Abstract
Background
Jellyfish contain diverse toxins and other bioactive components. However, large-scale identification of novel toxins and bioactive components from jellyfish has been hampered by the low efficiency of traditional isolation and purification methods.
Results
We performed de novo transcriptome sequencing of the tentacle tissue of the jellyfish Cyanea capillata. A total of 51,304,108 reads were obtained and assembled into 50,536 unigenes. Of these, 21,357 unigenes had homologues in public databases, but the remaining unigenes had no significant matches due to the limited sequence information available and species-specific novel sequences. Functional annotation of the unigenes also revealed general gene expression profile characteristics in the tentacle of C. capillata. A primary goal of this study was to identify putative toxin transcripts. As expected, we screened many transcripts encoding proteins similar to several well-known toxin families including phospholipases, metalloproteases, serine proteases and serine protease inhibitors. In addition, some transcripts also resembled molecules with potential toxic activities, including cnidarian CfTX-like toxins with hemolytic activity, plancitoxin-1, venom toxin-like peptide-6, histamine-releasing factor, neprilysin, dipeptidyl peptidase 4, vascular endothelial growth factor A, angiotensin-converting enzyme-like and endothelin-converting enzyme 1-like proteins. Most of these molecules have not been previously reported in jellyfish. Interestingly, we also characterized a number of transcripts with similarities to proteins relevant to several degenerative diseases, including Huntington’s, Alzheimer’s and Parkinson’s diseases. This is the first description of degenerative disease-associated genes in jellyfish.
Conclusion
We obtained a well-categorized and annotated transcriptome of C. capillata tentacle that will be an important and valuable resource for further understanding of jellyfish at the molecular level and information on the underlying molecular mechanisms of jellyfish stinging. The findings of this study may also be used in comparative studies of gene expression profiling among different jellyfish species.
Introduction
In recent decades, frequent outbreaks of jellyfish have occurred in oceans, potentially due to overfishing by humans, nutrient pollution and global warming. Jellyfish outbreaks have a strong adverse impact on marine ecological balance. However, the large amount of jellyfish biomass could be considered a valuable source of bioactive compounds. Thus, the overall development and comprehensive utilization of jellyfish have also triggered interest among many scientists.
Jellyfish bodies contain a great variety of natural bioactive components, among which the most studied are jellyfish nematocyst toxins. Nematocysts are densely located on the tentacles, and each contains a tiny dose of venom. People stung by toxic jellyfish may develop severe pain, dyspnea or even cardiorespiratory failure [1]. Many studies have explored the physicochemical properties of nematocyst toxins, which are now believed to be a type of novel protein or peptide. Jellyfish nematocyst toxins exhibit various bioactivities, such as hemolytic, enzymatic, neurotoxic, myotoxic and cardiovascular activities [2–4]. In addition to nematocyst toxins, the jellyfish body contains a wide range of novel proteins or peptides that exhibit activities such as antioxidation, antibiosis and immune reinforcing. Antioxidant activity of the giant jellyfish Nemopilema nomurai was observed by Kazuki [5]. We previously reported the first peroxiredoxin (Prx) and thioredoxin (Trx) genes from the jellyfish Cyanea capillata; both of these genes exhibit general intracellular antioxidant activity [6–7]. The jellyfish body also contains abundant collagens, which have immunostimulatory effects without inducing allergic complications [8].
Recent research has investigated and identified the bioactive components of jellyfish, particularly their toxins. However, the low efficiency of traditional isolation and purification methods has hampered the large-scale identification of novel toxins and bioactive components from jellyfish. No complete genome sequencing of jellyfish has been reported. In the absence of genome sequencing, the transcriptome represents a valuable searchable database. However, of the nearly 250 jellyfish species, only three species, Stomolophus meleagris, Chironex fleckeri and Aurelia aurita, have been sequenced using the transcriptome approach [9–11]. Only a small number of jellyfish sequences (54,247 ESTs and 3,795 nucleotides, as of Mar 10, 2015) have been deposited in the National Center for Biotechnology Information (NCBI) database, seriously limiting our understanding of the diverse bioactivities of this abundant marine zooplankton. Therefore, more sequence data and comprehensive analysis of jellyfish species transcriptomes are desired to explore more toxins and other bioactive components.
High-throughput next-generation sequencing technologies provide platforms to perform deep sequence analysis, which has greatly boosted comprehensive genetic research on some relatively uncharacterized species. C. capillata is one of the most common venomous jellyfish in the East China Sea. We previously demonstrated that a tentacle extract from C. capillata exhibits diverse bioactivities, including hemolytic, proteolytic, cardiovascular, cytolytic and antioxidant activities [12–14]. However, the underlying mechanisms of these bioactivities at the molecular level remain unclear. In the present study, we performed de novo transcriptome sequencing of the tentacle tissue of C. capillata using the Illumina HiSeq™ 2000 platform. A systematic bioinformatics strategy was used to conduct an in-depth and integrated analysis of this transcriptome, explore the venom composition in detail, and identify other important molecules in C. capillata.
Material and Methods
Jellyfish sample collection and RNA isolation
Samples of the jellyfish C. capillata were collected in July 2013 in the Sanmen Bay, East China Sea. No specific permit was required to catch C. capillata. The tentacle tissues were quickly excised manually after capture and frozen immediately in liquid nitrogen. Total RNA was isolated using TRIzol reagent (Invitrogen, CA, USA) and treated with RNase-free DNase I (Takara Biotechnology, China). RNA integrity was validated with a 2100 Bioanalyzer (Agilent Technologies, CA, USA).
Illumina sequencing
Illumina sequencing analysis was performed according to the methods described previously [15–16]. Briefly, poly(A) mRNA was isolated using Oligo (dT) beads and interrupted to short fragments. These fragments were then transcribed into first-strand cDNA, followed by synthesis of the second strand. The synthesized cDNA products were purified using a QiaQuick PCR extraction kit (Qiagen, Valencia, CA, USA) and dissolved in EB buffer for end repair and poly(A) addition. Subsequently, the cDNA fragments were ligated to the sequencing adapters and subjected to size selection using agarose gel electrophoresis. Suitable fragments were amplified by PCR, and the cDNA library was sequenced using an Illumina HiSeq™ 2000 sequencer at the Beijing Genomics Institute (BGI; Shenzhen, China).
De novo assembly and functional annotation
The image data output from the sequencer was transformed into sequence data called raw reads. After filtering low-quality reads and reads containing more than 5% unknown nucleotides, the sequencing adaptors were removed from the raw reads. Subsequently, the raw reads were assembled into contigs and unigenes by de novo assembly, which was performed with the Trinity program [17]. Finally, unigenes were aligned by BLASTx (e-value ≤ 10−5) to protein databases, including the NCBI non-redundant protein (Nr) database (http://www.ncbi.nlm.nih.gov), Swiss-Prot protein database (http://www.expasy.ch/sprot), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg) and Cluster of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG). Proteins with the highest sequence similarity with the given unigenes were used to determine the sequence direction, functional annotation and protein coding region. A preferential order of Nr, Swiss-Prot, KEGG and COG was followed if the results from these databases were inconsistent. If no hits were obtained for a unigene in these databases, ESTScan software [18] was used to decide the sequence direction and protein coding region. Based on Nr annotations, the Blast2GO program [19] was then used to obtain the gene ontology (GO) annotations of the unigenes, followed by GO classification using WEGO software [20]. COG and KEGG were also used to obtain functional annotations for the unigenes and analyze gene products involved in metabolism.
Identification of toxin-like transcripts
According to our previous studies of C. capillata and other reports on various jellyfish, the toxic effects of jellyfish venom primarily include vasoconstriction, hemorrhage, and hemolytic and cardiovascular toxicities. To explore the underlying molecular mechanisms of these toxic actions and identify as many putative toxin transcripts in C. capillata as possible, three strategies were used. First, we compared the unigene sequences to a toxin database in Swiss-Prot, Tox-Prot (http://www.uniprot.org/program/Toxins), based on sequence homology. Second, to make the screening more complete, we also manually searched the annotations of the unigenes under the term ‘toxin’ or ‘venom’. Third, according to the symptoms after jellyfish envenomation, we referred to many previous reports on venomous components in different types of venomous animals, such as snakes, scorpions, spiders, wasps and sea anemones, to construct a reference guide of estimated toxin-like transcripts.
Analysis of transcripts related to degenerative diseases
Sequences encoding proteins associated with degenerative diseases, including Huntington’s disease (HD), Alzheimer's disease (AD) and Parkinson's disease (PD), were identified by BLAST results against the Nr database, with a cut-off value of e-value ≤ 10−5.
Bioinformatics analyses and alignments
Bioinformatics analyses were performed following methods we have described previously [7]. Briefly, the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) were used to search for the open reading frames and signal peptides, respectively, in the sequences. Sequence alignments were performed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Phylogenetic analysis was performed using MEGA 4 software.
Results and Discussion
Illumina sequencing and reads assembly
A total of 54,109,750 raw reads were obtained using the Illumina HiSeq™ 2000 platform. After cleaning and removing dirty reads containing adapters, unknown or low quality bases, a total of 51,304,108 clean reads corresponding to more than 4.61 billion clean nucleotides were generated (Table 1). The average length of the clean reads was 90 bp, consistent with the sequencing capacity of the Illumina device. The Q20 percentage, N percentage and GC percentage were 99.15%, 0.02% and 40.26%, respectively. The original sequencing data for the clean reads have been deposited in the NCBI Sequence Read Archive (SRA) database (accession number SRP056566).
Table 1. Summary of the C. capillata tentacle transcriptome.
| Sequencing statistics | |
| Number of raw reads | 54,109,750 |
| Number of clean reads | 51,304,108 |
| Total clean nucleotides (bp) | 4,617,369,720 |
| Average length of clean reads (bp) | 90 |
| Q20 (%) | 99.15 |
| N (%) | 0.02 |
| GC (%) | 40.26 |
| Assembly statistics | |
| Contigs | |
| Total number | 125,058 |
| Total length (bp) | 32,284,466 |
| Mean length (bp) | 258 |
| N50 length | 313 |
| Unigenes | |
| Total number | 50,536 |
| Total length (bp) | 25,414,791 |
| Mean length (bp) | 503 |
| N50 length | 644 |
| Total consensus sequences | 50,536 |
| Distinct clusters | 861 |
| Distinct singletons | 49,675 |
Using the Trinity program, a total of 125,058 contigs corresponding to more than 32 million nucleotides were assembled from the short reads. Among these assembled contigs, 90.50% (113,176) were between 100 and 500 bp in length, 5.96% (7,451) were between 500 and 1000 bp, 2.99% (3,740) were between 1000 and 2000 bp, and 0.55% (691) were more than 2000 bp. Finally, the contigs were connected, and 50,536 unigenes were generated, with a mean length of 503 bp. Although most unigenes (36,224, 71.68%) were between 100 and 500 bp, we obtained 14,312 unigenes that were greater than 500 bp in length. The length distributions of these assembled contigs and unigenes are shown in S1 Fig. Protein coding sequences (CDS) of all assembled unigenes were predicted. A total of 20,892 potential CDSs were identified by BLAST searches, and 5,817 CDSs were predicted by ESTScan.
Transcriptome analysis using Illumina sequencing technology is one of the most popular tools for gene discovery, and it has recently been applied to several species that lack genomic sequence information [21–23]. Therefore, the transcriptome data for C. capillata obtained here will enrich the sequence information previously available for jellyfish in public databases. In addition, this transcriptome could provide more detailed and general genetic data to facilitate large-scale discovery and rapid characterization of novel important genes from jellyfish.
Functional annotation and classification
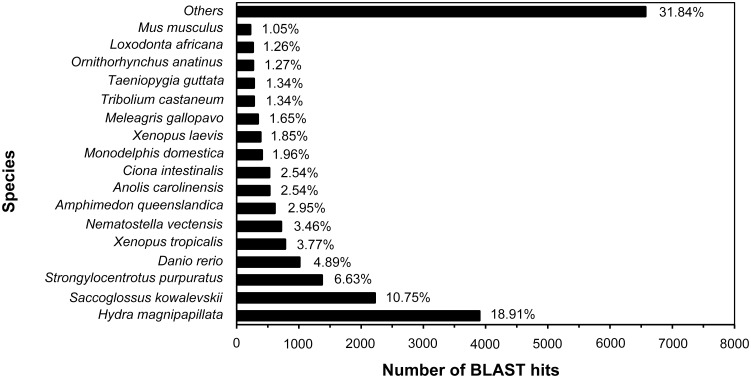
To obtain functional annotations of the predicted proteins, the assembled unigenes were used as a query for Blastx alignments to several public protein databases, including the Nr, Swiss-Prot, KEGG and COG databases. An e-value < 10−5 was used as a cut-off for confident homologue detection. Of a total of 50,536 unigenes, hits were obtained for 20,629 (40.8%) in the Nr database. This large percentage of hits was anticipated. In addition, hits were obtained for 17,006 (33.7%), 13,663 (27.0%) and 6,202 (12.3%) unigenes in the Swiss-Prot, KEGG and COG protein databases, respectively. We also aligned all of the unigenes by Blastn to the Nt nucleotide database, and 4,878 (9.7%) had significant hits. The functional annotations by BLAST searches of the 50,536 unigenes are presented in S1 Table. The e-value distributions were also calculated. Among the 20,629 unigenes that had homologous proteins in the Nr protein database, more than half of the matched sequences (11,793, 57.2%) had an e-value ranging between 1e-10 and 1e-50, and 6.6% (1,368) had homology with an e-value smaller than 1e-100, indicating strong reliability of the alignment. To sum up, a total of 21,357 (42.3%) unigenes were significantly similar to the unique protein accessions in each of the above databases. However, more than half of the unigenes had no significant matches due to the limited sequence information for jellyfish and their closely related species or partly due to a high number of species-specific transcripts and novel sequences. The species distributions of the top BLAST hits against the Nr database were also analyzed. The assembled unigenes had the greatest number (3,900, 18.91%) of matches with Hydra magnipapillata. Because jellyfish and hydra are both included in the phylum of Cnidaria, sequence similarities are likely to be highest with closely related species. The next species were Saccoglossus kowalevskii (2,218, 10.75%), Strongylocentrotus purpuratus (1.368, 6.63%), Danio rerio (1,008, 4.89%), Xenopus tropicalis (778, 3.77%) and another typical species of Cnidaria, Nematostella vectensis (713, 3.46%). Other species with proportions of greater than 1% are also shown in Fig 1. However, few matches corresponded to jellyfish species, which might also be due to the limited number of jellyfish protein sequences available in the database.
Fig 1. Top-hit species distribution of the BLASTx matches of the unigenes.
The species distributions of the top BLASTx hits against the Nr database were analyzed (cut-off e-value < 10−5). Species with proportions of greater than 1% are shown.
GO annotation based on sequence homology was used to determine the GO terms of the unigenes. A total of 6,657 (13.2%) unigenes were categorized into at least one group of 49 sub-categories of three independent ontology categories (S2 Fig). Among the 26 sub-categories of “biological process”, “cellular process” (3412 unigenes) was the most dominant group, followed by “metabolic process” (2578 unigenes) and “biological regulation” (1467 unigenes), indicating that many extensive metabolic activities and rapid growth may occur in the tentacle of C. capillata. For the “cellular component” category, the most representative of the assignments were “cell” (4373 unigenes), “cell part” (4041 unigenes) and “organelle” (2701 unigenes). Within the 12 groups corresponding to “molecular function”, the dominant distributions were from “binding” (2895 unigenes) and “catalytic activity” (2718 unigenes). These GO annotations represented the general gene expression profile characteristics for the tentacle of C. capillata. To further predict and classify possible functions of the unigenes, COG assignments were used (S3 Fig). The category of “general function prediction only”, which contained 2,079 unigenes (33.52%), was the largest group, followed by “replication, recombination and repair” (898, 14.48%), “translation, ribosomal structure and biogenesis” (841, 3.94%) and “posttranslational modification, protein turnover, chaperones” (769, 12.40%). The categories of “extracellular structures” (3, 0.05%) and “nuclear structure” (8, 0.13%) were the smallest groups. The GO and COG functional classifications thus provided valuable and detailed information for investigating specific processes and functions in jellyfish tentacle.
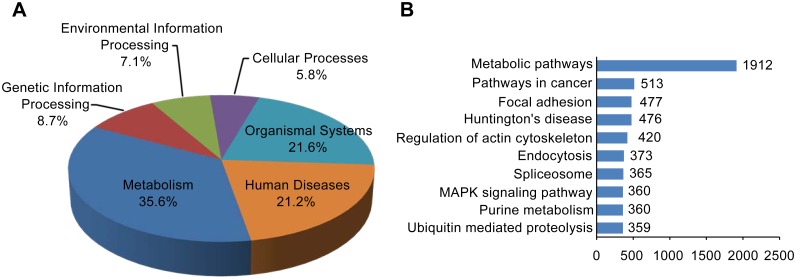
To identify the active metabolic pathways in the tentacle of C. capillata, the annotated unigenes were mapped to KEGG pathways. A total of 13,663 unigenes were assigned to 241 KEGG pathways consisting of the categories of “metabolism” (86 pathways), “genetic information processing” (21 pathways), “environmental information processing” (17 pathways), “cellular processes” (14 pathways), “organismal systems” (52 pathways) and “human diseases” (51 pathways) (Fig 2A). Among the mapped pathways, “metabolic pathways” contained 1,912 unigenes (13.99%) and was obviously larger than the other groups, such as “pathways in cancer” (513, 3.75%), “focal adhesion” (477, 3.49%), “‘Huntington’s disease” (476, 3.48%) and “regulation of actin cytoskeleton” (420, 3.07%). The top 10 pathways are shown in Fig 2B, and all pathways are summarized in S2 Table.
Fig 2. Categorization of unigenes into KEGG biochemical pathways.
A total of 13,663 unigenes were assigned to 241 KEGG pathways belonging to six categories. (A) The percentage of the pathway amount in each category is shown. (B) The ten largest groups with KEGG database annotation. The x-axis indicates the number of annotated unigenes.
In summary, based on de novo sequencing and in-depth analysis, we obtained a well-annotated transcriptome that could provide valuable information for identifying novel genes and investigating specific metabolic pathways in the tentacle of the jellyfish C. capillata.
Analysis of putative toxin transcripts in C. capillata
Interest in the biological activities of jellyfish toxins has increased considerably in recent years. However, due to the technical difficulties of obtaining toxins and their labile nature, the molecular mechanisms of jellyfish toxins remain largely unknown. Few molecules in jellyfish venom have been described, hampering the exploration of the pathophysiology of jellyfish envenomation and proper patient care. Many studies have demonstrated that jellyfish venom consists of a complex mixture of proteins and peptides with toxic or enzymatic actions [13,24–26]. Therefore, one of the main goals of the present study is to identify as many putative toxin transcripts in C. capillata as possible and build a reference database of toxin sequences to facilitate the future analysis of other jellyfish species.
1. Venom constituents similar to known toxin families
Based on the symptoms of jellyfish stings, we identified a large number of transcripts encoding proteins similar to several well-known toxin families that have been widely described in various venomous animals. These toxin transcripts are further classified and summarized in Table 2.
Table 2. Annotation of transcripts similar to known toxin families.
| Sequence ID | ORF a /Length(bp) | Signal peptide b | Annotation | Best matched species/Maximum identify (%) | E-value |
|---|---|---|---|---|---|
| Phospholipases | |||||
| Unigene7860 | F/784 | Y | Group 3 secretory phospholipase A2-like | Acyrthosiphon pisum/46 | 3E-19 |
| Unigene9375 | N/289 | U | Group 3 secretory phospholipase A2-like | Gallus gallus/45 | 2E-09 |
| Unigene17685 | F/1945 | Y | Group XV phospholipase A2-like | Aplysia californica/47 | 6E-104 |
| Unigene19288 | F/626 | Y | Phospholipase A2 | Rhopilema esculentum/43 | 2E-39 |
| Unigene27084 | F/662 | Y | Phospholipase A2 | Rhopilema esculentum/57 | 1E-52 |
| Unigene35028 | F/804 | Y | Phospholipase A2 | Rhopilema esculentum/36 | 6E-19 |
| Unigene37728 | F/562 | N | Phospholipase A2 | Rhopilema esculentum/49 | 2E-15 |
| Unigene28609 | N/284 | U | 85/88 kDa calcium-independent phospholipase A2 | Hydra vulgaris/59 | 1E-11 |
| Unigene47062 | N/546 | U | 85/88 kDa calcium-independent phospholipase A2 | Hydra vulgaris/54 | 3E-55 |
| Unigene41691 | N/967 | U | Calcium-independent phospholipase A2-gamma-like | Lepisosteus oculatus/52 | 3E-94 |
| Unigene4792 | N/305 | U | Phospholipase D1-like | Hydra vulgaris/62 | 5E-29 |
| Unigene23083 | N/277 | U | Phospholipase D1 | Ixodes scapularis/46 | 3E-17 |
| Unigene48846 | N/324 | U | Phospholipase D1-like | Metaseiulus occidentalis/45 | 3E-12 |
| Unigene30910 | N/435 | U | Phospholipase D3-like | Aplysia californica/54 | 9E-36 |
| Unigene48788 | N/255 | U | Phospholipase D3 | Ascaris suum/56 | 9E-23 |
| Metalloproteinases | |||||
| Unigene18144 | F/1437 | N | Matrix metalloproteinase-14 | Camelus ferus/33 | 2E-24 |
| Unigene34794 | N/670 | N | Matrix metalloproteinase-14 | Ascaris suum/43 | 1E-31 |
| Unigene39586 | N/1639 | N | Matrix metalloproteinase-1 | Drosophila melanogaster/43 | 2E-16 |
| Unigene9370 | N/215 | U | Matrix metalloproteinase-14-like | Saccoglossus kowalevskii/70 | 1E-13 |
| Unigene18671 | F/2463 | Y | Matrix metalloproteinase | Hydra vulgaris/38 | 8E-98 |
| Unigene19077 | F/2544 | Y | Matrix metalloproteinase | Hydra vulgaris/39 | 1E-98 |
| Unigene18967 | N/1127 | Y | Matrix metalloproteinase-24-like | Hydra vulgaris/44 | 9E-57 |
| Unigene21910 | N/836 | Y | Matrix metalloproteinase-9-like | Strongylocentrotus purpuratus/37 | 1E-12 |
| Unigene41786 | F/1953 | Y | Matrix metalloproteinase-25-like | Hydra vulgaris/36 | 6E-93 |
| Unigene6794 | F/2042 | N | Matrix metalloproteinase | Hydra vulgaris/42 | 1E-116 |
| Unigene13316 | N/413 | U | Matrix metalloproteinase-28 | Sarcophilus harrisii/48 | 3E-11 |
| Unigene8341 | F/1412 | Y | Zinc metalloproteinase nas-4 | Hydra vulgaris/53 | 6E-76 |
| Unigene19338 | F/1253 | Y | Zinc metalloproteinase nas-4 | Hydra vulgaris/39 | 3E-58 |
| Unigene17414 | F/1901 | Y | Zinc metalloproteinase nas-13-like | Hydra vulgaris/50 | 1E-59 |
| Unigene18943 | F/2096 | Y | Zinc metalloproteinase nas-13-like | Hydra vulgaris/40 | 7E-54 |
| Unigene7643 | F/1098 | Y | Zinc metalloproteinase nas-15-like | Hydra vulgaris/43 | 6E-66 |
| Unigene18427 | F/1440 | Y | Astacin 3 | Hydractinia echinata/49 | 6E-86 |
| Serine proteases | |||||
| Unigene17637 | N/1100 | U | Serine protease 1 | Aurelia aurita/52 | 4E-80 |
| Unigene8850 | F/1033 | Y | Serine protease 1 | Aurelia aurita/68 | 7E-134 |
| Unigene34706 | N/834 | U | Serine protease 1 | Aurelia aurita/70 | 7E-120 |
| Unigene10683 | N/459 | U | Serine protease 1 | Aurelia aurita/65 | 1E-66 |
| Unigene10980 | N/340 | U | Serine protease 2 | Aurelia aurita/59 | 1E-41 |
| Unigene23736 | N/324 | U | Serine protease 2 | Aurelia aurita/50 | 1E-32 |
| Unigene18480 | N/2005 | U | Transmembrane protease serine 6-like | Hydra vulgaris/37 | 8E-55 |
| Unigene19320 | F/2208 | Y | Chymotrypsin-like elastase family member 2A-like | Hydra vulgaris/35 | 3E-49 |
| Unigene41758 | F/1540 | Y | Chymotrypsin-like elastase family member 2A-like | Hydra vulgaris/39 | 6E-64 |
| Unigene40159 | N/262 | U | Chymotrypsin-like elastase family member 1-like | Hydra vulgaris/50 | 1E-23 |
| Unigene1406 | N/756 | U | Chymotrypsin-like elastase family member 3B-like | Hydra vulgaris/50 | 1E-70 |
| Unigene44510 | F/609 | N | Chymotrypsin-like elastase family member 3B-like | Hydra vulgaris/49 | 2E-48 |
| Unigene10156 | N/232 | U | Chymotrypsin-C-like | Meleagris gallopavo/45 | 5E-08 |
| Unigene15770 | F/789 | N | Chymotrypsin-like protease CTRL-1-like | Danio rerio/37 | 1E-35 |
| Unigene42233 | F/1362 | N | Serine proteinase stubble-like | Bombus impatiens/27 | 2E-17 |
| Serine proteinase inhibitor | |||||
| Unigene17288 | F/1137 | N | Kazal-type serine proteinase inhibitor 4 | Procambarus clarkii/51 | 1E-05 |
| Unigene23109 | F/1006 | N | Kazal-type serine proteinase inhibitor 1 | Fenneropenaeus chinensis/34 | 2E-31 |
| Unigene35150 | F/1748 | N | Kazal-type serine proteinase inhibitor 1 | Fenneropenaeus chinensis/35 | 4E-42 |
| Unigene38364 | F/1109 | Y | Kazal-type serine proteinase inhibitor | Pacifastacus leniusculus/48 | 2E-18 |
| Unigene40624 | F/420 | N | Kunitz-type serine protease inhibitor 3 | Chlorocebus sabaeus/59 | 4E-15 |
| Unigene18473 | F/546 | Y | Kunitz domain protein | Ixodes scapularis/53 | 9E-14 |
| Unigene23096 | F/764 | N | Papilin-like | Xiphophorus maculatus/34 | 4E-11 |
| Unigene40962 | N/1476 | U | Serine peptidase inhibitor, Kunitz-type 1 b | Danio rerio/36 | 8E-09 |
| Unigene42995 | F/1779 | Y | Serpin B4 | Lctidomy tridecemlineatus/33 | 1E-61 |
| Unigene17438 | F/2066 | N | Serine protease inhibitor | Cyanea capillata/66 | 2E-140 |
| Unigene18138 | F/1576 | Y | Leukocyte elastase inhibitor-like | Condylura cristata/39 | 4E-70 |
| Unigene18959 | F/1653 | Y | Proteinase inhibitor 14 serpin | Pedosphaera parvula/36 | 6E-69 |
| Unigene41584 | F/1356 | Y | Serpin B4 | Lctidomys tridecemlineatus/40 | 4E-71 |
a Full-length and not full-length open reading frames (ORFs) are indicated by “F” and “N”, respectively.
b Transcripts with or without a signal peptide are indicated by “Y” and “N”, respectively, whereas transcripts in which the presence of a signal peptide was unclear are indicated by “U” (Unknown).
Phospholipases: We identified many members and isoforms of phospholipase A2s (PLA2s) and phospholipase D (PLD) in the C. capillata tentacle transcriptome. PLA2s are the most common type of phospholipases identified in the venom of various toxic animals, such as snakes, scorpions and ants [27–30]. High levels of PLA2 activity have also been described in the tentacles of scyphozoan and cubozoan species [31]. PLA2s were also recently reported to be one of the most abundant toxins in the venom of the jellyfish S. meleagris [9]. In this study, we identified 10 unique transcripts encoding PLA2s in the tentacle of C. capillata (Table 2), and six of the 10 transcripts had clear open reading frames. Generally, hemolysis is considered to be the direct result of PLA2s and hemolysin, which can interact with ion channels, membrane proteins and membrane ion pumps. We also previously observed both in vitro and in vivo hemolysis of the tentacle extract from C. capillata [12]. Hemolytic activity is considered one of the most common biological activities of jellyfish venom [32–33]. In addition to hemolytic effects, venom PLA2s can also mediate several other toxic responses, such as cytotoxicity, cardiotoxicity, neurotoxicity, myotoxicity, edema and blood coagulation disturbance [34–35].
In addition to PLA2s, we also identified five transcripts for PLDs in the transcriptome. This is the first report of PLDs in jellyfish species. PLD has been reported to act as a dermonecrotic factor in the venom of brown spiders and plays a role in the necrotic effect and severe inflammatory response [36]. Interestingly, jellyfish venom exhibits an obvious dermonecrotic effect [37–38]. C. capillata venom can induce dermonecrotic lesions in the skins of rats and Guinea pigs [39]. Thus, the presence of PLDs in the C. capillata tentacle transcriptome is a significant finding that may help to advance the discovery of the dermonecrotic mechanism of jellyfish venom.
Metalloproteases: Two types of metalloproteases were identified: matrix metalloproteinases and astacin-like metalloproteases (Table 2). The identification of these diverse metalloprotease transcripts is not surprising, because metalloproteases have been described as the central toxic component in various venomous animals [40–42]. In snake venoms, metalloprotease toxins are predominantly responsible for local pathological effects, such as tissue damage, necrosis and hemorrhage [43]. In this study, 11 unique transcripts encoding matrix metalloproteinases families, including matrix metalloproteinase-14, -1, -9, -24 and -25, were identified, and three of the 11 transcripts aligned best with metalloproteinase-14. This identification supports a previous report describing metalloprotease-14 as the main venom-derived proteins in the jellyfish Nemopilema nomurai [44].
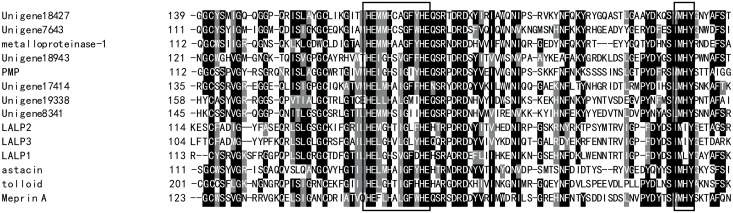
In addition, six transcripts for the astacin-like metalloproteases were also identified. Astacin-like metalloproteases have been reported to be the main components of Loxosceles toxins [45]. Multiple alignment analysis revealed that all of these deduced amino acid sequences contain the astacin family signatures HEXXHXXGXXHE (enzymatic catalytic domain) and MXY (Met-turn), similar to the astacin-like toxins in the Loxosceles genus and other astacin family-related members (Fig 3). Loxosceles astacin toxins can degrade extracellular matrix proteins and assist the spread of other venom components. However, the study of astacin-like metalloproteases in jellyfish venom remains in its infancy. Scyphozoan jellyfish venom has significant gelatinolytic, caseinolytic, and fibrinolytic activities [46]. Astacin-like metalloproteases were also recently identified as an important component of the venoms of S. meleagris and N. nomurai jellyfish [9,44]. Based on these findings, these astacin-like metalloproteases identified from C. capillata likely also play an important role in the pathological processes of C. capillata envenomation.
Fig 3. Multiple sequence alignment of astacin-family enzymes.
The deduced amino acid sequences of unigenes 18427, 7643, 18943, 17414, 19338 and 8341 were aligned with Loxosceles intermedia LALP 1, 2 and 3 (A0FKN6, C9D7R2 and C9D7R3, respectively), metalloproteinase 1 from H. vulgaris (AAA92361), astacin from A. astacus (CAA64981), PMP1 from Podocoryna carnea (CAA06314), tolloid from Drosophila melanogaster(AAF56329) and Meprin A from Homo sapiens (AAI36560). Sequence accession numbers are shown in brackets. The characteristic astacin family signatures are boxed. Black and grey indicate amino acids that are identical or highly conserved, respectively, across all aligned sequences. Absent amino acids are indicated by dashes (-) to improve the alignment.
Serine proteases: Serine proteases are the best-characterized venom component. In this study, we identified 15 unique sequences of this family (Table 2), but only six were confirmed to have a complete ORF due to their high molecular weights. Among the 15 transcripts, six sequences were significantly homologous to serine proteases from the jellyfish Aurelia aurita. Serine proteases have been described in the venoms of snakes, spiders and scorpions [28,36,47]. Generally, this toxin family affects a wide array of physiological functions, including platelet aggregation and fibrinolytic pathways [48]. They can also play a role in post-translation modification and spreading other toxins [49]. However, the exact role of serine proteases in jellyfish envenomation remains to be clarified.
Serine protease inhibitors: Several serine protease inhibitors were identified in this study, including Kazal-type (4 transcripts) and Kunitz-type (KUNs) (4 transcripts). The Serpin family (5 transcripts) was also identified (Table 2). Serine protease inhibitors have been widely found in the venoms of many well-known toxic animals [50–52]. However, few toxins of this type have been reported in jellyfish venom. Among this toxin family, Kunitz-type inhibitors have been commonly observed in snake venoms and inhibit both serine proteases and calcium ion channels. They are characterized by three disulfide bonds belonging to a highly conserved motif of C-8X-C-15X-C-4X-YGGC-12X-C-3X-C [50]. In this study, multiple alignment analysis demonstrated that compared with other typical Kunitz-type inhibitors, most of these identified Kunitz-type inhibitor transcripts possess a native Kunitz architecture (Fig 4). The function of serine protease inhibitors in the various venoms has been suggested to be primarily related to the protection of toxin integrity. Additionally, this toxin family may play a role in various physiological processes, such as blood coagulation, fibrinolysis and host defense. However, whether serine protease inhibitors in jellyfish venoms have a similar function remains to be explored, and further investigations are needed.
Fig 4. Alignment of the conserved sequence motifs of Kunitz-type inhibitors.
The deduced amino acid sequences of unigenes 18473, 23096, 40962 and 40624 were aligned with known Kunitz-type inhibitors, including Ixodes scapularis Kunitz-domain protein (XP_002435213), Crassostrea gigas putative Kunitz-type proteinase inhibitor (EKC39386), Latrodectus hesperus Kunitz-like protease inhibitor (ADV40132), Astyanax mexicanus Kunitz-type protease inhibitor 1-like (XP_007252976) and Danio rerio Kunitz-type protease inhibitor 1 (AAI63937). The six highly conserved cysteine residues are indicated by asterisks.
2. Other possible venom components
In addition to the well-known types of venom toxin families described above, some transcripts resembled putative molecules with potential toxic activities reported in other venomous animals. Moreover, most of these sequences have not yet been described in jellyfish species. Thus, these transcripts were classified as ‘other possible venom components’, and the main features of these molecules are presented in Table 3.
Table 3. Additional putative toxin transcripts identified by similarity searches.
| Sequence ID | ORF a /Length(bp) | Signal peptide b | Identification (best matched species) | E-value | Possible venom function |
|---|---|---|---|---|---|
| Unigene6213 | N/307 | U | Toxin TX2(Aurelia aurita) | 2E-06 | Hemolytic activity and pore-forming action |
| Unigene8172 | F/2294 | N | Venom dipeptidyl peptidase 4 isoform X2 (Nasonia vitripennis) | 2E-109 | Endopeptidases |
| Unigene41308 | F/961 | N | Venom toxin-like peptide-6 (Mesobuthus eupeus) | 4E-07 | Unknown |
| Unigene41678 | N/566 | N | Plancitoxin-1-like (Hydra vulgaris) | 5E-59 | Potently hepatotoxic |
| Unigene33521 | N/523 | Y | Plancitoxin-1-like (Hydra vulgaris) | 9E-17 | Potently hepatotoxic |
| Unigene4165 | N/327 | U | Venom protein Ci-120 (Chelonus inanitus) | 6E-30 | Proteoglycan metabolism |
| Unigene49242 | N/330 | N | Venom protein Ci-80a (Chelonus inanitus) | 4E-42 | Peptidase |
| Unigene17029 | F/734 | N | Translationally controlled tumor protein homology, TCTP (Ixodes scapularis) | 3E-23 | Histamine releasing factor |
| Unigene7297 | F/1784 | N | Angiotensin-converting enzyme-like (Hydra vulgaris) | 1E-178 | Releasing vasoconstrictive peptides |
| Unigene29942 | N/231 | U | Endothelin-converting enzyme 1-like (Hydra vulgaris) | 5E-06 | Releasing vasoconstrictive peptides |
| Unigene25995 | N/313 | U | Neprilysin-like (Myotis davidii) | 5E-30 | Endopeptidases and potent neurotoxicity |
| Unigene1506 | N/672 | U | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2-like (Hydra vulgaris) | 6E-22 | Hemostatic disturbance |
| Unigene21452 | N/1661 | N | Ectonucleotide pyrophosphatase/phosphodiesterase family member 5 (Zonotrichia albicollis) | 3E-101 | Hemostatic disturbance |
| Unigene28395 | N/221 | U | Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 (Columba livia) | 4E-05 | Hemostatic disturbance |
| Unigene39268 | N/1821 | U | Ectonucleotide pyrophosphatase/phosphodiesterase family member 4-like (Hydra vulgaris) | 6E-73 | Hemostatic disturbance |
| Unigene17070 | F/1758 | Y | Vascular endothelial growth factor A-like (Acyrthosiphon pisum) | 7E-09 | Inducing vasodilation and increasing vascular permeability |
| Unigene17673 | F/1989 | Y | Vascular endothelial growth factor A-like (Hydra vulgaris) | 2E-17 | Inducing vasodilation and increasing vascular permeability |
| Unigene39767 | F/1870 | N | Vascular endothelial growth factor toxin (Lepeophtheirus salmonis) | 2E-06 | Unknown |
| Unigene17310 | F/1239 | Y | Vascular endothelial growth factor D (Crassostrea gigas) | 3E-05 | Unknown |
| Unigene40990 | N/498 | U | Lysosomal acid lipase/cholesteryl ester hydrolase-like (Hydra vulgaris) | 2E-48 | Unknown |
| Unigene17402 | F/2567 | Y | Lysosomal acid lipase/cholesteryl ester hydrolase-like (Hydra vulgaris) | 1E-160 | Unknown |
| Unigene17303 | F/1764 | Y | Lysosomal acid lipase/cholesteryl ester hydrolase-like (Hydra vulgaris) | 5E-147 | Unknown |
| Unigene9684 | N/406 | U | Alkaline phosphatase D domain containing protein (Acanthamoeba castllanii) | 2E-36 | Unknown |
| Unigene41552 | F/1246 | N | Dipeptidyl peptidase 3-like (Saccoglossus kowalevskii) | 3E-97 | Inducing hypotension |
| Unigene8247 | F/1977 | N | Ectonucleoside triphosphate diphosphohydrolase 1 (Falco peregrinus) | 7E-65 | Potential inhibitor of platelet aggregation |
| Unigene19363 | F/1872 | N | Ectonucleoside triphosphate diphosphohydrolase6 (Danio rerio) | 5E-94 | Potential inhibitor of platelet aggregation |
a Full-length and not full-length open reading frames (ORFs) are indicated by “F” and “N”, respectively.
b Transcripts with or without a signal peptide are indicated by “Y” and “N”, respectively; transcripts in which the presence of a signal peptide was unclear are indicated by “U” (Unknown).
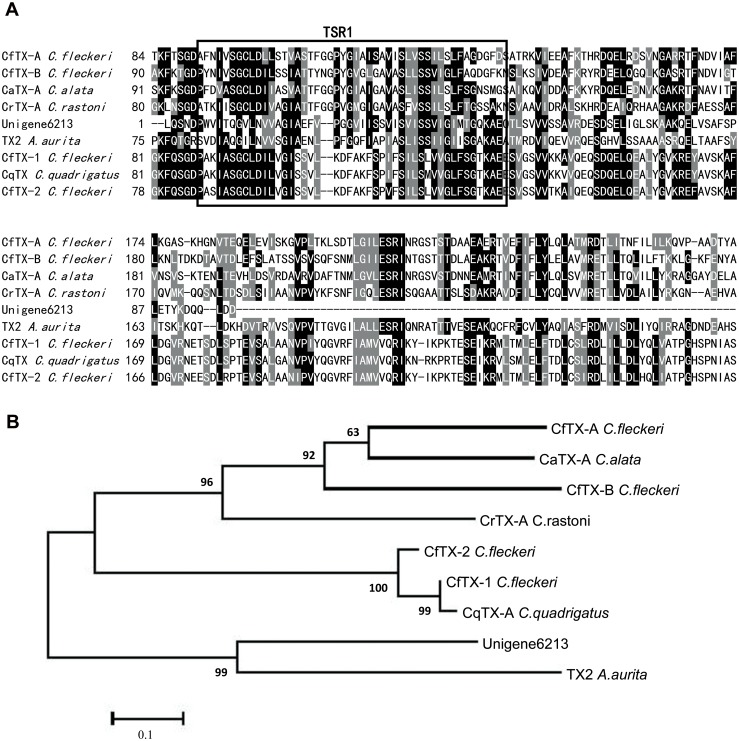
In this study, we identified a transcript (Unigene6213) with significant similarity to the N-terminal sequences of a family of known jellyfish toxins, including TX2 isolated from Aurelia aurita, CfTX-1,2 and CfTX-A,B from Chironex fleckeri, CqTX-A from C. quadrigatus, CrTX-A from C. rastoni and CaTX-A from C. alata [53–54]. The multiple sequence alignment and phylogenetic analysis of these similar jellyfish toxins are presented in Fig 5. This toxin family is primarily associated with potent hemolytic activity and pore-forming action [53,55]. As shown in Fig 5, a putative transmembrane spanning region (TSR1) that is highly conserved in the N-terminal sequences of this toxin family is also present in the predicted amino acid range (6–50) of Unigene6213. The presence of the transmembrane spanning region in these jellyfish toxins is very important because it may play a role in the pore-forming process [50]. Therefore, Unigene6213 is likely a new member of this family of jellyfish toxins, even though it does not contain a full-length ORF. Further research is needed to determine the full-length sequence, structure and biological role of this putative toxin in C. capillata.
Fig 5.
(A) Multiple sequence alignment of the amino acid sequence of unigene 6213 with other known CfTX-like cnidarian toxins. The abbreviations and sequence accession numbers for the aligned sequences are as follows: Aurelia aurita TX2 (AFK76349), Chiropsalmus quadrigatus CqTX-A (BAB82520), Carybdea rastonii CrTX-A (BAB12728), Carybdea alata CaTX-A (BAB12727), and Chironex fleckeri CfTX-A, CfTX-B, CfTX-1 and CfTX-2 (AFQ00676, AFQ00677, ABS30940 and ABS30941, respectively). The putative highly conserved transmembrane spanning region (TSR1) in the N-terminal sequences of this toxin family is boxed. (B) Phylogenetic relationships of the CfTX-like cnidarian toxins.
In addition to the homologues of known jellyfish toxins, we identified a number of toxin-like transcripts similar to some unique venom components isolated from other venomous animals. Two putative toxin transcripts with high similarity to plancitoxin-1-like, a gene closely related to plancitoxin-1 from the crown-of-thorns starfish Acanthaster planci, were also identified (Fig 6). Plancitoxin-1 is one of the few known toxic DNase II proteins and exhibits hepatotoxic properties in Acanthaster planci, the species from which this toxin was first described [56–57]. Additionally, plancitoxin-1 toxin was also previously identified in several nemertean species and the jellyfish S. meleagris [9,58]. A single transcript with a full-length ORF was identified that encoded a peptide exhibiting high identity with venom toxin-like peptide-6 (GenBank accession number ABR21046.1) and venom protein 2 (ABR21036.1), which were both isolated from the venom of the scorpion Mesobuthus eupeus (Fig 7). The sequence of the transcript comprises 71 amino acids and displays 45% and 44% identity, respectively, with these two scorpion toxins. However, the functions of these venom toxins in envenomation remain unknown. Two transcripts similar to Ci-120 and Ci-80a were also characterized (S4 Fig). Ci-120 and Ci-80a are both venom proteins identified from the parasitic wasp Chelonus inanitus [40]. The Ci-120 protein, which may affect proteoglycan metabolism, exhibits high sequence similarity to alpha-N-acetylglucosaminidase. Ci-80a is a member of the papain family, and its role in envenomation remains to be determined. In addition, we identified a transcript encoding a protein similar to venom dipeptidyl peptidase 4(DPP4) isoform X2 (S5 Fig). DPP4 has been widely identified in snake venoms [59], suggesting an important role in envenomation. Venom DPP4 is involved in the processing of venom peptides and may also play a role in cardiovascular disorders caused by the venom [60–61].
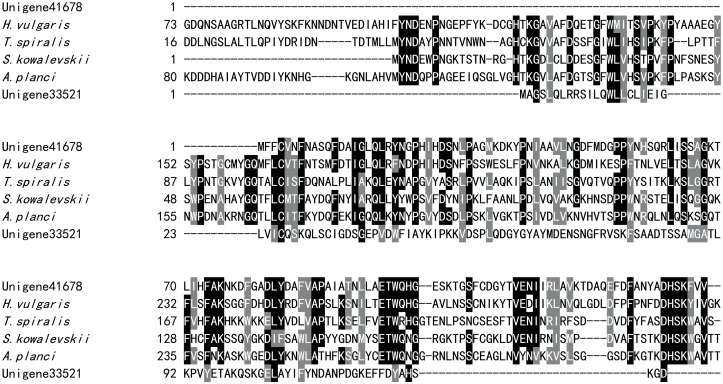
Fig 6. Alignment of plancitoxin-1-like proteins.
The aligned sequences are as follows: Acanthaster planci plancitoxin 1 (BAD12432), Saccoglossus kowalevskii plancitoxin 1-like (XP_006823536), T. spiralis plancitoxin1-like protein (AHM10158) and Hydra vulgaris plancitoxin 1-like (XP_004209299). Black and gray indicate amino acids that are identical or highly conserved, respectively, across all aligned sequences.
Fig 7. Alignment of the amino acid sequence of unigene 41308 with Mesobuthus eupeus venom toxin-like peptide-6 and venom protein 2.
The GenBank accession numbers for venom toxin-like peptide-6 and venom protein 2 are ABR21046 and ABR21036, representatively. Black and gray indicate amino acids that are identical or highly conserved across all aligned sequences, respectively.
In addition to these toxin-like candidates, some other putative venom components were also identified in the transcriptome of C. capillata tentacle by BLAST searches. Translationally controlled tumor protein (TCTP) has been identified in the gland secretions of ticks and mites [62–63]. TCTP is also called histamine-releasing factor (HRF) and has been reported as a venom toxin in several spider species [36]. In our transcriptome of C. capillata, a transcript exhibiting significant similarity with TCTP (or HRF) was identified (Fig 8). TCTP induces histamine release in basophil leukocytes and is one of the molecules responsible for histamine-associated symptoms. Interestingly, local tissue edema, the main effect of histamine release, always occurs after jellyfish stinging. Therefore, we suggest that TCTP in C. capillata tentacle may play a role in histamine release during jellyfish envenomation.
Fig 8. Alignment of translationally controlled tumor protein (or histamine-releasing factor).
The aligned sequences are as follows: Rhipicephalus microplus histamine release factor (AAY67698), Amblyomma americanum histamine release factor (AAY67700), Latrodectus hesperus histamine release factor (ADV40083), Saccoglossus kowalevskii TCTP (XP_002740226) and Ixodes scapularis TCTP (AAY66972). Black and gray indicate amino acids that are identical or highly conserved across all aligned sequences, respectively.
We also identified a transcript exhibiting significant similarity to various angiotensin-converting enzyme-like (ACE-like) proteins and a transcript similar to endothelin-converting enzyme 1-like (ECE 1-like) proteins (S6 Fig). ACE and ECE are both metalloproteases. ACE plays a role in converting angiotensin I to angiotensin II, which can induce vasoconstriction and elevate blood pressure. ECE is involved in the processing of ET-1, a potent vasoconstrictor, from its inactive precursor. Expression of ACE (or ACE-like) and ECE proteins in the venoms of several species of cone snails and wasps has been reported [40,64]. However, this is the first report of the presence of these molecules in jellyfish. Cardiovascular toxicity is the major bioactivity of jellyfish venoms. We previously demonstrate that C. capillata venom induces marked vasoconstriction [65]. Therefore, the identification of ACE and ECE-1 in C. capillata tentacle strongly suggests that these molecules might contribute to the disruption of cardiovascular function caused by jellyfish venoms.
Additionally, a transcript that exhibited significant sequence similarity to neprilysin (or neprilysin-like protein) was identified (Fig 9). Neprilysin is a zinc-dependent metalloprotease with affinity for a broad range of physiological targets, including natriuretic, vasodilatory and neuro peptides [66]. Notably, neprilysin has been identified in the venoms of several species of spiders and snakes and is likely correlated with neurotoxicity, potentially due to its activity in the inactivation of peptide transmitters and their modulators [66]. Interestingly, jellyfish venom comprises neurotoxins that immediately paralyze prey. We have also observed this neurotoxic activity in C. capillata venom. Therefore, neprilysin (or neprilysin-like protein) may also play a role in C. capillata envenomation, but its precise role requires further study.
Fig 9. Multiple sequence alignment of the amino acid sequence of unigene 25995 with other known neprilysin or neprilysin-like proteins.
The aligned sequences are as follows: Oryctolagus cuniculus neprilysin (NP_001095155), Equus przewalskii neprilysin isoform X2 (XP_008519227), Loxodonta africana neprilysin (XP_003416187), Cricetulus griseus neprilysin (XP_007625698), Peromyscus maniculatus bairdii neprilysin (XP_006972674), Myotis lucifugus neprilysin isoform X2 (XP_006090392), Myotis brandtii neprilysin (EPQ12789) and Myotis davidii neprilysin-like (XP_006769958).
We also identified four transcripts exhibiting similarity to the ectonucleotide pyrophosphatase/phosphodiesterase family (Table 3). Phosphodiesterase activity has been described in many snake venoms [67–68]. A phosphodiesterase family member has been identified as a toxin in the jellyfish S. meleagris [9]. However, the contributions of these proteins to the poisoning mechanism are poorly understood. Phosphodiesterases might exhibit inhibitory activity on ADP-induced platelet aggregation and contribute to hemostatic disturbances. Therefore, the discovery of an ectonucleotide pyrophosphatase/phosphodiesterase family in C. capillata tentacle implies that this superfamily might be constituents of jellyfish venom and their roles in envenomation deserve further investigation.
In addition, sequences with similarity to vascular endothelial growth factors (VEGFs) were also identified (Table 3). VEGFs, which contain several subclasses, have been described as minor venom constituents in the venom glands of several snake species [28–29]. Among these transcripts, two sequences exhibited similarity to VEGF-A, which has been reported to induce vasodilation and potently increase vascular permeability. It can also promote tachycardia and hypotension and diminish cardiac output [69]. We observed these symptoms when C. capillata venom was injected in rats. Therefore, this family might also play a role in C. capillata envenomation.
In addition to the molecules mentioned above, we also identified several other possible venom components, including lysosomal acid lipases (LALs), alkaline phosphatase, dipeptidyl peptidase 3 and ectonucleoside triphosphate diphosphohydrolase (Table 3). These transcripts have also been previously reported as atypical venom components in some venomous animals [26,67], and none of these transcripts have been previously described in jellyfish. However, their contributions to the bioactivities of jellyfish venom require experimental verification.
Transcripts relevant to degenerative diseases
In addition to putative toxin transcripts, disease-related transcripts were also identified (data not shown). Among them, the most surprising were transcripts relevant to three nervous system diseases, Huntington’s disease (HD), Alzheimer's disease (AD) and Parkinson's disease (PD).
We characterized 476 unigenes involved in the pathway of Huntington’s disease, the fourth largest group in KEGG annotation. HD is an inherited neurodegenerative disease [70]. In this study, transcripts homologous to genes closely related to HD, including Huntingtin, Huntingtin-interacting protein 1 and Huntingtin-interacting protein K, were all identified (S3 Table). Sequence analysis revealed that the Huntingtin genes were highly conserved in various species (Fig 10). We also identified two transcripts encoding proteins similar to presenilin-2 (S7 Fig). Mutations in the genes encoding presenilin-1 and presenilin-2 are responsible for early-onset autosomal dominant Alzheimer's disease, the most frequent degenerative dementia among the elderly [71]. There are no effective treatments for HD or AD. Transcripts encoding proteins homologous to Parkinson disease-related proteins, including Parkinson protein 7 (protein DJ-1) and Parkinson disease 7 domain-containing protein 1, were also identified in the transcriptome of C. capillata tentacle (S3 Table).
Fig 10. Multiple sequence alignment of the amino acid sequence of unigenes 5396 and 5817 with those of other known huntingtin proteins.
The aligned sequences are as follows: Hydra vulgaris huntingtin-like (XP_004206850), Saccoglossus kowalevskii huntingtin (XP_006822985), Poecilia reticulate huntingtin isoform X 5 (XP_008406720), Pongo abelii huntingtin (XP_002814571), Oryctolagus cuniculus huntingtin (XP_008248081), Python bivittatus huntingtin (XP_007432531) and Branchiostoma floridae huntingtin (ABP04240).
This is the first description of the expression of these nervous system disease-associated genes in jellyfish species. Interestingly, in a previous study of the genome of Hydra magnipapillata, genes associated with nervous system diseases, including HD and AD, were also identified [72]. Therefore, these results strongly suggest that degenerative disease-related genes are highly conserved from invertebrates to vertebrates. These findings also suggest the great potentials of these marine invertebrates as models in the study of degenerative diseases.
Conclusions
This study contributes to a more comprehensive view of the origin and functional diversity of venom proteins in jellyfish, provides a foundation and valuable resource for further investigations of bioactive components, and promotes the general development of jellyfish resources.
Supporting Information
(A) Length distribution of contigs. (B) Length distribution of unigenes. The number under the x-axis indicates the length range (e.g., ‘300’ indicates a length range of (200, 300), whereas‘>3000’ indicates a length range longer than 3,000 bp.). The y-axis is in logarithmic scale. The number above each bar indicates the total number of sequences falling in this length range.
(TIF)
The GO categories shown in the x-axis were grouped into three main categories: biological process, cellular component and molecular function. The right y-axis indicates the number of annotated unigenes in each sub-category, and the left y-axis indicates the percentage of total unigenes in that sub-category.
(TIF)
6,202 unigenes (12.3% of the total) were annotated and classified into 25 COG functional categories.
(TIF)
(A) Alignment of the amino acid sequence of unigene 4165 with parasitic wasp Chelonus inanitus venom protein Ci-120 (CBM69278). (B) Alignment of the amino acid sequence of unigene 49242 with parasitic wasp Chelonus inanitus venom protein Ci-80a (CBM69275).
(TIF)
The aligned sequences are as follows: Nasonia vitripennis venom dipeptidyl peptidase 4 isoform X2 (XP_008202161), Ceratosolen solmsi marchali venom dipeptidyl peptidase 4 isoform X2 (XP_011494919) and Cerapachys biroi venom dipeptidyl peptidase 4 isoform X2 (XP_011336230). Black and gray indicate amino acids that are identical or highly conserved across all aligned sequences, respectively.
(TIF)
(A) Multiple sequence alignment of the amino acid sequence of unigene 7297 with other known ACE and ACE-like proteins. The aligned sequences are as follows: Strongylocentrotus purpuratus ACE-like isoform 1(XP_003724612), Hydra vulgaris ACE-like (XP_004208490), Colius striatus ACE (XP_010200493), Acanthisitta chloris ACE (XP_009082259), Notothenia coriiceps ACE (XP_010793898), Pseudopodoces humilis ACE (XP_005532492) and Homo sapiens ACE 1(EAW94314). (B) Multiple sequence alignment of the amino acid sequence of unigene 29942 with other known ECE and ECE-like proteins. The aligned sequences are as follows: Hydra vulgaris ECE 1-like (XP_004211607), Apis florea ECE 2-like (XP_003693348), Hydra vulgaris ECE (AAD46624) and Metaseiulus occidentalis ECE 2-like (XP_003743930). Black and gray indicate amino acids that are identical or highly conserved across all aligned sequences, respectively.
(TIF)
The aligned sequences are as follows: Xenopus tropicalis presenilin 2 (NP_001017181), Lepisosteus oculatus presenilin-2-like (XP_006638781), Octodon degus presenilin-2-like isoform X2 (XP_004626898), Takifugu rubripes presenilin-2-like (XP_003972298), Stegastes partitus presenilin-2 (XP_008297105), Taeniopygia guttata presenilin-2 (XP_002197681) and Callorhinchus milii presenilin-2 (XP_007891888).
(TIF)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We thank the Beijing Genomics Institute (BGI; Shenzhen, China) for assistance with sequencing and Fang Wei at Second Military Medical University for editing this manuscript.
Data Availability
All the original sequencing data of clean reads' files are available from the NCBI Sequence Read Archive (SRA) database (accession number(s)SRP056566).
Funding Statement
This work was supported by the Young Scientists Fund of National Natural Science Foundation of China (41306136), the Natural Science Foundation of Shanghai Municipal Government (12ZR1437000) and the National High Technology Research and Development Program of China (863 Program) (2013AA092904). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Learmont SA. Chironex fleckeri (box jellyfish) envenomation: a case study. Australasian Emergency Nursing Journal 2006. May 9; 9(2). [Google Scholar]
- 2. Kim E, Lee S, Kim JS, Yoon WD, Lim D, Hart AJ, et al. Cardiovascular effects of Nemopilema nomurai (Scyphozoa: Rhizostomeae) jellyfish venom in rats. Toxicol Lett 2006. December 15; 167(3). [DOI] [PubMed] [Google Scholar]
- 3. Tibballs J. Australian venomous jellyfish, envenomation syndromes, toxins and therapy. Toxicon 2006. December 1; 48(7). [DOI] [PubMed] [Google Scholar]
- 4. Yu H, Li C, Li R, Xing R, Liu S, Li P. Factors influencing hemolytic activity of venom from the jellyfish Rhopilema esculentum Kishinouye. Food Chem Toxicol 2007. July; 45(7). [DOI] [PubMed] [Google Scholar]
- 5. Harada K, Maeda T, Hasegawa Y, Tokunaga T, Ogawa S, Fukuda K, et al. Antioxidant activity of the giant jellyfish Nemopilema nomurai measured by the oxygen radical absorbance capacity and hydroxyl radical averting capacity methods. Mol Med Rep 2011. Sep-Oct; 4(5). [DOI] [PubMed] [Google Scholar]
- 6. Ruan Z, Liu G, Guo Y, Zhou Y, Wang Q, Chang Y, et al. First report of a thioredoxin homologue in jellyfish: molecular cloning, expression and antioxidant activity of CcTrx1 from Cyanea capillata. PLoS One 2014. May 13; 9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruan Z, Liu G, Wang B, Zhou Y, Lu J, Wang Q, et al. First report of a peroxiredoxin homologue in jellyfish: molecular cloning, expression and functional characterization of CcPrx4 from Cyanea capillata. Mar Drugs 2014. January 9;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sugahara T, Ueno M, Goto Y, Shiraishi R, Doi M, Akiyama K, et al. Immunostimulation effect of jellyfish collagen. Biosci Biotechnol Biochem 2006. September;70(9). [DOI] [PubMed] [Google Scholar]
- 9. Li R, Yu H, Xue W, Yue Y, Liu S, Xing R, et al. Jellyfish venomics and venom gland transcriptomics analysis of Stomolophus meleagris to reveal the toxins associated with sting. J Proteomics 2014. June 25;106. [DOI] [PubMed] [Google Scholar]
- 10. Brinkman DL, Jia X, Potriquet J, Kumar D, Dash D, Kvaskoff D, et al. Transcriptome and venom proteome of the box jellyfish Chironex fleckeri . BMC Genomics 2015. May 27;16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brekhman V, Malik A, Haas B, Sher N, Lotan T. Transcriptome profiling of the dynamic life cycle of the scypohozoan jellyfish Aurelia aurita . BMC Genomics 2015. February 14;16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiao L, Zhang J, Wang QQ, He Q, Liu SH, Li Y, et al. In vitro and in vivo haemolytic studies of tentacle-only extract from jellyfish Cyanea capillata . Toxicol In Vitro 2010. June;24(4). [DOI] [PubMed] [Google Scholar]
- 13. Beilei W, Lin Z, Qian H, Qianqian W, Tao W, Jia L, et al. Direct cardiac toxicity of the tentacle-only extract from the jellyfish Cyanea capillata demonstrated in isolated rat heart. J Cardiovasc Pharmacol 2012. April;59(4). [DOI] [PubMed] [Google Scholar]
- 14. Liang X, Beilei W, Ying L, Qianqian W, Sihua L, Yang W, et al. Cardiovascular effect is independent of hemolytic toxicity of tentacle-only extract from the jellyfish Cyanea capillata. PLoS One 2012;7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Q, Sun P, Zhou X, Lei C. Characterization of head transcriptome and analysis of gene expression involved in caste differentiation and aggression in Odontotermes formosanus (Shiraki). PLoS One 2012;7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hao DC, Ge GB, Xiao PG, Zhang YY, Yang L. The First Insight into the Tissue SpecificTaxus Transcriptome via Illumina Second Generation Sequencing. PLOS ONE 2011;6(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011. May 15;29(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 1999. [PubMed] [Google Scholar]
- 19. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005. September 15; 21(18). [DOI] [PubMed] [Google Scholar]
- 20. Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 2006. July 1;34(Web Server issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen GM, Dou W, Niu JZ, Jiang HB, Yang WJ, Jia FX, et al. Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis). PLoS One 2011. December 15;6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang D, Fu Y, Wu X, Xie Y, Nie H, Chen L, et al. Annotation of the transcriptome from Taenia pisiformis and its comparative analysis with three Taeniidae species. PLoS One 2012. April 13; 7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tao X, Gu YH, Wang HY, Zheng W, Li X, Zhao CW, et al. Digital gene expression analysis based on integrated de novo transcriptome assembly of sweet potato [Ipomoea batatas (L.) Lam]. PLoS One 2012. April 27;7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Toom PM, Chan DS. Enzymatic activities of venom from the jellyfish Stomolophus meleagris . Comp Biochem Physiol B 1972. October 15;43(2). [DOI] [PubMed] [Google Scholar]
- 25. Nagai H, Takuwa K, Nakao M, Ito E, Miyake M, Noda M, et al. Novel proteinaceous toxins from the box jellyfish (sea wasp) Carybdea rastoni. Biochem Biophys Res Commun 2000. August 28;275(2). [DOI] [PubMed] [Google Scholar]
- 26. Turk T, Kem WR. The phylum Cnidaria and investigations of its toxins and venoms until 1990. Toxicon 2009. December 15;54(8). [DOI] [PubMed] [Google Scholar]
- 27. Schwartz EF, Diego-Garcia E, Rodriguez DLVR, Possani LD. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genomics 2007. May 16;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casewell NR, Harrison RA, Wuster W, Wagstaff SC. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 2009. November 30;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rokyta DR, Lemmon AR, Margres MJ, Aronow K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genomics 2012. July 16;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres AF, Huang C, Chong CM, Leung SW, Prieto-da-Silva AR, Havt A, et al. Transcriptome analysis in venom gland of the predatory giant ant Dinoponera quadriceps: insights into the polypeptide toxin arsenal of hymenopterans. PLoS One 2014. January 31;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nevalainen TJ, Peuravuori HJ, Quinn RJ, Llewellyn LE, Benzie JA, Fenner PJ, et al. Phospholipase A2 in cnidaria. Comp Biochem Physiol B Biochem Mol Biol 2004. December;139(4). [DOI] [PubMed] [Google Scholar]
- 32. Bailey PM, Bakker AJ, Seymour JE, Wilce JA. A functional comparison of the venom of three Australian jellyfish—Chironex fleckeri, Chiropsalmus sp., and Carybdea xaymacana—on cytosolic Ca2+, haemolysis and Artemia sp. lethality. Toxicon 2005. February;45(2). [DOI] [PubMed] [Google Scholar]
- 33. Marino A, Morabito R, Pizzata T, La Spada G. Effect of various factors on Pelagia noctiluca (Cnidaria, Scyphozoa) crude venom-induced haemolysis. Comp Biochem Physiol A Mol Integr Physiol 2008. September;151(1). [DOI] [PubMed] [Google Scholar]
- 34. Arni RK, Ward RJ. Phospholipase A2—a structural review. Toxicon 1996. August;34(8). [DOI] [PubMed] [Google Scholar]
- 35. Kini RM. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon 2005. June 15;45(8). [DOI] [PubMed] [Google Scholar]
- 36. Chaim OM, Trevisan-Silva D, Chaves-Moreira D, Wille AC, Ferrer VP, Matsubara FH, et al. Brown spider (Loxosceles genus) venom toxins: tools for biological purposes. Toxins (Basel) 2011. March 22;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burke WA. Cnidarians and human skin. Dermatologic Therapy volume 2002. March 1;15(1). [Google Scholar]
- 38. Uri S, Marina G, Liubov G. Severe delayed cutaneous reaction due to Mediterranean jellyfish (Rhopilema nomadica) envenomation. Contact Dermatitis 2005. May 17;52(5). [DOI] [PubMed] [Google Scholar]
- 39. Cegolon L, Heymann WC, Lange JH, Mastrangelo G. Jellyfish stings and their management: a review. Mar Drugs 2013. February 22;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vincent B, Kaeslin M, Roth T, Heller M, Poulain J, Cousserans F, et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genomics 2010. December 7;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Almeida DD, Scortecci KC, Kobashi LS, Agnez-Lima LF, Medeiros SR, Silva-Junior AA, et al. Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genomics 2012. August 1;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aird SD, Watanabe Y, Villar-Briones A, Roy MC, Terada K, Mikheyev AS. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics 2013. November 14;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gutierrez JM, Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie 2000. Sep-Oct;82(9–10). [DOI] [PubMed] [Google Scholar]
- 44. Kang C, Han DY, Park KI, Pyo MJ, Heo Y, Lee H, et al. Characterization and neutralization of Nemopilema nomurai (Scyphozoa: Rhizostomeae) jellyfish venom using polyclonal antibody. Toxicon 2014. August;86. [DOI] [PubMed] [Google Scholar]
- 45. Trevisan-Silva D, Gremski LH, Chaim OM, Da SR, Meissner GO, Mangili OC, et al. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 2010. January;92(1). doi: 10.1016/j.biochi.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 46. Lee H, Jung ES, Kang C, Yoon WD, Kim JS, Kim E. Scyphozoan jellyfish venom metalloproteinases and their role in the cytotoxicity. Toxicon 2011. September 1;58(3). [DOI] [PubMed] [Google Scholar]
- 47. Ma Y, Zhao R, He Y, Li S, Liu J, Wu Y, et al. Transcriptome analysis of the venom gland of the scorpion Scorpiops jendeki: implication for the evolution of the scorpion venom arsenal. BMC Genomics 2009. July 1;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serrano SM, Maroun RC. Snake venom serine proteinases: sequence homology vs. substrate specificity, a paradox to be solved. Toxicon 2005. June 15;45(8). [DOI] [PubMed] [Google Scholar]
- 49. Almeida FM, Pimenta AM, De Figueiredo SG, Santoro MM, Martin-Eauclaire MF, Diniz CR, et al. Enzymes with gelatinolytic activity can be found in Tityus bahiensis and Tityus serrulatus venoms. Toxicon 2002. July;40(7). [DOI] [PubMed] [Google Scholar]
- 50. Siang AS, Doley R, Vonk FJ, Kini RM. Transcriptomic analysis of the venom gland of the red-headed krait (Bungarus flaviceps) using expressed sequence tags. BMC Mol Biol 2010. March 29;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao R, Dai H, Qiu S, Li T, He Y, Ma Y, et al. SdPI, the first functionally characterized Kunitz-type trypsin inhibitor from scorpion venom. PLoS One 2011. November 8; 6(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Undheim EA, Sunagar K, Herzig V, Kely L, Low DH, Jackson TN, et al. A proteomics and transcriptomics investigation of the venom from the barychelid spider Trittame loki (brush-foot trapdoor). Toxins (Basel) 2013. December 13;5(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brinkman D, Burnell J. Identification, cloning and sequencing of two major venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 2007. November;50(6). [DOI] [PubMed] [Google Scholar]
- 54. Brinkman DL, Konstantakopoulos N, McInerney BV, Mulvenna J, Seymour JE, Isbister GK, et al. Chironex fleckeri (box jellyfish) venom proteins: expansion of a cnidarian toxin family that elicits variable cytolytic and cardiovascular effects. J Biol Chem 2014. February 21;289(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nagai H, Takuwa-Kuroda K, Nakao M, Oshiro N, Iwanaga S, Nakajima T. A novel protein toxin from the deadly box jellyfish (Sea Wasp, Habu-kurage) Chiropsalmus quadrigatus. Biosci Biotechnol Biochem 2002. January;66(1). [DOI] [PubMed] [Google Scholar]
- 56. Shiomi K, Midorikawa S, Ishida M, Nagashima Y, Nagai H. Plancitoxins, lethal factors from the crown-of-thorns starfish Acanthaster planci, are deoxyribonucleases II. Toxicon 2004. October;44(5). [DOI] [PubMed] [Google Scholar]
- 57. Ota E, Nagashima Y, Shiomi K, Sakurai T, Kojima C, Waalkes MP, et al. Caspase-independent apoptosis induced in rat liver cells by plancitoxin I, the major lethal factor from the crown-of-thorns starfish Acanthaster planci venom. Toxicon 2006. December 15;48(8). [DOI] [PubMed] [Google Scholar]
- 58. Whelan NV, Kocot KM, Santos SR, Halanych KM. Nemertean toxin genes revealed through transcriptome sequencing. Genome Biol Evol 2014. November 27;6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aird SD. Snake venom dipeptidyl peptidase IV: taxonomic distribution and quantitative variation. Comp Biochem Physiol B Biochem Mol Biol 2008. June;150(2). [DOI] [PubMed] [Google Scholar]
- 60. Lee VS, Tu WC, Jinn TR, Peng CC, Lin LJ, Tzen JT. Molecular cloning of the precursor polypeptide of mastoparan B and its putative processing enzyme, dipeptidyl peptidase IV, from the black-bellied hornet, Vespa basalis . Insect Mol Biol 2007. April;16(2). [DOI] [PubMed] [Google Scholar]
- 61. Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-like vesicles in Gloydius blomhoffii blomhoffii venom. Toxicon 2008. May;51(6). doi: 10.1016/j.toxicon.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 62. Mulenga A, Azad AF. The molecular and biological analysis of ixodid ticks histamine release factors. Exp Appl Acarol 2005. December; 37(3–4). [DOI] [PubMed] [Google Scholar]
- 63. Sade YB, Boia-Ferreira M, Gremski LH, Da SR, Gremski W, Senff-Ribeiro A, et al. Molecular cloning, heterologous expression and functional characterization of a novel translationally-controlled tumor protein (TCTP) family member from Loxosceles intermedia (brown spider) venom. Int J Biochem Cell Biol 2012. January;44(1). [DOI] [PubMed] [Google Scholar]
- 64. Safavi-Hemami H, Moller C, Mari F, Purcell AW. High molecular weight components of the injected venom of fish-hunting cone snails target the vascular system. J Proteomics 2013. October 8;91. [DOI] [PubMed] [Google Scholar]
- 65. Xiao L, He Q, Guo Y, Zhang J, Nie F, Li Y, et al. Cyanea capillata tentacle-only extract as a potential alternative of nematocyst venom: its cardiovascular toxicity and tolerance to isolation and purification procedures. Toxicon 2009. January;53(1). [DOI] [PubMed] [Google Scholar]
- 66. Turner AJ, Isaac RE, Coates D. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 2001. March;23(3). [DOI] [PubMed] [Google Scholar]
- 67. Santoro ML, Vaquero TS, Paes LA, Serrano SM. NPP-BJ, a nucleotide pyrophosphatase/phosphodiesterase from Bothrops jararaca snake venom, inhibits platelet aggregation. Toxicon 2009. September 15;54(4). [DOI] [PubMed] [Google Scholar]
- 68. Fox JW. A brief review of the scientific history of several lesser-known snake venom proteins: l-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon 2013. February;62. [DOI] [PubMed] [Google Scholar]
- 69. Yang R, Thomas GR, Bunting S, Ko A, Ferrara N, Miriuka S, et al. Effects of vascular endothelial growth factor on hemodynamics and cardiac performance. J Cardiovasc Pharmacol 2005. February;129(2). [DOI] [PubMed] [Google Scholar]
- 70. Benraiss A, Goldman SA . Cellular therapy and induced neuronal replacement for Huntington's disease. Neurotherapeutics 2011. October;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Armstrong RA. What causes alzheimer's disease? Folia Neuropathol 2013. March;51(3). [DOI] [PubMed] [Google Scholar]
- 72. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, et al. The dynamic genome of Hydra. Nature 2010. March 25;464(7288). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Length distribution of contigs. (B) Length distribution of unigenes. The number under the x-axis indicates the length range (e.g., ‘300’ indicates a length range of (200, 300), whereas‘>3000’ indicates a length range longer than 3,000 bp.). The y-axis is in logarithmic scale. The number above each bar indicates the total number of sequences falling in this length range.
(TIF)
The GO categories shown in the x-axis were grouped into three main categories: biological process, cellular component and molecular function. The right y-axis indicates the number of annotated unigenes in each sub-category, and the left y-axis indicates the percentage of total unigenes in that sub-category.
(TIF)
6,202 unigenes (12.3% of the total) were annotated and classified into 25 COG functional categories.
(TIF)
(A) Alignment of the amino acid sequence of unigene 4165 with parasitic wasp Chelonus inanitus venom protein Ci-120 (CBM69278). (B) Alignment of the amino acid sequence of unigene 49242 with parasitic wasp Chelonus inanitus venom protein Ci-80a (CBM69275).
(TIF)
The aligned sequences are as follows: Nasonia vitripennis venom dipeptidyl peptidase 4 isoform X2 (XP_008202161), Ceratosolen solmsi marchali venom dipeptidyl peptidase 4 isoform X2 (XP_011494919) and Cerapachys biroi venom dipeptidyl peptidase 4 isoform X2 (XP_011336230). Black and gray indicate amino acids that are identical or highly conserved across all aligned sequences, respectively.
(TIF)
(A) Multiple sequence alignment of the amino acid sequence of unigene 7297 with other known ACE and ACE-like proteins. The aligned sequences are as follows: Strongylocentrotus purpuratus ACE-like isoform 1(XP_003724612), Hydra vulgaris ACE-like (XP_004208490), Colius striatus ACE (XP_010200493), Acanthisitta chloris ACE (XP_009082259), Notothenia coriiceps ACE (XP_010793898), Pseudopodoces humilis ACE (XP_005532492) and Homo sapiens ACE 1(EAW94314). (B) Multiple sequence alignment of the amino acid sequence of unigene 29942 with other known ECE and ECE-like proteins. The aligned sequences are as follows: Hydra vulgaris ECE 1-like (XP_004211607), Apis florea ECE 2-like (XP_003693348), Hydra vulgaris ECE (AAD46624) and Metaseiulus occidentalis ECE 2-like (XP_003743930). Black and gray indicate amino acids that are identical or highly conserved across all aligned sequences, respectively.
(TIF)
The aligned sequences are as follows: Xenopus tropicalis presenilin 2 (NP_001017181), Lepisosteus oculatus presenilin-2-like (XP_006638781), Octodon degus presenilin-2-like isoform X2 (XP_004626898), Takifugu rubripes presenilin-2-like (XP_003972298), Stegastes partitus presenilin-2 (XP_008297105), Taeniopygia guttata presenilin-2 (XP_002197681) and Callorhinchus milii presenilin-2 (XP_007891888).
(TIF)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All the original sequencing data of clean reads' files are available from the NCBI Sequence Read Archive (SRA) database (accession number(s)SRP056566).