Abstract
The mononuclear phagocyte system (MPS) has historically been categorized into monocytes, dendritic cells and macrophages on the basis of functional and phenotypical characteristics. However, considering that these characteristics are often overlapping, the distinction between and classification of these cell types has been challenging. In this Opinion article, we propose a unified nomenclature for the MPS. We suggest that these cells can be classified primarily by their ontogeny and secondarily by their location, function and phenotype. We believe that this system permits a more robust classification during both steady-state and inflammatory conditions, with the benefit of spanning different tissues and across species.
Dendritic cells (DCs), monocytes and macrophages are members of the mononuclear phagocyte system (MPS) that exhibit multiple functions during immune responses. Historically, these cells have been grouped together because although monocytes have their unique functions as mononuclear phagocytic cells, they were also considered as the definitive precursors of macrophages and DCs1–3 (Box 1). Macrophages are distinguished as larger vacuolar cells that excel in the clearance of apoptotic cells, cellular debris and pathogens4,5, and have been phenotypically defined in mice as F4/80hi cells6. By contrast, DCs are usually defined as cells with a stellate morphology that can efficiently present antigens on MHC molecules and activate naive T cells7,8. In mice, DCs are defined as CD11chiMHC class II+ cells9–11.
Box 1. A historical perspective.
The preliminary studies on mononuclear phagocytes occurred at the same time as the publication of the histological accounts of von Recklinghausen (1863)125. Nonetheless, it was Ilya Metchnikoff (1892) — the father of cellular immunity — who established the phagocyte system4,5,126. Metchnikoff was the first to fully comprehend the capabilities of phagocytes, by carrying out a series of classical studies spanning from the echinoderm amoebocyte to the vertebrate. The phagocyte system comprised cells that he termed macrophages (from the Greek for ‘large eaters’) and microphages (‘small eaters’; now known as polymorphonuclear leukocytes). Remarkably, Metchnikoff appreciated that phagocytosis is more than the ability of a cell to engulf foreign microorganisms and that it is also an active defence mechanism — this gave rise to the concept of innate immunity.
By the turn of the twentieth century, the phagocyte system had undergone a number of amendments and the term macrophage had become synonymous with erythrophagocyte, pyrrhol cell, adventitia cell, rhagiocrine cell, polyblast, clasmatocyte and histiocyte. The many names that have been assigned to these cells reflected the divergence of opinion at the time as to the relationships between these cells. Ribbert (1904) restored order to the macrophage system when he discovered that diluted lithium carmine that is injected intravenously is specifically taken up by a group of cells, which became ‘vitally stained’ (Ref. 127). Aschoff128 coined the name ‘reticulo endothelial system’ (RES) to describe this group of cells. Shortly after the RES was introduced, a number of laboratories were in pursuit of the origin of these macrophages. Several in vitro studies that were published in close succession described the transformation of circulating monocytes into macrophages129–131. Carrel and Ebbing129 observed that, over time, blood cultures became primarily composed of monocyte-derived macrophages that had phagocytosed the relics of the other blood cells. However, it was the set of elegant experiments carried out by Ebert and Florey132, using the rabbit ear chamber, that first showed mammalian blood monocytes actively migrating towards sites of injury and differentiating into macrophages in vivo. Subsequently, Volkman and Gowans133 demonstrated, with the aid of thymidine autoradiography, that these infiltrating macrophages originate from the bone marrow. These new technologies (thymidine autoradiography, immunohistochemistry, parabiosis and electron microscopy) highlighted that the cells of the RES differ in morphology, function and origin134.
By the late 1960s, a group of leading scientists — including Ralph van Furth, James G. Hirsch and Zanvil A. Cohn — formulated the ‘mononuclear phagocyte system’ (MPS)1. The MPS constituted monocytes and macrophages with the premise that all macrophages are derived from blood monocytes. Nevertheless, scant evidence existed to suggest that monocytes differentiate into tissue-resident macrophage populations. On the contrary, it was acknowledged that macrophages exist in lower multicellular organisms, such as Porifera (sponges), in the absence of circulating monocytes135, 136. Furthermore, as early as 1907, Maximow137 concluded from embryonic studies in amphibians, rodents and larger mammals that macrophages and leukocytes arise from separate lineages.
While the MPS was being devised in the 1960s, scientists were in pursuit of the ‘third cell’ (Ref. 138) required for adaptive immune responses. In the 1970s, Steinman identified and characterized the dendritic cell (DC)78. This seminal discovery redefined our understanding of the immune response. Nevertheless, the identification of the DC has caused much debate among scientists about whether the DC is a constituent of the MPS or not. It should be noted that shortly after Steinman's discovery, van Furth incorporated DCs into the MPS139. Since then, monocytes, macrophages and DCs have been grouped together, and they are distinguished on the basis of their morphology, function and origin. Several attempts to formulate an inclusive system encompassing monocytes, macrophages and DCs have included the ‘custocyte system’ and the ‘mononuclear-phagocyte and immunoregulatory effector (M-PIRE) system’ (Refs 140,141).
Yet again, we have reached a crossroads in MPS nomenclature. Lineage-tracing studies have demonstrated that, under steady-state conditions, most macrophages in adults are maintained independently of blood monocytes and rely almost exclusively on self-renewal24–30,32,73. They have also shown that classical DCs arise from adult haematopoietic stem cell (HSC)-derived common DC precursors (CDPs) that are distinct from classical monocytes46,47. These findings highlight that the MPS is not a closed monocyte-macrophage system as originally proposed but instead that the MPS encompasses three broad families of cells — namely, CDP-derived DCs, embryonic-derived macrophages and monocyte-derived cells.
Since the original description of the MPS, the advent of polychromatic flow cytometry has enabled the assessment of different surface markers and allowed an unparalleled exploration of cellular phenotype and heterogeneity. This has facilitated the characterization of multiple distinct DC, monocyte and macrophage subsets in mice12–14. However, it has also revealed that many of the proposed unique markers and functions are, in fact, shared between cell types. Further complicating matters, markers of a particular cell subset are not always consistent between mice and humans. This has led to much confusion and debate regarding which subsets represent distinct cell types and which are simply modified versions of the same cell type15.
The complexity of the current mononuclear phagocyte nomenclature can be illustrated by examining the situation in the intestine. DCs have been divided into many different subsets on the basis of the expression of some of the following surface markers: CD103 (also known as integrin αE), CD11b (also known as integrin αM), CX3C-chemokine receptor 1 (CX3CR1), F4/80, CD8α, CD24, CD172a (also known as SIRPα and SHPS1), XC-chemokine receptor 1 (XCR1), CLEC9A (also known as DNGR1), E-cadherin (also known as cadherin 1) and CD64 (also known as FcγRI). Although some researchers define monocyte-derived cells in the intestine as DCs or macrophages on the basis of their respective expression or lack of expression of CD11c16,17, others consider many of these CD11c+ ‘DCs’ to, in fact, be ‘macrophages’ (Ref. 18). Morphological analysis is equally ambiguous, as intestinal monocyte-derived cells possess transepithelial dendrites19,20, leading some to consider these as DCs. Inflammation further complicates the picture, as mononuclear phagocytes in the inflamed intestine undergo phenotypical changes, and monocyte-derived cells that are not present in the steady-state intestine infiltrate during inflammation — for example, tumour necrosis factor (TNF)- and inducible nitric oxide synthase (iNOS)-producing DCs (TIP-DCs)21–23.
Consequently, interpreting the published literature is a minefield, as the same cell type is often given a different name on the basis of a prescribed functional or phenotypical characteristic. Although one can argue that naming is arbitrary and unimportant — as it has no bearing on the function of a cell — it becomes a concern when there is poor consistency between laboratories that leads to assumptions, bias, miscommunication and confusion. It is important for us to demarcate fundamentally novel subsets in the immune system, as opposed to simply identifying yet another marker for an existing subset. We believe that the issue has now moved beyond an etymological debate or trivial semantics because the name that is given to these cells often implies a functional specialization.
In this Opinion article, we propose a unifying nomenclature for cells of the MPS, in which the cellular origin forms the principal basis for their classification. Although primarily based on data from the mouse immune system, we suggest that this nomenclature could also be used in humans and other species on the basis of transcriptional, phenotypical and functional interspecies homology.
A new nomenclature for the MPS
In the sections below, we suggest and describe a new nomenclature for the MPS that could be adopted by researchers in the field in order to overcome the issues highlighted above. In devising this nomenclature, we have used historical terms where possible but have primarily based the terminology on a two-level system. We propose that mononuclear phagocytes should first be defined on the basis of their ontogeny (level one) and that these cells can subsequently be classified on the basis of their function, location and/or phenotype (level two). We think that this approach will yield a more robust nomenclature for mononuclear phagocytes, generating mutually exclusive level one selection criteria that are conserved across tissues and species. Of course, further discussion will be needed among the international research community before these — or an improved version of these — recommendations are accepted as an official nomenclature, and we propose to discuss this during round table sessions organized at upcoming international DC and macrophage meetings. Indeed, the aim of this article is not to be overly prescriptive but instead it is an attempt to propose a refined and less ambiguous MPS nomenclature in order to facilitate communication between different research groups.
Level one nomenclature
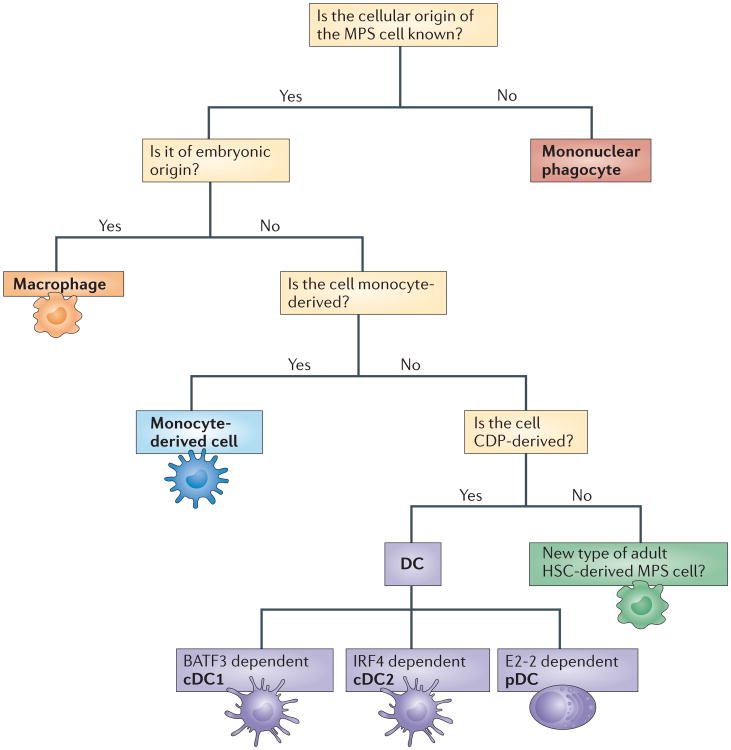
Terminally differentiated cells of the MPS were initially thought to derive exclusively from blood monocytes. However, the ontogeny of the MPS has undergone a conceptual revolution with three key recent findings. First, most adult macrophages are predominantly maintained through self-renewal, independently of adult haematopoiesis, and derive from precursors that arise during embryonic development24–30,73,142. Second, monocytes arise from precursor cells that are committed to the monocyte lineage — so-called common monocyte progenitors (cMoPs)31. Monocytes can traffic to tissues and maintain their phenotype in the steady state32,33 but they can also give rise to cells with a vast array of functions depending on the microenvironment in which they reside16,33,34. Third, conventional or classical DCs (cDCs) and plasmacytoid DCs (pDCs) — but not monocytes and macrophages — arise from a common DC precursor (CDP)35,36. We therefore propose the initial division of mononuclear phagocytes into three main categories — namely, macrophages, monocytes (and monocyte-derived cells) and DCs (Fig. 1).
Figure 1. A decision tree to facilitate nomenclature decisions for mononuclear phagocytes.
We propose that mononuclear phagocytes would primarily be categorized according to their ontog eny (level one nomenclature). Although the ontogenetic nomenclature aims to prevent the mis-categorization of cells, we fully acknowledge that studies pertaining to cellular origin are not always feasible. Furthermore, adopting an ontogeny-based nomenclature for mononuclear phagocytes in other species, such as humans, remains challenging. We suggest that when cell transfers are unfeasible and fate-mapping or genetic ablation models are not available, a parallel nomenclature should be used. This level two nomenclature will identify mononuclear phagocyte subsets on the basis of their expression of conserved phenotypical markers or transcripts (not shown). See Supplementary Information S1 (table) and S2 (table) for details of the markers that can be used to aid classification decisions in mice and humans. Cells of embryonic origin would be referred to as ‘macrophages’ (orange box). Note that some macrophages have historical names such as ‘Langerhans cells’, ‘Kupffer cells’ or ‘microglia’ and we propose to keep these terms. Others would be categorized as, for example, ‘peritoneal macrophages’ or ‘alveolar macrophages’. Mononuclear phagocyte system (MPS) cells of uncertain origin would be referred to as ‘mononuclear phagocytes’ (red box) and be further categorized on the basis of their functional or phenotypical properties and their tissue localization (level two classification). This aims to prevent the miscategorization of MPS cells and should facilitate scientific communication. MPS cells derived from monocytes would be referred to as ‘monocyte-derived cells’ (blue box). Note that these cells are very plastic and can acquire functional properties of both dendritic cells (DCs) and macrophages in some settings. Monocyte-derived cells would be further categorized according to functional specialization, phenotypical properties and transcriptional networks under level two nomenclature. Monocytes, DCs and macrophages were historically grouped in the MPS (see Box 1), and for continuity, we propose to maintain this classification. Hypothetically, if a new immune cell type is identified with a distinct cellular origin from monocytes, DCs and macrophages (green box), it is difficult to determine whether they should be incorporated into the MPS or not. This is because the current cells within the MPS are not develop-mentally linked and have no obvious functional property in common that would distinguish them from other immune cells. We suggest that transcriptional profiling could be used to determine whether any newly identified cell has important homology with monocytes, DCs or macrophages. This would open the possibility of incorporating a new cell type into the MPS. BATF3, basic leucine zipper transcriptional factor ATF-like 3; cDC1, classical type 1 DC; cDC2, classical type 2 DC; CDP, common DC precursor; HSC, haematopoietic stem cell; IRF4, interferon-regulatory factor 4.
Level two nomenclature
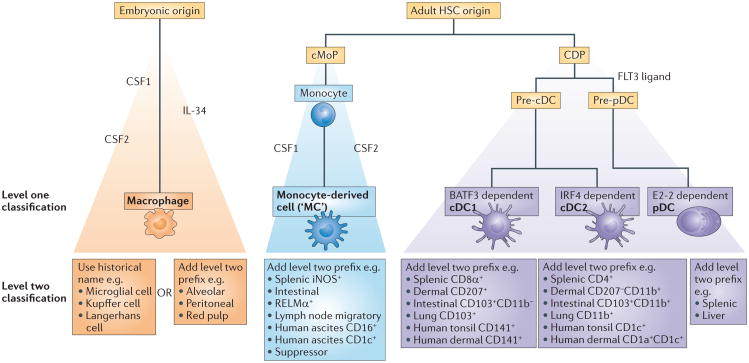
As MPS research mostly focuses on function, and not cellular origin, we supplement our level one nomenclature with a level two nomenclature by allowing the addition of a marker or functional property as an optional and flexible feature of the naming scheme. The overarching principal of this format is to allow scientists to freely describe the features of their cell of interest but also encourage them to place it in the context of its cellular origin. Although allowing flexibility in level two nomenclature could add confusion, we believe it allows the evolution of terminology, concomitant with evolving knowledge of function. The stringency of level one should provide sufficient structure in the nomenclature and provide the best compromise at this juncture. We propose that the level one nomenclature — which has a restricted set of options — trumps the level two nomen clature when classifying cells, as is illustrated by the examples shown in Fig. 2.
Figure 2. Two levels of nomenclature for classifying mononuclear phagocytes.
We suggest that mononuclear phagocytes should first be defined on the basis of their ontogeny (level one nomenclature; yellow boxes), followed by their function, location and/or morphology (level two nomenclature; blue boxes). This yields three main groups of cells — namely, common dendritic cell (DC) precursor (CDP)-derived DCs, embryonic-derived macrophages and monocyte-derived cells. We suggest that DCs should be further subdivided into ‘classical type 1 DCs (cDC1s)’, ‘cDC2s’ and plasmacytoid DCs (pDCs) because their development depends on distinct sets of transcription factors and because they arise from discrete committed precursors. In the lower part of the figure, we have added some examples to illustrate how our approach can yield a unifying nomenclature without losing flexibility. Level one nomenclature also includes unambiguous and widely accepted historical names (green box). Level two nomenclature can include surface markers that are used to identify the cells, the functional specialization studied or information on cell localization. Examples of level two nomenclature are provided, however, in many cases, level one should be sufficient to adequately define a population,, except when a novel function and/or relevant marker is required to discern a particular cell subset. We suggest the use of ‘MC’ as an abbreviation for monocyte-derived cells. However, this is not an officially accepted abbreviation and is incorporated here merely as a suggestion. BATF3, basic leucine zipper transcriptional factor ATF-like 3; cMoP, common monocyte progenitor; CSF1, colony-stimulating factor 1 (also known as M-CSF); CSF2, colony-stimulating factor 2 (also known as GM-CSF); FLT3, FMS-like tyrosine kinase 3; HSC, haematopoietic stem cell; IL-34, interleukin-34; iNOS, inducible nitric oxide synthase; IRF4, interferon-regulatory factor 4; RELMα, resistin-like molecule-α.
Classifying mouse dendritic cells
We propose that DCs should be classified as a separate lineage of mononuclear phagocytes on the basis of the fact that they arise from adult haematopoietic stem cell (HSC)-derived precursors that are distinct from the precursors of monocytes and macrophages (Fig. 1). We further propose to subdivide DCs into only three main subtypes — two main lineages of cDCs (which we propose should be called ‘classical type 1 DCs (cDC1s)’ for CD8α+ and CD103+ DCs, and ‘cDC2s’ for CD11b+ and CD172a+ DCs, on the basis of their distinct developmental pathways) and pDCs, which retain their original name.
When devising a nomenclature for the different DC subtypes, we were aware of similar previous attempts to achieve this using numbering systems that have not subsequently been adopted by the field37,38. However, we still believe that a numbering system helps to simplify the nomenclature across tissues and species, and we suggest that the inclusion of a ‘c’ for ‘conventional’ or ‘classical’ that has been in use for many years39 discriminates our proposed nomenclature sufficiently from prior attempts. Importantly, we do not mean to imply that cDC1s and cDC2s always regulate T helper 1 (TH1)-type and TH2-type immune responses, respectively, although a functional parallel has been observed40–45. In our proposed nomenclature, pDCs (also known as interferon (IFN)-producing cells or IPCs) would also keep their name. Although they are morphologically closer to plasma cells, their development correlates with that of cDCs35,36,46,48. Moreover, they can assume a dendritic appearance upon activation and can influence T cell fate, so we feel that this justifies their categorization as DCs.
At which point of their development DCs first branch from other cell lineages is still a matter of debate. There is evidence to suggest that this may occur at a relatively early stage of haematopoiesis46 and it is clear that DCs can develop from CDPs in the bone marrow35,36. CDPs give rise to distinct DC subtypes35,36 and cellular markers have been identified that help to delineate distinct CDP populations that are biased towards the generation of either cDCs or pDCs35,36,47,48. The intermediate stage between CDPs and cDCs is the precursor for cDCs (the pre-cDC). These pre-cDCs develop from CDPs in the bone marrow and then migrate to peripheral organs where they develop locally into cDCs49–51,58. On the other hand, pDCs terminally differentiate from CDPs in the bone marrow via a pre-pDC intermediate stage36,48,52,53. Note that it has been proposed that some pDCs may arise from a lymphoid precursor54,55.
Although the development of all DC subsets is mostly dependent on the cytokine FMS-like tyrosine kinase 3 ligand (FLT3L)56–58, differentiation into DC subtypes is specifically controlled by distinct sets of transcription factors. Mice lacking IFN-regulatory factor 8 (IRF8)59, DNA-binding protein inhibitor ID2 (Refs 58,60), basic leucine zipper transcriptional factor ATF-like 3 (BATF3)61 or nuclear factor interleukin (IL)-3-regulated protein (NFIL3)62 exhibit a severe defect in the development of cDC1s, whereas cDC2 development is strongly controlled by RELB63, PU.1 (Ref. 64), recombining binding protein suppressor of hairless (RBPJ)65–67 and IRF4 (Refs 42,68,69). Notably, under certain inflammatory settings a few splenic cDC1s are still able to develop in the absence of BATF3, ID2 and NFIL3 (Ref. 70). The development of pDCs is regulated by the transcription factor E2-2 (also known as TCF4)71,72, which counteracts the actions of ID2 that are required for cDC1 development. Several targets of E2-2 — such as SPIB, IRF7 and IRF8 — contribute to pDC lineage specification, and E2-2 is thus regarded as the ‘master regulator’ of pDCs71,72.
Mouse macrophages
Recent studies have shown that the majority of macrophages are derived from embryonic progenitors24–30,73,74, which include yolk sac-derived macrophages and fetal monocytes (as recently review in Ref. 13). When the circulation is established, these cells spread via the blood into peripheral tissues of the fetus, giving rise to tissue-resident macrophages that self-maintain throughout life. Their development is highly dependent on macrophage colony-stimulating factor 1 receptor (CSF1R; also known as M-CSFR), which is the receptor for the cytokines colony-stimulating factor 1 (CSF1; also known as M-CSF) and IL-34. These cytokines are crucial for the differentiation and survival of most macrophages75,76,143.
On the basis of their shared embryonic origins, we suggest that microglia, Kupffer cells, alveolar macrophages and splenic red pulp macrophages should be defined as part of the macrophage family but we propose to keep their historical names (Fig. 2). Although Langerhans cells share many functional properties with cDCs, we suggest that these cells should also be classified as macrophages on the basis of their embryonic origin. Importantly, despite grouping these cells as one family, gene expression analysis of macrophages from various tissues has demonstrated the astonishing diversity of these cells77, suggesting that each macrophage population is specifically adapted to its tissue of residence78,79. However, we suggest that any newly identified mononuclear phagocyte of embryonic origin should be classified as a macrophage.
Mouse monocyte-derived cells
Mouse monocytes consist of two subtypes — namely, LY6Chi classical monocytes and LY6Clow non-classical monocytes80. LY6Chi classical monocytes derive from the recently identified cMoP31. Undifferentiated LY6Chi classical monocytes are not only found in the blood but also in several steady-state tissues, including the spleen, lymph nodes, skin and lungs32,33,81. LY6Clow non-classical monocytes remain mostly within the blood vessels where they patrol the vascular wall82. Whether all LY6Clow blood monocytes differentiate from LY6Chi monocytes83,84,25 is still a matter of debate. Until this is firmly established, we retain the term LY6Clow non-classical monocytes. By contrast, LY6Chi classical monocytes are the definitive precursors of many mononuclear phagocytes and in certain adult tissues — including the gut, heart and dermis — these cells rely on continuous monocytic input for their maintenance in the steady state16,33,85. Strikingly, inflammation is often associated with such a substantial influx of monocytes80,86,87,144 that these cells can outnumber cDCs and macrophages. LY6Chi monocyte-derived cells have been classified as monocyte-derived DCs, monocyte-derived macrophages or myeloid-derived suppressor cells (MDSCs) on the basis of a set of restricted but non-exclusive functional properties that can be difficult to robustly assess in vivo.
Like cDCs, monocyte-derived cells can express CD11c and MHC class II, and they can present antigen to induce naive T cell activation44,88,89. However, similarly to macrophages, they can express F4/80, the tyrosine protein kinase MER (MERTK) and CD64, and they are efficient at phagocytosis and often poor at migration23,33,44,90. Monocyte-derived cells are often highly heterogeneous, even within a single organ or inflamed tissue. Therefore, it remains unclear whether monocyte-derived DCs and monocyte-derived macrophages constitute two ontogenically distinct lineages that are controlled by distinct sets of molecular regulators (as demonstrated for cDC1s and cDC2s) or if they are, instead, highly plastic cells that are able to acquire a multitude of functional modules in response to the cues they receive from their microenvironment.
Currently, there is a lack of suitable methods to accurately discriminate between different populations of monocyte-derived cells in adults. Therefore, we propose to regroup these cells under a single level one term — namely, ‘monocyte-derived cells’. Importantly, we do not deny that monocyte-derived cells can acquire functional properties that are very similar to cDCs (including migrating to the lymph nodes and activating naive T cells) or to macrophages (for example, they can participate in pathogen killing, phagocytosis or tissue repair responses) depending on the context in which they develop.
We therefore suggest that the level two nomenclature could be used to underline the functional heterogeneity of monocyte-derived cells (Fig. 2). As ‘monocyte-derived cell’ is quite long, we propose that this term could be abbreviated to ‘MC’. For instance, we suggest that TIP-DCs could be called ‘iNOS + MCs’ to underline both their monocytic origin21,91 (level one nomenclature) and their iNOS-mediated killing capabilities (level two nomenclature). We would term the monocyte-derived cells that are found in the steady-state intestine ‘intestinal MCs’. When monocyte-derived cells migrate to the lymph nodes, we would propose to call these cells ‘lymph node migratory MCs’. We would equally favour the use of ‘arginase+ MCs’ or ‘RELMα+ MCs’ (which express resistin-like molecule-α), for example. If monocyte-derived cells show suppressive activity, they could be called ‘suppressive MCs’ instead of MDSCs. Note that MC is not an officially accepted abbreviation and is incorporated here merely as a suggestion.
Translation to the human immune system
Although the MPS is well established in humans, determining cell ontogeny remains a challenge. As such, our proposed nomenclature scheme is predominantly based on evidence from mice. Nonetheless, we believe that a parallel nomenclature can be used in humans on the basis of transcriptomic and phenotypic profiling studies42,92–94,104 that have shown an important level of homology between mouse and human mono nuclear phagocyte populations. This has also been done in other species such as sheep, chicken, macaques and pigs95–100. Therefore, on the basis of this homology, we suggest that it could be feasible to apply the level one and level two nomenclature to the human immune system. To further facilitate the translation from mouse to humans, we have compiled tables that indicate the surface markers that are most commonly used to identify distinct mononuclear phagocyte populations in both species (see Supplementary Information S1 (table) and S2 (table)).
Dendritic cell populations in humans
Historically, human DCs found in lymphoid and non-lymphoid tissues were classified into two main groups — namely, pDCs and ‘classical’ or ‘myeloid’ DCs. Classical or myeloid DCs have been further subdivided into two subsets on the basis of their expression of CD141 (also known as BDCA3 and thrombomodulin) and CD1c (also known as BDCA1)37,42,101–105. It has been shown that the gene-expression profiles and functions of human CD141+ DCs and CD1c+ DCs resemble those of mouse cDC1s and cDC2s, respectively92,104,106–110. Accordingly, we propose that human CD141+ DCs could be referred to as cDC1s and human CD1c+ DCs referred to as cDC2s in a unifying nomenclature scheme. Further support for the equivalence of the mouse and human DC systems is that the injection of FLT3L into human volunteers dramatically increased the number of blood pDCs, CD141+ cDCs (cDC1s) and CD1c+ (cDC2s)111. In addition, E2-2, BATF3 and IRF4 have been proposed to act as master transcription factors for human pDCs72, CD141+ cDCs (cDC1s)112 and CD1c+ DCs (cDC2s)42,113, respectively.
Macrophages in humans
Human macrophages are found throughout the body. During HSC transplantation, dermal macrophages in the recipient show prolonged survival and slower replacement compared with dermal DCs, which is consistent with the idea that macrophages are also self-maintaining in humans114. Furthermore, patients harbouring a mutation in GATA2 (which encodes GATA-binding protein 2) lack blood monocytes and all cDC subsets, yet they have normal numbers of Langerhans cells and macrophages in the skin and lungs, respectively, suggesting that the development of these populations is independent of monocytes and DCs115. In a case of limb transplantation, Langerhans cells in the transplanted limb were still of donor origin 10 years after transplantation116, which also supports the idea that human Langerhans cells are self-renewing, as is the case in mice. Therefore, there is accumulating evidence that human macrophages show similar properties to mouse macrophages.
Human monocyte-derived cells
Human blood monocytes are defined as CD14+CD16- ‘classical’, CD14+CD16+ ‘intermediate’ and CD14lowCD16+ ‘non-classical’ monocytes37. Transcriptomic analyses have demonstrated that CD14lowCD16+ human monocytes are the counterparts of LY6Clow non-classical mouse monocytes and that CD14+CD16- human monocytes the counter parts of LY6C+ classical mouse monocytes117,118. However, it is not clear exactly which human cells are monocyte derived. Transcriptomic analyses suggest that dermal CD14+ DCs and intestinal CD103-CD172a+ DCs are related to monocytes104,113, and that they potentially represent populations of monocyte-derived cells. In inflamed tissues, the ‘inflammatory DCs’ expressing CD1c, CD1a and CD14 are also likely to be monocyte-derived cells119,120.
Devising a human MPS nomenclature
As exploring cell ontogeny in humans is challenging, we propose that human mono nuclear phagocyte subtypes could be classified on the basis of conserved phenotypic markers and transcriptomic analyses. Although some specialized functions of DC subsets (for example, the secretion of type I (IFN) by pDCs121 or type III IFNs by cDC1s122) are conserved between species, other functional specializations do not seem to be conserved (for example, cross-presentation and IL12p70 secretion by cDC1s123,124). We believe that this illustrates the strength of a nomenclature strategy that is based primarily on ontogeny, rather than on function, such that it can be applied across species.
Conclusion
In this Opinion article, we suggest that a new unified nomenclature for cells of the MPS will benefit the scientific community. We believe that distinguishing cells on the basis of their ontogeny will ensure a more robust classification of mononuclear phagocytes during steady-state and inflammatory conditions in different tissues across species. This should, in turn, allow for further studies of their functions in different contexts while avoiding some of the current confusion. In Fig. 2, we illustrate how our approach can yield a unifying nomenclature without losing flexibility. With ontogeny at the foundation of identifying mononuclear phagocytes, we also hope to avoid the ever-expanding number of mononuclear phagocyte subsets. Only when a cell type fulfils certain key criteria (Fig. 1) would a new level one name be ascribed.
We believe that applying a unified nomenclature across tissues and species will improve our understanding of mononuclear phagocyte function and benefit scientific communication both within and outside the field. However, we fully appreciate that this is a work in progress that will require further refinement as we gain a better understanding of the haematopoietic system. A final nomenclature system should be discussed with a wider panel of experts before acceptance by official nomenclature committees37. Nevertheless, we hope that the core principles that are described in this Opinion article will be helpful for finding a practical solution.
Supplementary Material
Acknowledgments
The opinions presented here are solely those of the authors but during the course of writing this piece, invaluable guidance and advice was sought from S. Jung, K. Shortman, K. Murphy S. Gordon, A. Mowat, G. Randolph, C. Reis e Sousa, S. Amigorena, B. Malissen, T. Ohteki, P. Henson, D. Riches, M. Merad, M. Manz, M. Dalod, S. Henri, B. Lambrecht, C. Scott, L. van de Laar, D. Metcalf, S. Zelenay and P. Whitney. The authors also thank M. Haniffa for reviewing the manuscript.
Footnotes
Contributor Information
Martin Guilliams, Email: Martin.guilliams@irc.ugent.be, Laboratory of Immunoregulation, VIB Inflammation Research Center, 9052 Ghent, Belgium, and the Department of Respiratory Medicine, Ghent University Hospital, 9000 Ghent, Belgium.
Florent Ginhoux, Email: Florent_ginhoux@immunol.a-star.edu.sg, Singapore Immunology Network (SIgN), 8A Biomedical Grove, Immunos Building, Level 3, BIOPOLIS, Singapore 138648.
Claudia Jakubzick, Email: jakubzickc@njhealth.org, Department of Pediatrics and Integrated Department of Immunology, National Jewish Health, 1400 Jackson Street, Denver, Colorado 80206, USA.
Shalin H. Naik, Email: naik.s@wehi.edu.au, Molecular Medicine Division, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia, and the Department of Medical Biology, The University of Melbourne, Parkville, Victoria 3010, Australia.
Nobuyuki Onai, Email: onai.bre@mri.tmd.ac.jp, Department of Biodefense Research, Medical Research Institute, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113–8510, Japan, and the Japan Science and Technology Agency, Core Research for Evolutional Science and Technology (CREST), Tokyo 102–0081, Japan.
Barbara U. Schraml, Email: barbara.schraml@tum.de, Institut für Medizinische Mikrobiologie, Immunologie und Hygiene, Trogerstr. 30, 81675 München, Germany.
Elodie Segura, Email: elodie.segura@curie.fr, Institut Curie, Centre de recherche and the INSERM U932, Pavillon Pasteur, 26 rue d'Ulm, 75248 Paris Cedex 05, France.
Roxane Tussiwand, Email: r.tussiwand@path.wustl.edu, Department of Pathology and Immunology, Washington University, St. Louis, Missouri 63110, USA, and the Department of Biomedicine, University of Basel, Mattenstrasse 28, 4058 Basel, Switzerland.
Simon Yona, Email: s.yona@ucl.ac.uk, Division of Medicine, University College London, 5 University Street, London WC1E 6JF, UK.
References
- 1.van Furth R, et al. Mononuclear phagocytic system: new classification of macrophages, monocytes and of their cell line. Bull World Health Organ. 1972;47:651–658. in French. [PMC free article] [PubMed] [Google Scholar]
- 2.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor-α. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chomarat P, Banchereau J, Davoust J, Palucka AK. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nature Immunol. 2000;1:510–514. doi: 10.1038/82763. [DOI] [PubMed] [Google Scholar]
- 4.Metchnikoff E. Ueber den Kampf der Zellen gegen Erypselkokken, ein Beitrag zur Phagocytenlehre. Arch Pathol Anat [Virchows' Arch] 1887;107:209–249. [Google Scholar]
- 5.Metchnikoff E. Leçons sur la Pathologie Comparée de l'Inflammation Faites à l'Institut Pasteur en Avril et Mai 1891(G Masson 1892) [Google Scholar]
- 6.Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 7.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinman RM, Witmer MD. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc Natl Acad Sci USA. 1978;75:5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metlay JP, et al. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J Exp Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nussenzweig MC, et al. Studies of the cell surface of mouse dendritic cells and other leukocytes. J Exp Med. 1981;154:168–187. doi: 10.1084/jem.154.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinman RM, Kaplan G, Witmer MD, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. V. Purification of spleen dendritic cells, new surface markers, and maintenance in vitro. J Exp Med. 1979;149:1–16. doi: 10.1084/jem.149.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nature Rev Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 14.Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol. 2008;181:5829–5835. doi: 10.4049/jimmunol.181.9.5829. [DOI] [PubMed] [Google Scholar]
- 16.Varol C, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Bogunovic M, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bain CC, Mowat AM. The monocyte-macrophage axis in the intestine. Cell Immunol. 2014 doi: 10.1016/j.cellimm.2014.03.012. http://dx.doi.org/10.1016/j.cellimm.2014.03.012. [DOI] [PMC free article] [PubMed]
- 19.Kim KW, et al. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood. 2011;118:e156–e167. doi: 10.1182/blood-2011-04-348946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rescigno M, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nature Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 21.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 22.Dunay IR, et al. Gr1+ inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29:306–317. doi: 10.1016/j.immuni.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamoutounour S, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 24.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 25.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito M, Hasegawa G, Takahashi K. Development, differentiation, and maturation of Kupffer cells. Microsc Res Tech. 1997;39:350–364. doi: 10.1002/(SICI)1097-0029(19971115)39:4<350::AID-JEMT5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Yamada M, Naito M, Takahashi K. Kupffer cell proliferation and glucan-induced granuloma formation in mice depleted of blood monocytes by strontium-89. J Leukoc Biol. 1990;47:195–205. [PubMed] [Google Scholar]
- 29.Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res. 1999;117:145–152. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 30.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hettinger J, et al. Origin of monocytes and macrophages in a committed progenitor. Nature Immunol. 2013;14:821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 32.Jakubzick C, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamoutounour S, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Avraham-Davidi I, et al. On-site education of VEGF-recruited monocytes improves their performance as angiogenic and arteriogenic accessory cells. J Exp Med. 2013;210:2611–2625. doi: 10.1084/jem.20120690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nature Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 36.Onai N, et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nature Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 37.Ziegler-Heitbrock L, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–e80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 38.Rissoan MC, et al. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 39.Vremec D, Shortman K. Dendritic cell subtypes in mouse lymphoid organs: cross-correlation of surface markers, changes with incubation, and differences among thymus, spleen, and lymph nodes. J Immunol. 1997;159:565–573. [PubMed] [Google Scholar]
- 40.Meredith MM, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Persson EK, et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Schlitzer A, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashayekhi M, et al. CD8α+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. 2011;35:249–259. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plantinga M, et al. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Desch AN, et al. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. 2011;208:1789–1797. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naik SH, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 47.Schraml BU, et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 48.Onai N, et al. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 2013;38:943–957. doi: 10.1016/j.immuni.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Naik SH, et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nature Immunol. 2006;7:663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 50.Diao J, Winter E, Chen W, Cantin C, Cattral MS. Characterization of distinct conventional and plasmacytoid dendritic cell-committed precursors in murine bone marrow. J Immunol. 2004;173:1826–1833. doi: 10.4049/jimmunol.173.3.1826. [DOI] [PubMed] [Google Scholar]
- 51.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyama-Sorimachi N, et al. Inhibitory NK receptor Ly49Q is expressed on subsets of dendritic cells in a cellular maturation- and cytokine stimulation-dependent manner. J Immunol. 2005;174:4621–4629. doi: 10.4049/jimmunol.174.8.4621. [DOI] [PubMed] [Google Scholar]
- 53.Jackson JT, et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 2011;30:2690–2704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corcoran L, et al. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol. 2003;170:4926–4932. doi: 10.4049/jimmunol.170.10.4926. [DOI] [PubMed] [Google Scholar]
- 55.Pelayo R, et al. Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood. 2005;105:4407–4415. doi: 10.1182/blood-2004-07-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 57.Waskow C, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nature Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiavoni G, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hacker C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nature Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 61.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 2011;117:6193–6197. doi: 10.1182/blood-2010-07-295873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L, et al. RelB is essential for the development of myeloid-related CD8α- dendritic cells but not of lymphoid-related CD8α+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- 64.Guerriero A, Langmuir PB, Spain LM, Scott EW. PU. 1 is required for myeloid-derived but not lymphoid-derived dendritic cells. Blood. 2000;95:879–885. [PubMed] [Google Scholar]
- 65.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis KL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satpathy AT, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nature Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki S, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8α- dendritic cell development. Proc Natl Acad Sci USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 70.Seillet C, et al. CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood. 2013;121:1574–1583. doi: 10.1182/blood-2012-07-445650. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoeffel G, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zigmond E, et al. Infiltrating monocyte-derived macrophages and resident Kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193:344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 75.Greter M, et al. Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nature Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosas M, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 81.Swirski FK, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–616. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Auffray C, et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 83.Sunderkotter C, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- 84.Varol C, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Epelman S, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mildner A, et al. CCR2+Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain. 2009;132:2487–2500. doi: 10.1093/brain/awp144. [DOI] [PubMed] [Google Scholar]
- 88.Kool M, et al. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J Exp Med. 2008;205:869–882. doi: 10.1084/jem.20071087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheong C, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209+ dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langlet C, et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- 91.Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 2007;179:7577–7584. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- 92.Robbins SH, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crozat K, et al. Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol Rev. 2010;234:177–198. doi: 10.1111/j.0105-2896.2009.00868.x. [DOI] [PubMed] [Google Scholar]
- 94.Guilliams M, et al. From skin dendritic cells to a simplified classification of human and mouse dendritic cell subsets. Eur J Immunol. 2010;40:2089–2094. doi: 10.1002/eji.201040498. [DOI] [PubMed] [Google Scholar]
- 95.Fairbairn L, et al. Comparative analysis of monocyte subsets in the pig. J Immunol. 2013;190:6389–6396. doi: 10.4049/jimmunol.1300365. [DOI] [PubMed] [Google Scholar]
- 96.Chamorro S, et al. Phenotypic characterization of monocyte subpopulations in the pig. Immunobiology. 2000;202:82–93. doi: 10.1016/S0171-2985(00)80055-8. [DOI] [PubMed] [Google Scholar]
- 97.Contreras V, et al. Existence of CD8α-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J Immunol. 2010;185:3313–3325. doi: 10.4049/jimmunol.1000824. [DOI] [PubMed] [Google Scholar]
- 98.Marquet F, et al. Characterization of dendritic cells subpopulations in skin and afferent lymph in the swine model. PLoS ONE. 2011;6:e16320. doi: 10.1371/journal.pone.0016320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vu Manh TP, et al. Existence of conventional dendritic cells in Gallus gallus revealed by comparative gene expression profiling. J Immunol. 2014;192:4510–4517. doi: 10.4049/jimmunol.1303405. [DOI] [PubMed] [Google Scholar]
- 100.Dutertre CA, et al. TLR3-responsive, XCR1+, CD141(BDCA-3)+/CD8α+-equivalent dendritic cells uncovered in healthy and simian immunodeficiency virus-infected rhesus macaques. J Immunol. 2014;192:4697–4708. doi: 10.4049/jimmunol.1302448. [DOI] [PubMed] [Google Scholar]
- 101.Segura E, et al. Characterization of resident and migratory dendritic cells in human lymph nodes. J Exp Med. 2012;209:653–660. doi: 10.1084/jem.20111457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Summers KL, Hock BD, McKenzie JL, Hart DN. Phenotypic characterization of five dendritic cell subsets in human tonsils. Am J Pathol. 2001;159:285–295. doi: 10.1016/S0002-9440(10)61694-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McIlroy D, et al. Investigation of human spleen dendritic cell phenotype and distribution reveals evidence of in vivo activation in a subset of organ donors. Blood. 2001;97:3470–3477. doi: 10.1182/blood.v97.11.3470. [DOI] [PubMed] [Google Scholar]
- 104.Haniffa M, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu CI, et al. Human CD1c+ dendritic cells drive the differentiation of CD103+ CD8+ mucosal effector t cells via the cytokine TGF-β. Immunity. 2013;38:818–830. doi: 10.1016/j.immuni.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jongbloed SL, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DCsubset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crozat K, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J Exp Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bachem A, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+ CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Poulin LF, et al. Characterization of human DNGR-1+BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells. J Exp Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Galibert L, et al. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J Biol Chem. 2005;280:21955–21964. doi: 10.1074/jbc.M502095200. [DOI] [PubMed] [Google Scholar]
- 111.Pulendran B, et al. Flt3-ligand and granulocyte colony-stimulating factor mobilize distinct human dendritic cell subsets in vivo. J Immunol. 2000;165:566–572. doi: 10.4049/jimmunol.165.1.566. [DOI] [PubMed] [Google Scholar]
- 112.Poulin LF, et al. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and non-lymphoid tissues. Blood. 2012;119:6052–6062. doi: 10.1182/blood-2012-01-406967. [DOI] [PubMed] [Google Scholar]
- 113.Watchmaker PB, et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nature Immunol. 2014;15:98–108. doi: 10.1038/ni.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haniffa M, et al. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J Exp Med. 2009;206:371–385. doi: 10.1084/jem.20081633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bigley V, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med. 2011;208:227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kanitakis J, Morelon E, Petruzzo P, Badet L, Dubernard JM. Self-renewal capacity of human epidermal Langerhans cells: observations made on a composite tissue allograft. Exp Dermatol. 2011;20:145–146. doi: 10.1111/j.1600-0625.2010.01146.x. [DOI] [PubMed] [Google Scholar]
- 117.Cros J, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ingersoll MA, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 2010;115:e10–e19. doi: 10.1182/blood-2009-07-235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Segura E, et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013;38:336–348. doi: 10.1016/j.immuni.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 120.Guttman-Yassky E, et al. Major differences ininflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J Allergy Clin Immunol. 2007;119:1210–1217. doi: 10.1016/j.jaci.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 121.Nakano H, Yanagita M, Gunn MD. CD11c+B220+Gr-1+ cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J Exp Med. 2001;194:1171–1178. doi: 10.1084/jem.194.8.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lauterbach H, et al. Mouse CD8α+ DCs and human BDCA3+ DCs are major producers of IFN-λ in response to poly IC. J Exp Med. 2010;207:2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nizzoli G, et al. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T cell responses. Blood. 2013;122:932–942. doi: 10.1182/blood-2013-04-495424. [DOI] [PubMed] [Google Scholar]
- 124.Segura E, Amigorena S. Cross-presentation by human dendritic cell subsets. Immunol Lett. 2014;158:73–78. doi: 10.1016/j.imlet.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 125.Tauber A, Chernyak L. Metchnikoff and the Origins of Immunology: From Metaphor to Theory. Oxford Univ. Press; 1991. [Google Scholar]
- 126.Gordon S. Elie Metchnikoff: father of natural immunity. Eur J Immunol. 2008;38:3257–3264. doi: 10.1002/eji.200838855. [DOI] [PubMed] [Google Scholar]
- 127.Florey H. General Pathology. Lloyd-Luke; 1970. [Google Scholar]
- 128.Aschoff L. Das reticuloendotheliale System. Erg Inn Med Kinderheilk. 1924;26:1–118. [Google Scholar]
- 129.Carrel A, Ebeling AH. The fundamental properties of the fibroblast and the macrophage: II the macrophage. J Exp Med. 1926;44:285–305. doi: 10.1084/jem.44.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Awrorow PP, Timofejewskij AD. Virchows Archiv Fur Pathologische Anatomie Und Physiologie Und Fur Klinische Medizin. Vol. 216. Springer; 1914. pp. 184–214. [Google Scholar]
- 131.Lewis MR, Lewis WH. The transformation of white blood cells into clasmatocytes (macrophages), epithelioid cells, and giant cells. J Am Med Associ. 1925;84:798–799. [Google Scholar]
- 132.Ebert RH, Florey HW. The extravascular development of the monocyte observed in vivo. Br J Exp Pathol. 1939;20:342–356. [Google Scholar]
- 133.Volkman A, Gowans JL. The origin of macrophages from bone marrow in the rat. Br J Exp Pathol. 1965;46:62–70. [PMC free article] [PubMed] [Google Scholar]
- 134.Gall EA. The cytological identity and interrelation of mesenchymal cells of lymphoid tissue. Ann NY Acad Sci. 1958;73:120–130. doi: 10.1111/j.1749-6632.1959.tb40796.x. [DOI] [PubMed] [Google Scholar]
- 135.Ratcliffe NA, Rowley AF. Invertebrate Blood Cells. Academic Press; 1981. [Google Scholar]
- 136.George WC. Comparative hematology and the functions of the leucocytes. Quarterly Rev Biol. 1941;16:426–439. [Google Scholar]
- 137.Maximow A. Uber die Entwicklung der blut - und Bindegewebszellen beim Saugetierembryo. Folia Haematol. 1907;4:16. [Google Scholar]
- 138.Mosier DE, Coppleson LW. A three-cell interaction required for the induction of the primary immune response in vitro. Proc Natl Acad Sci USA. 1968;61:542–547. doi: 10.1073/pnas.61.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.van Furth R. In: Methods for Studying Mononuclear Phagocytes. Adams DO, Edelson PJ, Koren H, editors. Academic Press; 1980. pp. 243–252. [Google Scholar]
- 140.Foucar K, Foucar E. The mononuclear phagocyte and immunoregulatory effector (M-PIRE) system: evolving concepts. Semin Diagn Pathol. 1990;7:4–18. [PubMed] [Google Scholar]
- 141.Goerdt S, Kodelja V, Schmuth M, Orfanos CE, Sorg C. The mononuclear phagocyte-dendritic cell dichotomy: myths, facts, and a revised concept. Clin Exp Immunol. 1996;105:1–9. doi: 10.1046/j.1365-2249.1996.d01-740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytesy. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Witmer-Pack MD, et al. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. 1993;104:1021–1029. doi: 10.1242/jcs.104.4.1021. [DOI] [PubMed] [Google Scholar]
- 144.Hilgendorf I, et al. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res. 2014;114:1611–1622. doi: 10.1161/CIRCRESAHA.114.303204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.