Abstract
Genome-wide association studies (GWAS) have provided valuable insights into the genetic basis of complex traits, discovering >6000 variants associated with >500 quantitative traits and common complex diseases in humans. The associations identified so far represent only a fraction of those which influence phenotype, as there are likely to be very many variants across the entire frequency spectrum, each of which influences multiple traits, with only a small average contribution to the phenotypic variance. This presents a considerable challenge to further dissection of the remaining unexplained genetic variance within populations, which limits our ability to predict disease risk, identify new drug targets, improve and maintain food sources, and understand natural diversity. This challenge will be met within the current framework through larger sample size, better phenotyping including recording of non-genetic risk factors, focused study designs, and an integration of multiple sources of phenotypic and genetic information. The current evidence supports the application of quantitative genetic approaches, and we argue that one should retain simpler theories until simplicity can be traded for greater explanatory power.
The search for genetic variants
The majority of biological phenotypes and many of the characters of interest to humans are complex in that they are determined by many mutations at multiple loci [1–8], as well as by many non-genetic factors. Some phenotypes show classical Mendelian patterns of inheritance and segregate within families [9–12]. However, for most traits, there is evidence that rare Mendelian mutations, low frequency segregating variants, copy number variants, and common variants all contribute toward complex phenotypes. Furthermore, across all species there is evidence of widespread pleiotropy across common diseases [13], quantitative phenotypes and Mendelian traits [14], meaning that each variant is likely to influence multiple phenotypes. The majority of current evidence is from humans (but see [8] for stature across a number of organisms), where the data for psychiatric disorders [4,15–18], diabetes [5], cardiovascular disease [19,20], obesity [21,22], and height [1] are consistent with a model where a large number of loci contribute predominantly additively to the phenotypic variation observed within populations [23–29].
Such a large mutational target in the genome has consequences for identifying new variants and for explaining the heritable genetic effects observed for the majority of phenotypes. As a large number of loci are likely to influence complex traits, then on average their contribution to the population-level variance will be small. The associations identified so far represent only a small fraction of those that influence a given phenotype, as evidenced by the fact that studies of increased range of allele frequency and sample size continue to detect additional variants, i.e. [4]. Here, we discuss how further dissection of the genetic variation for many complex traits will require larger sample size, better phenotyping including recording of non-genetic risk factors, focused study designs, and an integration of multiple sources of phenotypic and genetic information. Rather than continuing to evoke ever more complex esoteric arguments for the as yet unexplained heritable effects, we believe that current approaches in quantitative genetics coupled with gathering adequate data will dissect additional genetic variance within populations. The goal of explaining heritable effects is not purely academic. Until more of the variation expected from family studies is explained by direct analysis of the genome, there remains the possibility that we have a fundamental misunderstanding in our knowledge and conceptual framework. Identification of specific genomic variants that underpin individual differences provides the foundation for prediction, risk profiling and personalized medicine; for identifying pathways and new potential drug targets; for classifying disease subtypes; for improving and maintaining food sources; and for understanding the influence of selection and the maintenance of diversity in the natural world.
Complex trait variation
The genomic variation that we observe within a population is the result of the evolutionary forces of mutation, genetic drift, recombination, and natural selection in the evolutionary past [24], which is something that we do not know, particularly given the extent of pleiotropy across traits (Box 1). A wide range of genetic architectures, in terms of the exact number, effect size and frequency of causal variants may be consistent with current findings in humans [30]. Linkage studies and GWAS have identified many thousands of significant associations across more than 500 human phenotypes (Box 1), and it is clear that for any given trait, genetic variance is likely contributed from a large number of loci across the entire allele frequency spectrum.
BOX 1. The distribution of genetic variants across allele frequency.
The variance explained by a single causal variant depends upon its effect size and its frequency within the population. Under neutrality and random mating, the allele frequency distribution is approximately proportional to 1/[p(1−p)] [23] and the genetic variance contributed by a single variant is 2p(1−p)a2, where p is the frequency of the causal variant and a is the effect size on an arbitrary scale. Under a neutral model, this implies that most variants are rare, but most of the genetic variance is due to common variants [24].
The effect of directional selection is to increase the amount of variation explained by rare variants, because natural selection should minimize the frequency of deleterious variants in the population [24]. Therefore, for any phenotype, many causal variants will be rare, and the proportion of population-level genetic variance in complex phenotypes attributable to variants across the allele frequency spectrum will depend upon the strength of selection in our evolutionary past. The problem is that this is something that we do not know. Additionally, newly arising mutations can have pleiotropic effects on multiple phenotypes and the effect (size and / or direction) of a given mutation may not be the same for all traits. Moreover, each of the traits affected may be associated with fitness in different ways, and thus held at frequencies that are intermediate between two phenotypes (e.g. balancing selection).
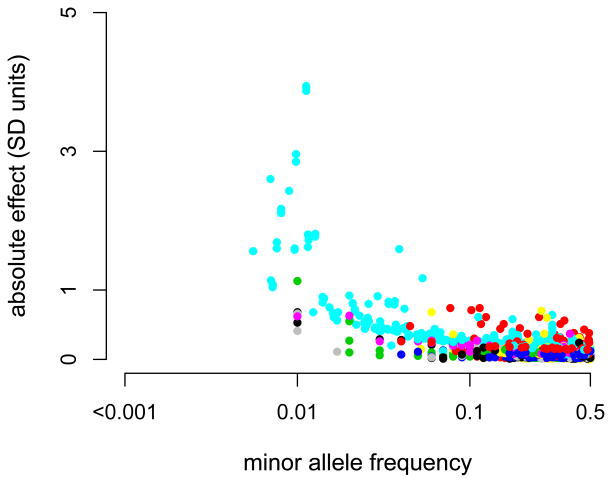
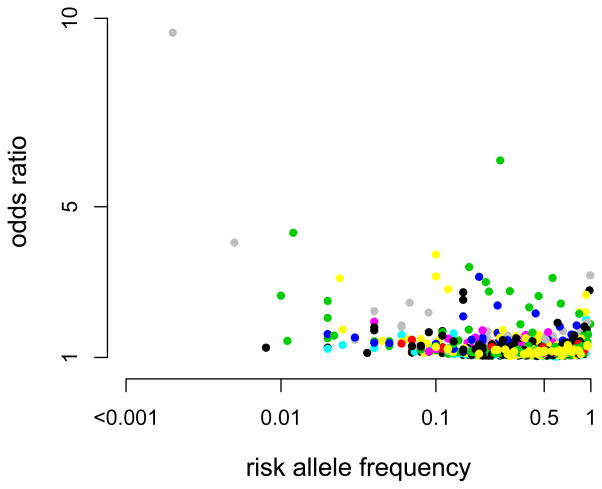
The distribution of GWAS findings to date, obtained from the Published GWAS Catalogue, across allele frequency is shown below for studies from 2008 on a selection of traits each of which is given a different color. For quantitative traits (Figure Ia) the absolute effect is plotted against the minor allele frequency, and for complex common diseases (Figure Ib) the odds ratio is plotted against the risk allele frequency. Each of the 38 quantitative traits and 43 disease traits are represented by different colors. There an ascertainment bias in that the power of detection is proportional to pa2, but it is clear that for each complex trait variance is contributed from the entire allele frequency spectrum. This highlights the scarcity of low frequency variants identified by GWAS for quantitative traits and complex disease in humans. Detecting these variants will require a combination of greater sample size, better genotyping and improved phenotyping.
BOX FIGURE I.
For quantitative traits (a) the absolute effect is plotted against the minor allele frequency, and for complex common diseases (b) the odds ratio is plotted against the risk allele frequency. Each of the 38 quantitative traits and 43 disease traits are represented by different colors.
Some researchers suggest that ‘synthetic associations’, where associations at common SNPs reflect LD with multiple rare variants, underlie many GWAS results and that drawing conclusions regarding genetic architecture from GWAS is not justified [31,32]. Although there are examples of ‘synthetic associations’ [33], they cannot explain all GWAS results [3,34,35]. Converging lines of evidence suggest a contribution from variants of >5% frequency: (i) conditional and joint analyses dissect allelic heterogeneity and distinguish among independent association signals at common SNPs [1,33,35–38]; (ii) common variants have been functionally validated [39]; (iii) associations have been replicated across distinct populations [40]; and (iv) there is some evidence for polygenic adaptation, meaning that selection has acted to alter the frequency of many common variants [41]. As sample sizes increase, the number of identified genomic regions and the amount of variation explained by association studies has increased. For example, a recent GWAS meta-analysis for rheumatoid arthritis (RA) in a total of >100,000 subjects discovered 42 novel risk loci bringing the total at the time of writing to 101 [42]. Functional annotation, the overlapping of GWAS hits and cis-action eQTL, and pathway analysis identified 98 biological candidate genes at the 101 risk loci. This GWAS study, as well as others, identifies a myriad of drug targets, which if verified, may be hugely effective because these regions are associated with RA across the majority of cases, rather than just a small number of families. GWAS has also identified new mechanisms involved in a range of diseases, such as autophagy of Crohn’s disease [43] and the role of lipid metabolism in Alzheimer’s [44]. It is clear that with sufficient sample size, large-scale association studies will shed light on fundamental genes, pathways and cell types involved in disease, and provides important information for drug discovery for treatments that are likely to be effective across many cases.
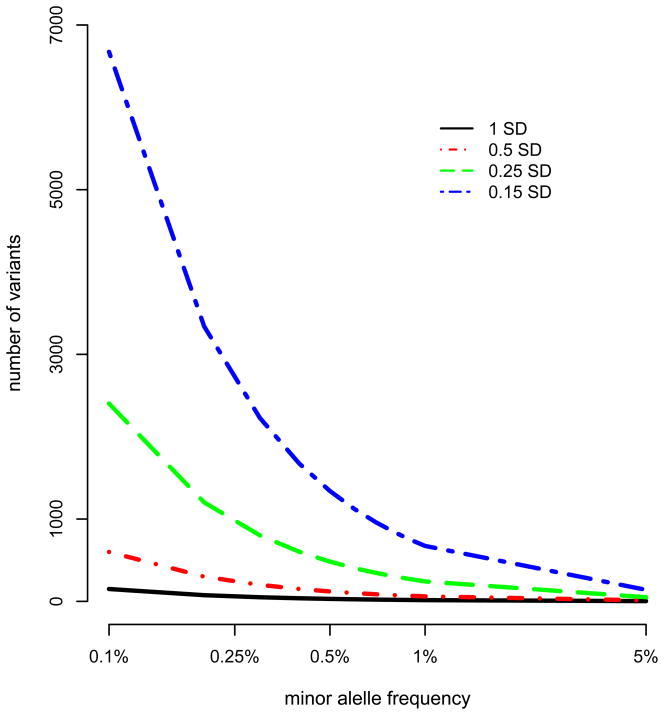
For common disorders and complex phenotypes, variation will be attributable to both rare and commonly varying regions of the genome. Individual effect sizes at common loci are modest and each SNP explains little of the phenotypic variance (Box 1). When we take the effects of all common SNPs collectively, the narrow sense heritability expected from family studies is not captured through linkage disequilibrium (LD) with currently tagged common SNPs [28]. For height, where 45% phenotypic variance is tagged by common SNPs, ~30% of genetic variation is still unexplained, and for many complex traits and diseases it appears that ½ to ⅔ of the genetic variance is not tagged by current and past SNP chips [1– 3,28,45]. These findings suggest that very many lower frequency variants are also needed to explain the genetic variance that is not tagged by current SNP chips. Using height as an example, we can model the expected number of variants that would be required to explain the remaining 30% of genetic variation, across a spectrum of low allele frequency and a range of effect sizes. Figure 1a shows that if the unexplained genetic variation for height can be attributed to low frequency variants, then a large number of segregating variants will exist even if their effect sizes on average are large.
FIGURE 1.
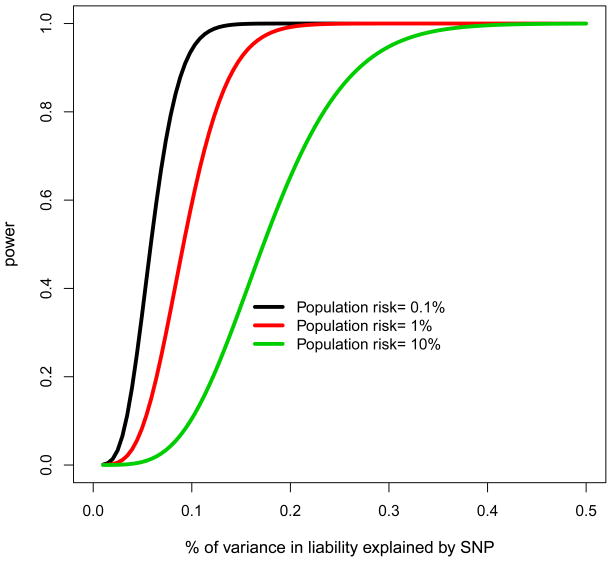
(a) The number of low frequency variants required to explain the remaining missing heritability for human height and (b) the power to detect variants that underlie complex common disease with 10,000 cases and 10,000 matched controls. For (a) the heritability remaining for human height that is not explained by associations with common SNPs was taken to be 30% and the number of variants was estimated by , where a is the effect size in SD (0.15, 0.25, 0.5, or 1) and p is the minor allele frequency of the causal variants. For (b) the power to detect variants for complex diseases of different prevalence with 10,000 cases and 10,000 matched controls.
The combined contribution of multiple rare loci to the population-level genetic variance remains an open question because association studies that focus on rare (<1% MAF) variants remain underpowered. Mutation rates have been estimated as ~1.2 × 10−8 bp−1 generation−1 in humans [46–49], meaning that individuals will posses ~60–70 novel SNP alleles, one of which (on average) will be a coding variant. Therefore, within an expanding human population most segregating variants will be rare [50]. Lower frequency coding variants that have yet to be identified are predicted to include functional variants with larger effects on risk (Box 1) and may also be key targets for new drug therapies, such as with LDL cholesterol [51,52]. Rare mutations have been identified which influence complex traits [53–57]. For example, mutations in the FBN1 gene gives a 10–20cm increase in height [58], and for schizophrenia a deletion at chr22q11 give an odds ratio of ~20 [59,60]. However, large effect sizes or odds ratios do not equate to a large contribution to the variance explained at the population-level. Even at a frequency of 10−4 (1 in 10,000) a mutation within a gene that has a large effect size of 2SD will only explain 8 × 10−4 of the phenotypic variance of a complex trait within a population. Correspondingly, FBN1 and the chr22q11 deletion explain 0.2 and 0.1% of the variation of height and schizophrenia respectively. Even more powerful recent whole exome sequencing (WES) studies of schizophrenia at the population-level [61], and Alzheimer’s disease within families [62], identified enrichment for rare variants, but this early work suggests a large polygenic burden of rare coding variants which alone may not account for the unexplained variation. Generally, rare variant association studies have found variants with large odds ratios which each explain only a tiny proportion of the phenotypic variance [63,64].
Given the likely large mutational target size for complex traits, with variation contributed across the entire allele frequency spectrum, it will be a significant challenge to identify all of the variants involved. Many researchers have questioned the need to explain genetic variation at the population-level, debated the usefulness of association studies, or even challenged the general application of quantitative genetic approaches to understanding complex trait variation [31,54,65–67], which is in our view is unjustified. Firstly, with the exception of very rare Mendelian traits, it makes little sense to focus only on affected families. The majority of complex traits will have a highly polygenic architecture which is consistent with (i) the existence of Mendelian forms in a small number of cases within a population, (ii) with common occurrences in families with no previous history [68], and (iii) with any two individuals carrying different sets of risk alleles (often termed ‘genetic heterogeneity’). High polygenicity and a large number of non-Mendelian mutations imply that variants segregate across families and thus nuclear families are no longer a natural unit. Family studies will compliment GWAS and will identify rare and de novo mutations in specific cases, but they will explain little of the variation in cases across the population as a whole, because there will be very many rare variants involved. Secondly, quantitative genetic theory does not make any assumptions about genetic architecture, which makes it a useful statistical description of the phenotypic data in pedigrees and populations. The evidence supports its application, it can accommodate non-additive effects, estimate interactions of higher orders, and it makes predictions that can be tested empirically (Box 2). Therefore this is not an either-or debate [69], and advocating a focus on solely rare or common variants will not be a productive way forward. Explaining genetic variance for complex traits will require a combination of large-scale GWAS and large-scale more targeted approaches at both the population and family-level to identify the remaining genetic variants.
Box 2. Do we need a new paradigm to dissect complex trait variation?
“We consider it a good principle to explain the phenomena by the simplest hypothesis possible.” – Ptolemy circa AD 90
Simpler explanations are, other things being equal, generally better than more complex ones. The field of quantitative genetics uses a long-standing polygenic model. Fisher’s infinitesimal model is, per definition, a simplification because there are not an infinite number of loci, each with a small effect. However, it allows a statistical treatment and description of the resemblance between relatives, partitioning of sources of variation and the response to natural or artificial selection. It may be a century after Fischer, but this paradigm has stood the test of time. Predictions from this statistical model can be tested empirically and, by-and-large empirical data are remarkably consistent with this model [25,106,107].
We do wish to know the genotypic values at interacting loci (termed by some the genotype-phenotype map [67]), the differences they create in environmental variance, and the influence of dominance and epistasis. However, while this ‘genotype-phenotype map’ is conceptually tractable, statistically determining these effects within a population at thousands of loci is a far from trivial task. Trading the GWAS assumptions of additivity for more complex assumptions worsens the problem, as interactive models lose power and there will be a huge sampling variance on the interactive effects of multiple loci. Most gene-gene and gene-environment interactions are undetected, but this is a limitation of sample size rather than the method of analysis. For example, for two loci of MAF 0.2, one homozygote has a frequency of 16 in 10,000 so estimating its effects relative to other genotypes will require large sample sizes. Given the large mutational target for complex traits, suggestions for the collection of datasets that “contain phenotypes associated with as many genotype combinations from the common and/or known allele variants as possible” [67], would likely require greater sample size than the entire planet to estimate all possible two and higher order interaction terms, be computationally non-trivial given a finite data set, and would be an impractical task for gaining conclusions.
More tractable are the improvements that we outline here, which will then enable identified regions and their potential pathways to be studied in further depth. Statistical methods fitting multiple markers [99], gene-set analyses [98], and machine learning approaches [108] will all complement GWAS findings. Through fine mapping, target gene identification, and functional identification in laboratory models [91] we can then better understand the underlying biology of complex phenotypes.
Dissecting more of the genetic variance
Epistasis, de novo mutations, or epigenetic effects are unlikely to be the explanation for the ‘missing heritability’. Appreciable epistatic variance is unlikely because most alleles will be rare and allelic substitutions have near additive effects, meaning that additivity in quantitative genetic statistical models is not inconsistent with epistasis commonly observed at the functional cellular level [25,70]. De novo mutations are not inherited by definition and so do not contribute to heritability [71] - in family studies their effects would be partitioned into a unique environmental variance component. Finally, inherited epigenetic effects would behave the same as a SNP in GWAS. Thus, a number of alternative explanations are more likely.
Firstly, as we have seen with rare variants, although effect sizes may be larger than for common variants, the variance explained at the population level by alleles of frequency <5% will be small, meaning that very large sample sizes are likely to be needed. Even when considering transmission within-families, which are likely to complement association studies, large numbers of families will be required [72]. Second, these variants are less likely to be in strong LD with common variants that are tagged on current SNP chips because (i) they may be under stronger selection and therefore be younger polymorphisms with lower minor allele frequency; and (ii) many may be deletions or duplications (i.e. CNVs) which interfere with the ability to assay SNPs near enough to be in strong LD [73]. Third, our phenotyping may be inaccurate such that we are combining phenotypes or diseases that have partially or even completely distinct underlying causal variants. This will average effect sizes across groups of individuals, who could be better separated on the basis of better phenotyping or a combination of information from different sources. Addressing these three issues is a far more pragmatic approach that will contribute significantly to identifying additional variants and explaining a larger portion of the genetic variance.
Power, sample size, and study design
The first step is increasing the number of individuals within a sample. The number of well-characterized phenotypic samples often limits sample sizes in GWAS. Power to link genotype to phenotype is a function of the set of SNPs on a chip, effect size, and sample size, and can be assessed analytically or through simulation. For ascertained case control disease studies, under a liability threshold model the expected chi-squared test statistic χ2 has a known analytic relationship 𝔼 [χ2] ∝ Nγ2 p(1 − p)r2 where N is the sample size, γ is the effect size, p is the allele frequency, and r2 is the correlation between the marker and the causal SNP [30]. Given the potentially large number of causal variants of frequency <5% that each explain little of the variance, then even if the variant is in complete LD with a genotyped SNP, and sample sizes are large there is currently low power to dissect additional genetic variation (Fig 1b). Figure 1b shows the power to detect variants that explain a small percentage of disease liability, which decreases rapidly for variants that explain <0.2% of the variance even with 10,000 cases and 10,000 matched controls.
Increasing sample size will have the greatest effect on power. Replacing high density SNP chips with full sequencing will tag low frequency loci, but it will not be enough alone to capture the effects of rare variants, because many rare variants will be at such low number that very large data-sets are is required for their detection. A recent whole-exome sequencing study for schizophrenia [61] provides an example, as it suggests evidence for a polygenic burden of rare variants, but was not successful in identifying individually significantly enriched genes. As large-scale parallel-sequencing studies of many thousands of individuals becomes common-place then sufficient power is likely to be gained, allowing both rare and common variants to be dissected to a far greater extent. This leaves only a choice of experimental design. Focused study designs will help in the identification of additional variants; using densely affected families will identify additional rare variants which can be followed-up with a combination of genotyping and deep re-sequencing of the variants or genes of interest in large numbers of cases and controls. However, as evidence by a recent Alzheimer’s study which identified only a single region [62], this study design also requires large sample size to distinguish signal from chance co-segregation.
Improvements to phenotyping
For all genetic studies, more samples have to go hand-in-hand with better phenotyping, but this is easier said than done. In general, genome analyses of a wider range of phenotypes across a wide range of species is required if we are to improve our understanding of the relationship between selection and genetic architecture. Many of our current phenotypes are subjectively measured and may represent many underlying biological processes. For example, many psychiatric disorders are diagnosed on a complex range of overlapping clinical characteristics [74], type-2 diabetes is diagnosed using a blood glucose threshold [75], metabolic syndrome is based on observing three of five criteria [76], and even many quantitative traits are arbitrary metrics or defined as functions of other characters. Misclassifying a phenotype, especially when multiple distinct phenotypes are influenced by different sets of underlying causal variants, can reduce power in GWAS relative to expectation based on power calculations of idealized homogeneous populations. Strong genotypic effects important in a small homogeneous sub-group could have a very small or even negligible effect within an entire population. These effects are prominent in cancers in which molecular subtypes have been identified such as ER +ve/−ve status HER2 expression in breast-cancer [77,78], or K-ras mutations in colorectal cancer and EGFR mutations in lung cancer, reviewed in [79]. If true for other complex phenotypes, then a single univariate measure may be unrepresentative of the biological etiology, and breaking the phenotype down into sub-phenotypes may reveal additional variants.
One approach to this may be to use additional phenotypic information collected on the same subjects. For many disease phenotypes, age-at-onset varies across subjects and could be used as a classifying term. Modeling can then be done by dividing cases into sub-groups, or by estimating genetic effects as a function across age-at-onset (for an example see [80]). Additionally, phenotypes could be stratified across the values of another associated phenotype, as has been done with T2D and BMI [81]. Insight may also be gained from adopting a multivariate approach, where jointly modeling multiple traits can give higher power than standard univariate GWAS [82]. There may be differing underlying effect sizes, or even different causal variants, depending upon the onset of a disease, or upon the values of another component of phenotype, which are more likely to be detected with these approaches. When only a single phenotypic measure is available for a given sample, mixture models may enable causal variants associated with phenotypic heterogeneity to be identified [83], but these models remain little explored and have yet to be applied in any great detail in GWAS.
Using clinical classifications and finer scale diagnoses rather than a simple case-control status can also serve the same purpose, as reviewed in [84]. The use of endophenotypes - intermediate or underlying phenotypic components that form the expression of a trait or disorder - may also be useful for dissecting additional genetic variation. For example in psychiatric disorders, different causal variants may influence multiple neural systems differentially [85–87]. Provided these endophenotypes are heritable, numerous, vary continuously, and are associated with the cause rather than the effect of the disease, GWAS on these more direct physiological or anatomical assessments may dissect additional genetic variation (for example [88]). Endophenotypes may be equally as complex as the complex traits they aim to reflect, but the use of endophenotypes within a multivariate analysis framework may enable genetic covariance to be partitioned across different shared and independent underlying pathways as a mechanism to dissect the etiology of complex diseases. Functional genomic profiling of serum or tissue, etiology-specific functional assays, and improved phenotypic assays maximizing information content will enable more rigorous genetics provided that candidate phenotypes are readily scalable and robust enough to provide accurate measures under routine data collection. There is a general perception that the study of endophenotypes has delivered less than it promised, but this may be because samples sizes are often very limited. As larger samples are collected it seems inappropriate to not accompany this with more detailed phenotyping to allow fully powered interrogation of clinical phenotypic heterogeneity.
Linking component information together
Overlapping GWAS results with other genomic sources of information is likely to explain additional variation and identify novel pathways. Studies have shown that ‘all SNPs are not created equal’, with functional SNPs being more frequently associated with phenotypes [89]. For example, chromatin marks (modifications of proteins that package DNA) were once dismissed as ‘junk DNA’, but are now thought to fulfill regulatory functions. GWAS results cluster near chromatin marks more frequently in certain cell types, enabling genetic variation to be apportioned to different cell types and regulatory pathways involved in disease expression [90]. The targeting of expression SNPs and the linking of GWAS, gene expression, and methylation data has uncovered additional variants and provided direct information on the underlying biology of complex phenotypes [91].
These approaches may also help us to understand whether genetic associations among traits represents gene expression that is shared, or whether the same variant contributes to variation in expression in different tissues that affect different traits. Additionally, leveraging this information to inform prior probability of SNP association within a Bayesian GWAS framework may also enable additional variants to be detected [92]. Thus far, integrative analyses within a systems genetics approach have largely focused on validated SNPs. Using a broader set of variants by integrating genome sequencing and cellular phenotype data will help to pinpoint putative causal genetic variants underlying GWAS associations and enable a better understanding of the biological basis of phenotype.
Using additional genomic variation
Copy number variations (CNVs) represent a significant source of genetic variation affecting ~12% of the human genome. CNVs may influence gene function and thus complex phenotypes through gene dosage imbalances, altered messenger RNA (mRNA) expression levels, or through the expression of truncated proteins [93]. Studies have demonstrated associations with both rare and common CNVs with several complex phenotypes including schizophrenia and autism [94,95] and a focus on intermediate frequency CNVs may yield additional associations.
In most GWAS, analysis is also largely confined to the nuclear genome, with much less attention paid to the organellar genome (mitochondrial DNA). This is in contrast to the central role that the organellar genome plays in controlling organismal metabolism and function, and increasing evidence from other non-human organisms that mitochondrial genomic variation can modulate the effects of nuclear genomic variation (although see [96]). Genomic variation in human mitochondria has been linked to several severe diseases, and more recently quantitative studies of common human diseases have suggested that genetic variation in organellar genomes may modify the effects of nuclear loci [97]. Including imputed mtDNA in GWAS may also yield additional variants especially if interactions between nuclear and cytoplasmic effects are estimated.
Improved analysis methods
In standard analysis of GWAS effects are estimated one marker at a time, but fitting multiple SNPs together may improve ability to dissect additive genetic variation across the genome. Multiple SNP effects can be fitted at the discovery stage to estimate genome-wide heritability across SNPs of different frequency, across segments of the genome (termed ‘regional heritability’), or in gene sets [98], and these methods have been shown to capture additional genetic variance [2,13,29], and even multiple independent variants within genomic regions [99]. At the meta-analysis stage, conditional analyses and multi-SNP association methods can also be used [100,101]. There are many known examples of multiple semi-independent associations at individual loci; such associations might arise either because of true allelic heterogeneity or because of imperfect tagging of an unobserved causal variant, and these approaches have used GWAS summary statistics to estimate the effect of loci harboring multiple association signals [100,101], which has explained additional genetic variation for many phenotypes.
In additional to the power gained [82], adopting a multivariate approach allows an estimation of the amount co-heritability, or pleiotropy across traits. Associations among multiple morphological phenotypes and among psychiatric diseases at common SNPs have been identified which further supports the role of common SNPs in complex trait variation [13,27]. At the phenotypic level there is evidence of associations between Mendelian disorders and between Mendelian and complex diseases [14], which can be better understood by examining genetic correlations among phenotypes across the genome.
Additional extensions to current models may also include the estimation of non-additive effects such as dominance and epistasis [102]; estimation of maternal effects in data where maternal genotypes are known [103]; and genotype-environment interactions [104]. Although each of these sources may only contribute to the variance of complex traits to a small degree, the variance attributed to these effects across all SNPs can be estimated. Ultimately with the plummeting costs of DNA sequencing, GWAS will employ direct DNA sequencing. Even though this will allow tests of association for low frequency variants, rare variants occur too infrequently to allow for individual associations to be tested and require aggregating variants into sets and comparing frequencies [105]. All genetic studies, whether common and rare variant association studies, or within family studies, require large samples size and well-defined phenotypes if we are to fully dissect heritable genetic variation.
Concluding remarks
The evidence to date shows that complex trait variation is due to very many loci contributed throughout the genome and across the allele frequency spectrum, each of which influences multiple phenotypes, and makes a small average contribution to the variance. Many authors, both in the early days of GWAS and more recently, have argued that GWAS has yet to dissect all of the expected genetic variance and that because many genes in a large number of distinct genomic regions have been detected, which are likely to show functional epistasis, a paradigm shift is required in order to link genotype to phenotype and dissect genetic variation. We feel that this is unnecessary (Box 2), and we believe that in humans, as well as in other species, the current framework, coupled with studies designed to identify rare variants will dissect the genetic variation of a wide range of complex traits. These steps will improve our ability to predict disease risk, identify new drug targets, improve and maintain food sources, and to understand diversity of the natural world.
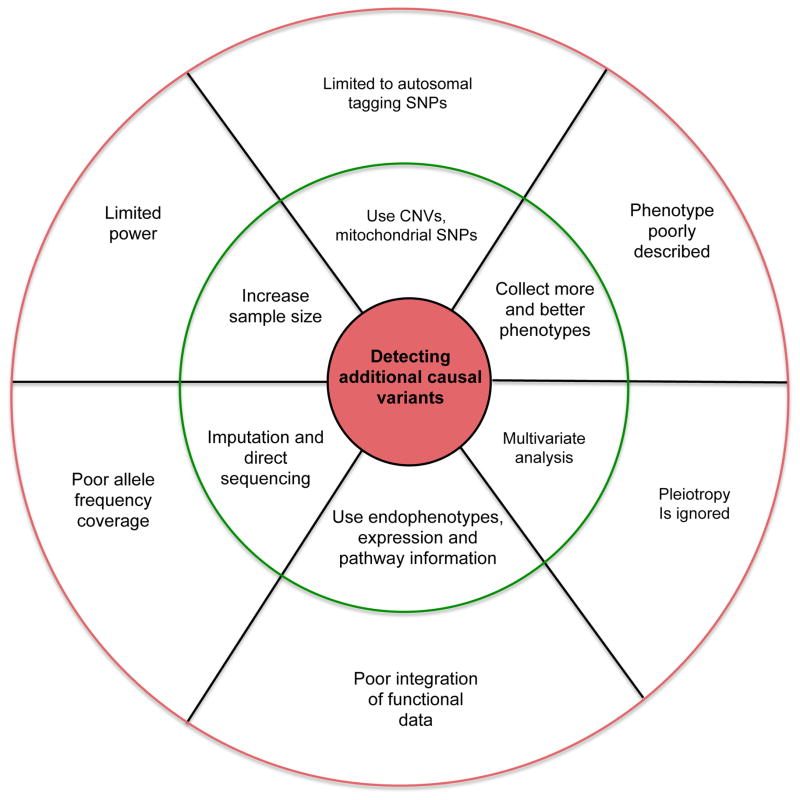
FIGURE 2. Identifying additional causal variants and dissecting additional genetic variation for complex traits.
Current limitations (outer circle) and potential solutions (inner circle) to targeting additional causal variants using whole genome studies.
Acknowledgments
We acknowledge support from the Australian Research Council (FT0991360, DP130102666), the Australian National Health and Medical Research Council (APP1011506, APP1047956, APP1048853, APP1050218, APP1047956, APP613601 APP613602) and the National Institutes of Health (GM099568, GM075091, MH100141).
Glossary
- Additive genetic variance
the total variance contributed by the additive effects of each causal variant
- Copy number variant (CNV)
a form of structural variation where there are alteration in the genome that result in variation in the number of copies of one or more sections of DNA
- De novo mutation
a genetic mutation that neither parent possessed nor transmitted
- Epigenetic inheritance
mitotically or meiotically heritable changes in gene expression or cellular phenotype caused by mechanisms other than changes in DNA sequence
- Epistasis
the interaction of genes, where the expression of one gene depends on the presence of one or more other genes
- Fitness
an organism’s ability to survive and reproduce in a particular environment
- Genetic drift
variation in the frequency of genotypes within a population due to chance events
- Heritability
proportion of observable differences in a trait among individuals within a population that is due to genetic differences
- Linkage disequilibrium (LD)
the occurrence in members of a population of combinations of linked loci in non-random proportions
- Minor allele frequency (MAF)
the frequency at which the least common allele occurs within a given population
- Mutation
the changing of the structure of a gene, resulting in a variant form which may be transmitted to subsequent generations, that is created by the alteration of a single base unit of DNA
- Odds ratio
Ratio of the odds of an event occurring in one group to the odds of it occurring in another group, i.e. the joint probability distribution of two binary random variables
- Pleiotropy
the production by a single gene of two or more apparently unrelated effects
- Single nucleotide polymorphism (SNP)
a DNA sequence variation occurring when a single nucleotide in the genome differs between members of a species
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official view of the funding bodies.
References
- 1.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SH, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–50. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visscher PM, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–9. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemper KE, Goddard ME. Understanding and predicting complex traits: knowledge from cattle. Hum Mol Genet. 2012;21:R45–51. doi: 10.1093/hmg/dds332. [DOI] [PubMed] [Google Scholar]
- 9.Scriver CR. The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat. 2007;28:831–45. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- 10.Sosnay PR, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–7. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacDonald ME, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg MH, Rodgers GP. Pathophysiology of sickle cell disease: role of cellular and genetic modifiers. Semin Hematol. 2001;38:299–306. doi: 10.1016/s0037-1963(01)90023-x. [DOI] [PubMed] [Google Scholar]
- 13.Lee SH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blair DR, et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sklar P, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connolly JJ, et al. A genome-wide association study of autism incorporating autism diagnostic interview-revised, autism diagnostic observation schedule, and social responsiveness scale. Child Dev. 2013;84:17–33. doi: 10.1111/j.1467-8624.2012.01838.x. [DOI] [PubMed] [Google Scholar]
- 17.Neale BM, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–97. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ripke S, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordell HJ, et al. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013;45:822–4. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lettre G, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–72. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom JS, et al. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–7. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eyre-Walker A. Evolution in health and medicine Sackler colloquium: Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc Natl Acad Sci U S A. 2010;107(Suppl):1752–6. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill WG, et al. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–9. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vattikuti S, et al. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8:e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. Ubiquitous polygenicity of human complex traits: genome-wide analysis of 49 traits in Koreans. PLoS Genet. 2013;9:e1003355. doi: 10.1371/journal.pgen.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwala V, et al. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet. 2013 doi: 10.1038/ng.2804. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson SP, et al. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell KJ. What is complex about complex disorders? Genome Biol. 2012;13:237. doi: 10.1186/gb-2012-13-1-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, et al. Interpretation of association signals and identification of causal variants from genome-wide association studies. Am J Hum Genet. 2010;86:730– 42. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wray NR, et al. Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biol. 2011;9:e1000579. doi: 10.1371/journal.pbio.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson CA, et al. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9:e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina-Gomez C, et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8:e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J-H, et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A. 2011;108:18026–31. doi: 10.1073/pnas.1114759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flister MJ, et al. Identifying multiple causative genes at a single GWAS locus. Genome Res. 2013 doi: 10.1101/gr.160283.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stranger BE, et al. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–83. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson CS, et al. Generalization and Dilution of Association Results from European GWAS in Populations of Non-European Ancestry: The PAGE Study. PLoS Biol. 2013;11:e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pritchard JK, et al. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–15. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2013 doi: 10.1038/nature12873. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Paolo G, Kim T-W. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–96. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Awadalla P, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316–24. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gazave E, et al. Population growth inflates the per-individual number of deleterious mutations and reduces their mean effect. Genetics. 2013;195:969–78. doi: 10.1534/genetics.113.153973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–5. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 52.Cohen JC, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264– 72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 53.Dauber A, et al. Genome-wide association of copy-number variation reveals an association between short stature and the presence of low-frequency genomic deletions. Am J Hum Genet. 2011;89:751–9. doi: 10.1016/j.ajhg.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malhotra D, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–63. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worthey EA, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 58.Nijbroek G, et al. Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am J Hum Genet. 1995;57:8–21. [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy KC, et al. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–5. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 60.Williams HJ, et al. Schizophrenia two-hit hypothesis in velo-cardio facial syndrome. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:177–82. doi: 10.1002/ajmg.b.32129. [DOI] [PubMed] [Google Scholar]
- 61.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014 doi: 10.1038/nature12975. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruchaga C, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2013;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonnefond A, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–9. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitchell KJ, Porteous DJ. Rethinking the genetic architecture of schizophrenia. Psychol Med. 2011;41:19–32. doi: 10.1017/S003329171000070X. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell KJ. What is complex about complex disorders? Genome Biol. 2012;13:237. doi: 10.1186/gb-2012-13-1-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson RM, et al. A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 2013;29:669–76. doi: 10.1016/j.tig.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Yang J, et al. Sporadic cases are the norm for complex disease. Eur J Hum Genet. 2010;18:1039–43. doi: 10.1038/ejhg.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Visscher PM, et al. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Mol Psychiatry. 2012;17:474–85. doi: 10.1038/mp.2011.65. [DOI] [PubMed] [Google Scholar]
- 70.Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002;3:11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- 71.Gratten J, et al. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat Genet. 2013;45:234–8. doi: 10.1038/ng.2555. [DOI] [PubMed] [Google Scholar]
- 72.Cheng KF, Chen JH. Detecting rare variants in case-parents association studies. PLoS One. 2013;8:e74310. doi: 10.1371/journal.pone.0074310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hannan AJ. TRPing Up the Genome: Tandem Repeat Polymorphisms as Dynamic Sources of Genetic Variability in Health and Disease. Discov Med. 2010;10:314–321. [PubMed] [Google Scholar]
- 74.Kapur S, et al. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol Psychiatry. 2012;17:1174–9. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- 75.Xu M, et al. The New Perspectives on Genetic Studies of Type 2 Diabetes and Thyroid Diseases. Bentham Science Publishers; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol Cell Endocrinol. 2012;382:740–57. doi: 10.1016/j.mce.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 78.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 79.Ferraldeschi R, Newman WG. Pharmacogenetics and pharmacogenomics: a clinical reality. Ann Clin Biochem. 2011;48:410–7. doi: 10.1258/acb.2011.011084. [DOI] [PubMed] [Google Scholar]
- 80.Traylor M, et al. Using phenotypic heterogeneity to increase the power of genome-wide association studies: application to age at onset of ischaemic stroke subphenotypes. Genet Epidemiol. 2013;37:495–503. doi: 10.1002/gepi.21729. [DOI] [PubMed] [Google Scholar]
- 81.Perry JRB, et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012;8:e1002741. doi: 10.1371/journal.pgen.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Reilly PF, et al. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS One. 2012;7:e34861. doi: 10.1371/journal.pone.0034861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Warde-Farley D, et al. Mixture model for sub-phenotyping in GWAS. Pac Symp Biocomput. 2012 at < http://www.ncbi.nlm.nih.gov/pubmed/22174291>. [PubMed]
- 84.Hall M-H, Smoller JW. A new role for endophenotypes in the GWAS era: functional characterization of risk variants. Harv Rev Psychiatry. 18:67–74. doi: 10.3109/10673220903523532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greenwood TA, et al. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2013;170:521–32. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Braff DL, et al. Deconstructing schizophrenia: an overview of the use of endophenotypes in order to understand a complex disorder. Schizophr Bull. 2007;33:21–32. doi: 10.1093/schbul/sbl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Dongen J, Boomsma DI. The evolutionary paradox and the missing heritability of schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:122–36. doi: 10.1002/ajmg.b.32135. [DOI] [PubMed] [Google Scholar]
- 88.Cruchaga C, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–68. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schork AJ, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–30. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edwards SL, et al. Beyond GWASs: Illuminating the Dark Road from Association to Function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Knight J, et al. Using functional annotation for the empirical determination of Bayes Factors for genome-wide association study analysis. PLoS One. 2011;6:e14808. doi: 10.1371/journal.pone.0014808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Henrichsen CN, et al. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 94.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–72. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Levinson DF, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–16. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Joseph B, et al. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. Elife. 2013;2:e00776. doi: 10.7554/eLife.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Samuels DC, et al. Recurrent Tissue-Specific mtDNA Mutations Are Common in Humans. PLoS Genet. 2013;9:e1003929. doi: 10.1371/journal.pgen.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, et al. Gene set analysis of genome-wide association studies: Methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uemoto Y, et al. The power of regional heritability analysis for rare and common variant detection: simulations and application to eye biometrical traits. Front Genet. 2013;4:232. doi: 10.3389/fgene.2013.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ehret GB, et al. A multi-SNP locus-association method reveals a substantial fraction of the missing heritability. Am J Hum Genet. 2012;91:863–71. doi: 10.1016/j.ajhg.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hemani G, et al. An evolutionary perspective on epistasis and the missing heritability. PLoS Genet. 2013;9:e1003295. doi: 10.1371/journal.pgen.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buyske S. Maternal genotype effects can alias case genotype effects in case-control studies. Eur J Hum Genet. 2008;16:783–5. doi: 10.1038/ejhg.2008.74. [DOI] [PubMed] [Google Scholar]
- 104.Thomas D. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu Rev Public Health. 2010;31:21–36. doi: 10.1146/annurev.publhealth.012809.103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sinauer Associates; 1998. [Google Scholar]
- 107.Hill WG, Zhang XS. On the pleiotropic structure of the genotype-phenotype map and the evolvability of complex organisms. Genetics. 2012;190:1131–7. doi: 10.1534/genetics.111.135681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szymczak S, et al. Machine learning in genome-wide association studies. Genet Epidemiol. 2009;33(Suppl 1):S51–7. doi: 10.1002/gepi.20473. [DOI] [PubMed] [Google Scholar]