zhang JP, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]
- 170.Austin JW, Sanders G, Kay WW, Collinson SK. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 1998;162:295–301. doi: 10.1111/j.1574-6968.1998.tb13012.x. [DOI] [PubMed] [Google Scholar]
- 171.Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gophna U, Barlev M, Seijffers R, Oelschlager T, Hacker J, Ron E. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli . Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 174.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli . Mol. Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 175.Ben Nasr A, Olsen A, Sjobring U, Muller-Esterl W, Bjorck L. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli . Mol. Microbiol. 1996;20:927–935. doi: 10.1111/j.1365-2958.1996.tb02534.x. [DOI] [PubMed] [Google Scholar]
- 176.Olsen A, Herwald H, Wikstrom M, Persson K, Mattsson E, Bjorck L. Identification of two protein-binding and functional regions of curli, a surface organelle and virulence determinant of Escherichia coli . J. Biol. Chem. 2002;277:34568–34572. doi: 10.1074/jbc.M206353200. [DOI] [PubMed] [Google Scholar]
- 177.Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 178.Kai-Larsen Y, Luthje P, Chromek M, Peters V, Wang X, Holm A, Kadas L, Hedlund KO, Johansson J, Chapman MR, Jacobson SH, Romling U, Agerberth B, Brauner A. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010;6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tukel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Baumler AJ. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol. 2010;12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 181.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid--from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 184.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, Wickner RB. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J. Biol. Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis . J. Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 188.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J. Mol. Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli . Proc. Natl. Acad. Sci. U. S. A. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang X, Zhou Y, Ren JJ, Hammer ND, Chapman MR. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Barak J, Gorski L, Naraghi-Arani P, Charkowski A. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 2005;71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 193.Taylor JD, Zhou Y, Salgado PS, Patwardhan A, McGuffie M, Pape T, Grabe G, Ashman E, Constable SC, Simpson PJ, Lee WC, Cota E, Chapman MR, Matthews SJ. Atomic resolution insights into curli fiber biogenesis. Structure. 2011;19:1307–1316. doi: 10.1016/j.str.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Romling U, Rohde M, Olsen A, Normark S, Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 195.Hammer N, Schmidt J, Chapman M. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J. Mol. Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 198.Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 2011;81:486–499. doi: 10.1111/j.1365-2958.2011.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U. S. A. 2009;106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tukel C, Raffatellu M, Humphries A, Wilson R, Andrews-Polymenis H, Gull T, Figueiredo J, Wong M, Michelsen K, Akcelik M, Adams L, Baumler A. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 201.Hung C, Marschall J, Burnham CA, Byun AS, Henderson JP. The bacterial amyloid curli is associated with urinary source bloodstream infection. PLoS One. 2014;9:e86009. doi: 10.1371/journal.pone.0086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Olsen A, Wick M, Morgelin M, Bjorck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli Fibers Are Required for Development of Biofilm Architecture in Escherichia coli K-12 and Enhance Bacterial Adherence to Human Uroepithelial Cells. Microbiol. Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 205.Cherny I, Rockah L, Levy-Nissenbaum O, Gophna U, Ron EZ, Gazit E. The formation of Escherichia coli curli amyloid fibrils is mediated by prion-like peptide repeats. J. Mol. Biol. 2005;352:245–252. doi: 10.1016/j.jmb.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 206.Leo JC, Grin I, Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat. Rev. Microbiol. 2012;10:213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 208.Jose J, Jahnig F, Meyer TF. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol. Microbiol. 1995;18:378–380. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 209.Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 210.Oomen CJ, Van Ulsen P, Van Gelder P, Feijen M, Tommassen J, Gros P. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Phadnis SH, Ilver D, Janzon L, Normark S, Westblom TU. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori . Infect. Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Emsley P, Charles IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 213.Benz I, Schmidt MA. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 214.Charbonneau ME, Janvore J, Mourez M. Autoprocessing of the Escherichia coli AIDA-I autotransporter: a new mechanism involving acidic residues in the junction region. J. Biol. Chem. 2009;284:17340–17351. doi: 10.1074/jbc.M109.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schembri MA, Dalsgaard D, Klemm P. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 2004;186:1249–1257. doi: 10.1128/JB.186.5.1249-1257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hasman H, Chakraborty T, Klemm P. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 1999;181:4834–4841. doi: 10.1128/jb.181.16.4834-4841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 2010;78:1659–1669. doi: 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Parham NJ, Srinivasan U, Desvaux M, Foxman B, Marrs CF, Henderson IR. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli . FEMS Microbiol. Lett. 2004;230:73–83. doi: 10.1016/S0378-1097(03)00862-0. [DOI] [PubMed] [Google Scholar]
- 219.Guyer DM, Henderson IR, Nataro JP, Mobley HL. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli . Mol. Microbiol. 2000;38:53–66. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 220.Alamuri P, Lower M, Hiss JA, Himpsl SD, Schneider G, Mobley HL. Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis . Infect. Immun. 2010;78:4882–4894. doi: 10.1128/IAI.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 222.van den Berg B. Crystal structure of a full-length autotransporter. J. Mol. Biol. 2010;396:627–633. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 223.Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nature Struct. Mol. Biol. 2007;14:1214–1220. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Klauser T, Kramer J, Otzelberger K, Pohlner J, Meyer TF. Characterization of the Neisseria Iga beta-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol. 1993;234:579–593. doi: 10.1006/jmbi.1993.1613. [DOI] [PubMed] [Google Scholar]
- 225.Oliver DC, Huang G, Fernandez RC. Identification of secretion determinants of the Bordetella pertussis BrkA autotransporter. J. Bacteriol. 2003;185:489–495. doi: 10.1128/JB.185.2.489-495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Maurer J, Jose J, Meyer TF. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 1999;181:7014–7020. doi: 10.1128/jb.181.22.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Roggenkamp A, Ackermann N, Jacobi CA, Truelzsch K, Hoffmann H, Heesemann J. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 2003;185:3735–3744. doi: 10.1128/JB.185.13.3735-3744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Surana NK, Cutter D, Barenkamp SJ, St Geme JW., 3rd The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 2004;279:14679–14685. doi: 10.1074/jbc.M311496200. [DOI] [PubMed] [Google Scholar]
- 229.Meng G, Surana NK, St Geme JW, 3rd, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 2006;25:2297–2304. doi: 10.1038/sj.emboj.7601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yeo HJ, Cotter SE, Laarmann S, Juehne T, St Geme JW, Waksman G. Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter. EMBO J. 2004;23:1245–1256. doi: 10.1038/sj.emboj.7600142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M, Goldman A. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 2004;23:701–711. doi: 10.1038/sj.emboj.7600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli . J. Bacteriol. 2008;190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW., 3rd Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 2011;30:3864–3874. doi: 10.1038/emboj.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Khan S, Mian HS, Sandercock LE, Chirgadze NY, Pai EF. Crystal structure of the passenger domain of the Escherichia coli autotransporter EspP. J. Mol. Biol. 2011;413:985–1000. doi: 10.1016/j.jmb.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Szczesny P, Lupas A. Domain annotation of trimeric autotransporter adhesins--daTAA. Bioinformatics. 2008;24:1251–1256. doi: 10.1093/bioinformatics/btn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hartmann MD, Grin I, Dunin-Horkawicz S, Deiss S, Linke D, Lupas AN, Hernandez Alvarez B. Complete fiber structures of complex trimeric autotransporter adhesins conserved in enterobacteria. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20907–20912. doi: 10.1073/pnas.1211872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Leo JC, Lyskowski A, Hattula K, Hartmann MD, Schwarz H, Butcher SJ, Linke D, Lupas AN, Goldman A. The structure of E. coli IgG-binding protein D suggests a general model for bending and binding in trimeric autotransporter adhesins. Structure. 2011;19:1021–1030. doi: 10.1016/j.str.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 239.Biedzka-Sarek M, Salmenlinna S, Gruber M, Lupas AN, Meri S, Skurnik M. Functional mapping of YadA- and Ail-mediated binding of human factor H to Yersinia enterocolitica serotype O:3. Infect. Immun. 2008;76:5016–5027. doi: 10.1128/IAI.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae . J. Bacteriol. 2009;191:6571–6583. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Leyton DL, Sevastsyanovich YR, Browning DF, Rossiter AE, Wells TJ, Fitzpatrick RE, Overduin M, Cunningham AF, Henderson IR. Size and conformation limits to secretion of disulfide-bonded loops in autotransporter proteins. J. Biol. Chem. 2011;286:42283–42291. doi: 10.1074/jbc.M111.306118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, Henderson IR. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology. 2007;153:59–70. doi: 10.1099/mic.0.29091-0. [DOI] [PubMed] [Google Scholar]
- 243.Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc. Natl. Acad. Sci. U. S. A. 2005;102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hagan CL, Silhavy TJ, Kahne D. beta-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 245.Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E383–E391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jain S, Goldberg MB. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 2007;189:5393–5398. doi: 10.1128/JB.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rossiter AE, Leyton DL, Tveen-Jensen K, Browning DF, Sevastsyanovich Y, Knowles TJ, Nichols KB, Cunningham AF, Overduin M, Schembri MA, Henderson IR. The essential beta-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis. J. Bacteriol. 2011;193:4250–4253. doi: 10.1128/JB.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol. Microbiol. 2009;71:1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 249.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17739–17744. doi: 10.1073/pnas.1009491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jose J, Kramer J, Klauser T, Pohlner J, Meyer TF. Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga beta autotransporter pathway. Gene. 1996;178:107–110. doi: 10.1016/0378-1119(96)00343-5. [DOI] [PubMed] [Google Scholar]
- 251.Jong WS, ten Hagen-Jongman CM, den Blaauwen T, Slotboom DJ, Tame JR, Wickstrom D, de Gier JW, Otto BR, Luirink J. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol. Microbiol. 2007;63:1524–1536. doi: 10.1111/j.1365-2958.2007.05605.x. [DOI] [PubMed] [Google Scholar]
- 252.Ohnishi Y, Nishiyama M, Horinouchi S, Beppu T. Involvement of the COOH-terminal pro-sequence of Serratia marcescens serine protease in the folding of the mature enzyme. J. Biol. Chem. 1994;269:32800–32806. [PubMed] [Google Scholar]
- 253.Oliver DC, Huang G, Nodel E, Pleasance S, Fernandez RC. A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol. Microbiol. 2003;47:1367–1383. doi: 10.1046/j.1365-2958.2003.03377.x. [DOI] [PubMed] [Google Scholar]
- 254.Soprova Z, Sauri A, van Ulsen P, Tame JR, den Blaauwen T, Jong WS, Luirink J. A conserved aromatic residue in the autochaperone domain of the autotransporter Hbp is critical for initiation of outer membrane translocation. J. Biol. Chem. 2010;285:38224–38233. doi: 10.1074/jbc.M110.180505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Junker M, Schuster CC, McDonnell AV, Sorg KA, Finn MC, Berger B, Clark PL. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Veiga E, de Lorenzo V, Fernandez LA. Structural tolerance of bacterial autotransporters for folded passenger protein domains. Mol. Microbiol. 2004;52:1069–1080. doi: 10.1111/j.1365-2958.2004.04014.x. [DOI] [PubMed] [Google Scholar]
- 257.Skillman KM, Barnard TJ, Peterson JH, Ghirlando R, Bernstein HD. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol. Microbiol. 2005;58:945–958. doi: 10.1111/j.1365-2958.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- 258.Sherlock O, Dobrindt U, Jensen JB, Munk Vejborg R, Klemm P. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J. Bacteriol. 2006;188:1798–1807. doi: 10.1128/JB.188.5.1798-1807.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sauri A, Soprova Z, Wickstrom D, de Gier JW, Van der Schors RC, Smit AB, Jong WS, Luirink J. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology. 2009;155:3982–3991. doi: 10.1099/mic.0.034991-0. [DOI] [PubMed] [Google Scholar]
- 260.Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the beta-domain pore during the assembly of a bacterial autotransporter. Mol. Microbiol. 2008;67:188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 261.Klemm P, Hjerrild L, Gjermansen M, Schembri MA. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli . Mol. Microbiol. 2004;51:283–296. doi: 10.1046/j.1365-2958.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 262.Henderson IR, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 263.Reidl S, Lehmann A, Schiller R, Salam Khan A, Dobrindt U. Impact of O-glycosylation on the molecular and cellular adhesion properties of the Escherichia coli autotransporter protein Ag43. Int. J. Med. Microbiol. 2009;299:389–401. doi: 10.1016/j.ijmm.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 264.van der Woude MW, Henderson IR. Regulation and function of Ag43 (flu) Annu. Rev. Microbiol. 2008;62:153–169. doi: 10.1146/annurev.micro.62.081307.162938. [DOI] [PubMed] [Google Scholar]
- 265.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 2007;75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Schembri MA, Klemm P. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 2001;20:3074–3081. doi: 10.1093/emboj/20.12.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc. Natl. Acad. Sci. U S A. 2014;111:457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Vejborg RM, Klemm P. Cellular chain formation in Escherichia coli biofilms. Microbiology. 2009;155:1407–1417. doi: 10.1099/mic.0.026419-0. [DOI] [PubMed] [Google Scholar]
- 270.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 2012;80:321–332. doi: 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Durant L, Metais A, Soulama-Mouze C, Genevard JM, Nassif X, Escaich S. Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli . Infect. Immun. 2007;75:1916–1925. doi: 10.1128/IAI.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Allsopp LP, Beloin C, Moriel DG, Totsika M, Ghigo JM, Schembri MA. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli . J. Bacteriol. 2012;194:5769–5782. doi: 10.1128/JB.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VA, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli . Proc. Natl. Acad. Sci. U. S. A. 2010;107:9072–9077. doi: 10.1073/pnas.0915077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HL, Rappuoli R, Pizza M, Serino L, Soriani M. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. MBio. 2012;3 doi: 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Isberg RR, Voorhis DL, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50:769–778. doi: 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- 276.Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, Finlay BB, Strynadka NC. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature. 2000;405:1073–1077. doi: 10.1038/35016618. [DOI] [PubMed] [Google Scholar]
- 277.Oberhettinger P, Schutz M, Leo JC, Heinz N, Berger J, Autenrieth IB, Linke D. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS One. 2012;7:e47069. doi: 10.1371/journal.pone.0047069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Leveille S, Caza M, Johnson JR, Clabots C, Sabri M, Dozois CM. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 2006;74:3427–3436. doi: 10.1128/IAI.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HL, Tarr PI. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Tarr PI, Bilge SS, Vary JC, Jr, Jelacic S, Habeeb RL, Ward TR, Baylor MR, Besser TE. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli . Nature Struct. Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 282.Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 2001;345:1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- 283.Burall LS, Harro JM, Li X, Lockatell CV, Himpsl SD, Hebel JR, Johnson DE, Mobley HL. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 2004;72:2922–2938. doi: 10.1128/IAI.72.5.2922-2938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 2010;34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 286.Delepelaire P. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 287.Vigil PD, Wiles TJ, Engstrom MD, Prasov L, Mulvey MA, Mobley HL. The repeat-in-toxin family member TosA mediates adherence of uropathogenic Escherichia coli and survival during bacteremia. Infect. Immun. 2012;80:493–505. doi: 10.1128/IAI.05713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Parham NJ, Pollard SJ, Chaudhuri RR, Beatson SA, Desvaux M, Russell MA, Ruiz J, Fivian A, Vila J, Henderson IR. Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli . J. Clin. Microbiol. 2005;43:2425–2434. doi: 10.1128/JCM.43.5.2425-2434.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Vigil PD, Alteri CJ, Mobley HL. Identification of in vivo-induced antigens including an RTX family exoprotein required for uropathogenic Escherichia coli virulence. Infect. Immun. 2011;79:2335–2344. doi: 10.1128/IAI.00110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ronald A. The etiology of urinary tract infection: traditional and emerging pathogens. Dis. Mon. 2003;49:71–82. doi: 10.1067/mda.2003.8. [DOI] [PubMed] [Google Scholar]
- 291.Koch S, Hufnagel M, Theilacker C, Huebner J. Enterococcal infections: host response, therapeutic, and prophylactic possibilities. Vaccine. 2004;22:822–830. doi: 10.1016/j.vaccine.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 292.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Chagnot C, Listrat A, Astruc T, Desvaux M. Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell Microbiol. 2012;14:1687–1696. doi: 10.1111/cmi.12002. [DOI] [PubMed] [Google Scholar]
- 294.Patti JM, Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 1994;6:752–758. doi: 10.1016/0955-0674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 295.Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, Weinstock GM, Murray BE, Hook M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis . J. Biol. Chem. 1999;274:26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 296.Sakinc T, Kleine B, Michalski N, Kaase M, Gatermann SG. SdrI of Staphylococcus saprophyticus is a multifunctional protein: localization of the fibronectin-binding site. FEMS Microbiol. Lett. 2009;301:28–34. doi: 10.1111/j.1574-6968.2009.01798.x. [DOI] [PubMed] [Google Scholar]
- 297.Nallapareddy SR, Qin X, Weinstock GM, Hook M, Murray BE. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect. Immun. 2000;68:5218–5224. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Bowden MG, Heuck AP, Ponnuraj K, Kolosova E, Choe D, Gurusiddappa S, Narayana SV, Johnson AE, Hook M. Evidence for the "dock, lock, and latch" ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J. Biol. Chem. 2008;283:638–647. doi: 10.1074/jbc.M706252200. [DOI] [PubMed] [Google Scholar]
- 299.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. A "dock, lock, and latch" structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 300.Ponnuraj K, Narayana SV. Crystal structure of ACE19, the collagen binding subdomain of Enterococus faecalis surface protein ACE. Proteins. 2007;69:199–203. doi: 10.1002/prot.21464. [DOI] [PubMed] [Google Scholar]
- 301.Liu Q, Ponnuraj K, Xu Y, Ganesh VK, Sillanpaa J, Murray BE, Narayana SV, Hook M. The Enterococcus faecalis MSCRAMM ACE binds its ligand by the Collagen Hug model. J. Biol. Chem. 2007;282:19629–19637. doi: 10.1074/jbc.M611137200. [DOI] [PubMed] [Google Scholar]
- 302.Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV. A 'Collagen Hug' model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, Bice T, Hook M, Narayana SV. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 304.Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Hook M. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008;4:e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hook M, Narayana SV. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 2002;21:6660–6672. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, Hartke A, Auffray Y, Giard JC. ace, Which encodes an adhesin in Enterococcus faecalis is regulated by Ers and is involved in virulence. Infect. Immun. 2009;77:2832–2839. doi: 10.1128/IAI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, Kuhara S, Hattori M, Ohta T. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.King NP, Beatson SA, Totsika M, Ulett GC, Alm RA, Manning PA, Schembri MA. UafB is a serine-rich repeat adhesin of Staphylococcus saprophyticus that mediates binding to fibronectin, fibrinogen and human uroepithelial cells. Microbiology. 2011;157:1161–1175. doi: 10.1099/mic.0.047639-0. [DOI] [PubMed] [Google Scholar]
- 309.Kline KA, Ingersoll MA, Nielsen HV, Sakinc T, Henriques-Normark B, Gatermann S, Caparon MG, Hultgren SJ. Characterization of a novel murine model of Staphylococcus saprophyticus urinary tract infection reveals roles for Ssp and SdrI in virulence. Infect. Immun. 2010;78:1943–1951. doi: 10.1128/IAI.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Symersky J, Patti JM, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas LJ, Hook M, Narayana SV. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 312.Nallapareddy SR, Singh KV, Sillanpaa J, Zhao M, Murray BE. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect. Immun. 2011;79:2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Matsuoka E, Tanaka Y, Kuroda M, Shouji Y, Ohta T, Tanaka I, Yao M. Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding. Protein Sci. 2011;20:406–416. doi: 10.1002/pro.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Model P, Russel M. Prokaryotic secretion. Cell. 1990;61:739–741. doi: 10.1016/0092-8674(90)90180-m. [DOI] [PubMed] [Google Scholar]
- 316.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 317.Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 319.Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol. Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 320.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 322.Ruzin A, Severin A, Ritacco F, Tabei K, Singh G, Bradford PA, Siegel MM, Projan SJ, Shlaes DM. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J. Bacteriol. 2002;184:2141–2147. doi: 10.1128/JB.184.8.2141-2147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus . Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 324.Murray BE. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Raz R, Colodner R, Kunin CM. Who are you--Staphylococcus saprophyticus? Clin. Infect. Dis. 2005;40:896–898. doi: 10.1086/428353. [DOI] [PubMed] [Google Scholar]
- 327.Szabados F, Kleine B, Anders A, Kaase M, Sakinc T, Schmitz I, Gatermann S. Staphylococcus saprophyticus ATCC 15305 is internalized into human urinary bladder carcinoma cell line 5637. FEMS Microbiol. Lett. 2008;285:163–169. doi: 10.1111/j.1574-6968.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 328.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol. 2008;190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Sakinc T, Kleine B, Gatermann SG. SdrI, a serine-aspartate repeat protein identified in Staphylococcus saprophyticus strain 7108, is a collagen-binding protein. Infect. Immun. 2006;74:4615–4623. doi: 10.1128/IAI.01885-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Holmes AR, McNab R, Millsap KW, Rohde M, Hammerschmidt S, Mawdsley JL, Jenkinson HF. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 2001;41:1395–1408. doi: 10.1046/j.1365-2958.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- 331.Torelli R, Serror P, Bugli F, Paroni Sterbini F, Florio AR, Stringaro A, Colone M, De Carolis E, Martini C, Giard JC, Sanguinetti M, Posteraro B. The PavA-like Fibronectin-Binding Protein of Enterococcus faecalis EfbA, Is Important for Virulence in a Mouse Model of Ascending Urinary Tract Infection. J. Infect. Dis. 2012 doi: 10.1093/infdis/jis440. [DOI] [PubMed] [Google Scholar]
- 332.Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 2001;69:4366–4372. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Tendolkar PM, Baghdayan AS, Shankar N. The N-terminal domain of enterococcal surface protein, Esp, is sufficient for Esp-mediated biofilm enhancement in Enterococcus faecalis . J. Bacteriol. 2005;187:6213–6222. doi: 10.1128/JB.187.17.6213-6222.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Spraggon G, Koesema E, Scarselli M, Malito E, Biagini M, Norais N, Emolo C, Barocchi MA, Giusti F, Hilleringmann M, Rappuoli R, Lesley S, Covacci A, Masignani V, Ferlenghi I. Supramolecular organization of the repetitive backbone unit of the Streptococcus pneumoniae pilus. PLoS One. 2010;5:e10919. doi: 10.1371/journal.pone.0010919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Linke C, Young PG, Kang HJ, Bunker RD, Middleditch MJ, Caradoc-Davies TT, Proft T, Baker EN. Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring. J. Biol. Chem. 2010;285:20381–20389. doi: 10.1074/jbc.M109.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes . J. Biol. Chem. 2009;284:20729–20737. doi: 10.1074/jbc.M109.014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of-Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Sillanpaa J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, Murray BE. Characterization of the ebp(fm) pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence. 2010;1:236–246. doi: 10.4161/viru.1.4.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis . J. Clin. Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Budzik JM, Poor CB, Faull KF, Whitelegge JP, He C, Schneewind O. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19992–19997. doi: 10.1073/pnas.0910887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Izore T, Contreras-Martel C, El Mortaji L, Manzano C, Terrasse R, Vernet T, Di Guilmi AM, Dessen A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae . Structure. 2010;18:106–115. doi: 10.1016/j.str.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 343.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 344.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Nielsen HV, Guiton PS, Kline KA, Port GC, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. MBio. 2012;3:e00177–e00112. doi: 10.1128/mBio.00177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sillanpaa J, Chang C, Singh KV, Montealegre MC, Nallapareddy SR, Harvey BR, Ton-That H, Murray BE. Contribution of Individual Ebp Pilus Subunits of Enterococcus faecalis OG1RF to Pilus Biogenesis, Biofilm Formation and Urinary Tract Infection. PLoS One. 2013;8:e68813. doi: 10.1371/journal.pone.0068813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Sillanpaa J, Nallapareddy SR, Prakash VP, Qin X, Hook M, Weinstock GM, Murray BE. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium . Microbiology. 2008;154:3199–3211. doi: 10.1099/mic.0.2008/017319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 350.Hilleringmann M, Giusti F, Baudner BC, Masignani V, Covacci A, Rappuoli R, Barocchi MA, Ferlenghi I. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog. 2008;4:e1000026. doi: 10.1371/journal.ppat.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. Amide bonds assemble pili on the surface of bacilli. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J. Infect. Dis. 2007;195:1671–1677. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Sillanpaa J, Xu Y, Nallapareddy SR, Murray BE, Hook M. A family of putative MSCRAMMs from Enterococcus faecalis . Microbiology. 2004;150:2069–2078. doi: 10.1099/mic.0.27074-0. [DOI] [PubMed] [Google Scholar]
- 354.Roberts JA, Kaack MB, Baskin G, Chapman MR, Hunstad DA, Pinkner JS, Hultgren SJ. Antibody responses and protection from pyelonephritis following vaccination with purified Escherichia coli PapDG protein. J. Urol. 2004;171:1682–1685. doi: 10.1097/01.ju.0000116123.05160.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Tchesnokova V, Aprikian P, Kisiela D, Gowey S, Korotkova N, Thomas W, Sokurenko E. Type 1 fimbrial adhesin FimH elicits an immune response that enhances cell adhesion of Escherichia coli . Infect. Immun. 2011;79:3895–3904. doi: 10.1128/IAI.05169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Otto M. Targeted immunotherapy for staphylococcal infections : focus on anti-MSCRAMM antibodies. BioDrugs. 2008;22:27–36. doi: 10.2165/00063030-200822010-00003. [DOI] [PubMed] [Google Scholar]
- 357.Patti JM. A humanized monoclonal antibody targeting Staphylococcus aureus . Vaccine. 2004;22(Suppl 1):S39–S43. doi: 10.1016/j.vaccine.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 358.Salminen A, Loimaranta V, Joosten JA, Khan AS, Hacker J, Pieters RJ, Finne J. Inhibition of P-fimbriated Escherichia coli adhesion by multivalent galabiose derivatives studied by a live-bacteria application of surface plasmon resonance. J. Antimicrob. Chemother. 2007;60:495–501. doi: 10.1093/jac/dkm251. [DOI] [PubMed] [Google Scholar]
- 359.Wellens A, Garofalo C, Nguyen H, Van Gerven N, Slattegard R, Hernalsteens JP, Wyns L, Oscarson S, De Greve H, Hultgren S, Bouckaert J. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One. 2008;3:e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Han Z, Pinkner JS, Ford B, Obermann R, Nolan W, Wildman SA, Hobbs D, Ellenberger T, Cusumano CK, Hultgren SJ, Janetka JW. Structure-based drug design and optimization of mannoside bacterial FimH antagonists. J. Med. Chem. 2010;53:4779–4792. doi: 10.1021/jm100438s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Schwardt O, Rabbani S, Hartmann M, Abgottspon D, Wittwer M, Kleeb S, Zalewski A, Smiesko M, Cutting B, Ernst B. Design, synthesis and biological evaluation of mannosyl triazoles as FimH antagonists. Bioorg. Med. Chem. 2011;19:6454–6473. doi: 10.1016/j.bmc.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 362.Pinkner JS, Remaut H, Buelens F, Miller E, Aberg V, Pemberton N, Hedenstrom M, Larsson A, Seed P, Waksman G, Hultgren SJ, Almqvist F. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17897–17902. doi: 10.1073/pnas.0606795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Dobrindt U, Hacker J. Targeting virulence traits: potential strategies to combat extraintestinal pathogenic E. coli infections. Curr. Opin. Microbiol. 2008;11:409–413. doi: 10.1016/j.mib.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 365.Jones CH, Dodson K, Hultgren SJ. Structure, function, and assembly of adhesive P pili p. 175–219. In: Mobley HLT, Warren JW, editors. Urinary tract infections: molecular pathogenesis and clinical management. 1 ed. Washington, DC: ASM Press; 1996. [Google Scholar]
- 366.Leffler H, Svanborg-Eden C. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect. Immun. 1981;34:920–929. doi: 10.1128/iai.34.3.920-929.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wu XR, Sun TT, Medina JJ. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9630–9635. doi: 10.1073/pnas.93.18.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Korhonen TK, Valtonen MV, Parkkinen J, Vaisanen-Rhen V, Finne J, Orskov F, Orskov I, Svenson SB, Makela PH. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Castelain M, Sjostrom AE, Fallman E, Uhlin BE, Andersson M. Unfolding and refolding properties of S pili on extraintestinal pathogenic Escherichia coli . Eur. Biophys. J. 2010;39:1105–1115. doi: 10.1007/s00249-009-0552-8. [DOI] [PubMed] [Google Scholar]
- 370.Khan AS, Kniep B, Oelschlaeger TA, Van Die I, Korhonen T, Hacker J. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli . Infect. Immun. 2000;68:3541–3547. doi: 10.1128/iai.68.6.3541-3547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Ong CL, Ulett GC, Mabbett AN, Beatson SA, Webb RI, Monaghan W, Nimmo GR, Looke DF, McEwan AG, Schembri MA. Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J. Bacteriol. 2008;190:1054–1063. doi: 10.1128/JB.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 2008;76:4055–4065. doi: 10.1128/IAI.00494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Gerlach GF, Clegg S, Allen BL. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae . J. Bacteriol. 1989;171:1262–1270. doi: 10.1128/jb.171.3.1262-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Stahlhut SG, Chattopadhyay S, Struve C, Weissman SJ, Aprikian P, Libby SJ, Fang FC, Krogfelt KA, Sokurenko EV. Population variability of the FimH type 1 fimbrial adhesin in Klebsiella pneumoniae . J. Bacteriol. 2009;191:1941–1950. doi: 10.1128/JB.00601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Tarkkanen AM, Allen BL, Westerlund B, Holthofer H, Kuusela P, Risteli L, Clegg S, Korhonen TK. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol. Microbiol. 1990;4:1353–1361. doi: 10.1111/j.1365-2958.1990.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 376.Stahlhut SG, Struve C, Krogfelt KA. Klebsiella pneumoniae type 3 fimbriae agglutinate yeast in a mannose-resistant manner. J. Med. Microbiol. 2012;61:317–322. doi: 10.1099/jmm.0.036350-0. [DOI] [PubMed] [Google Scholar]
- 377.Ong CL, Beatson SA, Totsika M, Forestier C, McEwan AG, Schembri MA. Molecular analysis of type 3 fimbrial genes from Escherichia coli Klebsiella and Citrobacter species. BMC Microbiol. 2010;10:183. doi: 10.1186/1471-2180-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Zunino P, Geymonat L, Allen AG, Preston A, Sosa V, Maskell DJ. New aspects of the role of MR/P fimbriae in Proteus mirabilis urinary tract infection. FEMS Immunol. Med. Microbiol. 2001;31:113–120. doi: 10.1111/j.1574-695X.2001.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 379.Li X, Johnson DE, Mobley HL. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis . Infect. Immun. 1999;67:2822–2833. doi: 10.1128/iai.67.6.2822-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Bahrani FK, Mobley HL. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J. Bacteriol. 1994;176:3412–3419. doi: 10.1128/jb.176.11.3412-3419.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Zunino P, Sosa V, Allen AG, Preston A, Schlapp G, Maskell DJ. Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiology. 2003;149:3231–3237. doi: 10.1099/mic.0.26534-0. [DOI] [PubMed] [Google Scholar]
- 382.Massad G, Lockatell CV, Johnson DE, Mobley HLT. Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infect. Immmun. 1994;62:536–542. doi: 10.1128/iai.62.2.536-542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Pellegrino R, Scavone P, Umpierrez A, Maskell DJ, Zunino P. Proteus mirabilis uroepithelial cell adhesin (UCA) fimbria plays a role in the colonization of the urinary tract. Pathog. Dis. 2013;67:104–107. doi: 10.1111/2049-632X.12027. [DOI] [PubMed] [Google Scholar]
- 384.Cook SW, Mody N, Valle J, Hull R. Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect. Immun. 1995;63:2082–2086. doi: 10.1128/iai.63.5.2082-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Lee KK, Harrison BA, Latta R, Altman E. The binding of Proteus mirabilis nonagglutinating fimbriae to ganglio-series asialoglycolipids and lactosyl ceramide. Can. J. Microbiol. 2000;46:961–966. [PubMed] [Google Scholar]
- 386.Massad G, Fulkerson JJF, Watson DC, Mobley HLT. Proteus mirabilis ambient-temperature fimbriae: cloning and nucleotide sequence of the aft gene cluster. Infect. Immun. 1996;64:4390–4395. doi: 10.1128/iai.64.10.4390-4395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Zunino P, Geymonat L, Allen AG, Legnani-Fajardo C, Maskell DJ. Virulence of a Proteus mirabilis ATF isogenic mutant is not impaired in a mouse model of ascending urinary tract infection. FEMS Immunol. Med. Microbiol. 2000;29:137–143. doi: 10.1111/j.1574-695X.2000.tb01516.x. [DOI] [PubMed] [Google Scholar]