Summary
Objective
To determine whether altered IL8 methylation status is associated with increased expression of IL8 in human osteoarthritic (OA) chondrocytes.
Methods
IL8 expression levels and the percentage CpG methylation in human chondrocytes were quantified by qRT-PCR and pyrosequencing in OA patients and in non-OA osteoporotic controls. The effect of CpG methylation on IL8 promoter activity was determined using a CpG-free vector; co-transfections with expression vectors encoding nuclear factor-kappa B (NF-κB), AP-1 and C/EBP were subsequently undertaken to analyse for IL8 promoter activity in response to changes in methylation status.
Results
IL8 expression in OA patients was 37-fold higher than in osteoporotic controls. Three CpG sites in the IL8 promoter were significantly demethylated in OA patients. Multiple regression analysis revealed that the degree of methylation of the CpG site located at −116-bp was the strongest predictor of IL8 expression. In vitro DNA methylation was noted to decrease IL8 promoter basal activity. Furthermore, NF-κB, AP-1 and C/EBP strongly enhanced IL8 promoter activity whilst DNA methylation inhibited the effects of these three transcription factors.
Conclusions
The present study demonstrates the key role of DNA methylation status on the expression of IL8 in human chondrocytes. We demonstrate a quantitative relationship between percentage methylation and gene expression within clinical samples. These studies provide direct evidence linking the activation of IL8, DNA demethylation and the induction of the OA process with important therapeutic implications therein for patients with this debilitating disease.
Keywords: Interleukin-8, Chemokine, DNA methylation, Epigenetics, Inflammation, Osteoarthritis
Introduction
Osteoarthritis (OA) remains, currently, the most frequent cause of pain, deformity and dysfunction in the elderly population1, 2. Although the pathogenesis of OA is far from clear, involvement of inflammation in the development and progression of OA has been implicated3, 4. Indeed, epidemiological studies indicate a significant association between OA disease progression and the presence of inflammatory synovium5, 6. Furthermore, a notable finding of inflammation in the tissue is the recruitment of neutrophils from the blood to the affected site; a process mediated by chemokines, small 8–12 kD chemotactic proteins7.
Interleukin 8 (IL-8), also named CXCL-8, is an inflammatory chemokine present under pathological conditions. IL-8, produced by human OA chondrocytes, is an important mediator in the pathophysiology of OA8, 9, 10, 11 including promotion of a number of pathogenic processes such as; (1) release of matrix metalloproteinase-13 (MMP-13), (2) neutrophil accumulation and, (3) activation and leukocyte homing to the synovium12, 13, 14. Furthermore, IL-8 and other chemokines are known to induce chondrocyte hypertrophy and differentiation9, 15, 16. Pierzchala et al. reported that synovial fluid from OA patients displayed significantly increased levels of IL-8 compared to controls17.
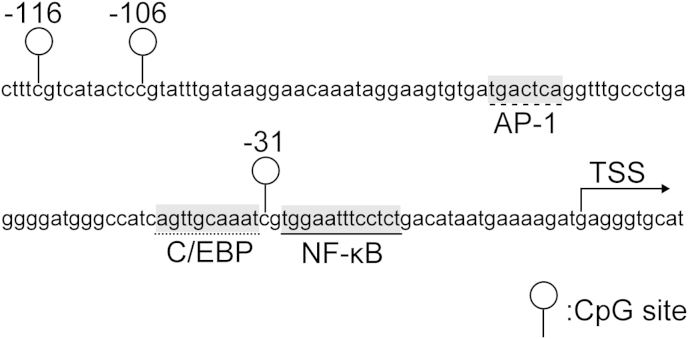
Gene expression is regulated by epigenetic and non-epigenetic mechanisms. Epigenetics refers to heritable or stable, long-term changes in gene activity without changes in the DNA sequence. DNA methylation at CpG sites is a central epigenetic mechanisms conferring long-term regulation of set genes in contrast to regulation observed by histone modifications18, 19, 20. While DNA methylation of the so-called CpG islands have been predominantly examined, a few studies have shown that single or a few specific CpG sites can dominate the promoter activities of a particular gene21, 22, 23, 24. DNA methylation at CpG sites has been shown as a critical mediator in human OA chondrocytes for a number of key genes implicated in OA including IL1B, MMP13, iNOS and COL9A121, 24, 25, 26. Furthermore, CpG methylation status can directly affect the binding of transcriptional factors resulting in altered transcriptional activity24. In previous studies, transcriptional regulation of IL8 by nuclear factor-kappa B (NF-κB), activator protein-1 (AP-1) and CCAAT/enhancer-binding protein (C/EBP) has been reported in a number of tissues and cell types including colonic epithelial cells, ovarian cancer cells and myometrial cells27, 28, 29, 30, 31, 32. Critically, sequences spanning nucleotides −1 to −133 within the IL8 proximal promoter were observed to be essential and sufficient for transcriptional regulation of the IL8 gene. This sequence (−1 to −133) includes binding sites for NF-κB, AP-1 and C/EBP28, 30, 33. In addition, within IL8, the 1000-bp of the proximal promoter region contains only three CpG sites all located close to transcriptional binding sites (Fig. 1). However, the methylation status of these CpG sites and subsequent involvement in the regulation of IL8 regulation remains, to date, unknown. The current study set out to examine whether the increased expression of IL8 in human OA is a consequence of epigenetic regulation, specifically DNA hypomethylation.
Fig. 1.
Diagrammatic representation of CpG sites within the proximal IL8 promoter with CpG sites and location of transcription factors indicated.
Materials and methods
Chondrocyte isolation
Human articular cartilage was obtained from the femoral heads of patients undergoing hemiarthroplasty following a fracture of the neck of femur (#NOF) or after total hip arthroplasty for OA (OARSI score for OA grade34 in all OA patients was 3–4). Samples were derived with full patient consent and prior approval of the local Institutional Review Board. Given patients with a #NOF are likely to be suffering from osteoporosis, and the accepted inverse relation between OA and osteoporosis [17], cartilage from these patients served as control samples35. Cartilage tissue was dissected within 6 h of surgery from OA and non-OA samples and primary chondrocytes isolated as previously detailed in Refs. 26, 36, 37. Briefly, non-OA/healthy chondrocytes were isolated from the cartilage deep zone of patients with #NOF, whereas cartilage pieces adjacent to weight-bearing areas of OA femoral heads (lacking surface zones) were harvested for OA chondrocytes. Cartilage samples were dissected and cut into small fragments and digested with 10% trypsin (Lonza) in PBS for 30 min, followed by sequential digestion using 1 mg/ml of hyaluronidase (Sigma–Aldrich) in PBS for 15 min, and in 10 mg/ml of collagenase B (Roche Applied Science) in DMEM/F12 medium (Life Technologies) for 12–15 h at 37°C. Isolated chondrocytes from 15 #NOF samples (controls, 5 men and 10 women with a mean ± SD age of 84.5 ± 5.3) and 15 OA samples (OA, seven men and eight women with a mean ± SD age of 66.7 ± 12.5) were directly used for extraction of genomic DNA and total RNA. The chondrocytes from seven #NOF patients were cultured for culture experiments.
Chondrocyte culture
Following cell isolation, non-OA chondrocytes were divided into three groups: (1) control culture, (2) IL-1β culture, and (3) 5-aza culture. Chondrocytes were cultured at a density of 2–4 × 105 cells/25-cm2 flask in 5 ml of DMEM/F12 supplemented with 5% fetal calf serum (FCS; Invitrogen), 1% insulin–transferrin–selenium (Sigma–Aldrich), 100 units/ml of penicillin and 100 μg/ml of streptomycin (Lonza), and 100 μg/ml of ascorbic acid (Sigma–Aldrich) in the atmosphere of 5% CO2 at 37°C. IL-1β (10 ng/ml) (Sigma–Aldrich) and oncostatin M (10 ng/ml) (Sigma–Aldrich) were added to cultures based on the observations these inflammatory cytokines are elevated in OA synovial fluid and known to induce the short-term induction of catabolic genes and to alter DNA methylation status following long-term stimulation4, 26, 38. The primary cultures were maintained for 5 weeks until cells reached confluence.
DNA and RNA extraction and molecular analysis (qRT-PCR)
Total RNA and genomic DNA were extracted simultaneously from the harvested chondrocytes using AllPrep DNA/RNA Mini kit (Qiagen). RNA was immediately reverse transcribed with SuperScript VILO cDNA Synthesis Kit (Life Technologies). Relative quantification of gene expression was performed with an ABI Prism 7500 detection system (Applied Biosystems). The 20-μl reaction mixture was prepared in triplicate, containing 1 μl of complementary DNA, 10 μl of Power SYBR Green PCR Master Mix (Applied Biosystems), and 250 nM of each primer. Thermal cycler conditions included an initial activation step at 95°C for 10 min, followed by a two-step PCR program of 95°C for 15 s and 60°C for 60 s for 40 cycles. The 2−ΔΔCt method was used for relative quantification of gene expression. Reactions were performed in triplicate and samples normalised against GAPDH gene expression as control. GAPDH primers were designed using Primer Express software (version 3.0; Applied Biosystems). IL8 primers were obtained from the PrimerBank database39 (PrimerBank ID: 10834978a2) and the primers used for quantitative reverse transcription PCR (qRT-PCR) are illustrated in Table I.
Table I.
Primer sequences for (a) RT-PCR, (b) pyrosequencing, and (c) site directed mutagenesis
| Name (length, bp) | Sequence (5′–3′) |
|---|---|
| (a) | |
| GAPDH (108) | F (CCAGGTGGTCTCCTCTGACTTC) |
| R (TCATACCAGGAAATGAGCTTGACA) | |
| IL8 (112) | F (ACTGAGAGTGATTGAGAGTGGAC) |
| R (AACCCTCTGCACCCAGTTTTC) | |
| (b) | |
| IL8-Pyro-1 (70) | F (AGGGGATGGGTTATTAGTTG) |
| R (ACTTATACACCCTCATCTTTTCATT) | |
| S (GGATGGGTTATTAGTTGTA) | |
| IL8-Pyro-2 (148) | F (GGTTTATTTTTTTAGGGTAAATTTGAGTTA) |
| R (ATTCACCAAATTATAAAACTTCAATATT) | |
| S (ATTATATTTTTTATTTGTTTTTTATTAA) | |
| (c) | |
| IL8-Mut1 (−116∗)† | F (AATTAAATTATTTTAAAGATCAAAGAAAACTTTtGTCATACTCCGTATTTGATAAGGAAC) |
| R (GTTCCTTATCAAATACGGAGTATGACaAAAGTTTTCTTTGATCTTTAAAATAATTTAATT) | |
| IL8-Mut2 (−106∗)† | F (CAAAGAAAACTTTCGTCATACTCtGTATTTGATAAGGAACAAATAGG) |
| R (CCTATTTGTTCCTTATCAAATaCAGAGTATGACGAAAGTTTTCTTTG) | |
| IL8-Mut3 (−31∗)† | F (GATGGGCCATCAGTTGCAAATtGTGGAATTTCCTCT) |
| R (AGAGGAAATTCCACaATTTGCAACTGATGGCCCATC) | |
| IL8-Mut4 (−106∗)† | F (GATCAAAGAAAACTTTTGTCATACTCtGTATTTGATAAGGAACAAATAGGAAG) |
| R (CTTCCTATTTGTTCCTTATCAAATACaGAGTATGACAAAAGTTTTCTTTGATC) | |
F: forward; R: reverse; S: sequencing.
Location of mutated CpG.
Lowercase letters indicate a mutated base.
Bisulfite pyrosequencing
Genomic DNA extracted from each sample was treated with sodium bisulfite to convert unmethylated cytosine in CpG sites to uracil using the EZ DNA Methylation-Gold Kit (Zymo Research Corporation). Following bisulfite treatment, PCR was performed with Premium PCR Supermix High Fidelity (Invitrogen). The percentage DNA methylation in the IL8 promoter was quantified using PyroMark MD (Qiagen). All primers were designed with Pyrosequencing Assay Design Software (Qiagen) (Table I).
Plasmid constructions
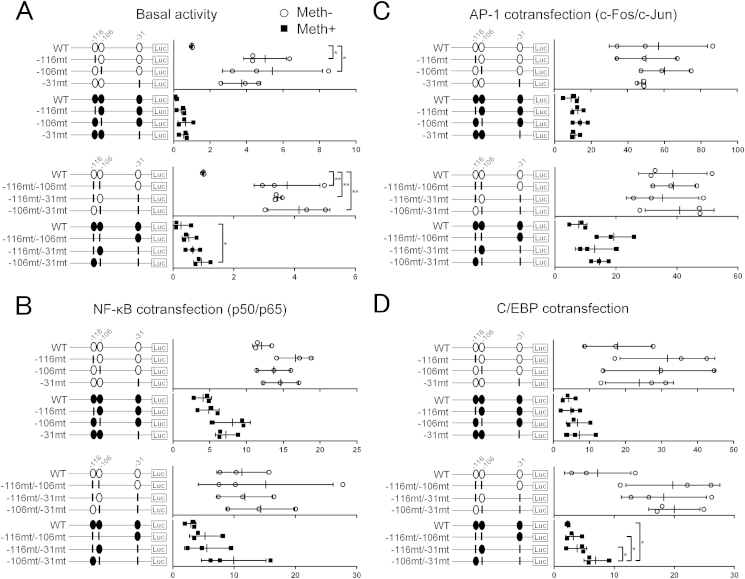
The IL8 promoter constructs were generated by PCR amplification using genomic DNA from human articular chondrocytes as a template. The following PCR primers were used: 5′-ATAGGATCCGCCTTGCTCCAACTGCCTTT-3′ (forward) and 5′-AATCCATGGTGGTTTCTTCCTGGCTCTTGT-3′ (reverse). Underlined letters indicate BamHI and NcoI recognition sequences, respectively. The resultant PCR products were digested with BamHI and NcoI (Thermo Scientific) and transferred into the multiple cloning site of a pCpGfree-Luc vector using Rapid DNA Ligation Kit (Thermo Scientific). The vector lacks CpG sites within the whole vector sequence and was generated as detailed by Klug and Rehli40. Point mutations at CpG sites were generated by converting CG to TG using QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies). Primers for mutagenesis were designed using QuikChange Primer Design (Agilent Technologies) (Table I). Promoter constructs with a mutation at CpG sites located at −31-bp, −106-bp and −116-bp from the transcription start site (TSS) were generated according to the manufacturer's instructions. Promoter constructs with two mutations at the CpG sites were produced by two-step mutagenesis. In total, six mutation patterns were generated (Fig. 6). The sequences of all constructs were confirmed by DNA sequencing using SmartSeq system (Eurofins Genomics).
Fig. 6.
Assessment of promoter activity of methylated or non-methylated IL8 promoter constructs containing different mutations analysed by luciferase assay. Point mutations (CG to TG) were created at CpG sites located at −31-bp, −106-bp, −116-bp, −116-bp and −106-bp, −116-bp and −31-bp, or −106-bp and −31-bp. (A) Basal IL8 promoter activity in C28/I2 cell line transfected with methylated or non-methylated CpG-free vector containing wild-type or mutated IL8 promoter constructs. (B) Co-transfection with the NF-κB p50 and p65 subunit expression vectors. (C) Co-transfection with the AP-1 c-Fos and c-Jun subunit expression vectors. (D) Co-transfection with the C/EBPβ expression vector. Values are the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
In vitro methylation, transfection and luciferase assay
Plasmids were methylated using CpG Methylase M.Sssl (New England Biolabs). Complete methylation was verified by plasmid DNA bisulfite modification and pyrosequencing with specific primers. The immortalized human chondrocytes, C28/I2, were seeded at a density of 30,000 cells per well in 24-well plates, cultured in DMEM/F12 overnight, and transfected with a mixture of 300 ng luciferase reporter vector and 1 ng pRL-TK Vector (Promega), using FuGENE HD in vitro Transfection Reagent (Promega). Transfected C28/I2 cells were cultured for 48 h prior to harvest. Cell lysates were assayed for firefly and renilla luciferase activity using a Dual-Luciferase Reporter Assay System on a Varioskan Flash (Thermo Scientific). Firefly luciferase activity of each transfection was normalized against renilla luciferase activity. Reactions were performed in duplicate, and each experiment was repeated at least three times.
The expression vectors for NF-κB (p50, p65, or p50/p65), AP-1(c-Fos, c-Jun, or c-Fos/c-Jun) and C/EBPβ were used (60 ng) for co-transfections. Blank expression vector pCMV4, pcDNA3.1 and pcDNA3.1(+) served as controls, respectively. Total DNA was normalized with empty vectors in the transfection mixture.
Statistical analysis
Statistical analysis was performed using SPSS Statistics (version 21.0; IBM). Cartilage samples were obtained from individual subjects. The Mann–Whitney U test was used to compare gene expression, CpG percentage methylation and age between two groups. Female to male ratios were compared using chi-square test. Spearman's rank correlation coefficient was used to analyse the relationship between percentage methylation and IL8 expression and multiple regression analysis using least-squares method was applied to determine the relationship between IL8 expression and patients' background data. Kruskal–Wallis test and Newman–Kuels multiple comparisons test were used to analyse the luciferase reporter assays. P values less than 0.05 were considered significant.
Results
IL8 proximal promoter CpG sites are demethylated in OA chondrocytes and correlate with higher IL8 gene expression
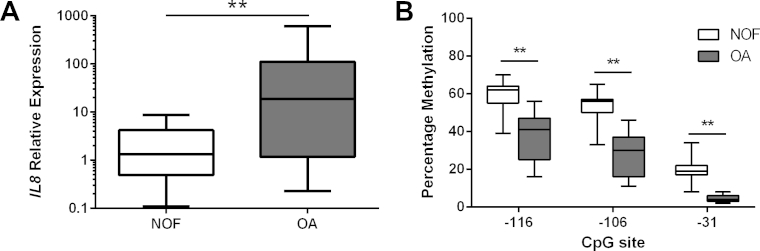
Initial studies centred on quantification of the CpG methylation status of the IL8 proximal promoter in human primary chondrocytes isolated from articular cartilage obtained from non-OA (#NOF) donors (n = 15) and patients with OA (n = 15). IL8 expression in OA chondrocytes was observed to be 37-fold higher than in #NOF controls [Fig. 2(A)]. Pyrosequencing analysis of the IL8 promoter in the same subjects revealed that all three CpG sites in the promoter region were significantly demethylated in OA chondrocytes in contrast to #NOF chondrocytes. OA chondrocytes displayed a 22%, 26% and 15% statistically significant (P < 0.01) reduction in methylation status at the −116, −106 and −31 CpG sites, respectively [Fig. 2(A)].
Fig. 2.
(A) Relative mRNA expression of IL8 in non-cultured primary human chondrocytes obtained from patients with femoral neck fracture (NOF) and OA patients. mRNA was analyzed by quantitative RT-PCR and normalized against GAPDH. (B) Percentage methylation of the indicated CpG sites in the IL8 proximal promoter in the same samples analysed by bisulfite pyrosequencing. Y-axis shows non-adjusted percentage methylation. Values are the mean ± SD of 15 independent samples from each group. **P < 0.01.
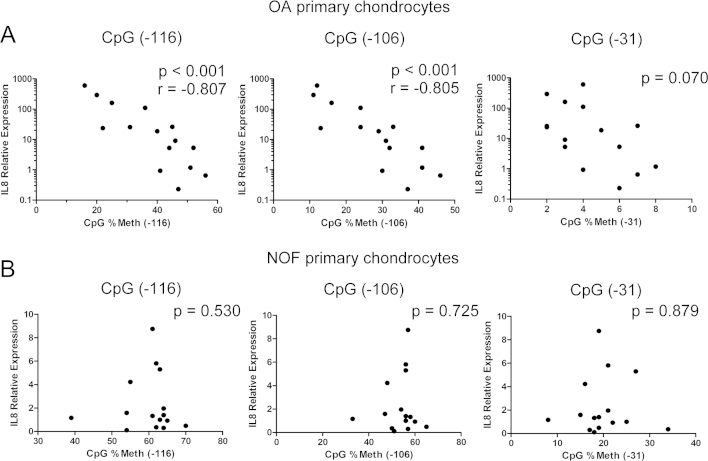
A significant negative correlation was observed between IL8 gene expression and the percentage methylation of the CpG sites located at −116-bp and −106-bp in OA chondrocytes [Fig. 3(A)]. The percentage methylation of the CpG site located at −31-bp displayed a correlation trend with IL8 expression although this was not statistically significant (P = 0.069). In contrast, no correlation was observed between IL8 expression and the percentage methylation in #NOF chondrocytes [Fig. 3(B)]. Importantly, multiple regression analysis revealed that the percentage methylation of the CpG site located at −116-bp was the strongest predictor of IL8 expression (P < 0.01). Furthermore, advanced age and OA were also associated with higher IL8 expression (Table II).
Fig. 3.
Results of Spearman's rank correlation coefficient comparing relative mRNA expression of IL8 and methylation status of the indicated CpG sites in the IL8 proximal promoter in OA chondrocytes (A) and controls (B).
Table II.
Factors associated with IL8 relative expression in human chondrocytes
| t value | P value | R2 | P value | |
|---|---|---|---|---|
| #NOF/OA [OA] | 2.71 | 0.012 | ||
| Female/Male [Female] | 0.81 | 0.427 | ||
| Age | 2.42 | 0.024 | ||
| IL8 %Methylation (−31) | 2.4 | 0.025 | ||
| IL8 %Methylation (−106) | 2.59 | 0.016 | ||
| IL8 %Methylation (−116) | −3.64 | 0.001 | ||
| Total model | 0.696 | <0.0001 | ||
#NOF: a fracture of the neck of femur, OA: osteoarthritis.
Demethylation of CpG sites in the IL8 promoter following long-term culture does not result in induction of IL8 gene expression
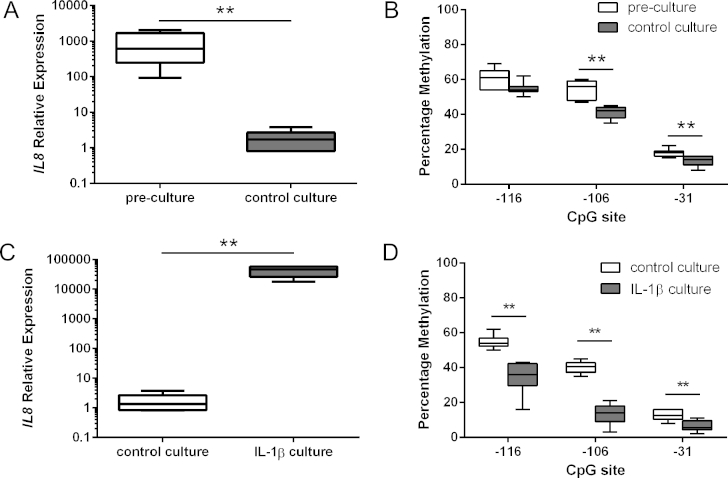
Given monolayer culture is known to affect the gene expression profile of chondrocytes41, IL8 mRNA levels were analysed in pre-culture control chondrocytes compared with cultured chondrocytes over for 5 weeks. The culture of chondrocytes resulted in a significant loss of IL8 expression [Fig. 4(A)]. CpG sites located at −106-bp and −31-bp of the IL8 promoter in the cultured chondrocytes showed significant demethylation compared with pre-culture chondrocytes [Fig. 4(B)].
Fig. 4.
Relative mRNA expression of IL8 was analyzed by quantitative RT-PCR and normalized against GAPDH in (A) pre-culture NOF chondrocytes and control culture chondrocytes, and (C) control culture and IL-1β culture. Percentage methylation of the indicated CpG sites in the IL8 proximal promoter was analysed using bisulfite pyrosequencing in (B) pre-culture NOF chondrocytes and control culture chondrocytes, and (D) control culture and IL-1β culture. Values are the mean ± SD of seven independent experiments. *P < 0.05, **P < 0.01.
Long-term exposure to IL-1β and oncostatin M results in enhanced expression of IL8 and loss of DNA methylation
Healthy chondrocytes were cultured for 5 weeks in IL-1β/OSM. Long-term treatment with IL-1β/OSM induced a 24,000-fold increase in IL8 expression compared to control cultures [Fig. 4(C)]. Pyrosequence analysis of the IL8 promoter revealed that #NOF chondrocytes cultured using IL-1β/OSM displayed a 22%, 25% and 2.3% reduction in methylation status at the −116, −106 and −31 CpG sites, respectively, in comparison to control cultures [Fig. 4(D)].
Methylation decreases IL8 promoter activity in vitro
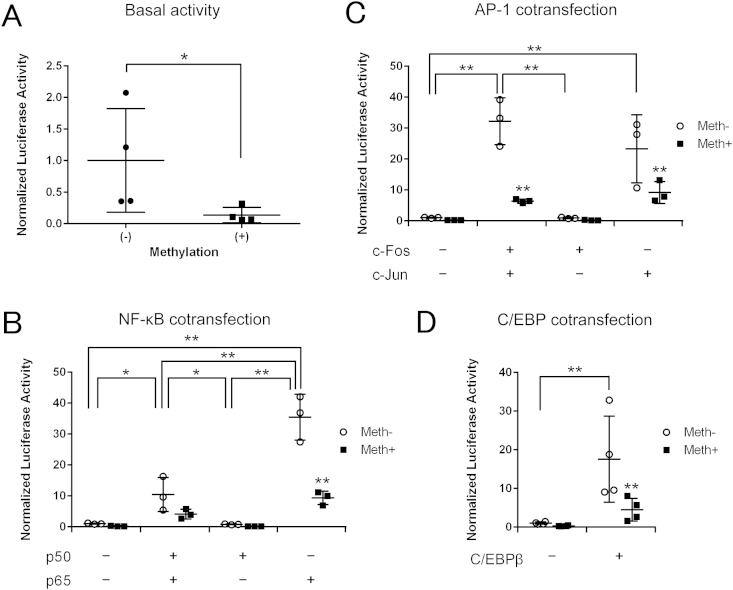
To determine the effects of DNA methylation on IL8 promoter activity, dual-luciferase reporter assays were performed. The C28/I2 chondrocyte cell line was transfected with the wild type IL8 promoter construct using a CpG free vector and pRL-TK vector as an internal control. The luciferase assay was performed 48 h after transfection. Methylation treatment significantly decreased the activities of the promoter constructs by seven fold [Fig. 5(A)].
Fig. 5.
IL8 promoter activity analysed by a luciferase assay in C28/I2 cell line transfected with non-methylated or methylated CpG-free vector containing wild-type IL8 promoter construct. (A) Basal activities without treatment. (B) Co-transfection with the empty control vector (pCMV4) or with the NF-κB p50 subunit expression vector, p65 subunit expression vector or both. (C) Co-transfection with the empty control vector (pcDNA3.1) or with the AP-1 c-Fos subunit expression vector, c-Jun subunit expression vector or both. (D) Co-transfection with the empty control vector (pcDNA3.1(+)) or with the C/EBPβ expression vector. Values are the mean ± SD of three independent experiments (B and C) or four (A and D). *P < 0.05, **P < 0.01.
NF-κB, AP-1 and C/EBPβ mediate IL8 transactivation in human chondrocytes and CpG methylation impairs IL8 promoter transactivation
To determine the effects of DNA methylation and the transcription factors NF-κB, AP-1 and C/EBP and on IL8 promoter activity, the expression vector encoding each transcription factor and the control empty vectors were co-transfected with wild type IL8 promoter construct using a CpG free vector. IL8 promoter activity was significantly enhanced (35-fold) with the NF-κB p65 subunit whilst, in contrast, DNA methylation suppressed the effect of NF-κB on IL8 activity [Fig. 5(B)]. The AP-1 c-Jun subunit significantly transactivated IL8 by 23-fold and the enhanced activity was higher with overexpression of c-Fos and c-Jun combined (32-fold) [Fig. 5(C)]. C/EBPβ significantly enhanced IL8-driven reporter activity 17-fold while DNA methylation significantly reduced the effect of C/EBPβ [Fig. 5(D)].
Mutations at three CpG sites proximal to the TSS increase IL8 promoter basal activity
To determine the CpG sites critical for IL8 promoter activity, we compared IL8 wild type promoter construct activity using a CpG free vector against six vectors containing mutations at different CpG sites [Fig. 6(A)]. Point mutations created on any single CpG site or two CpG sites resulted in a significant increase in IL8 promoter activity by 3.4–5.4 fold [Fig. 6(A)]. Furthermore, non-methylated −106/−31 mutant constructs showed a significant increase in promoter activity (2.7–4.5 fold).
NF-κB, AP-1 and C/EBPβ mediate IL8 transactivation in cooperation with specific CpG sites within the proximal promoter
Sequences spanning nucleotides −1 to −133 within the IL8 proximal promoter were observed to be essential and sufficient for transcriptional regulation of the IL8 gene30. This sequence includes binding sites for NF-κB, AP-1 and C/EBP, and the three CpG sites [Fig. 1].
To evaluate the role of each CpG site for transcription factor-mediated IL8 transactivation, the IL8 wild type promoter construct and the six vectors with point mutations were co-transfected with the expression vectors encoding NF-κB, AP-1 and C/EBPβ. NF-κB overexpression increased the activity of wild type and mutated non-methylated promoter constructs. Point mutations created at −31-bp CpG or −106-bp CpG or both displayed a trend for increased NF-κB-driven IL8 promoter transactivation in methylated plasmids [Fig. 6(B)]. In contrast, following AP-1 overexpression, methylated −116/−106 mutant constructs were observed to enhance IL8 promoter activity [Fig. 6(C)]. Promoter activity pattern under C/EBP overexpression was similar to that observed following NF-κB overexpression. Non-methylated −106/−31 mutant constructs showed a significant increase in the promoter activity (2.8–3.1 fold) [Fig. 6(D)].
Discussion
The current study demonstrates that the increased expression of IL8 in human OA chondrocytes is regulated by DNA demethylation in cooperation with transcription factors. We show for the first time that the percentage methylation of specific CpG sites correlates with IL8 gene expression level in clinical OA samples. Furthermore, long-term stimulation with IL-1β, a key pro-inflammatory cytokine involved in the pathophysiology of OA, resulted in the marked induction of IL8 with decreased CpG methylation in the IL8 promoter in human chondrocytes. Furthermore, 5-aza-dC treatment induced hypomethylation at the CpG site located at −116-bp in the IL8 promoter and enhanced IL8 expression (data not shown). Moreover, dual-luciferase reporter assays using a CpG free vector revealed that methylation treatment significantly decreased the activity of the IL8 promoter constructs, thus the methylation status of CpG sites is one of the key transcriptional regulators of IL-8 in human chondrocytes. Interestingly, methylation status of the CpG sites did not correlate with IL8 expression level in #NOF chondrocytes. Furthermore, a significant DNA demethylation of the IL8 promoter in cultured chondrocytes did not result in increased IL8 gene expression compared with pre-culture chondrocytes. These results suggest that methylation status alone was not sufficient to regulate IL8 transcription. However, in pathological situations such as OA, methylation status of specific CpG sites appears crucial to the regulation of IL8 expression. Critically, multiple regression analysis indicated that the methylation status of the CpG sites located at −116-bp from the TSS provided a strong association with IL8 expression. Interestingly, this CpG is the most distal to the transcriptional binding sites and, thus should be the least affected. However, to date, there is a paucity of information concerning the three-dimensional configuration of the chromatin in this region in combination with transcription factors and indeed DNA methylation status and thus this remains an area for further study. Future studies with enhanced patient numbers including different grades of clinical OA, would be necessary to confirm and reveal the relationship of DNA methylation status with disease progression.
The primary functions of IL-8 are chemotaxis and angiogenesis and IL-8 has been shown to play an essential role in acute inflammation7, 42, 43. In general, OA is considered to be a “non-inflammatory arthritis”. However, growing evidence indicates the involvement of inflammation in the development and progression of OA4, 5. A recent methylome study revealed an enrichment of several pathways involved in inflammation including IL2, IL3, IL4 and IL644. Furthermore, SOCS2 and CIS-1, inhibitors of cytokine signalling, have been shown to be suppressed in OA45. Enhanced expression of IL8 in OA chondrocytes in the present study supports an association, in part, of OA with inflammation. Thus, disease progression and joint symptoms could potentially be modified by modulation and control of IL8 expression. The current findings illustrating that DNA demethylation accounts for an increase in IL8 expression in OA suggesting a potential target for OA modulation and warranting further (clinical and in vivo) examination.
Interestingly, IL8 expression is typically, low or undetectable in normal non-inflammatory tissue. This is partly a result of transcriptional repression of the IL8 promoter30. The IL8 promoter contains a negative regulatory element (NRE) to which the NF-κB-repressing factor (NRF) binds. Reduction of cellular NRF by expressing NRF-antisense RNA results in spontaneous IL8 gene expression27. Additionally, mutation of the NRE site results in loss of NRF binding and increased basal IL8 expression27. The existence of a basal repression mechanism offers an explanation for the mutagenesis results observed in the current study. Point mutations created at the CpG sites in the IL8 promoter could interrupt basal repression and result in increased promoter activity. In addition, the present studies indicate methylated IL8 promoter constructs display low promoter activity indicating DNA methylation is an additionally basal repression mechanism of IL8 expression.
The transcription factors, NFκB, CEBP, and AP-1, have all been implicated in IL8 expression in a number of cell types27, 28, 29, 30, 32. NF-κB is a dimeric transcription factor composed of five different subunits27, 30, 46. We recently showed the NF-κB p65 subunit played a critical role in induction of iNOS in OA human chondrocytes in coordination with DNA demethylation of the enhancer elements25. NF-κB has also been shown to regulate the expression of a number of cytokines and chemokines, and several matrix degrading enzymes in OA pathogenesis47. A significant increase in IL8 promoter activity by the NF-κB p65 subunit, AP-1 c-Jun subunit and C/EBPβ27, 28, 29, 30, 31, 32 was demonstrated in this study. Furthermore, the current study demonstrates that CpG methylation impairs IL8 promoter transactivation by the transcription factors NF-κB, AP-1 and C/EBP. Interestingly, IL8 transactivation by NF-κB and C/EBP were predominantly regulated by the CpG site located at −31-bp. In contrast, AP-1 was predominantly regulated by the CpG site located at −106-bp explained by the location of the CpG sites and the binding sites of the transcription factors [Fig. 1].
In conclusion, the present study demonstrates the key role of DNA methylation status on the expression of IL8 in OA chondrocytes. This study demonstrates the quantitative relationship between percentage methylation of a CpG site and gene expression in clinical OA cartilage samples with evidence linking the activation of IL-8, DNA demethylation and, critically, induction of the OA process. These findings suggest, tentatively, a potential predictive marker, although additional in vivo and clinical studies are required before confirmation of this inflammatory chemokine, for pharmacological intervention in the treatment of OA and, potentially, other arthritic diseases.
Author contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. In detail: AT—conception, design, acquisition of data, analysis and interpretation of data, drafting the manuscript; MCA—conception, design, acquisition of data, analysis and interpretation of data, critical revision of the manuscript; KH—conception, design, analysis and interpretation of data, critical revision of the manuscript; EI—analysis and interpretation of data, critical revision of the manuscript; ROCO—conception, design, analysis and interpretation of data, critical revision of the manuscript.
Ethics approval
This study was approved by the Southampton & South West Hampshire Local Research Ethics Committee and informed consent was obtained from each patient.
Role of the funding source
Funding from the Leverhulme Trust and Biotechnology and Biological Sciences Research Council (BB/G010579/1) to RO is gratefully acknowledged. The study sponsors had no direct involvement in the study, in writing of the manuscript or the decision to submit.
Conflict of interest
The authors have no conflict of interest to declare.
Acknowledgements
The authors acknowledge Dr Miguel Otero and Dr Mary B. Goldring (Hospital for Special Surgery) for provision of the expression vectors and the chondrocytic cell line C28/I2, as well as the orthopaedic surgeons at Southampton General Hospital for provision of femoral head samples.
References
- 1.Loeser R.F., Goldring S.R., Scanzello C.R., Goldring M.B. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moskowitz R.W. The burden of osteoarthritis: clinical and quality-of-life issues. Am J Manag Care. 2009;15:S223–S229. [PubMed] [Google Scholar]
- 3.Blagojevic M., Jinks C., Jeffery A., Jordan K.P. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2010;18:24–33. doi: 10.1016/j.joca.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J.P., Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 5.Pozgan U., Caglic D., Rozman B., Nagase H., Turk V., Turk B. Expression and activity profiling of selected cysteine cathepsins and matrix metalloproteinases in synovial fluids from patients with rheumatoid arthritis and osteoarthritis. Biol Chem. 2010;391:571–579. doi: 10.1515/BC.2010.035. [DOI] [PubMed] [Google Scholar]
- 6.Ayral X., Pickering E.H., Woodworth T.G., Mackillop N., Dougados M. Synovitis: a potential predictive factor of structural progression of medial tibiofemoral knee osteoarthritis – results of a 1 year longitudinal arthroscopic study in 422 patients. Osteoarthritis Cartilage. 2005;13:361–367. doi: 10.1016/j.joca.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Rosenkilde M.M., Schwartz T.W. The chemokine system – a major regulator of angiogenesis in health and disease. APMIS. 2004;112:481–495. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- 8.Alaaeddine N., Di Battista J.A., Pelletier J.P., Kiansa K., Cloutier J.M., Martel-Pelletier J. Differential effects of IL-8, LIF (pro-inflammatory) and IL-11 (anti-inflammatory) on TNF-alpha-induced PGE(2)release and on signalling pathways in human OA synovial fibroblasts. Cytokine. 1999;11:1020–1030. doi: 10.1006/cyto.1999.0505. [DOI] [PubMed] [Google Scholar]
- 9.Merz D., Liu R., Johnson K., Terkeltaub R. IL-8/CXCL8 and growth-related oncogene alpha/CXCL1 induce chondrocyte hypertrophic differentiation. J Immunol. 2003;171:4406–4415. doi: 10.4049/jimmunol.171.8.4406. [DOI] [PubMed] [Google Scholar]
- 10.Chauffier K., Laiguillon M.C., Bougault C., Gosset M., Priam S., Salvat C. Induction of the chemokine IL-8/Kc by the articular cartilage: possible influence on osteoarthritis. Jt Bone Spine. 2012;79:604–609. doi: 10.1016/j.jbspin.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Yuan G.H., Masuko-Hongo K., Sakata M., Tsuruha J., Onuma H., Nakamura H. The role of C-C chemokines and their receptors in osteoarthritis. Arthritis Rheum. 2001;44:1056–1070. doi: 10.1002/1529-0131(200105)44:5<1056::AID-ANR186>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 12.Olson T.S., Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am J Physiol Regul Integr Comp Physiol. 2002;283:R7–R28. doi: 10.1152/ajpregu.00738.2001. [DOI] [PubMed] [Google Scholar]
- 13.Borzi R.M., Mazzetti I., Marcu K.B., Facchini A. Chemokines in cartilage degradation. Clin Orthop Relat Res. 2004:S53–S61. doi: 10.1097/01.blo.0000143805.64755.4f. [DOI] [PubMed] [Google Scholar]
- 14.Matsukawa A., Yoshimura T., Maeda T., Ohkawara S., Takagi K., Yoshinaga M. Neutrophil accumulation and activation by homologous IL-8 in rabbits. IL-8 induces destruction of cartilage and production of IL-1 and IL-1 receptor antagonist in vivo. J Immunol. 1995;154:5418–5425. [PubMed] [Google Scholar]
- 15.Borzi R.M., Mazzetti I., Macor S., Silvestri T., Bassi A., Cattini L. Flow cytometric analysis of intracellular chemokines in chondrocytes in vivo: constitutive expression and enhancement in osteoarthritis and rheumatoid arthritis. FEBS Lett. 1999;455:238–242. doi: 10.1016/s0014-5793(99)00886-8. [DOI] [PubMed] [Google Scholar]
- 16.Borzi R.M., Mazzetti I., Magagnoli G., Paoletti S., Uguccioni M., Gatti R. Growth-related oncogene alpha induction of apoptosis in osteoarthritis chondrocytes. Arthritis Rheum. 2002;46:3201–3211. doi: 10.1002/art.10650. [DOI] [PubMed] [Google Scholar]
- 17.Pierzchala A.W., Kusz D.J., Hajduk G. CXCL8 and CCL5 expression in synovial fluid and blood serum in patients with osteoarthritis of the knee. Arch Immunol Ther Exp (Warsz) 2011;59:151–155. doi: 10.1007/s00005-011-0115-4. [DOI] [PubMed] [Google Scholar]
- 18.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 19.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 20.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 21.Imagawa K., de Andres M.C., Hashimoto K., Itoi E., Otero M., Roach H.I. Association of reduced type IX collagen gene expression in human osteoarthritic chondrocytes with epigenetic silencing by DNA hypermethylation. Arthritis Rheumatol. 2014;66:3040–3051. doi: 10.1002/art.38774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murayama A., Sakura K., Nakama M., Yasuzawa-Tanaka K., Fujita E., Tateishi Y. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bui C., Barter M.J., Scott J.L., Xu Y., Galler M., Reynard L.N. cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J. 2012;26:3000–3011. doi: 10.1096/fj.12-206367. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto K., Otero M., Imagawa K., de Andres M.C., Coico J.M., Roach H.I. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem. 2013;288:10061–10072. doi: 10.1074/jbc.M112.421156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Andres M.C., Imagawa K., Hashimoto K., Gonzalez A., Roach H.I., Goldring M.B. Loss of methylation in CpG sites in the NF-kappaB enhancer elements of inducible nitric oxide synthase is responsible for gene induction in human articular chondrocytes. Arthritis Rheum. 2013;65:732–742. doi: 10.1002/art.37806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto K., Oreffo R.O., Gibson M.B., Goldring M.B., Roach H.I. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nourbakhsh M., Kalble S., Dorrie A., Hauser H., Resch K., Kracht M. The NF-kappa b repressing factor is involved in basal repression and interleukin (IL)-1-induced activation of IL-8 transcription by binding to a conserved NF-kappa b-flanking sequence element. J Biol Chem. 2001;276:4501–4508. doi: 10.1074/jbc.M007532200. [DOI] [PubMed] [Google Scholar]
- 28.Khanjani S., Terzidou V., Johnson M.R., Bennett P.R. NFkappaB and AP-1 drive human myometrial IL8 expression. Mediat Inflamm. 2012;2012:504952. doi: 10.1155/2012/504952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann E., Thiefes A., Buhrow D., Dittrich-Breiholz O., Schneider H., Resch K. MEK1-dependent delayed expression of Fos-related antigen-1 counteracts c-Fos and p65 NF-kappaB-mediated interleukin-8 transcription in response to cytokines or growth factors. J Biol Chem. 2005;280:9706–9718. doi: 10.1074/jbc.M407071200. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 31.Elliott C.L., Allport V.C., Loudon J.A., Wu G.D., Bennett P.R. Nuclear factor-kappa B is essential for up-regulation of interleukin-8 expression in human amnion and cervical epithelial cells. Mol Hum Reprod. 2001;7:787–790. doi: 10.1093/molehr/7.8.787. [DOI] [PubMed] [Google Scholar]
- 32.Edwards M.R., Mukaida N., Johnson M., Johnston S.L. IL-1beta induces IL-8 in bronchial cells via NF-kappaB and NF-IL6 transcription factors and can be suppressed by glucocorticoids. Pulm Pharmacol Ther. 2005;18:337–345. doi: 10.1016/j.pupt.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Mukaida N., Okamoto S., Ishikawa Y., Matsushima K. Molecular mechanism of interleukin-8 gene expression. J Leukoc Biol. 1994;56:554–558. [PubMed] [Google Scholar]
- 34.Pritzker K.P., Gay S., Jimenez S.A., Ostergaard K., Pelletier J.P., Revell P.A. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Dequeker J. The inverse relationship between osteoporosis and osteoarthritis. Adv Exp Med Biol. 1999;455:419–422. doi: 10.1007/978-1-4615-4857-7_63. [DOI] [PubMed] [Google Scholar]
- 36.da Silva M.A., Yamada N., Clarke N.M., Roach H.I. Cellular and epigenetic features of a young healthy and a young osteoarthritic cartilage compared with aged control and OA cartilage. J Orthop Res. 2009;27:593–601. doi: 10.1002/jor.20799. [DOI] [PubMed] [Google Scholar]
- 37.Roach H.I., Yamada N., Cheung K.S., Tilley S., Clarke N.M., Oreffo R.O. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 38.Imagawa K., de Andres M.C., Hashimoto K., Pitt D., Itoi E., Goldring M.B. The epigenetic effect of glucosamine and a nuclear factor-kappa B (NF-kB) inhibitor on primary human chondrocytes – implications for osteoarthritis. Biochem Biophys Res Commun. 2011;405:362–367. doi: 10.1016/j.bbrc.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spandidos A., Wang X., Wang H., Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klug M., Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 41.Ma B., Leijten J.C., Wu L., Kip M., van Blitterswijk C.A., Post J.N. Gene expression profiling of dedifferentiated human articular chondrocytes in monolayer culture. Osteoarthritis Cartilage. 2013;21:599–603. doi: 10.1016/j.joca.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Harada A., Sekido N., Akahoshi T., Wada T., Mukaida N., Matsushima K. Essential involvement of interleukin-8 (IL-8) in acute inflammation. J Leukoc Biol. 1994;56:559–564. [PubMed] [Google Scholar]
- 43.Hoch R.C., Schraufstatter I.U., Cochrane C.G. In vivo, in vitro, and molecular aspects of interleukin-8 and the interleukin-8 receptors. J Lab Clin Med. 1996;128:134–145. doi: 10.1016/s0022-2143(96)90005-0. [DOI] [PubMed] [Google Scholar]
- 44.Rushton M.D., Reynard L.N., Barter M.J., Refaie R., Rankin K.S., Young D.A. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66:2450–2460. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Andres M.C., Imagawa K., Hashimoto K., Gonzalez A., Goldring M.B., Roach H.I. Suppressors of cytokine signalling (SOCS) are reduced in osteoarthritis. Biochem Biophys Res Commun. 2011;407:54–59. doi: 10.1016/j.bbrc.2011.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner B.P., Westerheide S.D., Baldwin A.S., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcu K.B., Otero M., Olivotto E., Borzi R.M., Goldring M.B. NF-kappaB signaling: multiple angles to target OA. Curr Drug Targets. 2010;11:599–613. doi: 10.2174/138945010791011938. [DOI] [PMC free article] [PubMed] [Google Scholar]