SUMMARY
Chemical cross-linking and DNA sequencing have revealed regions of intra-chromosomal interaction, referred to as topologically associating domains (TADs), interspersed with regions of little or no interaction, in interphase nuclei. We find that TADs and the regions between them correspond with the bands and interbands of polytene chromosomes of Drosophila. We further establish the conservation of TADs between polytene and diploid cells of Drosophila. From direct measurements on light micrographs of polytene chromosomes, we then deduce the states of chromatin folding in the diploid cell nucleus. Two states of folding, fully extended fibers containing regulatory regions and promoters, and fibers condensed up to ten-fold containing coding regions of active genes, constitute the euchromatin of the nuclear interior. Chromatin fibers condensed up to 30-fold, containing coding regions of inactive genes, represent the heterochromatin of the nuclear periphery. A convergence of molecular analysis with direct observation thus reveals the architecture of interphase chromosomes.
INTRODUCTION
The basis of DNA folding and compaction in nuclei and chromosomes is one of the great mysteries of biology. How are two meters of DNA packaged in an interphase nucleus approximately ten micrometers in diameter? At the molecular level, folding begins with wrapping of DNA around histones in the nucleosome (Kornberg, 1974; Luger et al., 1997). At a cytological level, chromatin appears as darkly staining heterochromatin, most abundant at the periphery of the interphase nucleus, and lightly staining euchromatin, in the nuclear interior. The folding of chromatin fibers in heterochromatin and their organization in euchromatin have not been determined. We have gained insight into chromosome condensation from molecular analysis of polytene chromosomes of Drosophila.
Polytene chromosomes occur in cells of dipteran larvae that undergo as many as ten rounds of DNA replication without division, while retaining close alignment of sister chromatids and pairing of homologous chromosomes (Urata et al., 1995). Polytene chromosomes are visible by light microscopy (Agard and Sedat, 1983; Urata et al., 1995), and display an alternation of dense bands with less dense interbands (Balbiani, 1881; Flemming, 1882), which has been exploited for genetic analysis (Bridges, 1935). The increased density in bands reflects elevated DNA content and a stable state of chromatin condensation (Beermann, 1972). Two types of bands have been distinguished, loosely compacted grey bands and dense, tightly compacted intercalary heterochromatin (IH) bands (Vatolina et al., 2011; Zhimulev et al., 2014). Median expression levels of genes within grey bands is 27 times greater than that of genes within IH bands (Zhimulev et al., 2014).
Chromatin folding has been detected at the molecular level, by “chromosome conformation capture,” in which the chemical cross-linking of chromosomal material is followed by fragmentation, ligation, and DNA sequence analysis (Dekker et al., 2002). Extension of this approach by high-throughput sequencing (Hi-C) revealed, at a resolution of 1 megabase (Mb) pair, a genome-wide interaction network of the human genome (Lieberman-Aiden et al., 2009). Computational analysis of the Hi-C results identified two sets of chromosomal regions, termed compartments, within which very distant interactions occur more frequently than expected for the random coil configuration of a polymer. The two compartments correlate with regions of transcriptional activity and inactivity.
Increased sequencing depth revealed finer details of chromosome folding, at a resolution of 40 kilobase pairs or better. So-called topologically associating domains [“TADs” (Nora et al., 2012), also referred to as “physical domains” (Hou et al., 2012; Sexton et al., 2012) or “topological domains” (Dixon et al., 2012)], in which nucleotide sequences far apart along the DNA come in close proximity to one another are observed at a length scale of a few megabase pairs or less. Hi-C also revealed loops, frequently bridging between enhancers and promoters, that correlate with gene activation. Loops are apparently transient, greater than 250 nm apart in three quarters of a cell population (Rao et al., 2014), and though important for gene regulation, are unlikely to provide a structural basis for heterochromatin and euchromatin. TADs, however, represent a consistent feature among cell types (Dixon et al., 2012; 2015), and may therefore relate to stably folded states of chromatin. In studies performed to date, however, TADs have only been revealed by DNA sequencing and not directly related to chromosome condensation. A hierarchical relationship between TADs and compartments has been suggested by computational models (Gibcus and Dekker, 2013; Sexton et al., 2012). We report here on the direct visualization of TADs in polytene chromosomes of Drosophila, from which the relationship between TADs and compartments, as well as structural correlates of chromatin condensation in the interphase nucleus, may be derived.
RESULTS
Hi-C on Drosophila Polytene Chromosomes
We performed Hi-C on the salivary glands of wandering third instar larvae of Drosophila melanogaster (Figures 1A, 1C, S1, and S2). The limited amount of material available from manually-dissected, primary polytene tissue necessitated a Hi-C approach with improved signal-to-noise, and limited the resolution of our analysis to 15 kb (Figure S2 and Extended Experimental Procedures). To make reliable comparisons between the polytene Hi-C data, other Hi-C datasets, and cytological observations, we only considered chromosomal features of 75 kb or larger. The highly underreplicated and repetitive nature of pericentromeric heterochromatin also precluded reliable Hi-C analysis in these regions. Differences in copy number across the arms of polytene chromosomes due to incomplete DNA replication do not affect Hi-C results (Figure S3 and Extended Experimental Procedures).
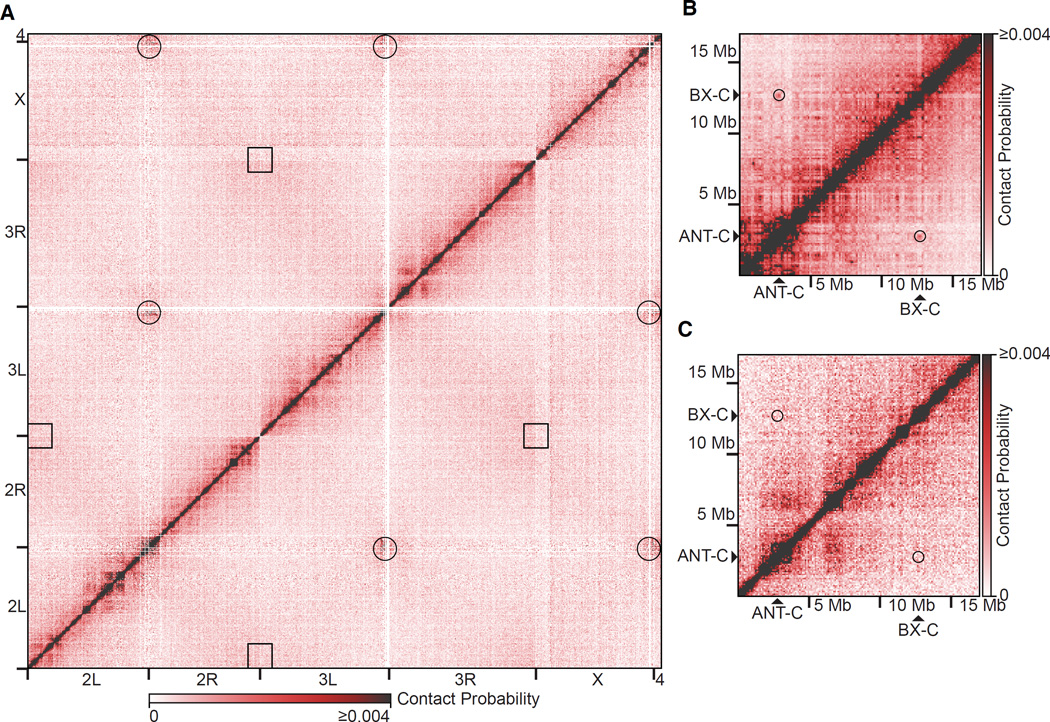
Figure 1. Lack of Regular, Long-range Contacts in Polytene Chromosomes.
(A) Genome-wide Hi-C heatmap from polytene cells. Black circles and squares represent where centromeres and telomeres intersect, respectively.
(B) Hi-C heatmap from Drosophila embryos (Sexton et al., 2012) of a 17 Mb region of chromosome 3R encompassing the ANT-C and BX-C loci. Black circles represent where the ANT-C and BX-C loci intersect.
(C) Hi-C heatmap from polytene cells of a 17 Mb region of chromosome 3R encompassing the ANT-C and BX-C loci. Black circles represent where the ANT-C and BX-C loci intersect. Heatmaps were normalized and divided into 100 kb bins.
See also Figures S1 and S2.
Our genome-wide, polytene Hi-C heatmaps revealed interactions between centromeres, but a lack of contact between telomeres (Figure 1A). These results are consistent with three-dimensional reconstructions of polytene chromosomes by light microscopy, in which centromeres were observed to cluster at the chromocenter at one pole of the nucleus with telomeres spread in the opposite hemisphere in a Rabl orientation (Hochstrasser et al., 1986). Centromeres are also clustered together in diploid Drosophila Kc167 cultured cells (Hou et al., 2012) and late-stage embryos (Sexton et al., 2012). Telomeres do not interact with one another in diploid cells (Hou et al., 2012), but do interact in embryos (Sexton et al., 2012).
Distant or long-range interactions (separated by greater than 1 Mb), such as those previously observed between the ANT-C and BX-C loci in Hi-C heatmaps of late-stage (mixed cell-type) Drosophila embryos (Sexton et al., 2012) (Figure 1B) were not detected in our polytene Hi-C heatmaps (Figure 1C). The lack of long-range interactions in our heatmaps was consistent with previously described light microscopy of polytene chromosomes in situ, which revealed only regular, short-range contacts and no reproducible, long-range interactions (Hochstrasser et al., 1986). Occasionally polytene bands interact with one another, resulting in ectopic pairing. The frequency of cells exhibiting ectopic pairing is low (Zhimulev et al., 1982) and complete mixing of bands is not observed, which explains why, at present, such pairing falls below our limit of detection by Hi-C.
We examined the proportion of paired-end reads that mapped to the same restriction fragment and pointed in the same direction, reflecting interactions between paired and aligned chromatids or homologous chromosomes (Sexton et al., 2012). A similar proportion of paired-end reads fell into this class for our polytene chromosome dataset (0.0328%) and the reported embryonic dataset (0.0553%). These values are much less than the proportion of paired-end reads representing intramolecular self-ligation (cyclization) events (0.117% and 0.132% in the polytene and embryo datasets, respectively) indicating that reads dues to interactions between paired and aligned chromatids or homologous chromosomes are very infrequent. A higher proportion of reads in the reported diploid chromosome (Kc167 cell line) dataset fall in this class (22.5% compared to 8.35% of paired-end reads representing intramolecular self-ligation events), but the chromatin was fragmented to a median size of 2377 bp, compared with 193 bp in the polytene and embryonic experiments. Homologous chromosome pairing is prevalent in many Drosophila primary tissues and culture cells (Fung et al., 1998; Williams et al., 2007), but the analysis of paired-end reads indicates that paired homologs or chromatids are not perfectly aligned at the level of a few hundred base pairs, even in polytene chromosomes, and the degree of alignment in polytene chromosomes is about the same as that in late-stage embryos.
Equivalence of Polytene Bands with TADs
Our polytene Hi-C heatmaps showed the presence of TADs, regions of high self-interaction, visible as boxes centered on the diagonal (Figures 2A, 5A, and S3). Our data revealed 346 polytene TADs. The median TAD size was 165 kb; the mean TAD size was 195 kb (Figure S3 and Table S1). Superimposing the Hi-C heatmap onto the locations of polytene bands for which reliable DNA sequence coordinates have previously been determined by FISH and non-histone protein localization (Belyaeva et al., 2012; Vatolina et al., 2011) demonstrated a correspondence of TADs with bands (Figures 2A, 2B, and 5A).
Figure 2. Polytene Bands are TADs.
(A) Normalized Hi-C heatmap (15 kb bins) of a 3 Mb region of polytene chromosome 2L. In the left panel, areas bounded by black boxes represent locations of polytene bands for which reliable DNA sequence coordinates are available (Belyaeva et al., 2012; Vatolina et al., 2011). In the right panel, areas bounded by black boxes represent TADs.
(B) Photographic image (bottom) of the region of polytene chromosome 2L from (A) and the same region of Bridges’s chromosome map (top). Arrows indicate bands represented by black boxes in (A). The DNA sequence coordinates of other bands in this region are not known and therefore cannot be compared with the Hi-C data. Adapted from (Lefevre, 1976).
(C) Mean directionality index (DI) of polytene bands (upper panel; n = 61) and heatmap of the directionality index of each band along its length (lower panel). Bands were normalized to the same length and 50 kb of flanking DNA is shown next to each normalized band.
(D) Heatmap of the agreement between polytene bands (n = 61) and TADs. Each band is represented by a row. Bands were normalized to the same length and 50 kb of flanking DNA is shown next to each normalized band. Orange segments overlap with polytene TADs, black segments overlap with regions between TADs.
(E) Fraction of band boundaries (n = 122) at the distance indicated on the abscissa from the closest TAD boundary (calculated in 20 kb windows).
See also Figures S3, S4, and S5 and Table S1.
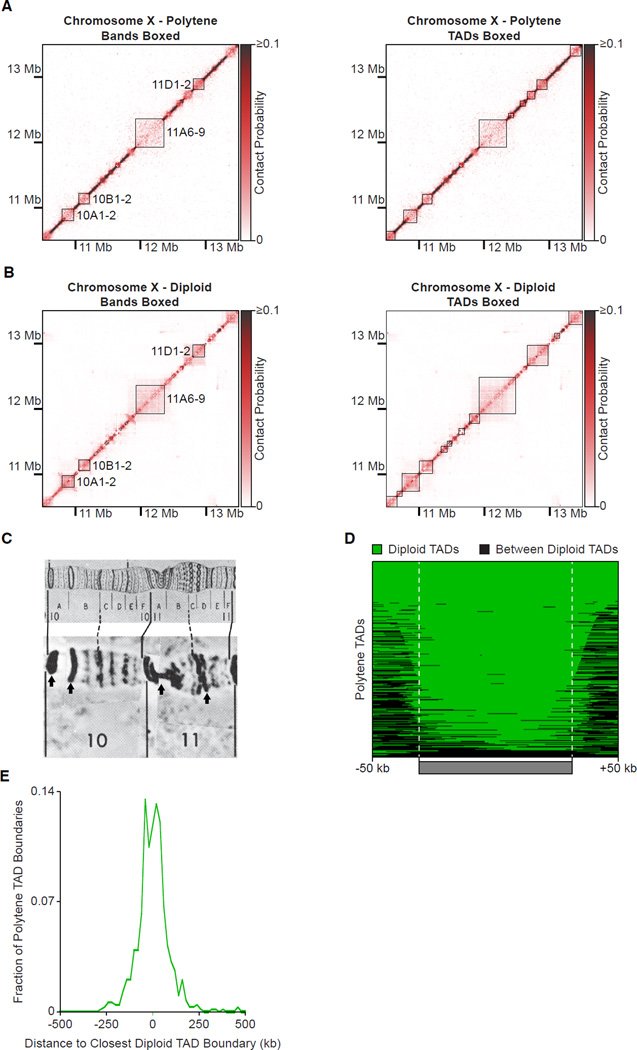
Figure 5. Conservation of Polytene and Diploid TADs.
Normalized Hi-C heatmaps (15 kb bins) of a 3 Mb region of the X chromosome. In the left panels, areas bounded by black boxes represent locations of polytene bands for which reliable DNA sequence coordinates are available (Belyaeva et al., 2012; Vatolina et al., 2011). In the right panel, areas bounded by black boxes represent TADs.
(A) Hi-C heatmap from polytene cells.
(B) Hi-C heatmap from diploid Kc167 cultured cells.
(C) Photographic image (bottom) of the region of polytene chromosome X from (A) and (B) and the same region of Bridges’s chromosome map (top). Arrows indicate cytological bands represented by black boxes in (A) and (B). The DNA sequence coordinates of other bands in this region are not known and therefore cannot be compared with the Hi-C data. Adapted from (Lefevre, 1976).
(D) Heatmap of the agreement between polytene TADs and diploid TADs. Each polytene TAD is represented by a row. Green segments overlap with diploid TADs, black segments overlap with regions between diploid TADs. TADs were normalized to the same length and 50 kb of flanking DNA is shown next to each normalized TAD.
(E) Fraction of polytene TAD boundaries (n = 692) at the distance indicated on the abscissa from the closest diploid TAD boundary (calculated in 20 kb windows).
See also Figures S4, S5, and S6.
For a quantitative assessment of the interaction pattern within polytene bands, we computed the genome-wide directionality index, a measure of the degree of bias of a locus for interaction with downstream (positive directionality index) or upstream loci (negative directionality index) (Dixon et al., 2012). Characteristically, the directionality index of a TAD starts positive, goes to zero at the middle of the TAD, and continues towards negative values at the end of a TAD (Dixon et al., 2012). Polytene bands exhibit this same trend, supporting their identification as TADs (Figure 2C).
The concordance between polytene bands and TADs was observed across all of the large chromosome arms (Figure 2D). At least 95% of polytene bands corresponded to uninterrupted TADs. The overlap between polytene bands and TADs was far greater than expected on a random basis (Z-score = 10.5, p = 3.01 × 10−25; Figure S4B and Extended Experimental Procedures). Differences were observed almost exclusively at band boundaries. Mismatches between polytene bands and TADs were far less than expected on a random basis (Z-score = −10.5, p = 2.78 × 10−25; Figure S4B and Extended Experimental Procedures). The majority (57.4%) of band boundaries were located within 20 kb of TAD boundaries (Figure 2E). Discrepancies at the boundaries were likely due to the limited resolution of the Hi-C experiment, limited accuracy of DNA sequence coordinates for polytene band borders, or both. Near identity of boundaries does not necessarily indicate equivalence of entire domains, but quantitative, unbiased examination of entire polytene bands revealed far greater equivalence with polytene TADs than expected on a random basis (Figure S5B and Extended Experimental Procedures).
Polytene Puffs Are Not TADs
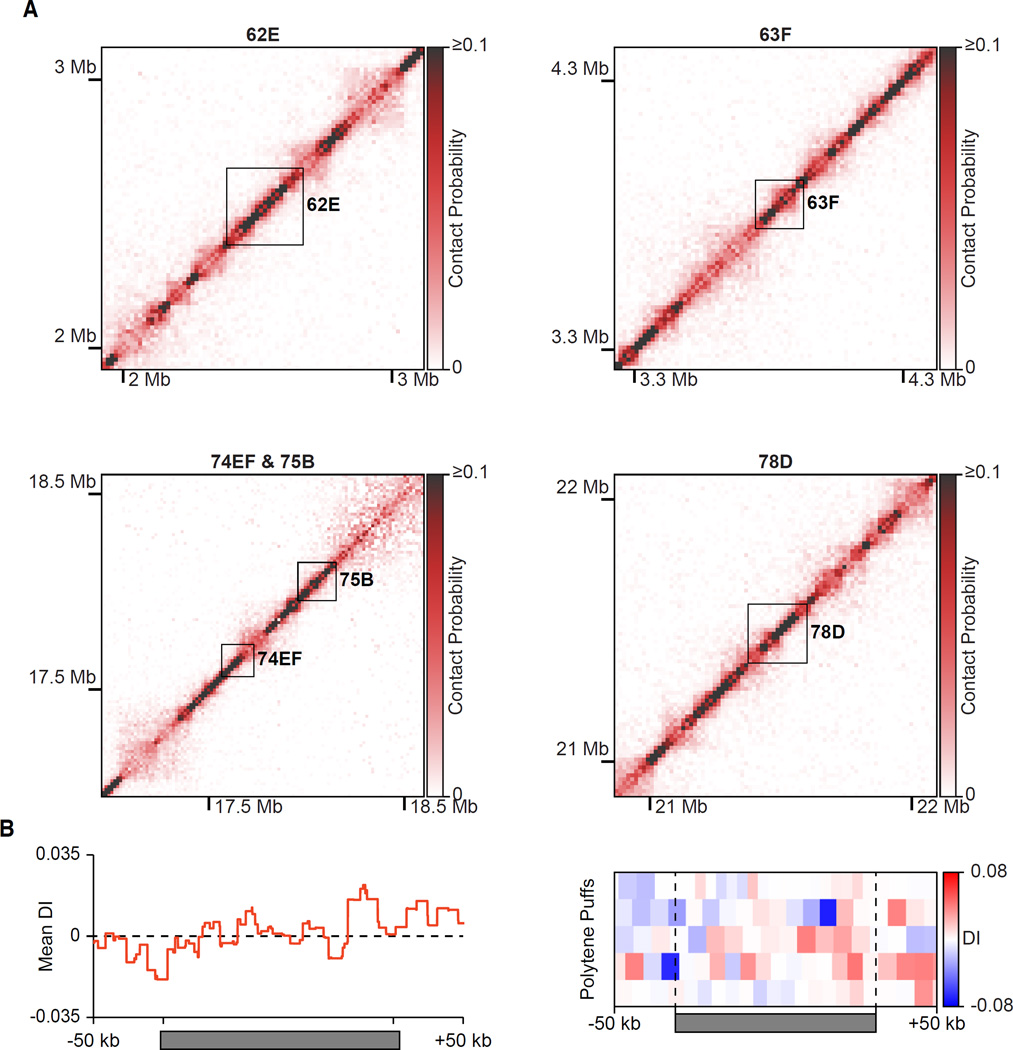
High transcriptional output of some genes at certain stages of development causes the conversion of polytene bands to puffs, regions of decondensed chromatin with a diameter wider than the rest of the chromosome (Ashburner, 1967). Loci that are known to convert from bands to puffs at the wandering third instar larval stage showed no evidence of TADs, but rather strong signals restricted to the diagonal (Figure 3A). Furthermore, quantitation of the interaction pattern of puffs showed a mostly uniform directionality index centered around zero, rather than a TAD-like bias in directionality (Figure 3B). The absence of TADs from puffed bands strengthens the connection between TADs and chromatin condensation.
Figure 3. Hi-C of Polytene Puffs.
(A) Normalized Hi-C heatmaps (15 kb bins) of puff stage 5–8 polytene puffs on chromosome 3L. Areas bounded by black boxes correspond to the indicated puff.
(B) Mean directionality index (DI) of polytene puffs (left) and heatmap of the directionality index of each puff along its length (right). Puffs were normalized to the same length and 50 kb of flanking DNA is shown next to each normalized puff.
Hi-C Predicts the Location of Bands and Interbands
As a further test of the relationship between TADs and bands, we asked whether the DNA sequence of a TAD could be used to predict the location of a band for which reliable DNA sequence information was not available. We designed fluorescent probes to the centers and borders of three TADs and hybridized them individually to polytene chromosomes (Figure 4). In every case, the FISH signal from a TAD center probe precisely overlapped with a polytene band. In five of six cases, the FISH signal from a TAD border probe overlapped with the adjacent polytene interband. In the one case where the TAD border probe overlapped with a band (Figure 4A; centromere proximal probe), the overlap occurred at the margin of the band immediately adjacent to a small interband (between bands 22A1–2 and 22A3 in Bridges’s map) (Bridges, 1935; Lefevre, 1976). Inasmuch as only 5% of DNA is located within interbands (Beermann, 1972), our success rate of 0.833 in identifying interbands is much greater than expected (p = 1.80 × 10−6, Binomial test). FISH thus confirms our Hi-C results and our identification of TADs with polytene bands.
Figure 4. Hi-C Predicts the Location of Polytene Bands.
The locations of TADs (normalized Hi-C heatmaps, 15 kb bins; left column) were used to generate FISH probes against TAD centers (red diamonds) or TAD borders (green diamonds), which were hybridized to polytene chromosome spreads counterstained with DAPI (middle-left column). The TAD border FISH signal and the TAD center FISH signal are pseudocolored green and red, respectively, in the merged images (rightmost two columns) and the identity of the polytene band is indicated (right column). White arrows indicate the polytene band of interest.
(A) FISH against a region from chromosome 2L. White arrowhead indicates a small interband between bands 22A1-2 and 22A3 in Bridges’s map.
(B) FISH against a region from chromosome 3L.
(C) FISH against a region from chromosome 3R.
Loci presented here are independent of those analyzed in Figure 1. Scale bars, 2 µm.
Conservation of TADs Between Polytene and Diploid Cells
Hi-C was previously performed on D. melanogaster diploid, Kc167 cultured cells (Hou et al., 2012) and late-stage embryos (Sexton et al., 2012). Both studies revealed TADs as an organizational feature of the Drosophila genome. The Hi-C heatmap from diploid cells and our Hi-C heatmap from polytene cells were closely similar (Figure 5A and 5B) and highly correlated (genome-wide Pearson’s r = 0.793, p < 2.2 × 10−16). Where comparisons could be made, diploid TADs could also be seen to correspond to polytene bands (Figure 5B and 5C). The overlap between polytene TADs and diploid TADs (Figure 5D) was far greater than expected on a random basis (Z-score = 20.5, p = 3.92 × 10−92; Figure S4B and Extended Experimental Procedures). Similarly, mismatches between polytene bands and TADs were far less than expected on a random basis (Z-score = −20.6, p = 6.89 × 10−93; Figure S4B and Extended Experimental Procedures). Approximately 50% of polytene TAD boundaries were located within 40 kb of diploid TAD boundaries (Figure 5E) and quantitative assessment of entire polytene TADs revealed far greater equivalence with Kc cell TADs than expected on a random basis (Figure S5C and Extended Experimental Procedures). We observed a similar correspondence in the Hi-C heatmaps between polytene cells and late-stage embryos (Figure S6; genome-wide Pearson’s r = 0.793, p < 2.2 × 10−16). TADs from polytene cells and Drosophila embryos significantly overlapped (Z-score = 15.2, p = 2.10 × 10−51; Figure S4B and Extended Experimental Procedures) and exhibited far fewer mismatches than expected on a random basis (Z-score = −15.3, p = 8.19 × 10−52; Figure S4B and Extended Experimental Procedures). The agreement was even more remarkable considering that late-stage embryos contain a mixture of cell-types, unlike the more homogenous composition of the salivary gland. Evidently, most TADs are conserved in their central regions across a range of cells types, while a fraction of TAD boundaries may exhibit some cell-type specificity.
Lack of Compartments in the Polytene Nucleus
Compartmentation refers to the preferential association of distant chromosomal loci in two groups, which correlate with transcriptional activity and inactivity. At large length scales, chromosomes exhibit polymeric behavior: loci separated by large linear distances are less likely to be in close spatial proximity than loci separated by small distances (Lieberman-Aiden et al., 2009). Associations of distant loci become apparent when a correction is applied for polymeric behavior: the observed number of interactions between two loci is divided by the number of interactions expected due to variations in polymer conformation. The resulting observed/expected heatmap (Figure 6, middle column) displays regions of more or less interaction than the expected genome-wide average. The delineation of such regions can be enhanced by correlation analysis, because neighboring loci along the genome in close spatial proximity should share the same interaction preferences and spatially distant loci should differ in their interaction preferences. A heatmap of the correlations between interaction profiles shows a “plaid” pattern if there are both shared and divergent interaction preferences (Figure 6, right column) (Lieberman-Aiden et al., 2009). Boxes in the plaid pattern are of two types, boxes along the diagonal, at the locations of TADs in the observed heatmap, and off-diagonal boxes, revealing enriched or depleted long-range interactions. The occurrence of off-diagonal boxes is indicative of compartments. Correlation heatmaps computed from published Hi-C data for Kc167 cells and late-stage Drosophila embryos (Figure 6, top two rows) exhibit a plaid pattern and thus compartments, as previously observed (Sexton et al., 2012). By contrast, a correlation heatmap computed from our Hi-C data for Drosophila polytene chromosomes shows no evidence of a plaid pattern (Figure 6, bottom row), indicating an absence of long-range interactions and a lack of compartments defined on that basis. A lack of compartments is not necessarily a consequence of homolog pairing, because homologs are paired in both embryonic (Fung et al., 1998) and Kc (Williams et al., 2007) cells.
Figure 6. Lack of Compartments in the Polytene Nucleus.
Normalized observed (left column), observed/expected (middle column), and Pearson correlation (right column) Hi-C heatmaps (100 kb bins) for embryonic (top row), diploid Kc167 cell (middle row), and polytene (bottom row) chromosome 3R. In the observed/expected heatmaps, interactions less than the expected genome-wide average are blue, those greater than the expected genome-wide average are red. A plaid pattern in a Pearson correlation heatmap indicates the presence of compartments.
See also Figure S7.
DISCUSSION
Our finding of an equivalence between polytene bands and TADs has two-fold significance. It complements the discovery of TADs by chemical cross-linking with their identification by a structural approach; and it shows that chromatin folding inferred from cross-linking corresponds to bona fide chromosome condensation, to a state so dense it can be seen by light microscopy. Polytene bands are stable from cell to cell (Painter, 1933), as well as when observed in real time (Hochstrasser et al., 1986); TADs are therefore similarly stable. Interbands correspond to regions between TADs, which thus reflect a stable state of chromosome decondensation. The band-interband pattern is similar between polytene nuclei of different tissues (Beermann, 1972). TADs evidently represent a largely invariant, or conserved, feature of chromosome structure. The equivalence of polytene, diploid, and embryonic TADs shows that chromosome structure manifest in the polytene state is general, applicable to the diploid interphase nucleus, and nearly constant among all cell types.
Although TADs are stable between embryonic stem (ES) cells and ES cell-derived lineages, interactions within and between TADs may change during ES cell differentiation (Dixon et al., 2015). So TADs themselves do not directly regulate transcription, but rather additional features, such as transient chromatin loops, are likely presumably involved. The equivalence of polytene bands with TADs indicates that the role of TADs is most likely for the compaction of DNA in the interphase nucleus.
Fine mapping of regions of polytene chromosomes (Zhimulev et al., 2014) has shown that interbands contain regulatory regions and promoters of genes expressed in most tissues, cell lines, developmental stages, and treatment conditions - so-called “housekeeping genes.”
Regulatory regions, transcription start sites, and 5’ transcribed regions are located in interbands, and the remaining coding portions of genes reside in the adjacent grey bands (Zhimulev et al., 2014). Only half of total RNA synthesis occurs in puffs (Zhimulev and Belyaeva, 1975); the other half may occur in grey bands. Although the moderate resolution of our Hi-C analysis and the lack of comprehensive epigenomic profiling in salivary gland tissues preclude the direct detection of active and inactive TADs in polytene chromosomes, our identification of bands as TADs, along with the characteristics of grey bands, implies a connection between loosely compact grey bands and “active TADs” reported by others (Rao et al., 2014; Sexton et al., 2012). TADs and the regions between them are conserved across cell types likely because transcription is regulated and initiated in interbands and many transcribed genes are active in all cell types.
Conserved interbands or regions between TADs may correspond not only to regulatory regions involved in gene activity but also to those needed for gene repression across cell types. The 5’-regulatory region of Notch, which is transcriptionally inactive in salivary glands, lies in an interband between bands 3C6 and 3C7, with the coding region in band 3C7 (Rykowski et al., 1988). The Notch mutation facet-strawberry (faswb) results in a small deletion harboring an insulator element (Vazquez and Schedl, 2000), which maps to the 3C6-3C7 interband (Rykowski et al., 1988). In the faswb mutation, the 3C6-3C7 interband disappears, resulting in fusion of bands 3C6 and 3C7 (Keppy and Welshons, 1977). Ectopic insertion of the DNA sequence comprising the faswb deletion was necessary and sufficient to split an endogenous band in two and form an interband at the ectopic site (Andreyenkov et al., 2010). Although a factor that binds to the faswb insulator has not yet been identified, maintaining the chromatin in a fully extended state would facilitate protein binding. If the same gene or set of genes is transcriptionally repressed across cell types, the state of chromatin and accessibility to transcriptional repressors may also be conserved, further accounting for the similarity in Hi-C results across cell types.
From the DNA sequences of bands and interbands, and measurements on micrographs of polytene chromosomes, the packing ratios (length of DNA to length of chromatin) of bands and interbands may be determined. The ratios range from 158:1 to 205:1 for IH bands, 12:1 to 73:1 for grey bands, and 5:1 for 12:1 in interbands (Rykowski et al., 1988; Vatolina et al., 2011). Conservation of Hi-C results between polytene and diploid cells allows us to deduce the structural states of chromatin, which we refer to as black, grey, and white, respectively, in the interphase, diploid nucleus. The packing ratio of a fully open chain of nucleosomes is expected to be 6.8:1 (Kornberg, 1974), so white chromatin evidently contains fully extended chromatin fibers. Approximately 5% of Drosophila DNA, containing promoters and regulatory regions, resides in this fully extended state (Beermann, 1972; Hou et al., 2012). Grey chromatin is up to 10-fold more compact, and contains approximately a quarter of the Drosophila genome (Filion et al., 2010; Sexton et al., 2012), within which are located the coding regions of active genes. Therefore, transcriptional regulation occurs in a fully extended state and elongation occurs in a partially compacted state. Black chromatin is up to 30-fold more condensed than fully extended chromatin fibers. It contains inactive genes and accounts for the remaining 70% of the genome.
The distribution of interactions within a TAD is informative about the pattern of chromatin condensation. The distribution is remarkably uniform, with every site in a TAD, particularly inactive TADs (Sexton et al., 2012), almost equally likely to become cross-linked with every other site (Figure 6B). Additionally, since homologs are paired in nearly all cell types (Fung et al., 1998; Williams et al., 2007), Drosophila chromosomes are particularly illuminating with regard to chromatin condensation. Although cytology clearly demonstrates homologous chromosome pairing, our Hi-C analysis indicates that, even for polytene chromosomes, at the molecular level they are not perfectly aligned. Nucleosome by nucleosome, the chromatin fibers from homologs and chromatids are not in lockstep – there is some variability in the path of each of the chromatin fibers. Although at the cytological level, bands are present at reproducible positions, there is some variation at the molecular level in the path of the DNA. Taken together, the interactions revealed by Hi-C cannot arise primarily from specific, stable chromatin loops, but rather the pattern of condensation within grey and black chromatin must vary from one cell to another within a population, consistent with conclusions of others (Gibcus and Dekker, 2013). There can be no unique pattern of condensation.
Studies of chromosomal protein distribution and histone modifications have revealed up to sixteen functional classes of chromatin (Ernst et al., 2011; Filion et al., 2010; Ho et al., 2014; Kharchenko et al., 2011; Ram et al., 2011; Rao et al., 2014; Sexton et al., 2012), and these classes are conserved between polytene and diploid cells (Zhimulev et al., 2014). One class corresponds closely with interbands, others classes with the flanking bands (Zhimulev et al., 2014). Structural information from polytene chromosomes identifies three structural states – black, grey, and white. A structural state may encompass multiple functional classes. For example, at the current level of resolution, we cannot distinguish between polycomb and HP1 repressed heterochromatin.
The equivalence of polytene bands with polytene TADs, and the virtual identity of polytene TADs with diploid TADs, implies a close correspondence of bands with diploid TADs. The structure of interphase chromosomes revealed by light microscopy of polytene nuclei therefore applies to the organization of the diploid cell nucleus. A subset of TADs in mammalian cells associates with the nuclear lamina (Dixon et al., 2012), and condensed chromatin is mostly located at the periphery of nuclei in electron micrographs of virtually all fixed, embedded, heavy metal-stained eukaryotic cell preparations (Figure 7, left). Polytene bands are also identified by heavy metal staining in electron micrographs and interactions of polytene chromosomes with the nuclear envelope, though infrequent, are almost entirely confined to the IH bands (Hochstrasser et al., 1986). If inactive TADs (black chromatin) are often located near the nuclear periphery, then the regions between inactive TADs, consisting of active TADs and fully extended chromatin fibers (white and grey chromatin), must loop into the interior of the nucleus (Figure 7, right). It follows that inactive TADs (black chromatin) correspond to classical heterochromatin (condensed chromatin at the nuclear periphery) and active TADs and the regions between TADs (white and grey chromatin) correspond to classical euchromatin (less dense chromatin in the nuclear interior). On the basis of the measurements mentioned above, heterochromatin at the nuclear periphery consists of up to 30-fold condensed chromatin fibers, while euchromatin in the nuclear interior contains both 10-fold condensed and fully extended fibers. The DNA sequences that reside in each of these regions are known from the results of Hi-C analysis. The identification of polytene TADs thus gives meaning to the familiar picture of the interphase nucleus.
Figure 7. Chromosome Condensation in the Interphase Nucleus.
At left, thin section electron micrograph of a nucleus (Cross and Mercer, 1993), with lightly staining euchromatin in the nuclear interior, surrounded by darkly staining heterochromatin, concentrated at the nuclear periphery. At right, cartoon representation of white, grey, and black chromatin, showing proposed relationships to heterochromatin, euchromatin, and the nuclear envelope (yellow). Active TADs in the euchromatin are nearby other active TADs and inactive TADs in the heterochromatin are nearby other inactive TADs, resulting in grey-grey and black-black TAD-TAD interactions. The actual pattern of chromatin folding is unknown and indicated only schematically.
Compartments can be understood in terms of the physical picture of the interphase nucleus. TADs and compartments are related, but differ in origin and significance. TADs are aligned with the off-diagonal boxes in the plaid pattern that defines compartments; the boundaries of TADs define the rows and columns of the plaid pattern (Figure 6, right column). Therefore the off-diagonal boxes arise from interactions between TADs. The difference is that TADs are due to persistent interactions. They reflect a stable state of axial condensation of the chromatin fiber.
By contrast, off-diagonal boxes are most apparent following correction for random polymer conformation and sharpening of the corrected heatmap. Off-diagonal boxes are therefore due to transient contacts between distant regions, for example short-lived contacts between TADs in neighboring euchromatic chromatin in the nuclear interior (Figure 7, right). This picture explains why off-diagonal boxes fall in two categories of interacting sequences, corresponding to active and inactive genes: active TADs (grey chromatin) tend to be in the vicinity of other active TADs, and inactive TADs (black chromatin) in the vicinity of other inactive TADs. In observed heatmaps, the off-diagonal boxes usually appear uniform in signal strength over their entire area, indicative of roughly equal likelihood of interaction of every sequence in one TAD with every sequence in another (Sexton et al., 2012). The reason is probably because the TADs themselves are condensed in a variable manner, as discussed above. All sequences have roughly equal probabilities of appearing on the surface of the condensed structure and interacting with adjacent condensed regions. Moreover, a single TAD may make many distant contacts, so there cannot be a unique pattern of TAD-TAD interactions, but rather the trajectory of a chromosome must vary from one cell to another.
Compartments refer to loci grouped on the basis of frequency of interaction. Compartments are not regions with boundaries, in the conventional sense of the term. They correspond to areas of the nucleus, for instance the interior and the periphery, but the areas themselves do not determine the state of gene activity (Therizols et al., 2014). The properties of polytene chromosomes are illustrative. Polytene chromosomes possess both active and inactive genes, but they exhibit no compartments, and they reside mostly in the nuclear interior, making little or no contact with the periphery. They lack compartments because they do not fold upon themselves to a significant extent, and therefore have no TAD-TAD interactions. TADs may occur without compartments, and chromosome condensation and gene regulation do not require compartments. Condensation along the chromosome axis is conserved, as shown by the equivalence of polytene, diploid, and embryonic TADs, whereas compartments may vary, depending on cell type and state of transcriptional activity.
The organization of the interphase nucleus in Drosophila is relevant to the mouse and to humans, where TADs organize chromosomes into spatial modules connected by short chromatin segments (Dixon et al., 2012). Futhermore, biochemical fractionation of open chromatin fibers from human cells revealed that the fibers are cytologically decondensed (Gilbert et al., 2004), and it is now apparent that these fibers are likely in the fully extended state. The packing ratios, DNA sequences, functional states, and chromosomal protein patterns of the differentially staining areas of the interphase nucleus are thus determined. Genome-wide amplification and alignment in the polytene state reveals interphase chromosome structure at the level of light microscopy, likely applicable to the diploid state in all monocentric metazoans.
EXPERIMENTAL PROCEDURES
Hi-C
Hi-C was performed using a tethering approach (Kalhor et al., 2012) to improve the signal-to-noise from a limited amount of manually-dissected, primary tissue. In brief, Drosophila melanogaster third instar larvae salivary glands were manually dissected and fixed with 2% EM grade paraformaldehyde. Cross-linked proteins were then biotinylated at cysteine residues and the DNA digested with DpnII. Digested chromatin was bound to streptavidin beads, thoroughly washed to remove uncross-linked DNA, DNA ends filled in with biotin-14-dATP, and free DNA ends ligated together. DNA-protein cross-links were reversed, DNA purified, biotinylated nucleotides marking unligated ends removed, and then DNA sheared to a mean size of approximately 200 bp. The biotinylated DNA was pulled down with streptavidin beads, prepared for and subjected to high-throughput Illumina sequencing. Further details provided in the Extended Experimental Procedures.
Hi-C Analysis
Hi-C reads were mapped to the dm3 reference genome using Bowtie 2 and assigned to DpnII restriction fragments. Reads mapping to the same restriction fragment, separated by less than the library insert size, within 4 bp of a restriction site, and duplicate reads were removed. Exceptionally large (> 100 kb) and small (<100 bp) restriction fragments and fragments with the highest 0.5% of counts were also removed. Filtered fragments were assigned to 15 kb genomic bins, unless otherwise indicated. Further filtering at the bin level removed bins where less than half the bin was sequenced, the lowest 1% of bins, and the highest 0.05% of interchromosomal bins. The resulting Hi-C heatmaps were normalized using a previously described iterative approach (Imakaev et al., 2012). All of the above steps were performed using a previously described pipeline (Imakaev et al., 2012).
The genome-wide directionality index (DI), a modified chi-squared statistic to measure the directional interaction bias of a locus, was determined as previously described (Dixon et al., 2012). TADs were identified by using the value midway between the mean of the values in the lowest two deciles of the contact probability along the diagonal and the mean of the values in the highest two deciles of the contact probability along the diagonal as a threshold for a low-pass filter. TADs were further required to have a minimum size of 75 kb to generously satisfy the Nyquist sampling criterion. Statistical comparisons between polytene bands and TADs or between TADs from different cell-types employed a bootstrapping approach to determine the significance of (1) the overlap between the features in the two lists, (2) the aggregate unmatched length of the features in the two lists, and (3) the Euclidean distance between the corners of the nearest features in the two lists.
The presence or absence of Hi-C compartments was determined for each chromosome as previously described (Lieberman-Aiden et al., 2009) by dividing the observed heatmap by the expected heatmap empirically determined by dividing the number of observed interactions at a given distance by the total number of loci separated by that same distance. The Pearson correlation coefficient between the ith row and jth column of the observed/expected heatmap gives the Pearson correlation heatmap.
Further details provided in the Extended Experimental Procedures.
FISH
FISH was performed on acid-fixed, squashed salivary glands as previously described (Kennison, 2000; Pardue, 2000) with further details provided in the Extended Experimental Procedures. Primers used to generate FISH probes are listed in the Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
A portion of this work was performed in the lab of Matthew P. Scott and we are grateful for his support of the project. We thank Michael Levitt for discussions and for advice and guidance regarding statistical analysis. We thank the Stanford Functional Genomics Facility and the Peter Parham lab for assistance with DNA sequencing and access to their Illumina MiSeq instrument (supported by NIH grant U01AI090905 to Peter Parham), Thomas Clandidin for use of fly facilities, and Ben A. Barres and Mariko L. Bennet for access to their fluorescence microscope and assistance with microscopy. We thank Shigeki Nagai for discussions and comments on the manuscript. Figures 2B and 5C adapted from (Lefevre, 1976), Copyright Elsevier. Figure 7, left panel adapted from (Cross and Mercer, 1993) is licensed under CC BY-NC-SA 3.0 US. This research was supported by NIH grants GM36659 and AI21144 to R.D.K. K.P.E. was supported by an American Heart Association Predoctoral Fellowship (11PRE7320067) and by the Stanford Medical Scientist Training Program NIH Training Grant (T32GM007365). T.A.H. was supported by an American Cancer Society Postdoctoral Fellowship (PF-13-191-01-DDC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The Hi-C data reported in this study are deposited in GEO: GSE72512.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures and one table and can be found with this article online.
AUTHOR CONTRIBUTIONS
K.P.E. and R.D.K. designed the study. K.P.E. performed Hi-C experiments and data analysis. T.A.H. prepared tissue for Hi-C and provided Drosophila expertise. K.P.E. performed FISH with guidance from T.A.H. All authors discussed the results. K.P.E. and R.D.K. wrote the manuscript with input from T.A.H.
REFERENCES
- Agard DA, Sedat JW. Three-dimensional architecture of a polytene nucleus. Nature. 1983;302:676–681. doi: 10.1038/302676a0. [DOI] [PubMed] [Google Scholar]
- Andreyenkov OV, Volkova EI, Demakov SA, Semeshin VF, Zhimulev IF. The decompact state of interchromomeric chromatin from the 3C6/C7 region of Drosophila melanogaster is determined by short DNA sequence. Dokl. Biochem. Biophys. 2010;431:57–59. doi: 10.1134/s1607672910020018. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. I. Autosomal puffing patterns in a laboratory stock of Drosophila melanogaster. Chromosoma. 1967;21:398–428. doi: 10.1007/BF00336950. [DOI] [PubMed] [Google Scholar]
- Balbiani EG. Sur la structure du noyau des cellules salivaires chez les larves de Chironomus. Zoologischer Anzeiger iv. 1881:637–641. [Google Scholar]
- Beermann W. Chromomeres and genes. Results Probl Cell Differ. 1972;4:1–33. doi: 10.1007/978-3-540-37164-9_1. [DOI] [PubMed] [Google Scholar]
- Belyaeva ES, Goncharov FP, Demakova OV, Kolesnikova TD, Boldyreva LV, Semeshin VF, Zhimulev IF. Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS ONE. 2012;7:e30035. doi: 10.1371/journal.pone.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. SALIVARY CHROMOSOME MAPS With a Key to the Banding of the Chromosomes of Drosophila Melanogaster. J Hered. 1935;26:60–64. [Google Scholar]
- Cross PC, Mercer KL. Cell and Tissue Ultrastructure: A Functional Perspective (W. H. Freeman) 1993 [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming W. Zellsubstanz, Kern und Zelltheilung (Leipzig: F. C. W. Vogel) 1882 [Google Scholar]
- Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The Hierarchy of the 3D Genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Ho JWK, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn K-A, Minoda A, Tolstorukov MY, Appert A, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512:449–452. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. Preparation and Analysis of Polytene Chromosomes. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophila Protocols. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. pp. 111–117. [Google Scholar]
- Keppy DO, Welshons WJ. The cytogenetics of a recessive visible mutant associated with a deficiency adjacent to the notch locus in Drosophila melanogaster. Genetics. 1977;85:497–506. doi: 10.1093/genetics/85.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lefevre G., Jr . A Photographic Representation and Interpretation of the Polytene Chromosomes of Drosophila melanogaster Salivary Glands. In: Ashburner M, Novitski E, editors. The Genetics and Biology of Drosophila. London, New York, San Francisco: Academic Press; 1976. pp. 31–66. [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter TS. A New Method for the Study of Chromosome Aberrations and the Plotting of Chromosome Maps. Science. 1933;78:585–586. doi: 10.1126/science.78.2034.585. [DOI] [PubMed] [Google Scholar]
- Pardue M-L. In Situ Hybridization to Polytene Chromosomes. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophila Protocols. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. pp. 119–129. [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial Patterning of Chromatin Regulators Uncovered by Genome-wide Location Analysis in Human Cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rykowski MC, Parmelee SJ, Agard DA, Sedat JW. Precise determination of the molecular limits of a polytene chromosome band: regulatory sequences for the Notch gene are in the interband. Cell. 1988;54:461–472. doi: 10.1016/0092-8674(88)90067-0. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata Y, Parmelee SJ, Agard DA, Sedat JW. A three-dimensional structural dissection of Drosophila polytene chromosomes. J Cell Biol. 1995;131:279–295. doi: 10.1083/jcb.131.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatolina TY, Boldyreva LV, Demakova OV, Demakov SA, Kokoza EB, Semeshin VF, Babenko VN, Goncharov FP, Belyaeva ES, Zhimulev IF. Identical functional organization of nonpolytene and polytene chromosomes in Drosophila melanogaster. PLoS ONE. 2011;6:e25960. doi: 10.1371/journal.pone.0025960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Schedl P. Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Genetics. 2000;155:1297–1311. doi: 10.1093/genetics/155.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Bateman JR, Novikov ND, Wu C-T. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics. 2007;177:31–46. doi: 10.1534/genetics.107.076356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev IF, Belyaeva ES. 3H–Uridine labelling patterns in the Drosophila melanogaster salivary gland chromosomes X, 2R and 3L. Chromosoma. 1975;49:219–231. [Google Scholar]
- Zhimulev IF, Semeshin VF, Kulichkov VA, Belyaeva ES. Intercalary heterochromatin in Drosophila. Chromosoma. 1982;87:197–228. [Google Scholar]
- Zhimulev IF, Zykova TY, Goncharov FP, Khoroshko VA, Demakova OV, Semeshin VF, Pokholkova GV, Boldyreva LV, Demidova DS, Babenko VN, et al. Genetic Organization of Interphase Chromosome Bands and Interbands in Drosophila melanogaster. PLoS ONE. 2014;9:e101631. doi: 10.1371/journal.pone.0101631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.