Abstract
Purpose
Fuchs endothelial corneal dystrophy (FECD) results in loss of vision associated with progressive corneal edema and loss of corneal transparency. The aim of the study was to evaluate changes in ZEB1, COL8A2, SLC4A11, and TCF4 rs613872 and correlate them with clinical findings.
Methods
Eighty-two patients with clinically diagnosed FECD and 143 controls were recruited during the period 2007–2012. Clinical details, pedigree information up to three generations, and 5 ml of blood samples were collected. Histopathological and transmission electron microscopy studies were performed on host corneal buttons from patients who underwent keratoplasty. Genomic DNA from blood was processed for PCR amplification followed by direct sequencing to screen genetic changes in the candidate genes. The pathogenic nature of the genetic variants was assessed using Sorting Intolerant From Tolerant (SIFT) and MutationTaster.
Results
The mean age at the onset of symptoms was 59.14±1.41years, the male to female ratio was 1:1.5, and the mean specular count (endothelial cell density) was 1629±93.62 cells/mm2 with a mean central corneal thickness (CCT) of 617.30±15.73 µm. ZEB1 showed a novel variant IVS2+276 C/T in 14% of the cases, a novel nonsense p.Leu947stop mutation in one patient, two novel missense mutations (p.Glu733Lys, p.Ala818Val) in one patient each, and one novel synonymous variation (p.Ser234Ser) in two patients. Reported mutation p.Gln840Pro and five polymorphisms were also identified. The TCF4 single nucleotide polymorphism (SNP) rs613872 was significantly higher in patients with FECD.
Conclusions
This is the first report of genetic variations in ZEB1 and TCF4 SNP rs613872 in patients with FECD from northern India that suggests a possible role in disease pathogenesis and the regulation of endothelial cell density.
Introduction
Fuchs endothelial corneal dystrophy (FECD) is characterized by the accumulation of focal excrescences called corneal guttae and the thickening of Descemet’s membrane accompanied by corneal edema [1]. Progressive endothelial cell loss causes failure of the endothelial pump allowing the relative influx of aqueous humor into the cornea, causing epithelial and stromal edema and resulting in distorted vision. FECD exists as early and late onset forms; the late form usually appears between the fourth and fifth decades of life. Most cases are sporadic, but autosomal dominant inheritance has also been reported [2].
Worldwide prevalence of FECD varies widely. It is estimated to affect about 5% of the population over the age of 40 years in the United States [3]. In the isolated population of Tangier Island, approximately 50% of the population over the age of 50 years is affected with FECD [4]. There is a lack of precise data on the prevalence rate in India and the exact underlying genetic basis is largely unknown, but reports have substantiated a genetic basis [5]. The role of genetic factors is well recognized in FECD, and different variants of genes such as COL8A2 (ID: 1296; OMIM: 120252), SLC4A11 (ID: 83959; OMIM: 610206), ZEB1 (ID: 6935; OMIM 613270), TCF4 (ID: 6925; OMIM 602272); AGBL1 (ID: 123624; OMIM 615496) show a disease association in a small proportion of patients with FECD.
Genome-wide linkage studies identified collagen type VIII alpha 2 (COL8A2; 1p34.3-p32; OMIM: 120252) as responsible for early onset FECD [6,7]. Type VIII collagen is the most important component of Descemet’s membrane and composed of two α-1 chains and one α-2 chain. The role of type VIII collagen is unknown, but its association with endothelial cells may help in determining the cell phenotype. Several studies have reported COL8A2 mutations in early onset FECD [6-8]. Late onset autosomal dominant- FECD (AD-FECD) has been mapped to three separate loci on chromosomes 13pTelq12 (FCD1), 18q21.2–21.3 (FCD2), and 5q33–35 (FCD3) [9-11], but no clear association has been established to date.
Involvement of SLC4A11 (20p13, OMIM: 610206), which encodes a solute carrier family 4, member 11, also known as BTR1-bicarbonate transporter related protein 1, or NaBC1-sodium-borate cotransporter 1 [11], was reported in patients with FECD and later confirmed in several studies [5,12]. The resulting mutant SLC4A11 proteins were also found to be defective in localization and turnover compared to the wild-type proteins [13].
Mutations in the TCF8 gene, a transcription factor (10p11.22; OMIM 613270), have been reported in patients with late onset FECD [14]. TCF8, also known as zinc finger E-box binding homeobox 1 (ZEB1), encodes a zinc finger transcription factor, a two-pronged homeodomain protein that functions as a transcriptional enhancer as well as a repressor [15]. Pathogenic ZEB1 mutations have been identified in posterior polymorphous corneal dystrophy-3 (PPCD3) [16], and later, they were also found to be associated with FECD [14].
The TCF4 (18q21.2, OMIM 602272) gene is a member of the E-protein family of class I basic helix–loop–helix (bHLH) transcription factors [17]. A genome-wide association study (GWAS) identified several single nucleotide polymorphisms (SNPs) in the TCF4 gene that included the SNP rs613872, the first major genetic risk factor for FECD. Several studies have shown genetic variations within TCF4 to be associated with FECD [17-19]. Of these variations, TCF4 rs613872 present in intron 3 was reported to be significantly associated with late onset FECD [17,18,20]. The present study was undertaken to identify known and unknown variations in the reported candidate and associated genes COL8A2, SLC4A11, and ZEB1 and the TCF4 SNP rs613872 by screening Indian patients with FECD.
Methods
Clinical examination
A total of 82 patients (27 males and 55 females) diagnosed with FECD were recruited from the outpatient department of Rajendra Prasad Centre for Ophthalmic sciences (RPC), AIIMS and 143 age and sex matched controls from the general population during the period 2007–2012. The subjects at the time of recruitment were healthy except for visual problems in the FECD patients and their age ranged between 41 years to 82 years. The study adhered to the Declaration of Helsinki and the ARVO statement on human subjects and was approved by Institute Ethics Committee. In addition to undergoing a complete ophthalmic examination, all patients were examined under a slit-lamp and evaluated clinically using specular microscopy and ultrasonic pachymetry. FECD was diagnosed clinically based on the following criteria: the presence of central guttae changes, frequent morning blurring of vision, progressive disease, increased central corneal thickness (CCT; >550 µm thickness), and decreased specular count (<2000 cells/mm2). Histopathology and transmission electron microscopy (TEM) studies were performed on the host corneal buttons obtained from patients who underwent keratoplasty. Corneal button specimens in the case of penetrating keratoplasty and Descemet’s membrane specimens in Descemet’s Stripping Automated Endothelial Keratoplasty (DSAEK) surgery preserved in formalin were obtained from the operation theater, cut into fine sections, stained with hematoxylin and eosin (H & E) stain, and evaluated under a light microscope (Nikon, Tokyo, Japan). The corneal tissues for TEM were immediately fixed in Karnovski fixative. Tissue samples were embedded in araldite after dehydration and clearing, and blocks were prepared. Sections were cut with a glass knife using an ultramicrotome (UC-6; Leica) and visualized under an electron microscope (Morgagni 268D; FEI Holland).
Study samples
A total of 82 patients diagnosed with FECD and 143 controls from the general population were recruited during the period 2007–2012. Five ml of peripheral blood samples were drawn by venipuncture in EDTA vials from all the participants after taking informed consent. Genomic DNA was extracted from the blood samples by the salting out method [21]. The DNA was stored at -20 degree centigrade till further use. Genomic DNA isolated from peripheral blood samples was processed for PCR amplification to screen the exons of COL8A2, SLC4A11, TCF8 (82 patients and 100 controls), and the TCF4 SNP (82 patients and 143 controls) using primers described previously [5,7]. The cycling conditions were 7 min initial denaturation at 95 °C, followed by 35 cycles of 94 °C for 30 s, 58 °C /60 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min. The reaction mixture of 25 μl was prepared using 200 ng genomic DNA, specific primers (0.5 μM each), MgCl2 (1.5 mM), deoxyribonucleotide triphosphates (dNTPs; 0.2 mM), 1× PCR buffer (Thermo, Wilmington, MA), and Taq polymerase (0.5 U; Roche) in a thermocycler (ABI 9700, Applied Biosystems [ABI], Foster City, CA). The PCR products were processed for gel purification using QIAmp gel extraction kits (Qiagen, GmBH, Hilden), and the purified PCR products were screened for sequence changes with Sanger sequencing. The amplified products were sequenced directly with BigDye Terminator Mix version 3.1 (ABI) according to the manufacturer’s instructions and were then analyzed on an ABI-3100 Genetic Analyzer (ABI). Nucleotide sequences for the coding regions were compared with the nucleotide sequence of the published SLC4A11 human cDNA (NM_032034), ZEB1 human cDNA (NM_030751), COL8A2 human cDNA (NM_0052021), and TCF4 human cDNA (NM_003199).
In silico analysis
Sorting Intolerant From Tolerant (SIFT) and MutationTaster tools were used to characterize the pathogenicity of the changes identified. SIFT predicts whether an amino acid substitution affects protein function, and the prediction is based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences.
It gives a value as a score, and a score <0.05 is considered potentially damaging. MutationTaster, a free, web-based application for rapid evaluation of the disease-causing potential of DNA sequence alterations, predicts the sequence variations to be disease causing. The probability of prediction, a value close to 1, indicates a high degree of accuracy for the prediction.
Statistical analysis
The genotype and allele frequency were calculated for the patient and control groups, and the significance was tested with chi-square and Fisher’s exact tests. Correlation of genotypes with parameters such as gender, age at onset, duration of the disease, CCT, and specular count was done through an unpaired Student t test using GraphPad Prism software. A ‘p’ value of less than 0.05 was considered significant.
Results
The study patients underwent clinical and histopathological examinations and genetic evaluation for variations in COL8A2, SLC4A11, ZEB1, and TCF4 (rs613872). The results obtained were correlated with gender, age at onset, presence of consanguinity, family history, CCT, and specular count.
Clinical profile
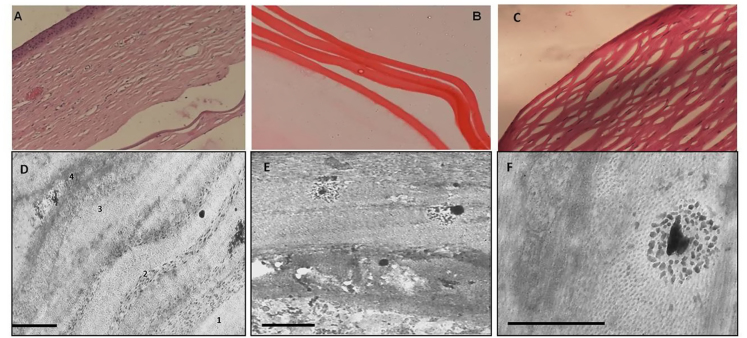
The patients in the study showed a mean age of onset of symptoms at 59.14±1.41 years and a male to female ratio of 1:1.5. Ultrasound pachymetry and specular microscopy showed increased corneal thickness (617.30±15.73 µm) and decreased endothelial count (1629±93.62 cells/mm2), respectively (Table 1). Histopathology confirmed FECD diagnosis in the patients who underwent keratoplasty. Light microscopy using H&E staining showed a marked increase in corneal and Descemet’s membrane thickness (Figure 1A,B) in addition to decreased endothelial cell counts. Electron microscopy study of the corneal buttons showed stromal thickening with severe disorganization and disruption of the lamellar pattern. Descemet’s membrane showed disorganized bundles of banded collagen. Spindle-shaped wide-space collagen, typical of late onset FECD, was found in the posterior banded layer (Figure 1D-F).
Table 1. Gender wise comparison of clinical parameters in the FECD patients.
| Clinical Parameters | Total (n=82) | Males (n=27) | Females (n=55) | P value |
|---|---|---|---|---|
| Specular count |
1629±93.62 |
1675±131.4 |
1588±134.7 |
0.651 |
| CCT |
617.30±15.73 |
593.2±21.19 |
634.5±21.96 |
0.200 |
| Age at Onset |
59.14±1.41 |
61.75±2.265 |
56.44±2.166 |
0.099 |
| Duration of Disease | 4.37±0.718 | 5.71±1.108 | 3.50±0.917 | 0.135 |
Values shown are Mean±SEM
Figure 1.
Representative images of histopathology and TEM in FECD. A: Photomicrograph of hematoxylin and eosin staining (20X) reveals stromal edema with thickened Descemet’s and retrocorneal membrane. B: Descemet’s membrane thickening of the DSAEK specimen (40X). C: Photomicrograph of hematoxylin and eosin staining (20X) of a control. D: Transmission electron microscopy (TEM) showing Descemet’s membrane of typical Fuchs endothelial corneal dystrophy (FECD). Four regions are distinguished: anterior banded, non-banded, posterior banded, and the fibrillar region (scale bar, 2 µm). E: Enlarged view of the posterior stroma with guttae-like deposits. F: Enlarged view of the stroma showing degenerative changes and guttae-like excrescences of spindle-shaped wide-space collagen (scale bar, 200 nm).
Genetic profile
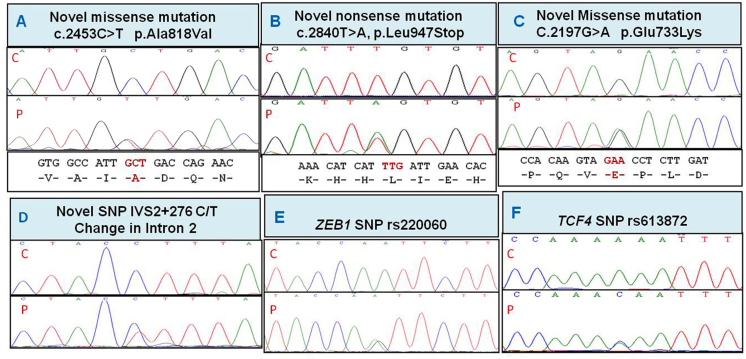
Consanguinity was seen in one family and a positive history in two families. The novel changes and variations associated with FECD are shown in Figure 2 and the in silico prediction in Table 2.
Figure 2.
Partial nucleotide sequence of the ZEB1 and TCF4 variations in FECD patients. Novel ZEB1 changes identified in the study are shown in panels A to D. The ZEB1 single nucleotide polymorphism (SNP) rs220060 and the TCF4 SNP rs613872 significantly associated with Fuchs endothelial corneal dystrophy (FECD) are shown in panels E and F. P=patient; C=control.
Table 2. Details of mutations and variations identified in FECD and their in-silico prediction.
| Changes | Gene | Exon/ Intron | Cases (n=82) | Control (n=100) | P-value | SIFT Prediction | SIFT Score** | Mutation taster Prediction* |
|---|---|---|---|---|---|---|---|---|
|
p.Glu733Lye |
ZEB1 |
Exon 7 |
1/82 |
NC |
NA |
DAMAGING |
0.01 |
D (0.9999) |
|
p.Ala818Val |
ZEB1 |
Exon 7 |
1/82 |
NC |
NA |
tolerated |
0.12 |
D (0.9998) |
|
p.Leu947stop |
ZEB1 |
Exon 8 |
1/82 |
NC |
NA |
DAMAGING |
0 |
D (1.0) |
|
p.Gln840Pro |
ZEB1 |
Exon 7 |
1/82 |
NC |
NA |
tolerated |
0.14 |
D (0.9998) |
|
p.Ser234Ser |
ZEB1 |
Exon 6 |
2/82 |
NC |
NA |
tolerated |
1 |
D (0.9998) |
|
rs220060 |
ZEB1 |
Intron 5 |
~24%(19/82) |
4% (4/100) |
0.000743 |
NA |
NA |
P (0.7116) |
|
rs77516068 |
ZEB1 |
Intron 3 |
~3.0% (2/82) |
NC |
NA |
NA |
NA |
P (0.9999) |
|
rs7918614 |
ZEB1 |
Exon 2 |
~10% (8/82) |
11% (11/100) |
0.8131 |
tolerated |
1 |
P (1.7600 ) |
|
rs161233 |
ZEB1 |
Intron3 |
17% (13/82) |
9% (9/100) |
0.087 |
NA |
NA |
P (0.9999) |
|
rs149166539 |
ZEB1 |
Exon 6 |
~3.0% (2/82) |
NC |
NA |
tolerated |
1 |
D (0.9999) |
|
IVS2+276 |
ZEB1 |
Intron 3 |
~14% (11/82) |
NC |
NA |
NA |
NA |
P (0.9999) |
|
rs613872 |
TCF4 |
Intron 9 |
~34% (28/82) |
~17% (25/143) |
0.0117 |
NA |
NA |
P (0.9999) |
| rs75864656 | COL8A2 | Exon 2 | ~17% (14/82) | 17% (17/100) | 1.000 | tolerated | 0.62 | NA |
** SIFT value scores of ‘0’ is most deleterious mutation, <0.05 is potentially damaging and ‘1’ is tolerated mutation. * value close to 1 indicates high security of prediction. D: Disease causing; P: Polymorphism; NC: None of the control; NA: Not applicable
Direct sequencing of the ZEB1 gene in the patients identified a novel nonsense p.Leu947stop mutation (1/82), two novel missense mutations, p.Glu733Lys (1/82) and p.Ala818Val (1/82), a novel synonymous mutation, p.Ser234Ser(2/82), and a novel variant at position IVS2+276 C/T in 11/82 of the patients. A reported nonsense mutation p.Gln840Pro was also found (1/82). In addition, SNPs p.Asp64Asp (rs7918614) in exon 2, p.T232T (rs149166539) in exon 6, and three intronic SNPs (rs77516068, rs220060, and rs161233) were also identified. The polymorphisms rs149166539 (2/82), rs77516068 (2/82; not identified in any of the controls), and rs161233 (13/82 in FECD; 9% controls) are being reported for the first time in FECD. The genotype frequency of AG in SNP rs220060 in the patients with FECD was significantly high (24%; 19/82) compared to the controls (4%; 4/100; p=7.43×10−4) showing an association with late onset FECD. The frequency of rs161233 genotype TG was 17% (13/82) in the patients with FECD and 9% (9/100) in the controls (p=0.087). Although a higher frequency was observed in patients for this SNP, a significant association was not observed.
The genotype frequency of the TCF4 rs613872 SNP represents a change A>C with genotypes CC, CA, and AA. A higher frequency of the allele C was seen in the patients with FECD (17.07%; 28/82) compared to the controls (10.14%; 29/143; p=0.0393). The frequency of genotype CA was found to be 34.15% (28/82) in the patients with FECD and 17.48% (25/143) in the controls, showing the CA genotype was significantly associated (p=0.0117) with the disease.
The rs613872 genotype was compared with CCT, age at onset, duration of disease, and endothelium cell density (Table 3). The CA genotype was found to be significantly associated with decreased endothelial cell density (p=0.0259) compared to the AA genotype (Table 3; Figure 3). No mutations were identified in COL8A2 and SLC4A11 except a previously reported SNP (rs75864656) in the COL8A2 gene that was present in equal frequency in the patients and the controls.
Table 3. Association of TCF SNP rs613872 with specular count in FECD.
| Clinical symptoms | Rs613872 AA genotype | Rs613872 AC genotype | p value |
|---|---|---|---|
|
Specular count |
1733±107.3 |
1239±181.6 |
0.0259* |
|
CCT |
619.1±16.06 |
606.1±37.45 |
0.7378 |
|
Age at Onset |
58.32±1.493 |
60.64±3.835 |
0.4994 |
| Duration of Disease | 3.488±0.634 | 6.300±1.756 | 0.0745 |
Values shown are Mean±S.E, * p<0.05.
Figure 3.
Difference in specular count between TCF rs613872 genotypes AA and AC as shown with an unpaired Student t test (p=0.0259, ≤0.05).
Discussion
FECD has a complex etiology with the involvement of genetic and environmental factors. Approximately 50% of FECD cases show familial clustering [22], and the disease generally follows an autosomal dominant pattern of inheritance [22,23].
The fundamental pathologic feature is the abnormal assembly of collagen within the extracellular matrix [6]. In particular, the accumulation in the anterior layer of the Descemet’s membrane during early fetal development [24] indicates that FECD is primarily a congenital disorder of collagen secretion and arrangement [6]. Final manifestation of the disease is variable and might depend on other genetic and environmental factors. Our TEM results are in accordance with those of a previous study [25] and suggest that the accumulation of large amounts of collagen in the anterior layer may be the primary pathologic attribute of the disease.
After COL8A2 was identified in early onset FECD, several studies reported the L450W [2,6,26] and Q455V [8] mutations confirming its role in early onset FECD. In the present study, we did not find any COL8A2 mutations in the patients from northern India, and our results are in agreement with a report on patients with FECD from southern India. Similarly, neither the present study nor a similar one from southern India documents any SLC4A11 changes in patients with late onset FECD [27].
ZEB1 was initially found to be associated with PPCD3 [16] and later with FECD [14] as well. ZEB1 is an E-box binding transcription factor involved in the differentiation and development of various tissues and is associated with hernias, hydroceles, and skeletal deformities [14,28]. To date, the exact role of ZEB1 in FECD pathogenesis has not been defined. Riazuddin et al. identified ZEB1 pathogenic mutations in FECD, but other studies did not show significant association of ZEB1 with the disease [29,30].
Chung et al. showed that truncating ZEB1 mutations due to a premature stop codon may lead to reduced protein production and impaired localization of the protein within the cell [31]. We found a novel nonsense mutation (p.Leu947 stop) in one patient in exon 8 that codes for the ZEB1 nuclear localization signal (NLS) domain. The NLS is required for import of ZEB1 into the nucleus [31]. The p.Leu947 stop mutant protein, although abundant, may lack the NLS domain and remain exclusively in the cytoplasm due to impaired nuclear translocation, thus leading to haploinsufficiency.
In contrast, ZEB1 missense mutations do not seem to affect the nuclear localization or alter the rate of protein production [31]. The FECD missense mutations documented to date lie in the conserved sites of the protein and are suggested to be hypomorphic; that is, they do not cause a severe disease phenotype as seen in PPCD [31]. The effects of these mutations are still unknown and may include protein regulation or signaling. The novel p.Glu733Lys and p.Ala818Val and the reported p.Gln840Pro missense mutations lie in the conserved regions and may be responsible for disease pathogenicity. In the mutation p.Ala818Val, replacement of alanine with valine might disrupt the alpha helix and affect the function of the protein. The p.Gln840Pro mutation identified in one patient in our study was earlier reported to be pathogenic in FECD [15]. SNPs found in the present study rs7918614 (equal frequency in patients and controls) and rs220060 (significantly associated with the disease) are reported to be associated with PPCD [19,28,32] and Fuchs dystrophy [29,30].
Substitutions involving amino acids or single nucleotide changes have the potential to affect protein function. Interpretation of the identified gene variations, whether they actually cause a disease or just confer susceptibility to it, is one of the greatest challenges encountered by investigators today. Many research tools are available but, unfortunately, may sometimes be misleading due to problems related to inconsistency and overall accuracy, especially in downstream experimental research [33].
ZEB1, which encodes a transcription factor, plays an important role in embryonic development as well as in disease-causing mechanisms such as tumor formation by downregulation of the genes involved in maintaining the epithelial phenotype of the cells [34,35]. ZEB1 has a critical role in epithelial-to-mesenchymal transition (EMT) and regulates several genes involved in EMT [36]. However, the function of ZEB1 in the endothelial cells of the cornea is not known. Based on the identification of ZEB1 mutations in the endothelial dystrophies, ZEB1 may be involved in maintaining endothelial cell density and corneal transparency.
TCF4 genetic variations are reported to be associated with FECD [17-19], of which TCF4 SNP rs613872 was reported to be significantly associated with late onset FECD [4,17,18,20,37]. Our results are in accordance with prior studies and show an association with increased risk of Fuchs dystrophy. Although in silico tools predict this change to be a polymorphism, our study, along with others, has identified this SNP to be associated with late onset FECD. Other TCF4 changes reported are a CTG intronic repeat expansion [38,39] and the SNP rs17089887, which have shown a significant association with late onset FECD [39].
Individuals with two risk alleles of TCF4 SNP rs613872 carry 30 times more chance of developing FECD compared to individuals who carry a single risk allele [17]. We did not identify any individuals with two risk alleles in our study, but the heterozygous genotype (CA) carrying a single risk allele showed significant association with FECD.
There is evidence the rs613872 SNP is associated with CCT in FECD [40]. We did not find any correlation of rs613872 with age at onset, gender, or CCT. However, endothelial cell density (specular count) was significantly associated with rs613872 in the patients.
TCF4 (also known as E2–2) is expressed in the developing corneal endothelium [17] and has a role in the downregulation of cell adhesion proteins such as E-cadherin that results in loss of cell polarity and may decrease endothelial cell density by inducing EMT [41]. The presence of the SNP rs613872 risk variant may alter the activity of TCF4 thus regulating the endothelial cell density. To summarize, E2–2 proteins under normal conditions may promote EMT during injury, age-related damage, and stress to the cornea by downregulation of E-cadherin, and possibly by increasing the expression of ZEB1, which is a direct repressor of the E-cadherin promoter. Dysregulation of E2–2 possibly due to the presence of polymorphisms may reduce ZEB1 expression and EMT in the cornea, thus decreasing the migration, proliferation, and replacement of the damaged corneal cells [42].
Novel mutations identified in the present study add to the repertoire of ZEB1 mutations that cause Fuchs corneal dystrophies. These findings also suggest that a single nucleotide change may be linked to other variants and alter the resulting protein in such a way that the presentation of the resulting phenotype becomes highly variable.
To the best of our knowledge, this is the first report on screening of ZEB1 mutations and TCF4 SNP in patients with late onset FECD from India in which novel ZEB1 variations and polymorphisms were identified. The TCF4 rs613872 was found to be associated with decreased endothelial cell density, thus envisaging a plausible role for the two in FECD pathogenesis.
Acknowledgments
The Authors thank the patients for their participation. The study was supported by a financial grant (No.BT/PR6376/MED/12/248/2005) provided by the Department of Biotechnology (DBT), India. None of the authors have any conflict of interest. The article was presented at the Asia ARVO, February 2015 in Yokohama, Japan. Dr Arundhati Sharma (arundhatisharma1@gmail.com) and Dr. Radhika Tandon (radhika_tan@yahoo.com) are co-corresponding authors for this paper.
References
- 1.Waring GO, III, Rodrigues MM, Laibson PR. Corneal dystrophies. II. Endothelial dystrophies. Surv Ophthalmol. 1978;23:147–68. doi: 10.1016/0039-6257(78)90151-0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=310583&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 2.Magovern M, Beauchamp GR, McTigue JW, Fine BS, Baumiller RC. Inheritance of Fuchs’ combined dystrophy. Ophthalmology. 1979;86:1897–923. doi: 10.1016/s0161-6420(79)35340-4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=399801&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Lorenzetti DW, Uotila MH, Parikh N, Kaufman HE. Central cornea guttata: incidence in the general population. Am J Ophthalmol. 1967;64:1155–8. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=6072991&dopt=Abstract [PubMed] [Google Scholar]
- 4.Eghrari AO, McGlumphy EJ, Iliff B, Wang J, Emmert D, Riazuddin SA, Katsanis N, Gottsch JD. Prevalence and severity of Fuchs Corneal Dystrophy in Tangier Island. Am J Ophthalmol. 2012;153:1067–72. doi: 10.1016/j.ajo.2011.11.033. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22321803&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vithana EN, Morgan PE, Ramprasad V, Tan DT, Yong VH, Venkataraman D, Venkatraman A, Yam GH, Nagasamy S, Law RW, Rajagopal R, Pang CP, Kumaramanickevel G, Casey JR, Aung T. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17:656–66. doi: 10.1093/hmg/ddm337. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18024964&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 6.Gottsch JD, Zhang C, Sundin OH, Bell WR, Stark WJ, Green WR. Fuchs corneal dystrophy: aberrant collagen distribution in an L450W mutant of the COL8A2 gene. Invest Ophthalmol Vis Sci. 2005;46:4504–11. doi: 10.1167/iovs.05-0497. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16303941&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 7.Biswas S, Munier FL, Yardley J, Hart-Holden N, Perveen R, Cousin P, Sutphin JE, Noble B, Batterbury M, Kielty C, Hackett A, Bonshek R, Ridgway A, McLeod D, Sheffield VC, Stone EM, Schorderet DF, Black GC. Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet. 2001;10:2415–23. doi: 10.1093/hmg/10.21.2415. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11689488&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 8.Mok JW, Kim HS, Joo CK. Q455V mutation in COL8A2 is associated with Fuchs' corneal dystrophy in Korean patients. Eye (Lond) 2009;23:895–903. doi: 10.1038/eye.2008.116. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18464802&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 9.Sundin OH, Broman KW, Chang HH, Vito EC, Stark WJ. b, Gottsch J. D. A common locus for late-onset Fuchs’ corneal dystrophy maps to 18q21.2-q21.32. Invest Ophthalmol Vis Sci. 2006;47:3919–26. doi: 10.1167/iovs.05-1619. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16936105&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Sundin OH, Jun AS, Broman KW, Liu SH, Sheehan SE, Vito ECL, Stark WJ, Gottsch JD. Linkage of late-onset Fuchs corneal dystrophy to a novel locus at 13ptel-13q12.13. Invest Ophthalmol Vis Sci. 2006;47:140–5. doi: 10.1167/iovs.05-0578. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16384955&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 11.Riazuddin SA, Eghrari AO, Al-Saif A, Davey L, Meadows DN, Katsanis N, Gottsch JD. Linkage of a mild late-onset phenotype of Fuchs’ corneal dystrophy to a novel locus at 5q33.1- q35.2. Invest Ophthalmol Vis Sci. 2009;50:5667–71. doi: 10.1167/iovs.09-3764. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19608540&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 12.Riazuddin SA, Vithana EN, Seet LF, Liu Y, Al-Saif A, Koh LW, Heng YM, Aung T, Meadows DN, Eghrari AO, Gottsch JD, Katsanis N. Missense mutations in the sodium borate cotransporter SLC4A11 cause late-onset Fuchs corneal dystrophya. Hum Mutat. 2010;31:1261–8. doi: 10.1002/humu.21356. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20848555&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vithana EN, Morgan P, Sundaresan P, Ebenezer ND, Tan DT, Mohamed MD, Anand S, Khine KO, Venkataraman D, Yong VH, Salto-Tellez M, Venkatraman A, Guo K, Hemadevi B, Srinivasan M, Prajna V, Khine M, Casey JR, Inglehearn CF, Aung T. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nat Genet. 2006;38:755–7. doi: 10.1038/ng1824. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16767101&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 14.Riazuddin SA, Zaghloul NA, Al-Saif A, Davey L, Diplas BH, Meadows DN, Eghrari AO, Minear MA, Li YJ, Klintworth GK, Afshari N, Gregory SG, Gottsch JD, Katsanis N. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010;86:45–53. doi: 10.1016/j.ajhg.2009.12.001. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20036349&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–87. doi: 10.1007/s00018-008-8465-8. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19011757&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krafchak CM, Pawar H, Moroi SE, Sugar A, Lichter PR, Mackey DA, Mian S, Nairus T, Elner V, Schteingart MT, Downs CA, Kijek TG, Johnson JM, Trager EH, Rozsa FW, Mandal MN, Epstein MP, Vollrath D, Ayyagari R, Boehnke M, Richards JE. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005;77:694–708. doi: 10.1086/497348. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16252232&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baratz KH, Tosakulwong N, Ryu E, Brown WL, Branham K, Chen W, Tran KD, Schmid-Kubista KE, Heckenlively JR, Swaroop A, Abecasis G, Bailey KR, Edwards AO. E2–2 protein and Fuchs's corneal dystrophy. N Engl J Med. 2010;363:1016–24. doi: 10.1056/NEJMoa1007064. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20825314&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 18.Riazuddin SA, McGlumphy EJ, Yeo WS, Wang J, Katsanis N, Gottsch JD. Replication of the TCF4 intronic variant in late-onset Fuchs corneal dystrophy and evidence of independence from the FCD2 locus. Invest Ophthalmol Vis Sci. 2011;52:2825–9. doi: 10.1167/iovs.10-6497. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21245398&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuot A, Hewitt AW, Griggs K, Klebe S, Mills R, Jhanji V, Craig JE, Sharma S, Burdon KP. Association of TCF4 and CLU polymorphisms with Fuchs' endothelial dystrophy and implication of CLU and TGFBI proteins in the disease process. Eur J Hum Genet. 2012;20:632–8. doi: 10.1038/ejhg.2011.248. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22234156&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YJ, Minear MA, Rimmler J, Zhao B, Balajonda E, Hauser MA, Allingham RR, Eghrari AO, Riazuddin SA, Katsanis N, Gottsch JD, Gregory SG, Klintworth GK, Afshari NA. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS ONE. 2011;6:e18044. doi: 10.1371/journal.pone.0018044. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21533127&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=3344216&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krachmer JH, Purcell JJ, Jr, Young CW, Bucher KD. Corneal endothelial dystrophy. A study of 64 families. Arch Ophthalmol. 1978;96:2036–9. doi: 10.1001/archopht.1978.03910060424004. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=309758&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 23.Rosenblum P, Stark WJ, Maumenee IH, Hirst LW, Maumenee AE. Hereditary Fuchs Dystrophy. Am J Ophthalmol. 1980;90:455–62. doi: 10.1016/s0002-9394(14)75011-1. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=6968504&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 24.Tamura Y, Konomi H, Sawada H, Takashima S, Nakajima A. Tissue distribution of type VIII collagen in human adult and fetal eyes. Invest Ophthalmol Vis Sci. 1991;32:2636–44. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=1869415&dopt=Abstract [PubMed] [Google Scholar]
- 25.Kayes J, Holmberg A. The fine structure of the cornea in fuch’s endothelial dystrophy. Invest Ophthalmol. 1964;3:47–67. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=14121146&dopt=Abstract [PubMed] [Google Scholar]
- 26.Meng H, Matthaei M, Ramanan N, Grebe R, Chakravarti S, Speck CL, Kimos M, Vij N, Eberhart CG, Jun AS. L450W and Q455K Col8a2 knock-in mouse models of Fuchs endothelial corneal dystrophy show distinct phenotypes and evidence for altered autophagy. Invest Ophthalmol Vis Sci. 2013;54:1887–97. doi: 10.1167/iovs.12-11021. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23422828&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemadevi B, Srinivasan M, Arunkumar J, Prajna NV, Sundaresan P. Genetic analysis of patients with Fuchs’ endothelial corneal dystrophy in India. BMC Ophthalmol. 2010;10:3. doi: 10.1186/1471-2415-10-3. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20144242&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aldave AJ, Yellore VS, Yu F, Bourla N, Sonmez B, Salem AK. Posterior polymorphous corneal dystrophy is associated with TCF8 gene mutations and abdominal hernia. Am J Med Genet. 2007;143A:2549–56. doi: 10.1002/ajmg.a.31978. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17935237&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 29.Mehta JS, Vithana EN, Tan DT, Yong VH, Yam GH, Law RW, Chong WG, Pang CP, Aung T. Analysis of the posterior polymorphous corneal dystrophy 3 gene, TCF8, in late-onset Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2008;49:184–8. doi: 10.1167/iovs.07-0847. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18172091&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 30.Minear MA, Li YJ, Rimmler J, Balajonda E, Watson S, Allingham RR, Hauser MA, Klintworth GK, Afshari NA, Gregory SG. Genetic screen of African Americans with Fuchs endothelial corneal dystrophy. Mol Vis. 2013;19:2508–16. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24348007&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 31.Chung DW, Frausto RF, Ann LB, Jang MS, Aldave AJ. Functional impact of ZEB1 mutations associated with posterior polymorphous and Fuchs' endothelial corneal dystrophies. Invest Ophthalmol Vis Sci. 2014;55:6159–66. doi: 10.1167/iovs.14-15247. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25190660&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechner J, Dash DP, Muszynska D, Hosseini M, Segev F, George S, Frazer DG, Moore JE, Kaye SB, Young T, Simpson DA, Churchill AJ, Héon E, Willoughby CE. Mutational spectrum of the ZEB1 gene in corneal dystrophies supports a genotype-phenotype correlation. Invest Ophthalmol Vis Sci. 2013;54:3215–23. doi: 10.1167/iovs.13-11781. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23599324&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 33.Castellana S, Mazza T. Congruency in the prediction of pathogenic missense mutations: state-of-the-art web based tool. Brief Bioinform. 2013;013:1–12. doi: 10.1093/bib/bbt013. [DOI] [PubMed] [Google Scholar]
- 34.Park SM, Gaur A, Peter M. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894. doi: 10.1101/gad.1640608. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18381893&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong TS, Gao W, Chan J. Transcription regulation of E-Cadherin by zinc finger E-box binding homeobox proteins in solid tumors. Biomed Res Int. 2014; 921564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurahara H, Takao S, Maemura K, Mataki Y, Kuwahata T, Maeda K, Ding Q, Sakoda M, Iino S, Ishigami S, Ueno S, Shinchi H, Natsugoe S. Epithelial-mesenchymal transition and mesenchymal-epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J Surg Oncol. 2012;105:655–61. doi: 10.1002/jso.23020. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22213144&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 37.Stamler JF, Roos BR, Wagoner MD, Goins KM, Kitzmann AS, Riley JB, Stone EM, Fingert JH. Confirmation of the association between the TCF4 risk allele and Fuchs endothelial corneal dystrophy in patients from the Midwestern United States. Ophthalmic Genet. 2013;34:32–4. doi: 10.3109/13816810.2012.726396. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22998502&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 38.Wieben ED, Aleff RA, Tosakulwong N, Butz ML, Highsmith WE, Edwards AO, Baratz KH. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2–2) gene predicts Fuchscorneal dystrophy. PLoS ONE. 2012;7:e49083. doi: 10.1371/journal.pone.0049083. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23185296&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanda GG, Padhy B, Samal S, Das S, Alone DP. Genetic association of TCF4 intronic polymorphisms, CTG18.1 and rs17089887, with Fuchs' endothelial corneal dystrophy in an Indian population. Invest Ophthalmol Vis Sci. 2014;55:7674–80. doi: 10.1167/iovs.14-15297. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25342617&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 40.Igo RP, Jr, Kopplin LJ, Joseph P, Truitt B, Fondran J, Bardenstein D, Aldave AJ, Croasdale CR, Price MO, Rosenwasser M, Lass JH, Iyengar SK. Differing roles for TCF4 and COL8A2 in central corneal thickness and fuchs endothelial corneal dystrophy. PLoS ONE. 2012;7:e46742. doi: 10.1371/journal.pone.0046742. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23110055&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16493418&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 42.Wright AF, Dhillon B. Major progress in Fuchs's corneal dystrophy. N Engl J Med. 2010;363:1072–5. doi: 10.1056/NEJMe1007495. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20825321&dopt=Abstract [DOI] [PubMed] [Google Scholar]