Aberrant kinase signaling is a hallmark of myeloid malignancies that has become the focus of targeted therapies due to its integral role in leukemia cell proliferation and survival.1 In acute myeloid leukemia (AML) in particular, aberrant kinase signaling results from activating mutations of FLT3, KIT and NRAS or mutational inactivation of their negative regulators CBL, PTEN and PTPN11. However, recent genome-wide surveys have revealed that no more than 50% of patients possess activating genetic mutations in these kinase signaling pathways.2 This suggests that other mechanisms of kinase activation may exist.
Ligand-dependent activation of receptor tyrosine kinases (RTKs) has long been recognized as an alternative mechanism of aberrant kinase signaling. Indeed, it is now over three decades since Sporn and Todaro3 proposed their ‘autocrine hypothesis’ that postulated the essential functions of secreted factors in the promotion of autonomous cell growth required for malignant transformation. We now recognize many examples of carcinogenic autocrine signaling, involving ligands of the epidermal growth factor (EGF), insulin-like growth factor, platelet-derived growth factor, fibroblast growth factor, EPH, vascular endothelial growth factor, and Fms-like tyrosine kinase 3 (FLT3) and KIT receptors in AML specifically.4–6 Recently, autocrine activation of the MET receptor kinase was found in almost 40% of patients with AML, causing enhanced leukemia cell survival owing to aberrant expression of the MET ligand hepatocyte growth factor (HGF) that could be blocked by the competitive RTK inhibitor crizotinib.7,8 However, translation of these findings into therapeutic clinical trials remains hindered by limited knowledge about the specific subtypes of AML in which MET activation occurs and by the development of resistance to MET kinase inhibitors due to the overexpression of HGF that adaptively reactivates MET signaling.8
Here, using reverse-phase protein arrays (RPPA) and mathematical modeling of ligand-dependent MET receptor activation, we determined signaling pathways associated with MET activation in primary AML cells and defined specific strategies to achieve durable inhibition of ligand-dependent kinase signaling in AML. Proteomic RPPA profiling of 511 AML patient specimens collected at the University of Texas M.D. Anderson Cancer Center was done using previously described methods, with experimental details and patient characteristics described in detail elsewhere.9,10 Matched peripheral blood and bone marrow samples were available for 140 patients. Paired primary and relapse samples were available for 48 of the AML and 1 of the acute promyelocytic leukemia patients. Outcome analysis was restricted to newly diagnosed patients. The majority of AML patients received arabinofuranosyl cytidine (Ara-C)-based therapy, while acute promyelocytic leukemia patients received combination all-trans retinoic acid therapy.9 The statistical methods used in this study are described elsewhere in detail,9 including the use of Super-Curve algorithms, paired t-tests, Bonferroni multiple-hypothesis corrections, mixed-effects linear models and Pearson and Spearman correlation analysis.
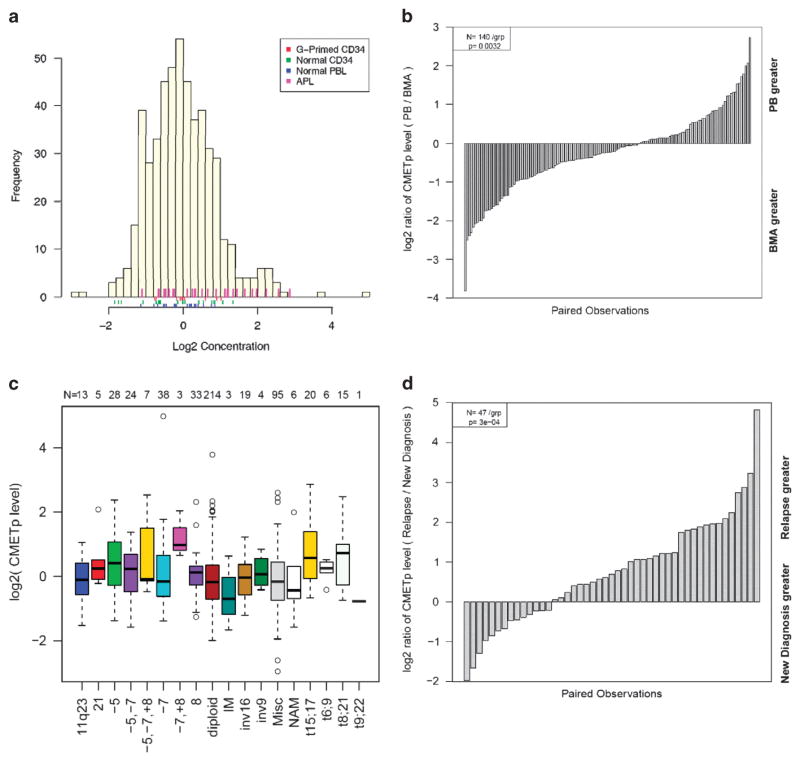
Using the RPPA approach, we found that 11% of all AML, 43% of acute promyelocytic leukemia, 9% of newly diagnosed, as well as 27% of relapsed AML specimens exhibit increased levels of MET phosphorylation, as compared with CD34+ controls (Figure 1a), which did not depend on cryo-preservation status (Supplementary Figure S1a). We found that MET phosphorylation was relatively higher in the bone marrow as compared with peripheral blood (P = 0.003, Figure 1b), consistent with ligand-dependent signaling that would be potentiated by restricted ligand diffusion of the bone marrow microenvironment.
Figure 1.
(a) Histogram of MET phosphorylation levels detected in primary AML cells relative to normal and stimulated controls. (b) Distribution of MET phosphorylation levels in pairs of samples from the peripheral blood and bone marrow showing significantly higher levels in bone marrow samples. (c) Analysis of MET phosphorylation as a function of AML cytogenetic subtype. (d) Analysis of MET phosphorylation in newly diagnosed versus relapsed AML specimens, demonstrating significantly increased levels at disease relapse.
Analysis of MET phosphorylation with respect to the specific AML molecular subtypes revealed increased MET activation associated with the t(15;17) and t(8;21) cytogenetic subtypes in addition to specimens harboring both chromosome 7 deletion (−7) and chromosome 8 trisomy (+8) (P<0.05 for all comparisons, Figure 1c). Importantly, we found that increased levels of MET activation were associated with the absence of FLT3 mutations (P = 0.0505; Supplementary Figure S1b). We did not identify an association between MET activation and primary chemoresistance (P = 0.89; Supplementary Figure S1c) but did find significantly increased MET activation in specimens collected at relapse as compared with diagnosis (P = 0.0003; Figure 1d). No association was observed between MET phosphorylation and overall or event-free survival, even when multivariate analysis was performed comparing MET activation with clinically relevant variables, including age, performance status, laboratory values and cytogenetics (Supplementary Figures S2a and b). Furthermore, MET phosphorylation had no apparent effect on the outcomes in the prognostically favorable t(15;17) and t(8;21) subgroups with which it had been associated (Supplementary Figures S2c and d).
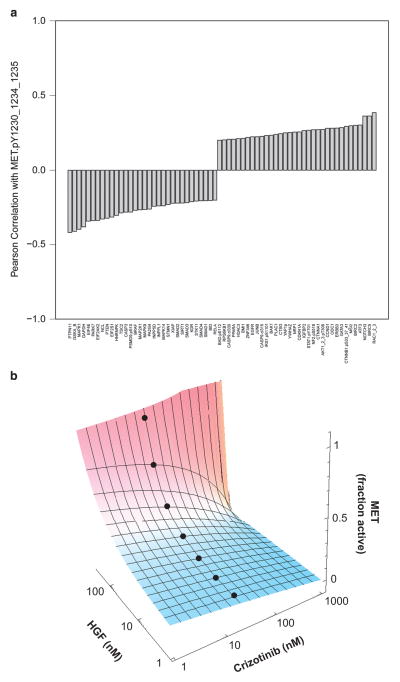
Analysis of MET phosphorylation in comparison with 210 additional RPPA protein markers revealed the expression levels of 34 and 32 proteins to be significantly positively and negatively correlated with MET phosphorylation, with the Pearson correlation coefficients of >0.2 and < − 0.2, respectively (Figure 2a). Among these proteins, we found phosphorylation of AKT1, signal transducer and activator of transcription factor 1, β-catenin, YAP1 and BAD, as well as increased expression of proto-oncogenes, such as MSI2, cyclin D1 and JUNB (Supplementary Figure S3a). These co-activated signaling pathways may constitute rational targets for combination therapy to achieve sustained MET inhibition.
Figure 2.
(a) Waterfall plot of proteins significantly associated with MET phosphorylation. (b) Dose–response surface of ligand-dependent MET kinase activation by HGF and competitive kinase inhibition by crizotinib. Blue-to-red color gradient indicates MET kinase activation. Black dots represent crizotinib plasma trough concentrations achievable in patients.
To understand the basis of resistance to competitive MET kinase inhibition, we developed and analyzed a mathematical model of the activated MET signaling complex (Supplementary Figures S3b and c). The mathematical model of ligand-dependent MET receptor activation is based on the mass action equilibria of the major molecular species. We assumed that the equilibrium of monomeric (M) and dimeric (M2) MET species is equal to its homolog EGF receptor (L2 = 0.8 M), with M2 being the active species.11,12 In the case of modeling receptor activation by HGF, we used high and low HGF-binding states (β2 = 100 pM, β1 = 90 nM), for the dimeric (M2) and monomeric (M) receptors, respectively.13,14 For competitive inhibition of MET kinase by crizotinib (C1 = 8 nM), we modeled its binding to the inactive monomeric (M) receptor.15,16 We then calculated total MET activity based on the fractional occupancy of the dimeric MET receptor, as a function of binding of HGF and inhibitor to the monomeric and dimeric MET receptors (β1 and β2 and C1 and C2), respectively. Dose–response surfaces were plotted using Mathematica version 8 (Wolfram Research, Champaign, IL, USA).
Using this mathematical analysis, we found that residual MET kinase activity can be maintained by increasing the levels of HGF in the presence of the competitive MET tyrosine kinase inhibitor crizotinib (Figure 2b).17 Even at supraphysiological crizotinib concentrations (1000 nM), sufficient HGF concentrations are available to maintain partial MET kinase activity. This indicates that the thermodynamic coupling between ligand-mediated receptor dimerization and inhibitor kinase binding can limit the therapeutic efficacy of competitive inhibitors, particularly those that preferentially stabilize the inactive kinase conformation.
In summary, our study establishes that autocrine MET signaling, as assessed by direct measurements of MET phosphorylation, is associated with distinct molecular abnormalities in AML such as t(8;21) and t(15;17) translocations, which trans-activate HGF expression by the expression of leukemogenic transcription factor fusion proteins to drive autocrine MET signaling.8 Notably, increased MET activation was found to be associated with the absence of FLT3 mutations in the examined patient cohort. This may indicate that autocrine MET signaling functions as an alternative pathway for leukemogenesis in cells lacking FLT3 activation or in functionally privileged subsets of cells, such as leukemia-initiating cells.18 Consistent with this notion, we found an increased prevalence of MET activation upon leukemia relapse, suggesting that MET signaling may contribute to enhanced cell survival or the emergence of chemotherapy-resistant cell subsets.
In the previous work, therapeutic targeting of autocrine MET activation using the competitive kinase inhibitor crizotinib led to the emergence of resistance owing to HGF upregulation.8 Similar ligand-dependent mechanisms of resistance were found to contribute to the resistance to inhibition of EGF receptor, BRAF and FLT3 RTKs.19 Here we found that residual MET kinase activity can be preserved in the presence of a wide range of concentrations of the competitive MET tyrosine kinase inhibitor crizotinib, even at inhibitor levels higher than those achievable clinically in patients. This resistance phenomenon may be exacerbated in vivo, where the dynamic and micro-environmental nature of autocrine signaling can facilitate adaptive ligand upregulation and local potentiation, respectively.
These findings suggest that therapeutic targeting of MET activation in AML, and other ligand-driven RTKs, will need to rely on strategies that can overcome the thermodynamic mass action coupling between ligand and competitive inhibitor binding. Possible approaches to overcome this barrier may include the use of ligand-neutralizing antibodies or non-competitive inhibitors, such as the recently developed covalent inhibitors of the EGF RTK.20 Likewise, therapies targeting adaptive or cooperating pathways themselves, as identified by the studies here, may be used to potentiate the durability of competitive kinase inhibition.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health grant K08CA160660, Gabrielle’s Angel Foundation and Alex’s Lemonade Stand Foundation (to AK) and the Leukemia and Lymphoma Society (to SMK).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Contributor Information
SM Kornblau, Email: skornblau@mdanderson.org.
A Kentsis, Email: kentsisresearchgroup@gmail.com.
References
- 1.Ferrara F. New agents for acute myeloid leukemia: is it time for targeted therapies? Expert Opin Invest Drugs. 2012;21:179–189. doi: 10.1517/13543784.2012.646082. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sporn MB, Todaro GJ. Autocrine secretion and malignant transformation of cells. N Engl J Med. 1980;303:878–880. doi: 10.1056/NEJM198010093031511. [DOI] [PubMed] [Google Scholar]
- 4.Graeber TG, Eisenberg D. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat Genet. 2001;29:295–300. doi: 10.1038/ng755. [DOI] [PubMed] [Google Scholar]
- 5.Zheng R, Klang K, Gorin NC, Small D. Lack of KIT or FMS internal tandem duplications but co-expression with ligands in AML. Leuk Res. 2004;28:121–126. doi: 10.1016/s0145-2126(03)00184-x. [DOI] [PubMed] [Google Scholar]
- 6.Pietsch T, Kyas U, Steffens U, Yakisan E, Hadam MR, Ludwig WD, et al. Effects of human stem cell factor (c-kit ligand) on proliferation of myeloid leukemia cells: heterogeneity in response and synergy with other hematopoietic growth factors. Blood. 1992;80:1199–1206. [PubMed] [Google Scholar]
- 7.Tyner JW, Fletcher LB, Wang EQ, Yang WF, Rutenberg-Schoenberg ML, Beadling C, et al. MET receptor sequence variants R970C and T992I lack transforming capacity. Cancer Res. 2010;70:6233–6237. doi: 10.1158/0008-5472.CAN-10-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18:1118–1122. doi: 10.1038/nm.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornblau SM, Qiu YH, Zhang N, Singh N, Faderl S, Ferrajoli A, et al. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–5612. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter BZ, Qiu Y, Huang X, Diao L, Zhang N, Coombes KR, et al. Survivin is highly expressed in CD34(+)38(-) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood. 2012;120:173–180. doi: 10.1182/blood-2012-02-409888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci USA. 1987;84:6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boni-Schnetzler M, Pilch PF. Mechanism of epidermal growth factor receptor autophosphorylation and high-affinity binding. Proc Natl Acad Sci USA. 1987;84:7832–7836. doi: 10.1073/pnas.84.22.7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gherardi E, Sandin S, Petoukhov MV, Finch J, Youles ME, Ofverstedt LG, et al. Structural basis of hepatocyte growth factor/scatter factor and MET signalling. Proc Natl Acad Sci USA. 2006;103:4046–4051. doi: 10.1073/pnas.0509040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazzone M, Basilico C, Cavassa S, Pennacchietti S, Risio M, Naldini L, et al. An uncleavable form of pro-scatter factor suppresses tumor growth and dissemination in mice. J Clin Invest. 2004;114:1418–1432. doi: 10.1172/JCI22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor. EMBO J. 2004;23:2325–2335. doi: 10.1038/sj.emboj.7600243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwall RH, Chang LY, Godowski PJ, Kahn DW, Hillan KJ, Bauer KD, et al. Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists NK1 and NK2. J Cell Biol. 1996;133:709–718. doi: 10.1083/jcb.133.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Alvey C, Bello A, Wilner KD, Tan W. Pharmacokinetics (PK) of crizotinib (PF-02341066) in patients with advanced non-small cell lung cancer (NSCLC) and other solid tumors. ASCO Meeting Abstr. 2011;29(15_suppl):e13065. [Google Scholar]
- 18.Bonardi F, Fusetti F, Deelen P, van Gosliga D, Vellenga E, Schuringa JJ. A proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Mol Cell Proteomics. 2013;12:626–637. doi: 10.1074/mcp.M112.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuckmann JM, Rauh D, Thomas RK. Epidermal growth factor receptor (EGFR) signaling and covalent EGFR inhibition in lung cancer. J Clin Oncol. 2012;30:3417–3420. doi: 10.1200/JCO.2012.43.1825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.