Abstract
Background
Human rhinoviruses (HRV) are now considered major respiratory pathogens. We sought to determine whether HRV are a cause of wheezing and/or hospitalization in children < 2 years old.
Methods
A PCR assay was used to screen for HRV infection in 4 categories of children < 2 years old: 1) with symptoms of respiratory tract disease without wheezing; 2) with wheezing with or without other symptoms; 3) who were asymptomatic and; 4) who had a respiratory specimen submitted to a diagnostic laboratory. All specimens were collected between January and December, 2004. Phylogenetic analyses were performed on most HRV isolates.
Results
Twenty-eight (17%) of 165 children with symptoms of respiratory infection without wheezing; 21 (26.3%) of 80 children with wheezing; 3 (3%) of 93 asymptomatic children and 47 (23.3%) of 202 children with specimens submitted to the diagnostic laboratory tested positive for HRV. The difference between the rates of infection in the asymptomatic group and in each of the three other categories was statistically significant (p≤0.01). Among HRV-positive children with samples submitted to the diagnostic laboratory, 55% were hospitalized which was similar to that observed for respiratory syncytial virus (52.7%) among children of a similar age group and time period (P=0.85). Diverse groups of HRV were circulating during the one-year study period.
Conclusions
HRV are important pathogens among children < 2 years old and are responsible for a significant proportion of wheezing this age group. The hospitalization rates of HRV-positive children appear to be similar to that of RSV.
Keywords: Human rhinovirus, respiratory tract infection, wheezing, infants
INTRODUCTION
Respiratory tract infections are a major cause of morbidity and mortality worldwide (1). Many viral pathogens have been associated with respiratory tract disease. Among the most common of these pathogens are the human rhinoviruses (HRV) which, until recently, were thought to cause mainly mild upper respiratory tract infection. HRV were found to be the cause of 80% of cases of the common cold in adults who had self-diagnosed colds (2). HRV belong to the picornavirus family of viruses that include the polioviruses, the enteroviruses and hepatitis A virus, among others. The first HRV was isolated in 1956 (3, 4) and since that time, more than 100 genotypically and serotypically diverse HRV have been identified. New HRV continue to be discovered (5–11), suggesting that this genus may be even more diverse than previously thought. The diversity of HRV and their slow growth in cell culture hampered the study and detection of these viruses, when they were thought to be of minor clinical significance. The development of more sensitive molecular diagnostic techniques such as PCR assays(12, 13), capable of detecting multiple HRV with a single reaction, has led to a greater appreciation of the clinical significance of these viruses (for review, see (14)).
HRV are now known to be associated with serious infections of the lower respiratory tract (15–17), including bronchiolitis and pneumonia in young children (7, 15, 17–24), and these viruses may be the cause of a small, but significant proportion of influenza-like illnesses (6). Moreover, HRV have been implicated in exacerbations of asthma among school-aged children (25, 26) and adults (27). Recently, Miller et al (28) reported an association between HRV infection and hospitalization in children < 5 years old. These studies suggest that HRV infections are common and are likely the cause of a substantial proportion of respiratory tract infections.
Young children may be particularly prone to HRV infection. In a cohort of 329 children, 79% had at least one HRV symptomatic infection by the age of 2 years (29). HRV was the most common virus detected among children with acute respiratory illness in the first year of life (30). Asymptomatic HRV infection occurs in infants (31), children(32–35) and adults(32, 33, 36) raising the question as to whether HRV are a major cause of lower respiratory tract infection.
To further investigate the association between HRV and lower respiratory tract disease, including wheezing, among young children, we screened specimens obtained from children < 2 years old between January and December 2004 for HRV and compared those results with results from a group of asymptomatic controls whose specimens were obtained during the same year. We also screened specimens submitted to a clinical diagnostic laboratory for HRV. Lastly, we defined the molecular epidemiology of HRV during a 1 year period in New Haven, Connecticut.
MATERIALS AND METHODS
Specimens
Nasopharyngeal aspirates were collected from children < 2 years old between January and December 2004 according to standard protocols (37). Respiratory specimens were stored at −20°C after addition of an equal volume of “viral freezing medium” (2X Dulbecco’s modified Eagle medium, 200 mmol/L MgSO4 and 100 mmol/L HEPES [pH 7.5]). (Both magnesium and HEPES buffer has been shown to preserve the infectivity of RSV (38).) Study subjects originated from four clinical categories. Categories 1–3 were patients identified prospectively in the Yale-New Haven Hospital Primary Care Center Pediatric Clinic (PC). Category 1 included children who had evidence of upper and/or lower respiratory tract infection without wheezing (PC non-wheezing); Category 2 included children who had evidence of wheezing (PC wheezing) with or without other symptoms of respiratory tract disease or infection; Category 3 included asymptomatic children who were seen for routine well-child visits and did not have any evidence of either upper or lower respiratory tract infection on the day of enrollment or in the 15 days prior to enrollment (Asymptomatic). Category 4 included children from whom a respiratory specimen was submitted to the Yale-New Haven Hospital Clinical Virology Laboratory (Clinical Virology) for testing for respiratory viruses. Specimens from patients in Category 4 were submitted at the discretion of the medical team from the Emergency Department, inpatient wards, intensive care units and hospital-affiliated outpatient urgent care clinics. Patients were included in Category 4 only if the results of the tests on the specimens were negative for adenovirus, parainfluenza virus 1–3, influenza A and B and respiratory syncytial virus (RSV) by direct immunofluorescence assay (DFA). The specimens screened in this study were not cultured for HRV. All of the specimens from Category 4 were screened for the human bocavirus (HBoV) by PCR as previously described (39). Collection of specimens and clinical data were approved by the Yale University Human Investigation Committee. Signed informed consent was obtained for the enrolled of children in the Yale-New Haven Hospital Primary Care Center.
RNA extraction, RT-PCR
RNA was extracted using a commercially-available nucleic acid purification kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. Reverse transcriptase (RT) was performed using random hexamer primers and MuMLV RT (New England Biolabs, Beverly, MA), according to the manufacturer’s specifications. PCR amplification was performed using the PICOF3 and PICO-R3 primers (40) which amplify a 115-bp region of the picornavirus 5’-untranslated region (5’-UTR). For sequencing purposes, a larger (394 bp) portion of the picornavirus 5’-UTR was amplified using the OL-26 and OL-27 primers (41) PCR amplification was performed using HotStar Taq polymerase (Qiagen) according to the manufacturer’s instructions. PCR amplification cycles were performed as follows: 95°C for 15 minutes followed by 40 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and completed by a final extension cycle at 72°C for 10 minutes. Each set of PCR included appropriate positive and negative controls. Amplicons were detected by agarose gel electrophoresis. All primers used in this study were synthesized by the Oligonucleotide Laboratory, Department of Pathology, Yale University School of Medicine.
Sequencing and Phylogenetic Analysis
To verify the identification of HRV, amplicons were sequenced using an Applied Biosystems 3730 XL DNA Analyzer at the W. M. Keck Biotechnology Resource Laboratory, Yale University School of Medicine. Alignments and phylogenetic analysis were performed using Lasergene MegAlign Software (version 5.05; DNAstar) with use of the clustal W alignment method. GenBank accessions number assignments are pending.
Clinical data
The clinical data for patients in Categories 1–3 were collected on enrollment. The medical records of all HRV-positive children from Category 4 (Clinical Virology) were reviewed. Demographic and clinical characteristics were recorded on a standardized collection form. Specimen collection and collection of clinical data were approved by the Yale University Human Investigation Committee and collection of data was compliant with HIPAA regulations.
Statistical analysis
Fisher’s exact tests were used to determine whether the differences in percentage of HRV-positive specimens among different categories were statistically significant.
RESULTS
Overall, 447 symptomatic children (Categories 1, 2 and 4) and 93 asymptomatic controls (Category 3) were screened for HRV (Table 1, Figure 1). Of the symptomatic children screened, 245 (55%) were seen in the PC with symptoms of respiratory tract disease and 202 (45%) had specimens submitted to the Clinical Virology Laboratory. A total of 102 samples tested positive for a picornavirus by initial PCR screening with the PICO-F3 and PICO-R3 primers. Based on sequence analysis, 3 of these isolates were non-HRV picornaviruses and were not included in the analyses (see below).
Table 1.
Screening of children < 2 years of age for HRV, by Category
| Category | Description | HRV-positive specimens/ Total no. of specimens screened (%) |
|---|---|---|
| 1 | PCa Non-Wheezing | 28/165 (17.0) |
| 2 | PCa Wheezing | 21/80 (26.3) |
| 3 | Asymptomatic | 3/93 (3.2) |
| 4 | Clinical Virology | 47/202 (23.3) |
Yale-New Haven Hospital Primary Care Center Pediatric Clinic (PC)
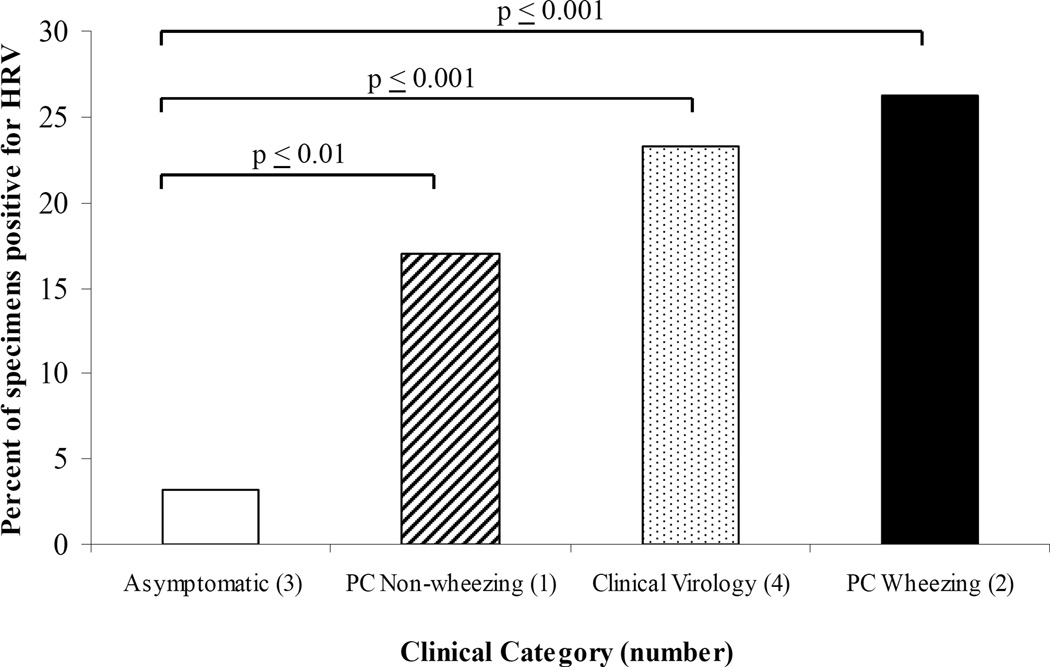
Figure 1.
Percent of HRV(+) specimens per category. P-values versus asymptomatic control are indicated for each category. Fisher’s exact test was used for each pair-wise comparison.
In all, 28 of 165 children with respiratory symptoms without wheezing (17.0%) (Category 1), 21 of 80 children with wheezing (26.3%) (Category 2), 47 of 202 samples obtained from the Clinical Virology Laboratory (23.3%) (Category 4) and 3 of 93 asymptomatic children (3.2%) (Category 3) had HRV identified in the samples. The differences among each of the three categories of symptomatic children (1, 2, 4) and the asymptomatic controls (Category 3) were statistically significant (p≤0.01). The difference in the proportion with HRV between the wheezing (Category 2) and non-wheezing (Category 1) (26.3% vs. 17.0) was not statistically significant (p≤0.09).
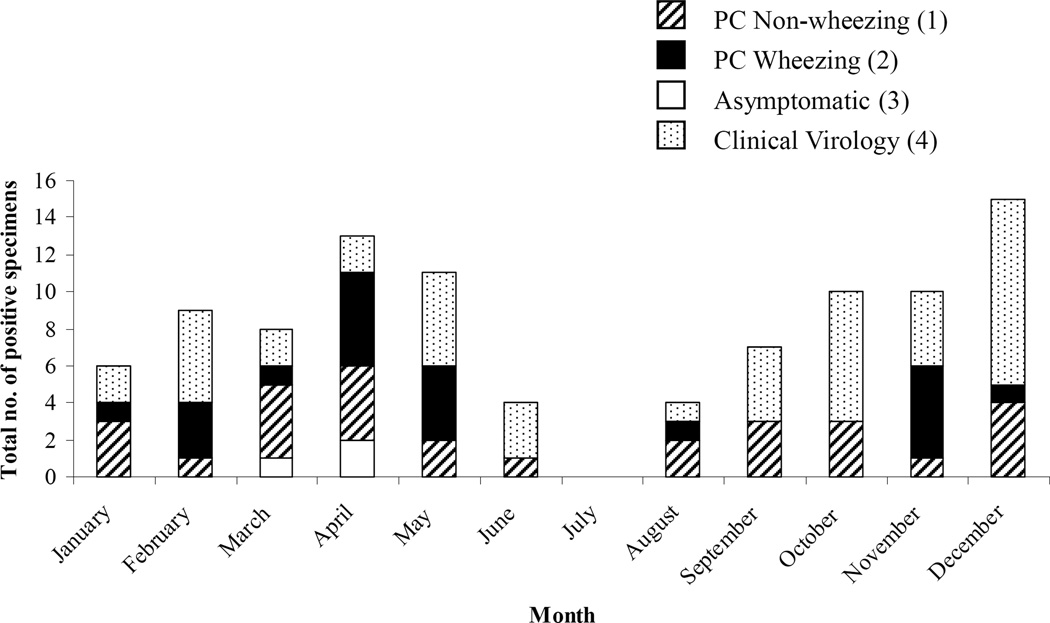
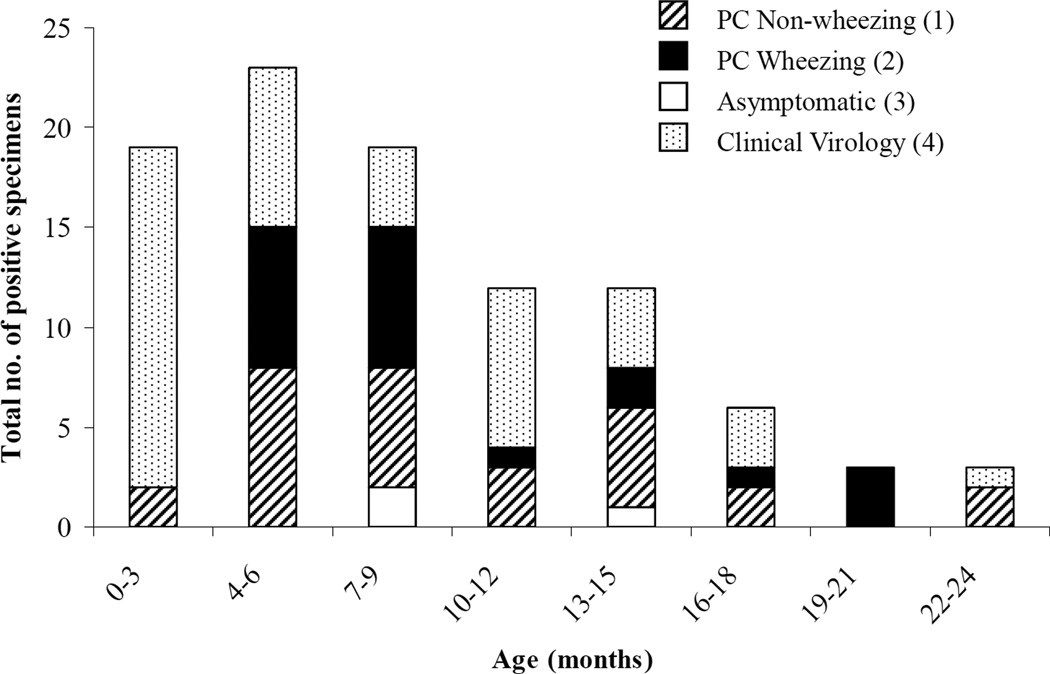
The monthly distribution of HRV-positive specimens is shown in figure 2. HRV-positive specimens were detected throughout the year with the exception of July. HRV-positive specimens were most frequently identified in the months of October–December and April–May. The three cases of asymptomatic HRV-infection occurred in March and April. The percentage of HRV-positive specimens for Categories 1, 2 and 4 (representing symptomatic children), by month, is displayed in Table 3. The distribution of HRV-positive specimens by age is shown in figure 3. All children enrolled in the study were < 2 years old. The youngest child from whom HRV was identified was 15 days old.
Figure 2.
Monthly distribution of HRV(+) specimens. The breakdown by category is indicated for each month.
Table 3.
Clinical features of HRV infection in the Clinical Virology Category
| Clinical Feature | Patients, no. (%)(a) |
|---|---|
| Fever(b) | 31 (77.5) |
| Cough | 24 (60.0) |
| Rhinorrhea | 24 (60.0) |
| Wheezing | 19 (47.5) |
| Retractions | 18 (45.0) |
| History of wheeze/asthma | 6 (15.0) |
| Hypoxia(c) | 6 (15.0) |
| Abnormal CXR (n=25)(d) | 5 (20.0) |
| Comorbidity(e) | 9 (22.5) |
| Hospitalized | 22 (55.0) |
| Male | 22 (55.0) |
Percent of total (40 patients)
Temperature > 38.0°C (Range 38.0°C–40.3°C, mean 38.7°C).
Oxygen saturation <90%.
Chest radiograph abnormalities included hyperinflation, infiltrates, peribronchial cuffing and atelectasis.
Comorbidities included prematurity, pulmonary abnormalities (hypoplastic left lung, primary pulmonary hypertension, bronchopulmonary dysplasia) tracheostomy, congenital heart abnormalities (double aortic arch, aortic stenosis, patent foramen ovale, patent ductus arteriosis), DiGeorge syndrome, Very Long Chain Acyl CoA Dehydrogenase Deficiency, and anoxic brain injury/seizure disorder.
Figure 3.
Distribution of HRV(+) individuals by age at time of specimen collection. The breakdown of each bar by study category is indicated.
The clinical features of 40 of the 47 HRV-positive children in the Clinical Virology group (Category 4) are shown in Table 3. Of the 7 children who were not included in the analyses, 2 were co-infected with HBoV and the medical records of the remaining 5 patients were not available for review. Of the 40 HRV-positive children in the Clinical Virology Category, 22 (55%) were hospitalized. In order to compare the rate of hospitalization associated with HRV with that caused by RSV in this age group, we reviewed the medical records of every 4th child < 2 years old who tested positive for RSV during the study period. Of the 93 RSV-positive children, 49 (52.7%) were hospitalized. The percentage of HRV-positive children who were hospitalized and the percentage of RSV-infected children who were hospitalized were not statistically significantly different (p=0.85).
All of the 102 picornavirus-positive specimens were screened with the HRV-specific primers and the resulting amplicons were sequenced. Of these, 87 (85.2%) yielded a full amplicon sequence that allowed for robust phylogenetic analyses (GenBank accession numbers EU699943-EU700028). Among these 87 isolates, 84 (96.6%) had sequences that matched known HRV sequences and 3 (3.4%) most closely resembled non-HRV picornaviruses: the sequence of 2 of these isolates were consistent with a human Coxsackie virus and the sequence of the third was consistent with a human enterovirus. As stated above, these 3 specimens were excluded. Of the 15 picornavirus-positive specimens that were not sequenced, it is likely that 3.4%, or about (less than) one isolate, was a non-HRV picornavirus. Therefore, these 15 specimens were considered to represent HRV infection and were included in further analyses. While it is possible, albeit unlikely, that one or more of these 15 isolates were non-HRV, we performed the statistical analyses with these 15 isolates omitted from the data set (data not shown). The results of statistical comparisons were unchanged.
Phylogenetic analysis of HRV-positive specimens indicated that a broad range of distinct genotypes of HRV circulated in New Haven throughout 2004 (figure 4, on line only). Most HRV sequences from New Haven showed close identity to previously described HRV serotypes and genotypes. Most isolates were HRV clade A, while a minority were HRV clade B or the newly identified clade C (6, 10, 11). Five isolates appeared to be distinct from other well-characterized HRV genotypes (figure 4, arrows). However, these isolates shared high sequence identity with short fragments of HRV genomes available in GenBank and may, in fact, be related to HRV genotypes which have not yet been fully characterized.
No association could be identified between particular genotypes and either specific clinical categories or clinical features within children from Category 4. Specifically, genotypes were not clustered among patients from the wheezing category or among patients with clinical features of wheezing in the clinical virology category. Likewise, there appears to be no association between particular genotypes and the month or season of specimen collection (data not shown).
DISCUSSION
We found a high rate of HRV infection (17–26%) among sick children in New Haven, CT compared with asymptomatic control patients (3%). The data support recent findings that HRV are a major cause of morbidity among young children and suggest that the detection of these viruses, at least in children < 2 years of age, does not simply represent asymptomatic infection.
We detected HRV in the respiratory specimens of 23.3% of children who had samples submitted to the Clinical Virology laboratory and who were negative for RSV, parainfluenza virus 1–3, influenza A and B, adenovirus and HBoV. This rate of infection is higher than the 8% rate of human metapneumovirus (hMPV) infection previously detected in a similar population in New Haven (42). Other researchers have found HRV to have a prevalence of 26% in sick children, higher than the prevalence of RSV, influenza, parainfluenza and enteroviruses (28). That study and ours strongly suggest that HRV may be a more common pathogen than other viruses traditionally associated with lower respiratory tract infection. Furthermore, 55% of HRV-infected patients in the Clinical Virology Category were hospitalized at the time of specimen collection, suggesting that HRV infection is a major cause of hospitalization among young children which is consistent with other studies (15, 22, 43). Indeed, the percentage of HRV-infected children who were hospitalized was similar to the percentage of RSV-infected children. The high prevalence of this pathogen, along with considerable morbidity as evidenced by high rates of hospitalizations, suggests that HRV are of major clinical significance and that a rapid sensitive assay capable of detecting most rhinovirus strains would be an important diagnostic tool.
Our study suggests that only a small percentage (3.3%) of isolates detected using picornavirus-specific primers represent non-rhinovirus picornavirus infection. In this study, we detected two cases of infection with coxsackie virus and one case of enterovirus infection. Miller et al (28) identified enterovirus infection in 2% of their study population. These data confirm that HRV are overwhelmingly the most common pathogen in the picornavirus family of the respiratory tract in young children with respiratory symptoms.
Few other studies have included sequence data and phylogenetic analyses of HRV strains collected from young children during a complete calendar year (11). Our phylogenetic analysis confirms the broad genetic diversity of HRV circulating in Connecticut during a one-year period. These viruses have a large number of nucleotide polymorphisms, suggesting that our current understanding of the diversity within this genus may underestimate the true genetic diversity of HRV strains in circulation. Many previously unidentified variants of HRV have been recently detected with the use of genome amplification technologies (5–11). Many of the New Haven isolates are closely related to several of these newly identified strains(10) though some may represent HRV strains that have yet to be fully characterized.
Based on our data, no particular HRV strains are more likely to cause wheezing and asthma exacerbation than others. Likewise, it does not appear that certain isolates are more likely to cause severe disease and hospitalization than others at least among those found in 2004. The previously-described seasonality of HRV infection, with peaks in spring and fall months (44), is not supported by our data. There appears to be no association between particular isolates and the seasonal distribution of this virus. While most cases of HRV infection occurred during spring and fall-winter months, there does not appear to have been any specific HRV isolates with peak activity during this period, suggesting that these viruses do not have a true seasonal distribution and circulate throughout the year. This is consistent with recent studies of HRV(22, 35).
We conclude that HRV are important pathogens among young children. The clinical significance of HRV infection may have previously been underestimated, as HRV appear to be more prevalent than other common respiratory viruses, such as hMPV, among children seeking medical care. Rapid diagnostic assays with a high degree of sensitivity for detecting this genotypically diverse group of viruses, as described by Lu et al (13), may prove to have an important role in the diagnosis of respiratory tract infection and of the cause of asthma exacerbations among young children.
Supplementary Material
Table 2.
HRV-positive specimens (percentage) for Categories 1, 2, and 4, by month
| Month | HRV-positive | Total screened | HRV-positive (%) |
|---|---|---|---|
| January | 6 | 63 | 9.5 |
| February | 10 | 72 | 13.9 |
| March | 6 | 45 | 13.3 |
| April | 11 | 51 | 21.6 |
| May | 11 | 34 | 32.4 |
| June | 4 | 19 | 21.1 |
| July | 0 | 10 | 0.0 |
| August | 4 | 8 | 50.0 |
| September | 7 | 20 | 35.0 |
| October | 11 | 34 | 32.4 |
| November | 11 | 44 | 25.0 |
| December | 15 | 47 | 31.9 |
| Total | 96(a) | 447 |
The 3 HRV-positive specimens from Category 3 (asymptomatic controls) and the 3 non-HRV picornavirus-positive specimens were not included.
ACKNOWLEDGEMENTS
We are indebted to George Miller, M.D., John F. Enders Professor of Pediatric Infectious Diseases for his continued support, intellectual and scientific input and exchange and critical review of the data. We thank the staff of the Clinical Virology Laboratory, Yale-New Haven Hospital for their assistance in this study. We thank Nancy Holabird, LPN for her assistance in enrollment and sampling of asymptomatic children. Support for this work is as follows: MV was supported by NIH grant AI68280 and EDS was supported by NIH grant AI001703. ZP was supported by a NIH-NCRR CTSA-T32 Medical Student Research Fellowship. This publication was made possible by Grant Number 5 T32 RR023261 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
Conflict of Interests: None.
References
- 1.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 2.Arruda E, Pitkaranta A, Witek TJ, Jr, Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–2868. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelon W, Mogabgab WJ, Phillips IA, Pierce WE. A cytopathogenic agent isolated from naval recruits with mild respiratory illnesses. Proc Soc Exp Biol Med. 1957;94:262–267. doi: 10.3181/00379727-94-22915. [DOI] [PubMed] [Google Scholar]
- 4.Price WH. The Isolation of a New Virus Associated with Respiratory Clinical Disease in Humans. Proc Natl Acad Sci U S A. 1956;42:892–896. doi: 10.1073/pnas.42.12.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McErlean P, Shackelton LA, Lambert SB, et al. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamson D, Renwick N, Kapoor V, et al. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renwick N, Schweiger B, Kapoor V, et al. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J Infect Dis. 2007;196:1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SK, Yip CC, Tsoi HW, et al. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiang D, Yagi S, Kantardjieff KA, et al. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–237. doi: 10.1016/j.jcv.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Lee WM, Kiesner C, Pappas T, et al. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loens K, Goossens H, de Laat C, et al. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. J Clin Microbiol. 2006;44:166–171. doi: 10.1128/JCM.44.1.166-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Holloway B, Dare RK, et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brownlee JW, Turner RB. New developments in the epidemiology and clinical spectrum of rhinovirus infections. Curr Opin Pediatr. 2008;20:67–71. doi: 10.1097/MOP.0b013e3282f41cb6. [DOI] [PubMed] [Google Scholar]
- 15.Juven T, Mertsola J, Waris M, et al. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J. 2000;19:293–298. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Michelow IC, Olsen K, Lozano J, et al. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics. 2004;113:701–707. doi: 10.1542/peds.113.4.701. [DOI] [PubMed] [Google Scholar]
- 17.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with "common cold" virus infections. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMillan JA, Weiner LB, Higgins AM, Macknight K. Rhinovirus infection associated with serious illness among pediatric patients. Pediatr Infect Dis J. 1993;12:321–325. doi: 10.1097/00006454-199304000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Papadopoulos NG, Moustaki M, Tsolia M, et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002;165:1285–1289. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- 20.Jacques J, Bouscambert-Duchamp M, Moret H, et al. Association of respiratory picornaviruses with acute bronchiolitis in French infants. J Clin Virol. 2006;35:463–466. doi: 10.1016/j.jcv.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Chidekel AS, Rosen CL, Bazzy AR. Rhinovirus infection associated with serious lower respiratory illness in patients with bronchopulmonary dysplasia. Pediatr Infect Dis J. 1997;16:43–47. doi: 10.1097/00006454-199701000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Cheuk DK, Tang IW, Chan KH, et al. Rhinovirus infection in hospitalized children in Hong Kong: a prospective study. Pediatr Infect Dis J. 2007;26:995–1000. doi: 10.1097/INF.0b013e3181586b63. [DOI] [PubMed] [Google Scholar]
- 23.Kim JO, Hodinka RL. Serious respiratory illness associated with rhinovirus infection in a pediatric population. Clin Diagn Virol. 1998;10:57–65. doi: 10.1016/s0928-0197(98)00004-x. [DOI] [PubMed] [Google Scholar]
- 24.Ong GM, Wyatt DE, O'Neill HJ, McCaughey C, Coyle PV. A comparison of nested polymerase chain reaction and immunofluorescence for the diagnosis of respiratory infections in children with bronchiolitis, and the implications for a cohorting strategy. J Hosp Infect. 2001;49:122–128. doi: 10.1053/jhin.2001.1044. [DOI] [PubMed] [Google Scholar]
- 25.Johnston SL, Pattemore PK, Sanderson G, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. Bmj. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakes GP, Arruda E, Ingram JM, et al. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care. IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999;159:785–790. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- 27.Bardin PG, Sanderson G, Robinson BS, Holgate ST, Tyrrell DA. Experimental rhinovirus infection in volunteers. Eur Respir J. 1996;9:2250–2255. doi: 10.1183/09031936.96.09112250. [DOI] [PubMed] [Google Scholar]
- 28.Miller EK, Lu X, Erdman DD, et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 2007;195:773–781. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blomqvist S, Roivainen M, Puhakka T, Kleemola M, Hovi T. Virological and serological analysis of rhinovirus infections during the first two years of life in a cohort of children. J Med Virol. 2002;66:263–268. doi: 10.1002/jmv.2140. [DOI] [PubMed] [Google Scholar]
- 30.Kusel MM, de Klerk NH, Holt PG, et al. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr Infect Dis J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- 31.van Benten I, Koopman L, Niesters B, et al. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14:363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston SL, Sanderson G, Pattemore PK, et al. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J Clin Microbiol. 1993;31:111–117. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright PF, Deatly AM, Karron RA, et al. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J Clin Microbiol. 2007;45:2126–2129. doi: 10.1128/JCM.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nokso-Koivisto J, Kinnari TJ, Lindahl P, Hovi T, Pitkaranta A. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J Med Virol. 2002;66:417–420. doi: 10.1002/jmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: Association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–650. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]
- 36.Peltola V, Waris M, Osterback R, et al. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 37.DeVincenzo JP, Hall CB, Kimberlin DW, et al. Surveillance of clinical isolates of respiratory syncytial virus for palivizumab (Synagis)-resistant mutants. J Infect Dis. 2004;190:975–978. doi: 10.1086/423213. [DOI] [PubMed] [Google Scholar]
- 38.Fernie BF, Gerin JL. The stabilization and purification of respiratory syncytial virus using MgSO4. Virology. 1980;106:141–144. doi: 10.1016/0042-6822(80)90229-9. [DOI] [PubMed] [Google Scholar]
- 39.Kesebir D, Vazquez M, Weibel C, et al. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006;194:1276–1282. doi: 10.1086/508213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roghmann M, Ball K, Erdman D, et al. Active surveillance for respiratory virus infections in adults who have undergone bone marrow and peripheral blood stem cell transplantation. Bone Marrow Transplant. 2003;32:1085–1088. doi: 10.1038/sj.bmt.1704257. [DOI] [PubMed] [Google Scholar]
- 41.Gama RE, Horsnell PR, Hughes PJ, et al. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. J Med Virol. 1989;28:73–77. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- 42.Esper F, Martinello RA, Boucher D, et al. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis. 2004;189:1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellner G, Popow-Kraupp T, Kundi M, et al. Contribution of rhinoviruses to respiratory viral infections in childhood: a prospective study in a mainly hospitalized infant population. J Med Virol. 1988;25:455–469. doi: 10.1002/jmv.1890250409. [DOI] [PubMed] [Google Scholar]
- 44.Monto AS. The seasonality of rhinovirus infections and its implications for clinical recognition. Clin Ther. 2002;24:1987–1997. doi: 10.1016/S0149-2918(02)80093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.