Abstract
High-throughput sequencing of microRNAs has revealed the diversity and variability of mature and functional short non-coding RNAs, including their genomic origins, biogenesis pathways, sequence variability, and newly identified products, such as microRNA-offset RNAs. Here, we review known cases of alternative mature miRNA-like RNA fragments and propose a revised definition of microRNAs to encompass this diversity. We then review nomenclature guidelines for microRNAs and propose to extend nomenclature conventions to align with those for protein-coding genes established by international consortia. Finally, we suggest a system to encompass the full complexity of sequence variations, i.e. isomiRs, in the analysis of small RNA sequencing experiments.
Keywords: non-coding RNA, miRNA-seq, moR, loop-origin miRNAs, isomiR, miRNA cluster
Small RNAs and their functions
At least three kinds of small RNAs help regulate animal gene expression: Piwi-interacting RNAs (piRNAs) and two RNA-interference (RNAi) systems, small interfering RNAs (siRNAs) and microRNAs (miRNAs) [1–4]. The animal-specific piRNA system involves RNA molecules about 24–30 nt (nucleotides) long that form RNA-protein complexes by interacting with Piwi proteins to regulate epigenetic and post-transcriptional gene silencing of retro-transposons and other genetic elements primarily in germ cells, especially during spermatogenesis [5]. Second, the siRNA pathway regulates genetic expression using about 21 nt long RNA molecules of either exogenous (often viral) or endogenous origin [6]. siRNAs can interact with a Argonaute protein and incorporate into an RNA-protein complex (RISC, RNA-Induced Silencing Complex) to alter chromatin structure, transcriptional regulation, and transposable element repression; siRNAs can also participate in post-transcriptional regulation by inducing messenger RNA degradation based on complementarity between the siRNA and specific mRNAs. siRNAs can act as an antiviral defense system that targets viral mRNAs using exogenous fragments of the target viral mRNA, or can act as a self-cell regulator of translation by the production of endogenous siRNA [6]. Finally, the miRNA pathway uses endogenous small RNA molecules about 22 nt long to modulate the translation of targeted mRNAs by acting on their stability or translation through interaction with the RISC [6].
miRNAs help control cell fate determination, cell differentiation, and organ development and physiology and are involved in pathology and disease in humans, other metazoans, and plants [7–11]. Recent deep sequencing experiments and powerful bioinformatic tools [12] have led to a dramatic increase in the number of known miRNA genes (miRNA genes), even in well-studied organisms such as mouse and human. Databases, such as miRBase [13], ZooMir [14], miRNEST [15] and miRMaid [16], have cataloged miRNA sequences across various organisms. miRBase, the most commonly used reference microRNA database, currently lists 1915 mouse and 2588 human mature miRNA sequences (miRNA), but dramatically fewer in most other species; for example, miRBase currently lists only 350 mature miRNAs in zebrafish (release 21; [13]). The extent to which the disparity in annotated miRNAs among species reflects biology with evolved differences in genome content or instead, simply variation in sequencing effort or in stringency of gene annotation (which seems more likely for closely related species), is an issue requiring investigation.
The constantly increasing number of reads delivered by massively parallel sequencing strategies reveals putative new miRNA genes, some of which are indeed novel but others of which are orthologous to miRNA genes previously described in other species. Curiously, many short reads containing some features of mature microRNAs appear to originate from other classes of RNA, such as snoRNAs (small nucleolar RNAs) or lncRNAs (long non-coding RNAs) [17]. Deep sequencing has also revealed isomiRs [18], which are variant miRNA sequences that all appear to derive from the same gene but vary in sequence due to post-transcriptional processing. Because isomiRs contain different sequences, they may have different targets and thus different cell functions [19]. Great sequencing depth and careful data mining also reveal moRs (microRNA-offset RNAs) [20–22] and single strand RNA fragments released from the loop RNA portion of the precursor miRNA hairpin [23,24] (which we call “loop-origin miRNAs”, or “loRs”). Although functional evidence remains scarce, these variants appear to be independently regulated and may play cooperative, complementary, or alternative roles in cell function and gene regulation compared to their associated miRNAs. This complexity in genomic origin and variation in sequence may impact cell biology but is so far under-studied; furthermore, current nomenclature conventions make annotation rather subjective depending on the emphasis of each researcher in accepting or rejecting putative new miRNA-related sequences.
A problem regarding short RNAs derived from non-microRNA genes is that transcription may be quite widespread across the genome and that many such perhaps randomly transcribed sequences may be without function, although they might be detected with deep sequencing. In addition, it is possible that some promiscuously transcribed sequences might be processed by Drosha/Dicer and with ever deeper sequencing, identifying this “noise” becomes more likely. Consistent lengths, 5′ processing sites, species conservation, and tissue-specific expression patterns provide evidence for control, and hence circumstantial evidence for function. A prudent course of action would seem to be to annotate apparent isomiRs and/or miRNA-like fragments derived from non-microRNA genes to facilitate testing for functional roles of individual sequences.
The complexity of miRNA origins and sequence variations inhibit optimal communication and connectivity among species. Here, we explore recent findings and opinions concerning miRNA variations and raise the question: What should define an animal miRNA: a single biogenesis pathway or a specific mode of action? To answer this question, we propose to define miRNAs from a functional rather than a biogenesis perspective. In addition, we suggest nomenclature guidelines that parallel international protein-coding gene nomenclature guidelines to improve connectivity in miRNA research across metazoans.
Canonical micoRNAs: products and biogenesis
Most miRNA genes are usually transcribed by RNA polymerase II into a long transcript called the “primary microRNA transcript” (pri-MiRNA) [19,17]. For miRNA genes organized in clusters, the pri-MiRNA can be monocistronic or polycistronic. Within the nucleus, the DROSHA and DGCR8 (known as Pasha in D. melanogaster and C. elegans) microprocessor complex cleaves the pri-MiRNA into a shorter sequence, called the “microRNA precursor” (pre-MiRNA), which displays a hairpin-like secondary structure [25]. The pre-MiRNA is exported to the cytoplasm by Exportin-5 in a complex with RAN-GTP, and then DICER1, an RNase III enzyme (hereafter called Dicer), cleaves off the loop of the pre-MiRNA and generates a mature miRNA duplex. The two strands of the miRNA duplex pair with a few mismatches, and each strand usually has a two-nucleotide overhang at its 3′ end. The duplex then loads onto an Argonaute protein, the effector of the RISC [25], where the unwinding of the duplex and the selection of a strand occur[26,27]. The mature miRNA, as part of the RISC, binds the 3′untranslated region (UTR) of specific messenger RNAs and mediates mRNA degradation, destabilization, or translational inhibition according to sequence complementarity to the target [19,28,29]. This general biogenesis pathway and the features of the intermediate transcripts define a so-called “canonical Mir gene” (Figure). Criteria for the annotation of canonical microRNA genes were first defined in 2003 [7] and rely on combined expression and biogenesis criteria (Box 1). Criteria have been revised over the years (Box 1) to fit with new knowledge often arising from technological improvements [13,30–33].
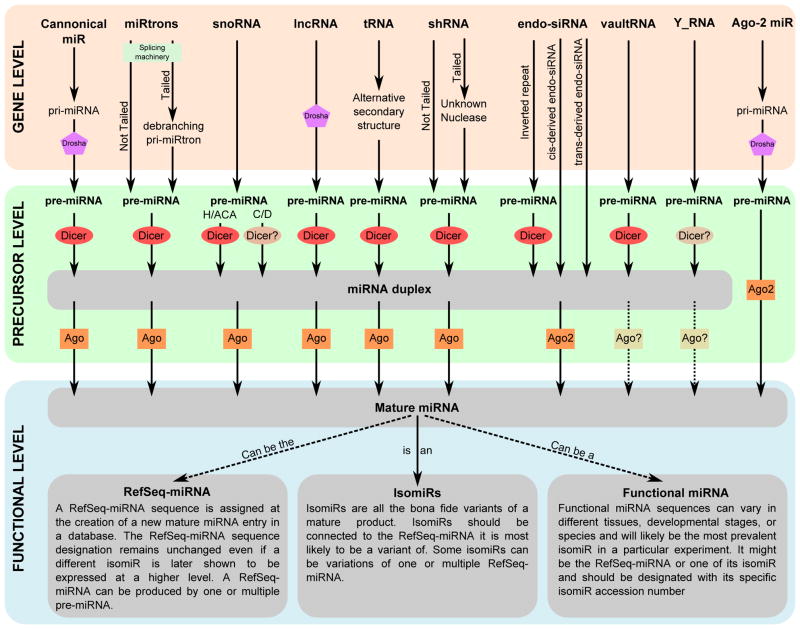
Figure 1. Diverse genetic origins and biogenesis pathways leading to functional microRNAs.
Many different types of non-coding RNA genes (e.g. canonical miRNAs, snoRNAs, lncRNAs, tRNAs) are transcribed, cleaved into precursor microRNAs (pre-MiRNAs) and eventually processed into miRNA duplexes from which one strand loads into an Argonaute protein (AGO) and represses mRNA translation through the RNA-Induced Silencing Complex (RISC). Canonical miRNAs, lncRNA and AGO2-processed miRNAs (i.e. MiR451) depend on DROSHA and DGCR8 for processing of the primary miRNA transcript into the precursor miRNA. miRtron precursors, on the other hand, are produced by the messenger RNA splicing machinery into simple miRtrons, in which intron splicing defines both ends of the hairpin, or tailed miRtrons, which contain templated regions 5′ or 3′ to a splice-derived terminal intron. snoRNA, shRNA, inverted-repeat derived endo-siRNA, vaultRNA and Y_RNA appear to be DROSHA-independent and are transcribed directly into an RNA fragment displaying the characteristics of a precursor miRNA. In some cases, tRNAs can also be cleaved to produce small RNA fragments by folding into an alternative secondary structure that shows characteristics of a precursor-miRNA. Dicer processes most precursor miRNAs (i.e. canonical miRNA, miRtron, some snoRNA, lncRNA, tRNA, shRNA, inverted-repeat derived endo-siRNA, and vaultRNA) to give rise to a miRNA duplex. Some snoRNA-derived miRNAs and Y_RNA-derived short fragments appear to be Dicer-independent but the enzyme producing the short fragments remains currently unknown. Cis- and trans-derived endo-siRNAs are by nature transcribed directly into a miRNA duplex. miRNA duplexes are then unwound and one side of the duplex, the mature miRNA, is loaded into an AGO protein that drives the assembly of the RISC and the repression of protein translation of the targeted transcript. An AGO2-processed miRNA is an exception to this biogenesis step because it is Dicer-independent and the catalytic activity of the AGO2 protein produces the mature miRNA. Functional experiments are critical to separate true microRNAs from degradation products of these other RNA types.
Box 1. A timeline for the evolution of miRNA annotation.
Criteria for annotating miRNA genes and miRNA transcripts have evolved since microRNAs were first discovered.
2003: An understanding of microRNA biology begins to emerge. microRNA identity requires a combination of at least one expression and one biogenesis criterion [109]:
-
Expression criteria:
-
A.1
Hybridization to a size-fractionated RNA sample detects an RNA transcript of about 22nt.
-
A.2
A cDNA library made from size-fractionated RNA identifies a sequence of about 22nt.
-
A.1
-
Biogenesis criteria:
-
B.1
The precursor sequence is predicted to fold into a hairpin-like structure with the lowest free energy and display the microRNA in one arm of the hairpin.
-
B.2
An approximately 22nt microRNA sequence and its predicted fold-back precursor is conserved across phylogenies.
-
B.3
The precursor accumulates in mutants with reduced Dicer function.
Special case: Candidates identified by sequence analysis can be annotated without further experimental evidence if they are homologous/orthologous to microRNAs from related organisms.
-
B.1
2011: Results begin to appear from deep sequencing technologies. Criteria revised to avoid mis-annotation of small RNA fragments originating from a variety of RNA species by focusing on microRNA biogenesis [13]:
Multiple reads support the sequence of the mature microRNA in several independent libraries.
Reads support mature sequences from both arms of the predicted hairpin and the putative mature sequences pair with correct two-nucleotide 3′-overhangs.
Reads map to genomic DNA and sequence flanking the putative mature microRNA is predicted to form a microRNA precursor-like hairpin with strong pairing between the mature microRNA and the opposite arm.
Reads mapping “very many times” to a genome are discarded.
Mapped reads do not overlap other annotated transcripts (mRNAs or other known RNA types).
Reads support a single 5′-end of the mature sequence, reflecting precise nucleotide processing, which contrasts to heterogeneous ends that characterize degraded RNAs processing of other RNA types.
2013: Increasing evidence suggests the presence of dubious miRNAs annotated in miRNA databases; criteria evolve toward defining “high confidence mirs” [33]:
At least 10 reads must map with no mismatches to each of the two possible mature miRs.
The two mature miRs must pair in a duplex with 0–4nt overhangs at their 3′ ends.
At least 50% of the reads for each arm of the pre-miRNA must have the same 5′ end.
The predicted hairpin structure must have a predicted folding free energy lower than −0.2kcal/mol/nt.
At least 60% of the bases in the mature miR should pair in the predicted hairpin structure.
2015: Special cases of miRNA-like sequences from non-canonical biogenesis pathways, like DROSHA-independent miRNAs (e.g. miRtrons), Dicer-independent miRNAs (e.g. Mir451), and pathways from other non-coding RNA genes (e.g. snoRNA or lncRNA) could be accepted as miRNAs.
Alternative sources of miR-like small RNAs
Although canonical miRNA products encompass the largest fraction of known mature miRNAs, several miRNA-like products originate by DROSHA-independent and/or Dicer-independent biogenesis pathways and from the processing of other non-coding RNA biotypes and associated with Argonaute proteins [34]. Examples include miRtrons, Argonaute RISC catalytic subunit 2 (Ago2) processing, and origins from various other non-coding RNA genes, including snoRNA, lincRNA, and tRNA genes (Figure).
miRtrons
The study of Drosha mutants identified miRtrons, miRNAs that are produced from an intron of a protein-coding gene by Drosha-independent pre-mRNA splicing machinery [35–37]. Expression of miRtrons depends on the host gene because at least one end of the pre-MiRNA hairpin aligns exactly to the pre-mRNA splice site junction; in contrast, other intronic miRNAs can be regulated independently of their host gene [38–40]. Because miRtrons are expressed with the host protein-coding gene and are processed by the mRNA splicing machinery rather than by the microprocessor complex, they are non-canonical miRNAs, although they follow other miRNA annotation criteria (Box 1).
DROSHA-dependent, Dicer-independent miRNAs
Another alternative mode of microRNA biogenesis came from the study of Dicer1 null mutant mice, which revealed miRNAs that are processed by DROSHA, but not Dicer [41,42]. For example, in mouse and zebrafish, pre-MiR451a is transcribed, along with its neighbor pre-MiR144, into a polycistronic pri-MiRNA that is processed by DROSHA, but its subsequent maturation into a unique strand, the 5p strand, occurs not by Dicer, but by AGO2, the only vertebrate Argonaute protein displaying catalytic activity [42–44].
miRNA-like small RNAs from other non-coding RNAs
Alternative miRNA origins have been identified due to recent high-throughput sequencing, which identified “miRNA-like” small RNAs that are loaded into the RISC [34]and mediate mRNA translation but derive from various non-coding RNA types, including small nucleolar RNA (snoRNA) [45–48], long non-coding RNA (lncRNA) [49], shRNA (Short hairpin RNA) [50] or transfer RNA (tRNA) [51]. For example, mouse MiR664 originates from Snora36b; mouse MiR320 is initially expressed as an shRNA [50]; mouse MiR1983 derives from the processing of an alternative secondary conformation of a tRNA(Ile) [50]; and mouse MiR205 comes from the lncRNA 4631405K08Rik [52] (although note that by definition, all canonical pri-miRNAs are lncRNAs). Several sequences originating from such non-coding RNAs, like MiR1274a, which is a fragment of a tRNA(Lys), have been removed from recent miRBase releases (present in release 16, but gone from release 17 and later), while others, like MiR1983, remain as valid entries (release 21). Short “miRNA-like” RNAs that derive from a variety of non-coding RNAs but load into the RISC and act like canonical miRNAs probably arise from a pre-miRNA-like hairpin alternative secondary structure of the precursor that Dicer processes to form a short fragment that subsequently loads into the RISC [50,53,54,34]. Although these sequences have a different genomic origin than canonical miRNAs, they show other key features of functional miRNAs, including origin from a hairpin-like precursor, processing by Dicer, evolutionary conservation, loading into the RISC, and functional repression of translation. These considerations call into question the current definitions of miRNA genes and mature miRNA products and the practice of rejecting microRNAs that emerge from non-microRNA non-coding RNA genes while emphasizing the need to distinguish bone fide microRNAs from degradation products.
endo-siRNAs
Some endogenous small interfering RNAs (endo-siRNAs) bind AGO2, load into the RISC, and repress translation [55]. Any of three different mechanisms can make endo-siRNAs: 1) transcription of inverted repeats producing a hairpin-derived endo-siRNA that Dicer processes into an RNA duplex; 2) transcription of short fragments from both DNA strands of a genomic locus giving a cis-derived endo-siRNA that forms a double-stranded RNA duplex; and 3) transcription of two different genomic loci producing RNA fragments that hybridize to assemble into a trans-derived endo-siRNA double-stranded RNA duplex [50,56]. At least some of these endo-siRNAs appear to have a function comparable to canonical miRNAs by cleaving mRNA targets when pairing is perfect [55]. endo-siRNAs, however, also play a role in chromatin regulation, silencing transposable elements in both germ line and somatic cell lines, and can act as co-transcriptional silencers of gene expression [57]. Although endo-siRNAs do not form by the canonical miRNA biosynthetic pathway, when loaded onto AGO2 and part of the RISC, they appear to function like miRNAs by inducing the decay of mRNA targets; the miRNA regulation system of animals and plants likely emerged from the siRNA machinery [58]. The finding that siRNAs and the miRNAs of both plants and cnidarians, the basally diverging sister group to bilateral animals, use near-perfect pairing to their targets and act by mRNA cleavage, suggests an independent origin of the miRNA systems of plants and animals and the evolution of new miRNA mechanisms in the bilateria [59]. Given the likely evolutionary origin of the miRNA system of gene regulation, it seems likely that some endo-siRNAs could be confused with micoRNAs, especially in basal metazoans.
Y_RNA and vaultRNA
Small RNAs derived from Y_RNA or vaultRNA genes can be found in small RNA sequencing and/or AGO pull-down sequencing datasets [34]. Interestingly, most of those miRNA-like reads map to genes orthologous to human or mouse Y_RNA or vaultRNA genes that also produce small RNA products from a similar position of the transcript, suggesting that they are evolutionarily conserved short products of those non-coding RNAs and not simply degradation fragments. For example, we identified reads from the mouse Y_RNA gene Rny3 (orthologous to human RNY3 [60]) or from the vaultRNA gene Vaultrc5 (co-orthologous to human VTRNA1 genes [61]) in small RNA sequencing and/or AGO pull-down sequencing datasets such as SRP010168 from [22] or datasets deposited at FaceBase (https://www.facebase.org/project/155/datasets). Sequences originating from orthologous regions of Y_RNA or vaultRNA genes were previously annotated in human as MIR1979 and MIR886, respectively, but have now been removed from the miRBase database [33]. Short reads originating from several Y_RNA and vaultRNA genes were also recently reported in zebrafish [62]; one, mir733, from the vault.4 gene and another one, mir735, from the Y_RNA.2 gene continue to appear in miRBase (MI0004778 and MI0004781, respectively). Even though those fragments appear in various datasets and show evolutionary conservation, a function in repressing translation has been shown only for vault fragments [63] and not yet for Y_RNA fragments [64,65], although the tests performed on Y_RNA fragments might have involved sub-optimal conditions [66]. More work is needed to decipher the putative function in translation repression of these evolutionary conserved miRNA-like fragments and hence whether they should be considered as miRNAs.
Clarifying the question: what is a microRNA?
Evidence reviewed in the previous section shows that many types of small non-coding RNA fragments can sometimes function in a manner similar to canonical miRNAs, i.e. they are small, they can load into the RISC, and they can act to mediate cleavage or to inhibit translation of targeted mRNAs even though they originate from other ncRNA biotypes and do not strictly follow the canonical miRNA biogenesis pathway. This situation raises the question of whether 1) miRNAs should be defined by mature sequence characteristics and their putative function, or 2) by their biosynthetic pathway. The biogenesis definition [31,32] would allow only canonical micoRNAs and possibly miRtrons to be annotated as microRNAs. The mature sequence characteristics definition emphasizes the structure, sequence conservation across species and samples, regulated expression, and putative action of the mature product, including short, single-stranded RNAs, effective Argonaute protein loading, and translation repression (Box 2).
Box 2. Expanding miRNA nomenclature to encompass alternative miRNA origins.
Many small non-coding RNA sequences are processed from hairpin-like precursors or RNA duplexes (like pre-MiRNAs), are functionally loaded into an Argonaute protein in the RISC (as are miRNAs), and are developmentally regulated, despite the fact that they are derived – non canonically –from non-Mir, non-coding RNA genes (such as snoRNA or lncRNA genes) (Figure). Many of those sequences were first annotated as miRNAs because of their function or conservation across samples and species, but some were subsequently removed from databases after their genomic origin was understood. A paradigm shift, however, that takes a functional rather than a biogenesis approach to those sequences suggests that some of these sequences should remain as valid entries in miRNA databases or in RNA-Induced Silencing Small RNA databases. To apprehend the full complexity and diversity of RNA-induced silencing mechanisms and to accommodate miRNA-like sequences emerging from non-miRNA genes, we propose three levels of miRNA annotation (Figure):
The “Functional sequence” level, which corresponds to the “mature sequence” level already implemented in miRBase and referenced with a “MIMAT” accession number. For a sequence to qualify, it must be an endogenous single-stranded RNA molecule ~22 nucleotides long, with a significant number of reads allowing statistical analysis and mapping to a ‘limited number’ of locations (maybe about ten) in the reference genome, preferentially found in several independent libraries, and displaying a consistent 5′-end of the mature sequence (to discriminate products originating from degradation or other modes of non-coding RNA processing).
The “Precursor miRNA” level, which corresponds to the actual miRBase “Stem-loop” level referenced with an “MI” accession number in miRBase. The sequence should be able to fold into a hairpin-like structure allowing its processing into a mature miRNA. If transcriptomic evidence exists, any identified moRs should be included into the precursor sequence because this additional sequence will influence the stability of secondary structure folding.
A new “Gene” level which denotes the genomic origin and encompasses miRNAs coming from canonical miRNA genes, miRtrons, snoRNA, lincRNA, tRNA, and others.
These revised criteria and additional hierarchical levels would accept new mature miRNAs if they originate from non-canonical miRNA genes (abandoning the criterion that they have to come exclusively from miRNA genes) and recognize miRNAs cleaved by AGO2 or originating from Dicer-independent pathways (abandoning the previous criterion accepting only Dicer-processed sequences). Revised criteria would continue to accept sequence orthology among closely related species as sufficient evidence for annotation. Special attention to sequence functionality should, however, be focused on novel miRNA-like sequences never previously described in any species. In addition, for species that can be readily studied in the laboratory, validation of dependence on RNase III, Argonaute loading and/or functional activity in translation and mRNA stability would reinforce the miRNA annotation.
We propose to distinguish miRNAs from other classes of RNA by dissociating the three notions of “miRNA gene” (Mir), “miRNA precursor” (pre-MiRNA), and “mature miRNA product” (MiRNA). The proposed new definition recognizes that miRNA genes produce miRNAs, but that some miRNAs may originate from genes transcribed into other types of non-coding RNAs. This revised definition distinguishes function from biogenesis and simplifies the nomenclature of miRNAs and miRNA-like short sequences (Box 2). These new relationships and connections are now implemented in the Sequence Ontology Database [67], which provides an ontology framework to human and mouse nomenclature consortia [68,69].
microRNA nomenclature evolution
New knowledge of miRNA origins, biogenesis, and function requires a revision of nomenclature guidelines to provide an unbiased naming protocol, consistent with coding-gene guidelines, that maximizes the connectivity of miRNA analyses across species (Table). International genome feature nomenclature guidelines for model animals are already in use (e.g. http://www.genenames.org/, [69]; http://www.informatics.jax.org/, http://rgd.mcw.edu/ [68,70], http://zfin.org/, [71]), but have not been applied consistently by researchers and repositories regarding miRNA nomenclature. Examples given here use human, mouse, and zebrafish guidelines. Other species, however, should use guidelines established for the most closely related model species.
Table 1.
Summary of nomenclature guidelines for microRNA genes and their various products in human, mouse, and zebrafish. In species lacking nomenclature conventions, symbols should follow nomenclature guidelines established by the most closely related reference model organism consortium as agreed on by researchers studying the species.
| Symbol
|
Signifies | ||
|---|---|---|---|
| Human | Mouse | Zebrafish | |
| MIRX | MirX | mirX | microRNA gene X |
| MIRXy | MirXy | mirXy | yth member of the mirX family (y can be a letter or a “dot-number”) |
| MIRXOS | MirXos | mirXos | mirror microRNA gene of the microRNA gene X |
|
| |||
| pri-MIRX | pri-MiRX | pri-miRX | primary microRNA transcript X |
| pre-MIRX | pre-MiRX | pre-miRX | microRNA precursor X |
|
| |||
| MIRX-5p | MIRX-5p | MiRX-5p | 5′ arm mature miRNA sequence |
| MIRX-3p | MIRX-3p | MiRX-3p | 3′ arm mature miRNA sequence |
| MORX-5p | MORX-5p | MoRX-5p | 5′ arm mature moR sequence |
| MORX-3p | MORX-3p | MoRX-3p | 3′ arm mature moR sequence |
| LORX | LORX | LoRX | loop fragment of the miRNA precursor |
|
| |||
| MIRCX | MircX | mircX | mir gene cluster X |
General case
Nomenclature guidelines should:
Respect species-specific gene nomenclature conventions. For example, MIR125a and Mir125a represent orthologous miRNA genes in human and mouse, respectively, and mir125aa and mir125ab symbolize the two zebrafish co-orthologs of the human MIR125a gene that were derived from the teleost genome duplication [72,73].
Retain the convention that MIR, Mir, mir (human, mouse, zebrafish), italicized and with a lower case “r” (except for human), refers to the gene or genomic locus; “MIR” (MiR, miR) italicized and with a capitalized “R”, refers to intermediate transcripts; and “MIR” (MIR, MiR) not-italicized and with a capitalized “R”, refers to the final products: the mature microRNAs.
Remove reference to species in the official gene name. For example, “mmu-” should be removed from the previous designator for the mouse gene “mmu-Mir-125a”. Reference to the organism should be clearly stated within the text to avoid confusion. Databases that cover multiple species might need to retain species designators.
Remove hyphens separating ‘Mir’ from the number designator. Hyphens are not generally used in human, rodent, and zebrafish gene designations. For example, the gene “hsa-MIR-125a” becomes MIR125A, “mmu-Mir-125a” becomes “Mir125a”, and “dre-mir-125aa” becomes “mir125aa”.
Specify various forms by unambiguous designators. Genomic locus, primary-miRNA transcript, miRNA precursor, and mature forms should use, for example, for the mouse gene, “Mir125a”; the primary miRNA transcript, “pri-MiR125a”; the miRNA precursor, “pre-MiR125a”; and the mature form, “MiR125a”. Hyphenation of the primary miRNA and the miRNA precursor (i.e., “pri-MiR” and “pre-MiR”, respectively) are retained in accordance with the common usage of the “pre-mRNA” symbol to refer to a precursor to a messenger RNA, and the italicization follows nomenclature guidelines for italicization of intermediate transcripts.
Annotation should inform without bias. Formerly, one of the two mature products of a Mir gene was called the “mature strand” or “guide strand”, and was considered to be the miRNA because it was more highly expressed, while the opposite strand was called the “passenger strand” or “star strand” (miR*), because it was less expressed and thought to be inactive [74]. Alternative strands, however, can be differentially expressed in different tissues, developmental stages, pathological states, or species, requiring inversion of the miR/miR* annotation to reflect expression variations if the miR* strand is functionally active [75–78]. For example, MIR140-3p is the most highly expressed miRNA strand in mouse ovaries and testes but MIR140-5p is the most highly expressed strand in cartilage [79–81]. Appropriately, miRBase has, since release 17, replaced the “miR/miR*” symbolism with the unbiased “5p/3p” strand annotation to indicate the position of the strand in the pre-MiRNA hairpin independently of its expression status in any given dataset [33].
Special cases
Mirror-miRNA genes provide another nomenclature problem. Mirror-MiRs are pri-MiRNAs independently transcribed from both DNA strands [34,62,82,83]. Although mirror-mir pairs are evolutionarily associated, they may be transcribed independently and are unlikely to have a conserved seed; thus, mirror-miRs may act on different target protein coding genes. The convention for protein-coding genes is to use the suffix “os”, for “opposite strand”, for a pair of overlapping genes transcribed from opposite strands. We propose to apply this convention to mirror-miRs: the first Mir gene described would be assigned a number, and a transcript documented to come from the opposite strand in the same genomic location would get the “os” suffix. For example, Mir3120, the mirror gene of Mir214 [83], should be called Mir214os and its respective mature sequences MIR214os-5p and MIR214os-3p (Table).
miRNA genes with identical or similar mature products can be confused. Because these genes come from gene duplication events, including genome duplication, segmental duplication, or tandem duplication, using the same miRNA number has the advantage of grouping these genes in a common gene family that reflects related structure and evolutionary origin. Adding an additional letter or number as a suffix should follow established species-specific conventions for coding genes. For example, in zebrafish, “a” and “b” designators reflect an origin by the teleost genome duplication and thus an ohnology relationship. Examples include mir125aa and mir125ab, which reside on zebrafish chromosomes LG16 and LG19, which are ohnologous chromosomes from the teleost genome duplication [72], and MIR125A and MIR125B1 in human chromosome bands 19q13.41 and 11q24.1, respectively, derived from the two rounds of early vertebrate genome duplications [84,85]. In accord with tandemly duplicated protein coding genes, the use of a “dot-number” suffix should indicate tandem mir gene duplicates; for example, the zebrafish tandem duplicates mir34b and mir34c should be named mir34b.1 and mir34b.2, respectively according to zebrafish nomenclature conventions. In mouse and human, the approved guideline is the use of letter designators to reflect duplication (Table).
Clustered miRNA genes, such as a group of six miRNA genes (Mir17, Mir18, Mir19a, Mir20a, Mir19b-1, and Mir92a-1) on the distal region of mouse chromosome 14, are defined by a possible common expression as a unique polycistronic pri-MiRNA. No clear guidelines have yet been established to refer to microRNA clusters; sometimes they have been referred to by the first and last genes of the cluster, in the sense of transcription, separated by a tilde “ ~ ” or a dash “-“ when all genes are transcribed in the same orientation; in this example, the cluster would be annotated Mir17~92 or Mir17-92 [86]. For database purposes, however, the use of a tilde is not appropriate because it usually has a predefined meaning. We suggest the use of the nomenclature symbol “Mirc”, standing for “microRNA cluster”, which is already in use in the Mouse Nomenclature System [68]. MicroRNA clusters can be named and referred to by following nomenclature conventions already in use for specific species using sequentially numbered clusters [57], as for example, the cluster above is called Mirc1 in mouse. Or, we suggest that, in other species, the number of the cluster refers to the first gene of the cluster in the sense of transcription, as is currently used for human microRNA clusters [58]; the MIR17-92 cluster in the example above, which is currently called MIR17HG in human (where HG stands for host gene, but in many cases, each MIR gene in the cluster would be its own transcriptional unit, questioning the use of the term ‘host gene’), would be called MIRC17 in Human and mirc17 in zebrafish (Table). It is noteworthy that, in some species or duplicated clusters within a species, the first gene of the cluster, which would give the cluster its name, can be lost. It would then be the responsibility of the researcher to establish orthology/paralogy relationships between clusters and give the cluster a name that preserves evolutionary relationships even if the first gene (or indeed other genes in the cluster) is lost, for example, if in zebrafish the ‘b’ copy of this cluster is missing mir17, the cluster would nevertheless be called ‘mirc17b’, in analogy with Hox clusters, which retain the cluster designation across organisms even when the exact gene content of each cluster may differ in different species.
Other relevant microRNA products
IsomiRs
High-throughput sequencing has revealed a variety of mature sequences apparently transcribed from a single Mir gene. These bona fide mature miRNA variants, distinguished from sequencing errors by their frequency, are called isomiRs and they can vary in length or contain untemplated nucleotide additions or post-transcriptional base edits [18,22]. Most commonly, variations involve length at the 3′ end. Variations at the 3′end (3′-isomiRs) can result in shorter sequences due presumably to exonuclease “nibbling” at the mature sequence, or in longer sequences due likely to inconsistent Dicer processing or post-transcriptional addition of a few bases that may or may not match the genomic locus thus resulting in templated or untemplated variants, respectively. 5′-isomiRs, which vary in length at the 5′ terminus, are rarer, but can still account for a significant proportion of the isomiR pool. Finally, post-transcriptionally edited isomiRs harbor different internal nucleotide sequences [87].
Even though the broad biological significance of isomiRs is yet to be fully understood, increasing evidence suggests that some may be functionally important because they can be expressed in a cell-specific manner and hence are regulated [88–94]. Studies also show that some isomiRs vary in stability, loading efficiency onto Argonaute proteins, and target selection [87,95]. Indeed, because miRNA repression of mRNA function is hypothesized to rely mainly on “seed pairing” (the perfect or near-perfect complementary match of nucleotides 2 to 8 of the mature miRNA product [96–98]), length modifications at the 5′ end of the mature product could relocate the seed position and thus change in the seed sequence (called “seed shifting”), thereby altering mRNA target recognition and function [92,93]. Furthermore, because non-canonical base pairing [28,29,99–102] can cause any portion of the miRNA to influence the pairing of the miRNA to its mRNA-UTR, any modification or edition in the mature sequence can putatively affect biological function.
These considerations raise the question: How should we annotate isomiRs that vary in length or nucleotide sequence? This question is important because tissue- or time-specific variation in the relative abundance of different isomiRs could contribute to differences in cell differentiation, proliferation, or physiology among tissues within a species or to the evolution of different features between species.
Currently, miRNA databases typically display only one mature sequence per strand, which is generally inferred from small RNA sequencing data and/or from calculated miRNA precursor folding probabilities. Although some databases list sequences of isomiRs, this information is not easily accessible and is often associated only with a pre-MiRNA sequence and not a mature miRNA sequence, meaning that an isomiR can’t be specifically assigned to any mature product of the precursor and can’t be directly annotated and thus conveniently designated in an investigation. Furthermore, no consistent nomenclature conventions for isomiRs have yet been established, which complicates communication. In databases, the annotated mature sequence usually corresponds to the most highly expressed isomiR across all deposited sequences. The sequencing of more tissues or more species could make the most highly expressed sequence a moving target, which does not facilitate precise communication. To overcome this problem, we propose a new view of miRNA sequences organized into three interconnected levels: 1) the reference mature miRNA sequence which would be an unchanging standard for reference purposes, 2) the isomiR sequences, which include all the miRNA variants observed for a reference miRNA with the application of an accession number to each isomiR and its assignment to one or multiple reference miRNA sequences, if appropriate, and 3) the functional mature sequence which would correspond to the most highly expressed isomiR in a given situation, tissue, and developmental stage, and thus would provide a functional perspective (Box 3) (Figure).
Box 3. Introducing a Reference sequence and isomiR annotation.
Current small RNA databases list one “mature sequence” per miRNA strand, which is usually the most frequent sequence in the database, while sometimes displaying isomiR sequences in a visual quantitative presentation in which each isomiR can be followed to an individualized page with a sequence specific accession number. Several factors, however, could help better capture the complexity of isomiRs because so far, those isomiRs are associated only with a stem-loop and not a mature sequence (Figure).
A “reference sequence miRNA” (RefSeq-miRNA) should replace the concept of a single universal “mature sequence”. The effective “mature sequence” would now be seen in a functional perspective, and could vary in different tissues or developmental stages, whereas the “reference sequence” would be a species-specific invariable standard for each miRNA-encoding gene strand. This convention has the advantage that even if the ranking of isomiR expression levels vary in different tissues or stages or situation and thus the “mature sequence” varies, the “reference sequence” remains unchanged and isomiR sequences and quantities can be described compared to an unchanging reference.
Similar to mature sequences that can originate from one or multiple stem-loops when they are closely related, isomiRs can potentially originate from one or multiple stem-loops and thus be isomiRs of one or multiple mature sequences. Each isomiR should thus be associated with all the mature sequences for which it is possible to be a variant of, as for mature sequences.
Finally, to facilitate communication, each isomiR should continue to receive its own accession number, like the “MR” annotation system currently used in miRBase, and corresponding sequences and annotations should be easily accessible for download.
moRs and loRs
Increased sequencing depth has identified other Mir gene-encoded fragments in addition to the 5′ and 3′ miRs. For example, moRs (microRNA offset RNAs), are encoded by sequences adjacent to the ends of the 5′ and 3′ miRNA-encoding sequences that abut the loop [103]. moRs were first considered to be non-functional fragments or processing by-products, but because the quantity of a moR relative to its neighboring miRNA can vary between tissues or developmental stages, they appear to be regulated and thus are either functional or represent tissue-specific, regulated, transcriptional noise [20,21,104–106]. Genomically, moRs are generally more frequent on the 5′ side of the hairpin, independent of strand-specific miRNA expression levels [21], consistent with a putative function, but also suggesting that they might actually be co-products of miRNA biogenesis [107] and thus provide a distinct functional class of miRNA-related sequences. moRs are frequently found in RISC pull-down libraries, suggesting function, and a study has verified that some viral moRs show a moderate, but still significant repression of mRNA translation in luciferase assays [108]. Together, data suggest that moRs can program the RISC and act on gene expression, and should be considered in small RNA studies.
moRs were initially annotated as a miRNA with a “dot-number” suffix referring to its relative position in the hairpin, e.g. the moR for the Mir125a gene was labeled MiR125a.1 and the original miRNA was called MiR125a.2 because it is further from the end of the hairpin. The symbol moR, introduced by Shi et al [20], however, coupled with designating origin from the 5′ or 3′ end, for example “MOR125a-5p” and “MOR125a-3p”, has the advantage that it allows clear and unambiguous identification of the corresponding RNA sequence (Table).
Small RNA sequencing libraries also identify sequences that derive from the loop of pre-MiRNAs and, like moRs, their relative quantities can vary according to tissue or developmental stage [23,24], suggesting independent regulation of expression and thus independent cell functions. Furthermore, recent studies reported the expression and incorporation of loop-derived short sequences into the RISC and repression of translation of targeted mRNA in in vitro luciferase assays [23,24]. We propose to use the nomenclature “loR” for “loop-origin miRNAs”; for example, the loop-origin fragment from the Mir125a gene would become “LOR125a” (Table).
Concluding Remarks
The increasing amount of small RNA sequencing data provides a more complete view of the putative regulatory small RNAome, but it is important to distinguish the functional miRNAome from the degradome, small RNA fragments resulting from RNA degradation. miRNAs account for the majority of reads in small RNA sequencing efforts, but many other miRNA-like small RNAs that originate from non-canonical miRNA genes, show consistent and significant expression across experiments and can act on message stability and translation repression as do canonical miRNAs. These miRNA-like sequences thus compose a new kind of miRNA for which we suggest nomenclature conventions. Furthermore, the depth of small RNA sequencing data demonstrates that a single Mir gene can potentially produce not just a single mature miRNA from each of its two complementary paired stem sequences, but up to five different and transcriptionally regulated, putatively functional mature sequences in the order: moR-5p, miR-5p, loR, miR-3p, and moR-3p. Finally, most of these sequences show variations (isomiRs) arising from both templated and untemplated variations. Appreciation of the variety of sequences that can arise from genes that encode miRNAs should help to unravel the regulatory roles they play in development and physiology, health and disease. The revised nomenclature conventions we suggest should help facilitate communication about the variety of miRNA-related sequences derived from different species and hence, the conservation evolution of miRNA functions.
Outstanding questions.
What fraction of small non-coding miRNA-like sequences that derive from genes other than traditional miRNA genes actually act on translational control in a fashion similar to most miRNAs? Although these miRNA-like fragments consistently appear in smallRNA sequencing and Argonaute pulldown experiments, evidence of a functional role in translational control remain scarce or absent. Are those short ncRNAs incorporated into the RISC and do they inhibit translation of targeted messenger RNAs? Are they evolutionary conserved? Are they merely degradation products of other ncRNA biotypes without function? Do they modulate the miRNA pathway by overwhelming the RISC and inhibiting the loading of bone fide miRNAs? The later possibility would add an additional layer of complexity to the pathway of miRNA regulation.
What are the functions of moRs and loRs, which arise from the ends of pri-miRNA stems or the loop? Evolutionary conservation, sequence conservation, and variable expression levels suggest a functional role in mRNA translation control, but conclusive evidence in direct protein translation is lacking or minimal. Do moRs and loRs function like miRNA by entering the RISC and regulating protein translation? Are they simple by-products of the miRNA biogenesis pathway? And if so, are they needed for proper pri-miRNA and pre-miRNA processing, thus explaining their evolutionary conservation? Answering these questions would provide a deeper understanding of the miRNA biogenesis pathway and the roles of miRNA genes in regulatory biology.
Trends Box.
Functional microRNAs can originate from various types of non-coding RNA genes
The definition of microRNAs should take a functional perspective
The full complexity of isomiR populations should be considered in micoRNA studies
microRNA nomenclature systems should encompass origins, function, and isomiRs
Acknowledgments
Authors would like to thank Eric C. Lai for valuable comments. The work was funded by the grants NIH R01HG004341 (KE), NIH 5P41HG000330 (JTE & MSM), NIH HG004838 (AS), NIH HG004834 (AS), The Research Council of Norway (FishmiR project, grant no. 213825, JHP and I. Babiak), NIH U01DE020076 (JHP, K. Artinger and D. Clouthier), NIH 5R240D011199 (JHP and W. Cresko), and NIH R01 OD011116 (JHP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Dr. T. Desvignes, Email: desvignes@uoneuro.uoregon.edu.
P. Batzel, Email: pbatzel@uoregon.edu.
Prof. E. Berezikov, Email: e.berezikov@umcg.nl.
Dr. K. Eilbeck, Email: keilbeck@genetics.utah.edu.
Prof. J.T. Eppig, Email: janan.eppig@jax.org.
M.S. McAndrews, Email: monica.mcandrews@jax.org.
Prof. A. Singer, Email: asinger@zfin.org.
J.H. Postlethwait, Email: jpostle@uoneuro.uoregon.edu.
References
- 1.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ketting RF. The Many Faces of RNAi. Dev Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Ishizu H, et al. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Kosik KS. MicroRNAs and Cellular Phenotypy. Cell. 2010;143:21–26. doi: 10.1016/j.cell.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Bizuayehu TT, Babiak I. MicroRNA in Teleost Fish. Genome Biol Evol. 2014;6:1911–1937. doi: 10.1093/gbe/evu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christodoulou F, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes CPC, et al. A review of computational tools in microRNA discovery. Front Bioinforma Comput Biol. 2013;4:81. doi: 10.3389/fgene.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li SC, et al. Identification of homologous microRNAs in 56 animal genomes. Genomics. 2010;96:1–9. doi: 10.1016/j.ygeno.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Szcześniak MW, Makałowska I. miRNEST 2.0: a database of plant and animal microRNAs. Nucleic Acids Res. 2014;42:D74–D77. doi: 10.1093/nar/gkt1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobsen A, et al. miRMaid: a unified programming interface for microRNA data resources. BMC Bioinformatics. 2010;11:29. doi: 10.1186/1471-2105-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 18.Morin RD, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 20.Shi W, et al. A distinct class of small RNAs arises from pre-miRNA–proximal regions in a simple chordate. Nat Struct Mol Biol. 2009;16:183–189. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langenberger D, et al. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25:2298–2301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H, et al. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012;40:5864–5875. doi: 10.1093/nar/gks247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamura K, et al. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev. 2013;27:778–792. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winter J, et al. Loop-miRs: active microRNAs generated from single-stranded loop regions. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter J, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 26.Kawamata T, et al. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 27.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin C, et al. Expanding the MicroRNA Targeting Code: Functional Sites with Centered Pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown M, et al. Mammalian miRNA curation through next-generation sequencing. Front Genet. 2013:4. doi: 10.3389/fgene.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen TB, et al. Enhancing miRNA annotation confidence in miRBase by continuous cross dataset analysis. RNA Biol. 2011;8:378–383. doi: 10.4161/rna.8.3.14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langenberger D, et al. MicroRNA or Not MicroRNA? In: de Souza ON, et al., editors. Advances in Bioinformatics and Computational Biology. Springer; Berlin Heidelberg: 2011. pp. 1–9. [Google Scholar]
- 33.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burroughs AM, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamura K, et al. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruby JG, et al. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berezikov E, et al. Mammalian Mirtron Genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isik M, et al. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence. 2010;1:5. doi: 10.1186/1758-907X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramalingam P, et al. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2013 doi: 10.1261/rna.041814.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desvignes T, et al. Evolution of the miR199-214 cluster and vertebrate skeletal development. RNA Biol. 2014;11:281–294. doi: 10.4161/rna.28141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheloufi S, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cifuentes D, et al. A Novel miRNA Processing Pathway Independent of Dicer Requires Argonaute2 Catalytic Activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, et al. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 44.Yang JS, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ender C, et al. A Human snoRNA with MicroRNA-Like Functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 46.Taft RJ, et al. Small RNAs derived from snoRNAs. RNA. 2009 doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brameier M, et al. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babiarz JE, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia J, et al. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Hum Mol Genet. 2013;22:737–748. doi: 10.1093/hmg/dds481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol. 2013;15:1153–1163. doi: 10.1038/ncb2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyoshi K, et al. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 54.Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol. 2012;24:333–340. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim VN, et al. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 57.Chen L, et al. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics. 2012;7:758–771. doi: 10.4161/epi.20706. [DOI] [PubMed] [Google Scholar]
- 58.Axtell MJ, et al. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moran Y, et al. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014 doi: 10.1101/gr.162503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perreault J, et al. Ro-Associated Y RNAs in Metazoans: Evolution and Diversification. Mol Biol Evol. 2007;24:1678–1689. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- 61.Stadler PF, et al. Evolution of vault RNAs. Mol Biol Evol. 2009;26:1975–1991. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 62.Desvignes T, et al. Expanding the annotation of zebrafish microRNAs based on small RNA sequencing. Gene. 2014;546:386–389. doi: 10.1016/j.gene.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Persson H, et al. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 64.Meiri E, et al. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res. 2010;38:6234–6246. doi: 10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolas FE, et al. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012;586:1226–1230. doi: 10.1016/j.febslet.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 66.Verhagen APM, Pruijn GJM. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. BioEssays. 2011;33:674–682. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- 67.Eilbeck K, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bult CJ, et al. The Mouse Genome Database: Genotypes, Phenotypes, and Models of Human Disease. Nucleic Acids Res. 2012;41:D885–D891. doi: 10.1093/nar/gks1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gray KA, et al. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laulederkind SJF, et al. The Rat Genome Database 2013--data, tools and users. Brief Bioinform. 2013;14:520–526. doi: 10.1093/bib/bbt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bradford Y, et al. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amores A, et al. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 73.Braasch I, Postlethwait JH. Polyploidy in Fish and the Teleost Genome Duplication. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Springer; Berlin Heidelberg: 2012. pp. 341–383. [Google Scholar]
- 74.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Czech B, et al. Hierarchical Rules for Argonaute Loading in Drosophila. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okamura K, et al. Distinct Mechanisms for MicroRNA Strand Selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang JS, et al. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuddenham L, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 80.Eberhart JK, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rakoczy J, et al. MicroRNAs-140-5p/140-3p Modulate Leydig Cell Numbers in the Developing Mouse Testis1. Biol Reprod. 2013;88:Article 143, 1–11. doi: 10.1095/biolreprod.113.107607. [DOI] [PubMed] [Google Scholar]
- 82.Tyler DM, et al. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scott H, et al. MiR-3120 Is a Mirror MicroRNA That Targets Heat Shock Cognate Protein 70 and Auxilin Messenger RNAs and Regulates Clathrin Vesicle Uncoating. J Biol Chem. 2012;287:14726–14733. doi: 10.1074/jbc.M111.326041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dehal P, Boore JL. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakatani Y, et al. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saini HK, et al. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neilsen CT, et al. IsomiRs – the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 88.Lee LW, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martí E, et al. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Llorens F, et al. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genomics. 2013;14:104. doi: 10.1186/1471-2164-14-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo L, Chen F. A challenge for miRNA: multiple isomiRs in miRNAomics. Gene. 2014;544:1–7. doi: 10.1016/j.gene.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 92.Tan GC, et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014;42:9424–9435. doi: 10.1093/nar/gku656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia J, Zhang W. A meta-analysis revealed insights into the sources, conservation and impact of microRNA 5′-isoforms in four model species. Nucleic Acids Res. 2014;42:1427–1441. doi: 10.1093/nar/gkt967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Warnefors M, et al. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 2014;15:R83. doi: 10.1186/gb-2014-15-6-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thornton JE, et al. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4) Nucleic Acids Res. 2014 doi: 10.1093/nar/gku805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brennecke J, et al. Principles of MicroRNA–Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schirle NT, et al. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li N, et al. Dispatched Homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 2008;36:4277–4285. doi: 10.1093/nar/gkn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas M, et al. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 101.Chi SW, et al. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clark PM, et al. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci Rep. 2014;4 doi: 10.1038/srep05947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bortoluzzi S, et al. MicroRNA–offset RNAs (moRNAs): by-product spectators or functional players? Trends Mol Med. 2011;17:473–474. doi: 10.1016/j.molmed.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 104.Bortoluzzi S, et al. Characterization and discovery of novel miRNAs and moRNAs in JAK2V617F-mutated SET2 cells. Blood. 2012;119:e120–e130. doi: 10.1182/blood-2011-07-368001. [DOI] [PubMed] [Google Scholar]
- 105.Asikainen S, et al. Selective MicroRNA-Offset RNA Expression in Human Embryonic Stem Cells. PLoS ONE. 2015;10:e0116668. doi: 10.1371/journal.pone.0116668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang W, et al. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010;11:R81. doi: 10.1186/gb-2010-11-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berezikov E, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Umbach JL, et al. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology. 2010;405:592–599. doi: 10.1016/j.virol.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ambros V, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]