Abstract
The transition from a non-invasive to an invasive phenotype is an essential step in tumor metastasis. The Snail family of transcription factors (TFs) is known to play a significant role in this transition. These TFs are zinc fingers that bind to the CAGGTG Ebox consensus sequence. CoIII-Ebox is a cobalt(III) complex attached to an Ebox oligonucleotide that confers specificity towards Snail TFs. CoIII-Ebox has been shown to inhibit Snail-mediated embryonic neural crest development in Xenopus laevis, but its efficacy in inhibiting Snail-induced cancer cell invasiveness has not been explored. Here, we describe the efficacy of CoIII-Ebox in inhibiting the invasive aspects of heregulin β1(HRG)-treated breast cancer cells. CoIII-Ebox was found to inhibit the capacity of Snail to repress target genes after HRG induction. Snail inhibition by CoIII-Ebox reduced the invasive propensity of cells in 2D and 3D, thereby demonstrating promise in inhibiting metastasis.
Keywords: cancer, cobalt, metastasis, Snail, transcription factor
Introduction
Tumor metastasis is the process whereby cells disseminate from a primary tumor and are established as secondary tumors at a distal site. This process is the leading cause of cancer-related deaths.[1] Because most tumors are of epithelial origin, metastasis requires that these cells lose their cell–cell adhesions and adopt a mobile, invasive phenotype. Although inhibition of this process could lead to successful treatment of many cancers, it is a complex, multistep process that has been difficult to target with therapy. Current strategies include inhibition of tumor cell survival in the bloodstream,[2] prevention of cell attachment at the secondary tumor site,[3] and inhibition of extracellular proteins such as matrix metalloproteinases to prevent degradation of the extracellular matrix.[4] Notably, none of these approaches effectively treat metastasis in vivo. Antimetastatic capacity could be improved if drugs were able to suppress multiple metastatic traits simultaneously.
The Snail TF has been associated with metastatic tumors, and it is believed that this is a result of Snail’s involvement in cellular adhesion and motility.[5] Snail is most commonly associated with a distinct phenotypic change called the epithelial-to-mesenchymal transition (EMT) and is integral to embryogenic morphogenesis and fibrosis.[6] It is widely recognized that there are significant similarities between EMT and the progression to an invasive phenotype that develops in cancer.[7] Furthermore, Snail is believed to be a key component of both phenotypes. Snail family TFs bind to a consensus CAGGTG sequence known as the Ebox sequence by a Cys2His2-type zinc-finger where they act as transcriptional repressors, inhibiting the expression of target genes such as E-cadherin and other epithelial markers.[8] This transcriptional repression leads to the development of a mobile phenotype[9] (Figure 1 A and B). It has been well documented that epithelial tumors reduce E-cadherin as they progress toward malignancy.[10] Beyond repression of E-cadherin, Snail expression results in a decrease in cell–cell adhesions, an increase in cellular motility, an upregulation of cell survival genes, and expression of extracellular matrix remodeling proteins.[5d]
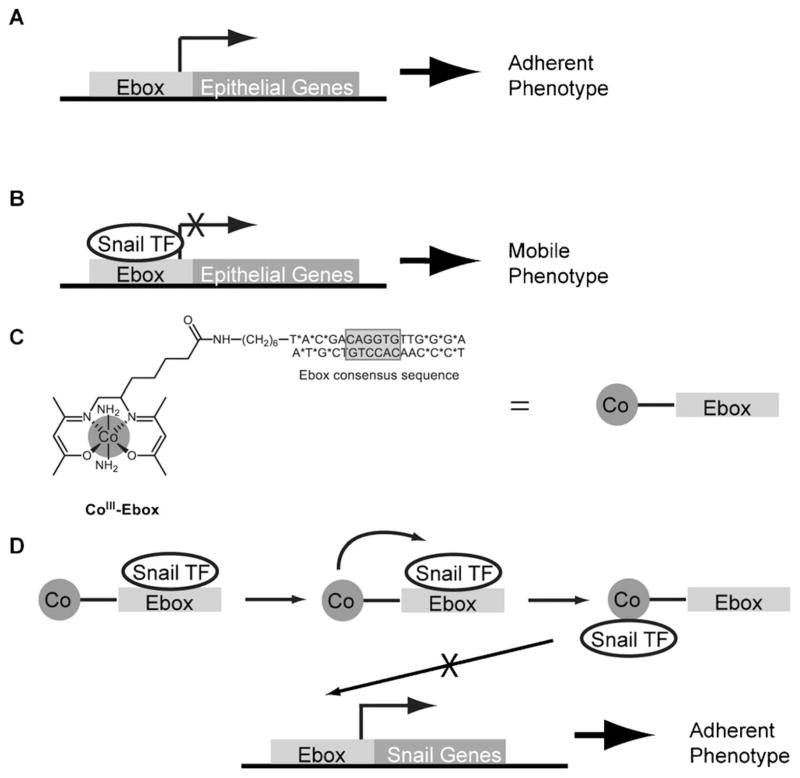
Figure 1.
Schematic showing the predicted mode by which CoIII-Ebox inhibits Snail family TF-mediated metastasis. A) The cells remain adherent when the Ebox consensus sequence is unoccupied.[9] B) The cells acquire a mobile phenotype when Snail family TFs bind to the Ebox consensus sequence and inhibit target gene transcription.[9] C) The structure of CoIII-Ebox. CoIII-Ebox is a conjugate of CoIII-sb peptide-coupled to an amine-modified Ebox oligonucleotide. * indicates the nucleotides with phosphorothioate bonds that prevent nuclease degradation. D) CoIII-Ebox inhibits Snail family TFs from binding to the Ebox consensus sequence. Snail family TFs are believed to reversibly bind to the targeting decoy oligonucleotide, bringing the TF into close proximity of the CoIII-sb inhibitor to elicit irreversible inhibition.
A critical barrier to inhibiting TFs is that pharmacological manipulation remains elusive.[11] Unlike enzyme active sites, TFs generally lack specific binding sites for small molecules to inhibit the function of the protein.[11] With the exception of drugs targeting TFs of the nuclear hormone receptor super family, TFs are generally considered undruggable.[11] For example, most TF-targeted drugs rely on reversible interactions to disrupt TF–DNA binding, necessitating the use of high concentrations of the drug to achieve effective modulation.[11–12] An alternate approach to targeting TFs is to use DNA as a decoy to bring TFs into proximity with an effector moiety capable of disabling the TF.[13]
CoIII-Schiff base complexes (CoIII-sb) consisting of the tetradentate bis(acetylacetone)ethylenediimine (acacen) as the equatorial ligand and amines as axial ligands have been shown to irreversibly inhibit the activity of histidine(His)-containing proteins such as thermolysin, α-thrombin, and matrix metalloproteinases.[14] The labile axial ligands of the CoIII-sb moiety undergo dissociative ligand exchange with the imidazole ring of essential His residues in the binding site of the protein, causing irreversible disruption of protein structure and loss of function.[15]
Specificity can be incorporated into CoIII-sb inhibitors by conjugation to a targeting moiety, such as a peptide or oligo-nucleotide.[14a,c,16] A CoIII-sb complex tethered to a decoy Ebox sequence oligonucleotide (CoIII-Ebox; Figure 1 C) has been previously reported and has shown remarkable specificity and efficacy in the inhibition of Snail family TFs involved in embryonic neural crest development of Xenopus laevis.[14a,16a] When CoIII-Ebox is administered to cells undergoing EMT, Snail family TFs are expected to reversibly bind to the decoy oligonucleotide. This binding event will bring the TF in close proximity to the inhibitor and allow for specific inhibition (Figure 1 D). Considering the high specificity and efficacy of the CoIII-Ebox conjugate, this strategy has the potential to overcome the shortcomings of previously studied TF inhibitors.
Based on the highly specific and efficient inhibition of EMT by CoIII-Ebox observed in embryonic neural crest development, this approach has strong potential as a TF inhibitor that could be used in the treatment of metastasis.[14a,16a] To further investigate this possibility, the capability of CoIII-Ebox to inhibit artificially induced EMT was studied in breast cancer cells. EMT induction was achieved in SKBR3 and MCF7 epithelial breast cancer cells by treatment with heregulin-β1 (HRG, or neuregulin), a known inducer of EMT in these cell lines.[17] Snail has explicitly been shown to be essential for this transition for SKBR3 cells, and in both cell lines Snail has been shown to localize to the nucleus after exposure to HRG.[17–18] It was hypothesized that inhibition of Snail activity with CoIII-Ebox would result in attenuation of multiple mesenchymal traits in these cells. The effects of CoIII-Ebox were analyzed from multiple aspects to characterize its in vitro efficacy as an antimetastatic agent with particular emphasis on studying the functional aspects of inhibition of Snail.
Results
The ability of CoIII-Ebox to selectively block Snail-induced motility in tumor-derived cells was examined. In previous studies, Snail-mediated transcriptional repression was shown by transfecting epithelial cells with murine Snai1 to achieve selective expression of Snail. This determined a clear link between the effects of CoIII-Ebox and the presence of its target.[14a,16a] In the present study, the ability of CoIII-Ebox to prevent phenotypic changes from endogenous Snail induced by HRG in cancer cells was explored.
HRG is a member of the EGF-like growth and differentiation factors that bind with high affinity to receptors ErbB3 and ErbB4.[19] HRG is overexpressed in breast cancers and is strongly associated with cancer progression and metastasis, an aggressive clinical course, and poor prognosis of the disease.[20] In vitro, HRG is known to transform MCF7[21] and SKBR3[18] epithelial breast cancer cell lines to a more invasive and aggressive phenotype (Figures S1 and S2 in the Supporting Information) and has been associated with induction of EMT.[17] As HRG is capable of inducing EMT in these cell lines within 48 h, it was used to test the efficacy of CoIII-Ebox to inhibit the effects of Snail.
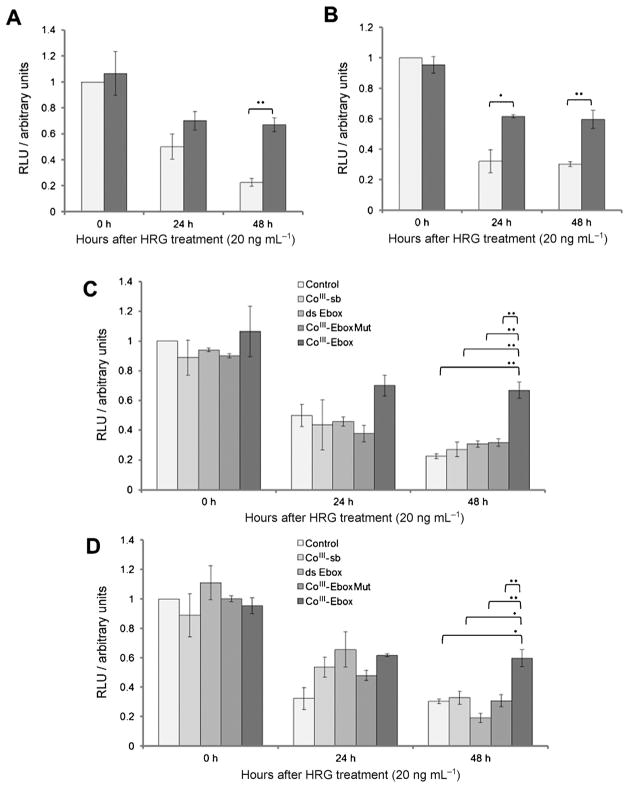
The ability of CoIII-Ebox to alleviate transcriptional repression of the E-cadherin promoter was examined. 20 ng mL−1 HRG causes a time-dependent decrease of E-cadherin expression in cells transfected with the wild-type luciferase reporter gene construct (Ecad-luc; Figure 2). To ensure that this was an effect mediated by Snail binding to the E-cadherin promoter, the experiment was repeated with a mutated luciferase reporter gene construct (EcadMut-luc) that does not bind Snail.[22] Cells transfected with EcadMut-luc did not show a decrease in E-cadherin expression in response to HRG (Figure S3). To test the inhibitory effect of CoIII-Ebox, the cells were cotransfected with 40 nM CoIII-Ebox and the Ecad-luc construct. As a result of having a nuclear export sequence, Snail resides in the cytosol in unstimulated cells.[23] Therefore, transfection agents which deposit cargo into the cytosol can effectively deliver CoIII-Ebox to Snail.[24] The concentrations of CoIII-Ebox and transfection agent used have previously been shown to be non-toxic to the cells (Table S1).[16a] The HRG-induced decrease in E-cadherin expression was inhibited, showing that CoIII-Ebox alleviates the repression of E-cadherin expression (Figure 2A and B).
Figure 2.
CoIII-Ebox treatment alleviated the HRG-induced decrease in E-cadherin expression in breast cancer cells. A) SKBR3 and B) MCF7 cells transfected with the luciferase reporter showed a time-dependent decrease in E-cadherin expression in response to HRG. This effect of HRG was alleviated when co-treated with CoIII-Ebox (■). Data are presented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance from control treatment groups (
 ) where *P <0.05 and **P<0.005. C) SKBR3 and D) MCF7 cells do not show the same inhibition of E-cadherin expression as CoIII-Ebox following treatment with CoIII-sb, ds-Ebox, or CoIII-EboxMut. Data are represented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance from control treatment groups where *P <0.05 and **P<0.005.
) where *P <0.05 and **P<0.005. C) SKBR3 and D) MCF7 cells do not show the same inhibition of E-cadherin expression as CoIII-Ebox following treatment with CoIII-sb, ds-Ebox, or CoIII-EboxMut. Data are represented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance from control treatment groups where *P <0.05 and **P<0.005.
To assess the specificity and efficacy of CoIII-Ebox, its effects were compared to treatments with the untargeted CoIII-sb, Ebox double-stranded oligonucleotide (ds-Ebox), and a mutated CoIII-DNA conjugate (CoIII-EboxMut; Figure 1). The Ebox sequence in CoIII-EboxMut has a two-base-pair substitution to diminish Snail protein binding. These three control derivatives were used to evaluate the specificity and efficacy of the binding interaction between Snail family TFs and CoIII-Ebox.[14a,16a] In all cases, the HRG-induced decrease in E-cadherin expression was not inhibited compared to CoIII-Ebox (Figure 2 C and D).
These results show that it is the cooperative effect between the sequence specificity of the targeting Ebox oligonucleotide and the inhibitory efficacy of the CoIII-sb that allows the potent inhibition of Snail family TFs by CoIII-Ebox, corroborating previously observed results.[14a,16a] Specific inhibition of the Snail transcription factor is particularly desirable because reducing Snail activity has the potential to simultaneously prevent several aspects of HRG-induced invasiveness. This prediction stems from the centrality of Snail in the induction of EMT.[8]
The extent of inhibition of E-cadherin repression by CoIII-Ebox was not complete, as a decrease in E-cadherin expression was observed over time. However, the alleviation of E-cadherin repression was specific and significant. This is particularly important, as Snail has a high turnover, with a t1/2 of less than 1 h.[25] Despite this high protein turnover, CoIII-Ebox was capable of inhibiting the Snail family TF-mediated repression of E-cadherin expression over 48 h, emphasizing the potency of this conjugate.
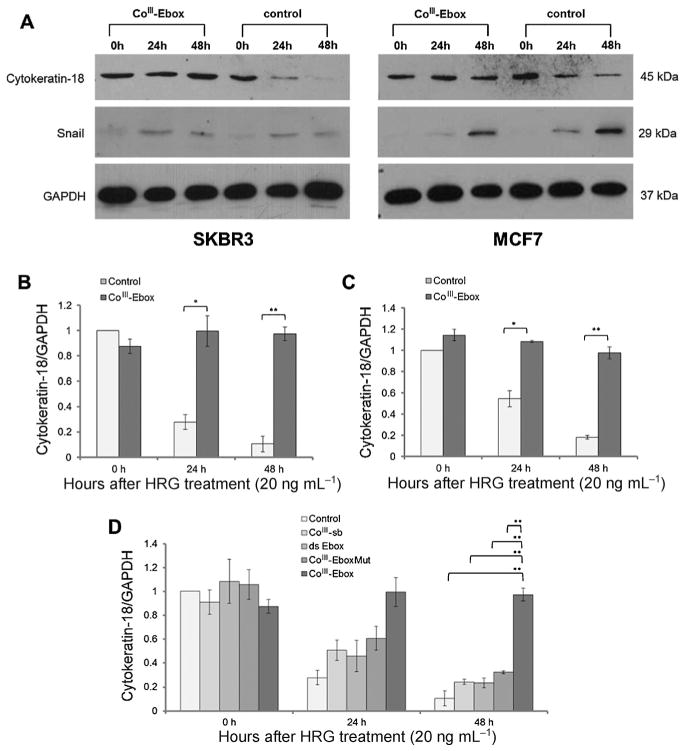
To correlate the results observed in the luciferase assay experiments, expression of cytokeratin-18 (a Snail target gene)[18] was monitored in the presence and absence of CoIII-Ebox by western blot analysis. Similar to E-cadherin, the cytokeratin-18 promoter includes Ebox sequences, and its expression is repressed by Snail.[26] GAPDH was used as a loading control and to normalize the cytokeratin-18 band intensities. HRG at 20 ng mL−1 caused a time-dependent decrease of cytokeratin-18 expression in both SKBR3 and MCF7 cells (Figure 3). When the same cells were treated with CoIII-Ebox, the HRG-induced decrease in cytokeratin-18 was not observed. Graphs showing the relative expression levels of cytokeratin-18 demonstrate that CoIII-Ebox effectively inhibits the Snail-induced decrease in this epithelial marker (Figure 3 B and C).
Figure 3.
CoIII-Ebox treatment alleviated the HRG-induced decrease in cytokeratin-18 expression in breast cancer cells. A) Western blot films showed a time-dependent decrease in cytokeratin-18 in response to HRG in SKBR3 and MCF7 cells. This effect of HRG was alleviated when co-treated with CoIII-Ebox. For both cell lines, the time-dependent increase in Snail in response to HRG was not affected by CoIII-Ebox treatment. GAPDH was used as a loading control. Relative quantification of cytokeratin-18 expression in B) SKBR3 and C) MCF7 cells. Data are represented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance from control treatment groups where *P <0.05 and **P<0.005. D) The inhibitory effect of CoIII-Ebox on cytokeratin-18 expression was not observed when SKBR3 cells were treated with CoIII-sb, ds-Ebox, or CoIII-EboxMut. Data are represented as means±S.E.M., n =3. Student’s t-tests determined statistical significance from control treatment groups where *P <0.05 and **P <0.005.
The expression levels of Snail were also probed by western blot. As the target of CoIII-Ebox is the Snail proteins themselves and not any of the upstream transducers, its expression was not expected to change in the presence or absence of CoIII-Ebox. Snail expression increased in cells treated with HRG (Figure 4 A). However, the increase in Snail expression was unaltered by CoIII-Ebox treatment. This showed that CoIII-Ebox - prevented the repression of E-cadherin and cytokeratin-18 through Snail inhibition and not through a reduction of Snail expression.
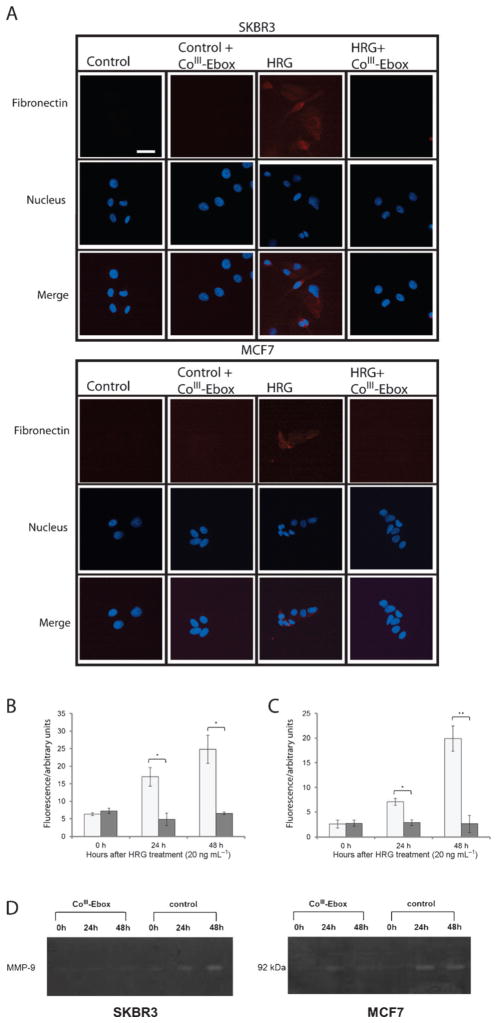
Figure 4.
CoIII-Ebox treatment alleviated the HRG-induced increase in mesenchymal marker expression in breast cancer cells. A) Fluorescence microscopy images of immunostained SKBR3 and MCF7 cells showing red (Cy5) fluorescence, indicating fibronectin expression in response to HRG. No fibronectin-associated fluorescence was observed following co-treatment with CoIII-Ebox. Scale bar: 200 μm. Relative quantification of B) SKBR3 cells and C) MCF7 cells showed a time-dependent increase in fibronectin in response to HRG. This effect was alleviated when co-treated with CoIII-Ebox (■). Data are represented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance from control treatment groups (
 ) where *P<0.05 and **P <0.005. D) Gel zymograms of SKBR3 and MCF7 cells showed a time-dependent increase in MMP-9 activity in response to HRG. The effects of HRG were alleviated by co-treatment with CoIII-Ebox.
) where *P<0.05 and **P <0.005. D) Gel zymograms of SKBR3 and MCF7 cells showed a time-dependent increase in MMP-9 activity in response to HRG. The effects of HRG were alleviated by co-treatment with CoIII-Ebox.
In a similar manner to the Ecad-luc experiment, the specificity and efficacy of CoIII-Ebox was tested by comparing its effects to control treatment with CoIII-sb, ds-Ebox oligonucleotide, and CoIII-EboxMut (Figure 3 D). In all cases, the HRG-induced decrease in cytokeratin-18 expression was not alleviated, which is in accordance with the Ecad-luc experiment. The requirement of having an Ebox sequence conjugated to the CoIII complex in order to prevent cytokeratin-18 repression further emphasizes the highly specific and effective nature of Snail inhibition by CoIII-Ebox.
The Snail-induced decrease in expression of epithelial markers allows for HRG signaling to cause a corresponding increase in mesenchymal markers, several of which are associated with metastasis. The expression of fibronectin is an indicator of the invasive phenotype associated with Snail.[27] For this reason, the ability of CoIII-Ebox to inhibit the HRG-induced increase in expression of fibronectin was investigated. This was accomplished by immunofluorescence staining of SKBR3 and MCF7 cells. Cells that were treated with HRG showed red fluorescence, corresponding to the Cy5-conjugated secondary antibody and indicative of the expression of fibronectin. In contrast, the cells that were treated with CoIII-Ebox or those that were not treated with HRG did not display red fluorescence. This observation shows that CoIII-Ebox is capable of inhibiting the downstream increase in fibronectin (Figure 4 A).
The level of fibronectin expression over time was compared by assessing the amount of red fluorescence corresponding to the Cy5-conjugated secondary antibody in a representative area of the image (Figure 4B and C). 20 ng mL−1 HRG was shown to cause a time-dependent increase in fibronectin expression, but when the same cells were treated with CoIII-Ebox, the HRG-induced increase in fibronectin was not observed. This result validates that CoIII-Ebox effectively inhibits the HRG-induced increase in fibronectin, potentially preventing the capacity to invade surrounding tissue.
MMP-9 has been associated with increased metastatic potential due to its ability to degrade the extracellular matrix and promote angiogenesis.[28] It has been shown that HRG increases MMP-9 protein, mRNA, and activity levels in MCF7 and SKBR3 cells.[29] Furthermore, in MDCK cells, Snail expression has been shown to directly increase expression of MMP-9.[30] Therefore, the capacity of CoIII-Ebox to inhibit HRG-induced expression of MMP-9 was explored. The activity of the enzyme was detected by gel zymography. Cells were grown in serum-free media and treated with vehicle or CoIII-Ebox, followed by HRG. Fractions of the media were collected 0, 24, and 48 h after treatment and analyzed for MMP-9 activity. The cells treated with HRG exhibited a time-dependent increase in MMP-9 activity (Figure 4 D). However, this time-dependent increase in MMP-9 activity was inhibited when the same cells were treated with CoIII-Ebox. This result further confirms that CoIII-Ebox inhibition of Snail is capable of attenuating the invasiveness caused by HRG exposure.
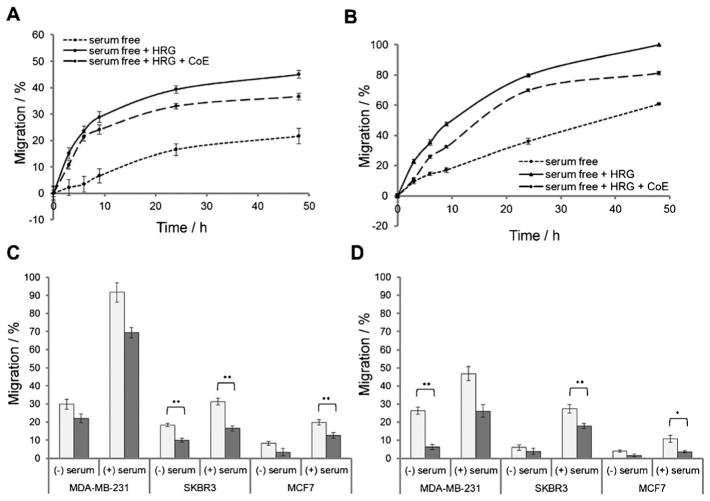
Snail expression increases the invasive and migratory properties of cells; hence, inhibition with CoIII-Ebox is expected to prevent these processes. To confirm this, a functional scratch wound assay was performed. Three scenarios were compared: cells grown with 1) no HRG, 2) HRG only, and 3) both HRG and CoIII-Ebox. Cells that were grown with no HRG showed little migration (Figure 5 A and B). However, cells treated with HRG showed much higher migration, and cells treated with CoIII-Ebox were shown to migrate slower than HRG-treated cells but faster than cells grown without HRG. Further, the cells treated with HRG were observed to migrate with a mesenchymal phenotype, in which the cells have a spindle-like appearance and migrate as individual cells rather than as clumps of cells (Figure S4). In contrast, cells treated with both HRG and CoIII-Ebox were observed to migrate less with a mesenchymal phenotype and more with an epithelial phenotype, further supporting that CoIII-Ebox is capable of inhibiting the HRG-induced invasiveness. These results show that CoIII-Ebox is capable of preventing the HRG-induced increase in migration by SKBR3 and MCF7 cells.
Figure 5.
CoIII-Ebox treatment alleviated the HRG-induced increase in breast cancer cell migration and invasion. Graphs comparing the percent migration of A) SKBR3 and B) MCF7 cells in a scratch wound assay in the presence and absence of HRG, and with or without CoIII-Ebox co-treatment. Data are represented as means±S.E.M., n =9. Comparison of C) the percent migration and D) the percent invasion of HRG-treated MDA-MB-231, SKBR3, and MCF7 cells with (■) or without (
 ) CoIII-Ebox co-treatment, and with or without chemo-attractant (serum). Data are represented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance between control and CoIII-Ebox treatment where *P<0.05 and **P <0.005.
) CoIII-Ebox co-treatment, and with or without chemo-attractant (serum). Data are represented as means ± S.E.M., n =3. Student’s t-tests determined statistical significance between control and CoIII-Ebox treatment where *P<0.05 and **P <0.005.
To corroborate the results of the scratch wound assay, trans-well migration and invasion assays were performed (Figure 5C and D). For these experiments, the migration and invasion of mesenchymal-like MDA-MB-231 cells[31] were compared to those of the more epithelial SKBR3 and MCF7 cells.[17] The transwell migration and invasion assays were both carried out by using Corning Transwell inserts which have 8 μm pores in a polycarbonate membrane.
For the migration assays, the cells were plated in wells and treated with HRG alone or with HRG and CoIII-Ebox. Serum was used as the chemo-attractant. For the invasion assay, basement membrane extract (BME) was plated into the inserts as an invasion substrate between the cells and the chemo-attractant. It was found that MDA-MB-231 cells had the highest migratory/invasive ability, followed by SKBR3 cells, and finally, MCF7 cells. For all cell lines (regardless of the availability of chemo-attractant), those treated with CoIII-Ebox displayed less migration and invasion compared to the untreated cells. In particular, SKBR3 and MCF7 cells treated with CoIII-Ebox migrated and invaded significantly less than the control in the presence of chemo-attractant. This demonstrates that CoIII-Ebox is capable of inhibiting the migration and invasion of cells undergoing HRG-induced EMT.
To study the effect of CoIII-Ebox in a more physiologically relevant environment, spheroids grown on a bed of agarose were used as a model of metastatic tumors.[32] Attempts were made to grow spheroids from MDA-MB-231, SKBR3, and MCF7 cells, but only MCF7 cells successfully formed spheroids. Consequently, spheroid outgrowth experiments were carried out in MCF7 cells.
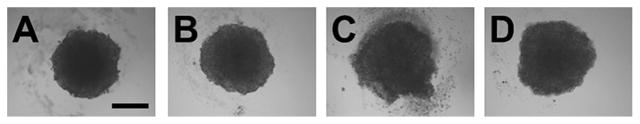
MCF7 cells were seeded at a density of 2.5 × 104 cells per well in a 96-well plate in serum-free medium, allowed to settle overnight, then transfected with vehicle or 40 nM CoIII-Ebox. After 24 h of incubation at 37 °C, the spheroids were treated with water or 20 ng mL−1 HRG. The cells were then allowed to aggregate for five days. Each spheroid was approximately 400 μm in diameter after the five day growth period. Figure 6 shows the transmittance images of spheroid morphology eight days after the cells were plated on the bed of agarose. Cells grown without HRG formed clearly defined spheroids. However, cells that were treated with HRG grew malformed spheroids with ill-defined perimeters. This is likely due to HRG causing the cells to acquire a more migratory and invasive phenotype, which causes the cells to break away from the spheroid. Spheroids that were treated with both HRG and CoIII-Ebox presented the same morphology as the spheroids untreated with HRG, showing that CoIII-Ebox is able to inhibit the effects of HRG, even in 3D culture.
Figure 6.
Transmittance images of MCF7 spheroids. A) Treated with vehicle only, B) treated with CoIII-Ebox only, C) treated with HRG only and D) treated with HRG and CoIII-Ebox. Scale bar: 200 μm.
To further assess the migratory and invasive behavior of spheroids, those grown in the same conditions as outlined in Figure 7 were embedded in BME, which mimics the extracellular matrix of cells. The purpose of this experiment was to observe the migration and invasion of cells originating from spheroids in a 3D setting, as this should be more representative of the actual behavior of cells metastasizing from tumors in vivo. To achieve efficient embedding in BME, the spheroids were harvested from the agarose bed and gently washed in PBS to remove any cell debris. They were then mixed with a BME solution in serum-free media supplemented with HRG, and the mixture was placed in a small well lined with pre-gelled BME such that the spheroids were suspended three-dimensionally in BME upon gelling at 37°C. Images of the spheroids were obtained every three days, and the spheroids were fixed and immunostained for fibronectin after a total of two weeks.
Figure 7.
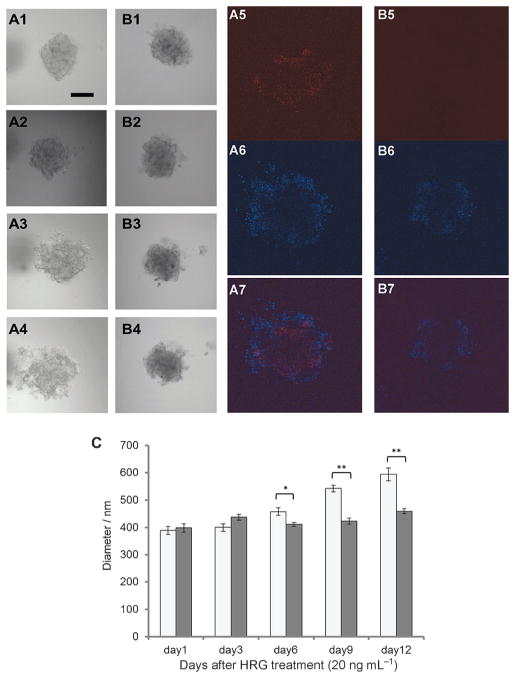
CoIII-Ebox treatment alleviated the HRG-induced increase in breast cancer spheroid invasion. MCF7 spheroids embedded in BME treated with A) HRG and B) HRG with CoIII-Ebox were imaged at 1) one day, 2) three days, 3) six days, and 4) nine days. After 12 days, the spheroids were fixed and immunostained for fibronectin. Fluorescence images of 5) fibronectin (Cy5), 6) nucleus (DAPI) and 7) merge of (5) and (6). Scale bar: 200 μm. C) Graph comparing the outgrowth of the spheroids as determined by measuring the diameter of the spheroids over time. Data are represented as mean ± S.E.M., n =3. Student’s t-tests determined statistical significance between control (
 ) and CoIII-Ebox treatment (■) where *P <0.05 and **P< 0.005.
) and CoIII-Ebox treatment (■) where *P <0.05 and **P< 0.005.
Spheroids that were treated with vehicle showed migration and invasion of cells disseminating from the spheroids over a 12-day period (Figure 7A). During this time, the spheroid was occupy a larger volume. The spheroid became less compact, with clusters of cells migrating away from the main body of the spheroid. This phenomenon was observed to occur in all directions, as captured by Z-stack images (Movies S1 and S2 in the Supporting Information). In contrast, the spheroid treated with CoIII-Ebox did not invade the BME to the same extent (Figure 7 B). Some alteration in the spheroid morphology was observed, as shown by reorganization of its shape over time, but the volume occupied by the spheroids appeared to remain unchanged. The spheroids treated with CoIII-Ebox remained dense, and no significant outgrowth or invasion into the BME was observed.
The visual observations of spheroid outgrowth were compared by measuring the spheroid density against the background using ImageJ software and determining the average diameter of the spheroids at each time point. The diameter of HRG-treated spheroids increased by approximately 200 μm after the 12-day period, indicating that the invasion of cells into the BME was extensive. In comparison, the diameter of the spheroids co-treated with CoIII-Ebox remained virtually unchanged. This corroborates the visual observations that CoIII-Ebox inhibits the invasion and migration of cells from spheroids. The difference in extent of outgrowth between the spheroids treated with HRG only and those that were co-treated with CoIII-Ebox highlight the efficacy of CoIII-Ebox at inhibiting the migratory and invasive propensity of cells in a 3D setting.
The expression of fibronectin (a mesenchymal marker) was probed in spheroids embedded in BME by immunostaining (Figure 7 A5–7 and B5–7). For the spheroids treated with vehicle only, the red fluorescence indicating fibronectin expression was found to be localized in the perimeter of the spheroids where the cells appeared to be disseminating away from the body of the spheroid. This suggests that fibronectin is being expressed by the cells that are located at the outermost part of the spheroid, where they are in direct contact with the BME. These cells are expected to be aggressively migrating and invading the matrix in response to HRG, which is in agreement with the high expression levels of fibronectin. In contrast, the spheroids that were treated with CoIII-Ebox did not show any red fluorescence, indicating that there was no fibronectin expression. This result further supports the observations that the outgrowth of HRG-treated spheroids requires Snail activity and that CoIII-Ebox is capable of inhibiting this process in 3D. To our knowledge, this is the first successful example of using spheroids to study inhibition of EMT using a 3D tumor model in vitro.
Conclusion
The results of the experiments described in this report show that CoIII-Ebox is capable of inhibiting multiple aspects of Snail activity during HRG-induced EMT in SKBR3 and MCF7 breast cancer cells. Specifically, CoIII-Ebox was observed to inhibit the Snail-induced decrease in epithelial markers such as E-cadherin and cytokeratin-18. CoIII-Ebox was also observed to inhibit the corresponding Snail-induced increase in mesenchymal markers such as fibronectin and MMP-9. Through Snail inhibition, CoIII-Ebox was capable of delaying the invasive phenotype induced by HRG, both in 2D and 3D cell culture. In particular, the observed inhibition of spheroid invasion into BME in 3D demonstrates that CoIII-Ebox has significant promise in preventing tumor metastasis. The success of the agent in vitro warrants further experiments in vivo and in the clinic as an effective treatment for metastasis with a novel mode of action.
Experimental Section
Materials
CoIII-Ebox conjugate was prepared by previously described methods.[14a,16b] HRG was purchased from Sigma–Aldrich and was used without further purification.
Assays and statistical analysis
Details of the cell culture conditions, cytotoxicity assay, reporter gene assay, Western blot analysis, immunofluorescence study, zymography, scratch wound assay, and transwell assays are described in the Supporting Information. Statistical analysis was performed on the means by using Student’s t-test where *P <0.05 and **P <0.005.
Confocal microscopy of spheroids
Spheroids were prepared by plating of a single cell suspension of MCF7 cells (100 μL; 2.5 × 105 cells mL−1) onto 96-well plates coated in each well with agarose (35 μL; 0.75 % w/v in PBS).[32] Following incubation overnight, the spheroids were treated with of CoIII-Ebox (40 nM) complexed to Turbofect Transfection Reagent (Thermo Scientific). After 24 h, the cells were treated with HRG (20 ng mL−1) and allowed to aggregate for five days without motion, resulting in the formation of a single spheroid per well.
On day 5, the spheroids were gently washed with PBS and embedded in Cultrex BME (75 μL; 0.25 μg μL−1; Trevigen) in medium with and without HRG (20 ng mL−1). The embedding was performed in a two-well insert attached to a 35 mm microscopy μ-dish (iBidi) lined with Cultrex BME (20 μL; 0.25 μg μL−1). The embedded spheroids were imaged on day 1 and subsequently every 3 days over 12 days. At the end of the 12 days, the spheroids were fixed in 3.7 % (w/v) formaldehyde solution in PBS for 2 h, then blocked overnight in 6 % (w/v) BSA with 0.25 % (v/v) TritonX-100 in PBS at room temperature. The wells were washed with 0.25 % (v/v) TritonX-100 in PBS (3 × 10 min) and incubated with anti-fibronectin mouse mAb (1:1000; Thermo Scientific) in BSA (3 %, w/v) with TritonX-100 (0.25 %, v/v) in PBS overnight at room temperature. The wells were washed in PBS (3 × 10 min) and incubated with Cy5-conjugated secondary antibody (1:500) in BSA (3 %, w/v) with TritonX-100 (0.25 %, v/v) in PBS overnight at room temperature. The wells were washed with TritonX-100 (0.25 %, v/v) in PBS (3 × 10 min), incubated with DAPI (300 nM) with TritonX-100 (0.25 %, v/v) in PBS overnight at room temperature, and imaged the following day.
Confocal fluorescence microscopy was performed on a LSM 510 inverted confocal scanning microscope (Zeiss) and an EC Plan-Neofluar 10 × 0.30 objective lens. Confocal images were acquired with a 700 nm Mai-Tai Ti-Sapphire crystal laser (Spectra-Physics), a 480 nm LGK7812ML4 Ar laser (Lasos), and a 633 nm LGK7628–1 He/Ne laser. The emission ranges were 390–465, 500–530, and 650–710 nm, respectively. A scan rate of 1.6 μs per pixel was used for all images, and an average of four scans per image were collected. Z-stack images were collected at increments of 15 μm. An incubator chamber (PeCon) was used to maintain the temperature at 37 °C during imaging. At least three spheroids were imaged on each occasion, and the experiments were repeated on at least three separate occasions. Images shown are a representative replicate.
Relative quantification of the spheroid outgrowth was carried out by using ImageJ (National Institutes of Health), by drawing an 800 μm line through the center of the spheroid in the Z-stack transmittance image and measuring the integrated density where the spheroid had the largest diameter. The diameter of the spheroid was determined from the plotted profile of the line. This procedure was repeated 16 times, rotating the line around the spheroid at approximately equal angle intervals, and the measured diameters were averaged. Measurements were taken from at least six different spheroids in each treatment group. Statistical analysis was performed on the means by using Student’s t-test where *P < 0.05 and **P <0.005.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute (grant RO3A167715), and the National Institute for Arthritis and Musculoskeletal and Skin Diseases (NIAMSD) (grant P30AR057216). The authors gratefully acknowledge Marie C. Heffern and Robert J. Holbrook for helpful discussions. Fluorescence and luminescence measurements were performed at the Northwestern University High Throughput Analysis Laboratory. Confocal fluorescence spectroscopy was performed at the Northwestern University Quantitative Bioelemental Imaging Center, generously supported by the National Science Foundation (grant CHE-9810378/005). This work was supported by the National Institutes of Health Centers of Cancer Nanotechnology Excellence initiative of the National Cancer Institute (grant U54A119341).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201500289.
References
- 1. [accessed August 14, 2015];Cancer Fact Sheet No. 297. http://www.who.int/mediacentre/factsheets/fs297/en/
- 2.Maraveyas A, Johnson JM, Yu PX, Noble S. Cancer Metastasis Rev. 2010;29:777–784. doi: 10.1007/s10555-010-9263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman ER, Lipton A, Roodman GD, Guise AT, Boyce FB, Brufsky MA, Clezardin P, Croucher IP, Gralow RJ, Hadji P, Holen I, Mundy RG, Smith RM, Suva JL. Cancer Treat Rev. 2010;36:615–620. doi: 10.1016/j.ctrv.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deryugina EI, Quigley JP. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 5.a) Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, Larsson O, Muller WJ, Miller WH, Sonenberg N. Oncogene. 2015;34:2032–2042;. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer Cell. 2009;15:195–206;. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]; c) Yang M-H, Chang S-Y, Chiou SH, Liu C-J, Chi C-W, Chen P-M, Teng S-C, Wu K-J. Oncogene. 2007;26:1459–1467;. doi: 10.1038/sj.onc.1209929. [DOI] [PubMed] [Google Scholar]; d) Barrallo-Gimeno A, Nieto MA. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 6.Baum B, Settleman J, Quinlan MP. Semin Cell Dev Biol. 2008;19:294–308. doi: 10.1016/j.semcdb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 7.a) Voulgari A, Pintzas A. Biochim Biophys Acta Rev Cancer. 2009;1796:75–90;. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]; b) Mego M, Mani SA, Cristofanilli M. Nat Rev Clin Oncol. 2010;7:693–701;. doi: 10.1038/nrclinonc.2010.171. [DOI] [PubMed] [Google Scholar]; c) Brabletz T. Nat Rev Cancer. 2012;12:425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 8.Nieto MA. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 9.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, Del Barrio MG, Portillo F, Nieto MA. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 10.a) Roy F, Berx G. Cell Mol Life Sci. 2008;65:3756–3788;. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Perl A-K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature. 1998;392:190–193;. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]; c) Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, Van Roy F. Cell. 1991;66:107–119;. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]; d) Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Loechner D, Birchmeier W. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konstantinopoulos PA, Papavassiliou AG. JAMA J Am Med Assoc. 2011;305:2349–2350. doi: 10.1001/jama.2011.727. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Martínez JA, Peterson-Kaufman KJ, Ansari AZ. Biochim Biophys Acta Gene Regul Mech. 2010;1799:768–774. doi: 10.1016/j.bbagrm.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Pazos E, Portela C, Penas C, Vazquez ME, Mascarenas JL. Org Biomol Chem. 2015;13:5385–5390;. doi: 10.1039/c5ob00318k. [DOI] [PubMed] [Google Scholar]; b) Portela C, Albericio F, Eritja R, Castedo L, Mascareñas JL. Chem Bio Chem. 2007;8:1110–1114. doi: 10.1002/cbic.200700115. [DOI] [PubMed] [Google Scholar]
- 14.a) Harney AS, Lee J, Manus LM, Wang P, Ballweg DM, La Bonne C, Meade TJ. Proc Natl Acad Sci USA. 2009;106:13667–13672;. doi: 10.1073/pnas.0906423106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Takeuchi T, Bottcher A, Quezada CM, Meade TJ, Gray HB. Bioorg Med Chem. 1999;7:815–819;. doi: 10.1016/s0968-0896(98)00272-7. [DOI] [PubMed] [Google Scholar]; c) Takeuchi T, Boettcher A, Quezada CM, Simon MI, Meade TJ, Gray HB. J Am Chem Soc. 1998;120:8555–8556;. [Google Scholar]; d) Louie AY, Meade TJ. Proc Natl Acad Sci USA. 1998;95:6663–6668. doi: 10.1073/pnas.95.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Manus LM, Holbrook RJ, Atesin TA, Heffern MC, Harney AS, Eckermann ALT. J Meade,Inorg Chem. 2013;52:1069–1076;. doi: 10.1021/ic302379j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heffern MC, Kurutz JW, Meade TJ. Chem Eur J. 2013;19:17043–17053;. doi: 10.1002/chem.201301659. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Heffern MC, Yamamoto N, Holbrook RJ, Eckermann ALT. J Meade, Curr Opin Chem Biol. 2013;17:189–196. doi: 10.1016/j.cbpa.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Harney AS, Meade TJ, Labonne C. PLoS One. 2012;7:e32318. doi: 10.1371/journal.pone.0032318. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hurtado RR, Harney AS, Heffern MC, Holbrook RJ, Holmgren RA, Meade TJ. Mol Pharm. 2012;9:325–333. doi: 10.1021/mp2005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Jeong H, Lee Y, Kim C, Kim H, Kim A. BMC Cancer. 2013;13:383. doi: 10.1186/1471-2407-13-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng LS, Zha Z, Lang B, Liu J, Yao XB. Cancer Lett. 2009;280:50–60. doi: 10.1016/j.canlet.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Breuleux M. Cell Mol Life Sci. 2007;64:2358–2377. doi: 10.1007/s00018-007-7120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Science. 1989;244:707–712;. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]; b) Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Science. 1987;235:177–182;. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]; c) Ross JS, Fletcher JA. Stem Cells. 1998;16:413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 21.Adam L, Vadlamudi R, Kondapaka SB, Chernoff J, Mendelsohn J, Kumar R. J Biol Chem. 1998;273:28238–28246. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- 22.Hajra KM, Chen DYS, Fearon ER. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 23.Domínguez D, Montserrat-Sentís B, Virgós-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Francí C, García de Herreros A. Mol Cell Biol. 2003;23:5078–5089. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas M, Klibanov AM. Appl Microbiol Biotechnol. 2003;62:27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 25.a) de Herreros Antonio G, Peiro S, Nassour M, Savagner P. J Mammary Gland Biol Neoplasia. 2010;15:135–147;. doi: 10.1007/s10911-010-9179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Franci C, Takkunen M, Dave N, Alameda F, Gomez S, Rodriguez R, Escriva M, Montserrat-Sentis B, Baro T, Garrido M, Bonilla F, Virtanen I, Garcia de Herreros A. Oncogene. 2006;25:5134–5144. doi: 10.1038/sj.onc.1209519. [DOI] [PubMed] [Google Scholar]
- 26.De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- 27.Moody SE, Perez D, Pan T-c, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Deryugina E, Quigley J. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 29.Yao J, Xiong S, Klos K, Nguyen N, Grijalva R, Li P, Yu D. Oncogene. 2001;20:8066–8074. doi: 10.1038/sj.onc.1204944. [DOI] [PubMed] [Google Scholar]
- 30.Jordà M, Olmeda D, Vinyals A, Valero E, Cubillo E, Llorens A, Cano A, Fabra À. J Cell Sci. 2005;118:3371–3385. doi: 10.1242/jcs.02465. [DOI] [PubMed] [Google Scholar]
- 31.Yin KB. In: Breast Cancer-Focusing Tumor Microenvironment, Stem Cells and Metastasis. Gunduz M, Gunduz E, editors. InTech Open Access Publisher Book Project; 2011. pp. 385–403. [Google Scholar]
- 32.Kim BJ, Hambley TW, Bryce NS. Chem Sci. 2011;2:2135–2142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.