Abstract
Background Centralized and federated models for sharing data in research networks currently exist. To build multivariate data analysis for centralized networks, transfer of patient-level data to a central computation resource is necessary. The authors implemented distributed multivariate models for federated networks in which patient-level data is kept at each site and data exchange policies are managed in a study-centric manner.
Objective The objective was to implement infrastructure that supports the functionality of some existing research networks (e.g., cohort discovery, workflow management, and estimation of multivariate analytic models on centralized data) while adding additional important new features, such as algorithms for distributed iterative multivariate models, a graphical interface for multivariate model specification, synchronous and asynchronous response to network queries, investigator-initiated studies, and study-based control of staff, protocols, and data sharing policies.
Materials and Methods Based on the requirements gathered from statisticians, administrators, and investigators from multiple institutions, the authors developed infrastructure and tools to support multisite comparative effectiveness studies using web services for multivariate statistical estimation in the SCANNER federated network.
Results The authors implemented massively parallel (map-reduce) computation methods and a new policy management system to enable each study initiated by network participants to define the ways in which data may be processed, managed, queried, and shared. The authors illustrated the use of these systems among institutions with highly different policies and operating under different state laws.
Discussion and Conclusion Federated research networks need not limit distributed query functionality to count queries, cohort discovery, or independently estimated analytic models. Multivariate analyses can be efficiently and securely conducted without patient-level data transport, allowing institutions with strict local data storage requirements to participate in sophisticated analyses based on federated research networks.
Keywords: distributed analytics, federated research network, privacy-preserving network infrastructure, comparative effectiveness research
BACKGROUND AND SIGNIFICANCE
Several electronic networks have been formed in the past two decades to provide support for public health surveillance, cohort discovery, and comparative effectiveness research (CER) studies, including the HMO Research Network,1 I2B2,2 caBIG,3 the Biomedical Informatics Research Network,4,5 SAFTINet,6 and more recently, PCORnet.7–18 Many of these electronic networks have also developed custom software tools as part of their implementation strategy. The majority of networks involve research data collection for a specific study (e.g., clinical trials), function (e.g., pharmacovigilance), or domain (e.g., disease registries), while a few involve the use of data collected for patient care (clinical data research networks (CDRNs)) to perform basic descriptive statistics about a selected sample cohort (e.g., counts, averages), or transmit selected observations. As interest in a learning healthcare system that can execute research studies on CDRNs increases19 it is important that CER studies support advanced descriptive and inferential statistics in distributed environments and allow study-based data governance.
This work addresses requirements and solutions that can be applied in scalable ways to manage some sociotechnical issues for managing analysis workflow in a distributed computing environment. Researchers interested in multi-institutional collaborations involving analysis of patient records face regulatory and ethical challenges that limit the scalability of research across projects and organizations. A widely endorsed architecture for addressing the legal and organizational barriers for using clinical data for research in US institutions has been the distributed research network. In such clinical data networks, data are maintained locally by each institution, and are coordinated via a common infrastructure that shares practices and software. Distributed networks still do not solve all problems, but they currently represent the most practical solution in cases where physical transfer of data is difficult (e.g., bandwidth limitations for big data, or international regulations20 against physical hosting of data outside geographical boundaries).
Simultaneously with the growing interest in CDRNs and in “big data,” there has been a resurgence in application of parallel processing algorithms based on “map-reduce”21 and related frameworks such as the Statistical Query Oracle22 whereby iterative algorithms that do not require row-level data transfer can be used to compute the same models that would normally be based on centrally pooled data. While it is not trivial to redevelop model estimation algorithms in these architectures, significant effort has been made in the computer science community to develop new algorithms that are available in open source software. This approach lends itself well to the policy requirements of federated research networks because it retains data control at each site. Without the ability to centralize analysis, most federated networks must independently estimate multivariate models for each site in the network.23 However, a single analytic model that can capture variation or adjust for confounders across the entire network is still desirable. Our approach supports a natural marriage of parallel computation algorithms and federated CDRN policy management infrastructure. By creating a platform that allows addition of novel methods to a repository by contributors both inside and outside the SCAlable National Network for Effectiveness Research (SCANNER) team, we enable scalability to new methods as well as new projects and research teams.
SCANNER design was informed by platforms adopted by several CDRNs, including PopMedNet, which has been adopted by the PCORnet Network and the MiniSentinel project,16 as well as the SHRINE system for distributing cohort discovery queries to harmonized I2B2 instances,24 which has been adopted by academic consortia such as UC-ReX.25 We refer readers to two recent reviews that cover governance and these (along with other) technical solutions in greater detail.26,27 While SCANNER does have a native system for query distribution, most of the study management services orchestrated at the portal, such as the study registry and library of data operations, are compatible with existing platforms for query federation (e.g., TRIAD,28 PopMedNet,29 or Hadoop30). That is, the SCANNER portal might be implemented with a plug-in interface to these distribution platforms while retaining functionalities of both. The intent of SCANNER was not to develop a query federation platform, which exists already in several commercial and academic contexts. Rather, SCANNER is a system for reusable web services for data operations and policy management that could be implemented in any framework, giving users the ability to form networks for research and cohort discovery on a reusable governance infrastructure. However, in order to support distributed analytic use cases, some features for request scheduling that are distinct to the SCANNER and other emerging platforms for REST-based parallel distributed processing were required (e.g., GridFactory31 and Apache Spark32). While we opted to develop our own portal interface, we could have adapted existing portal software for overlapping functionality.
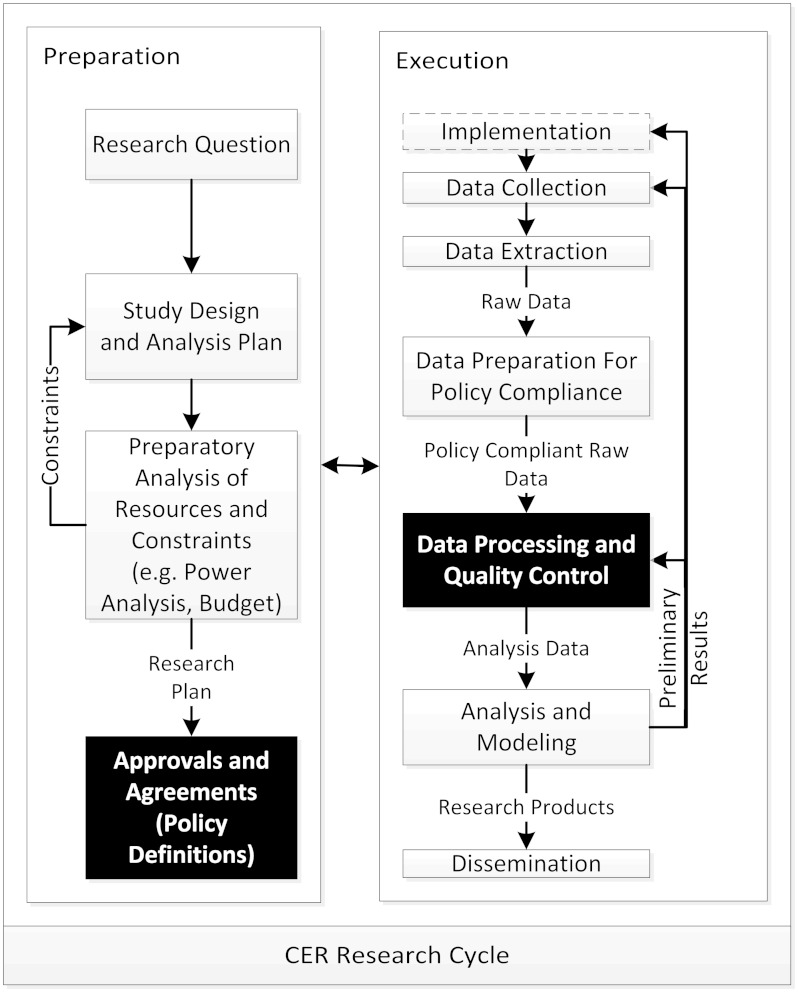
CDRNs need to flexibly support a variety of uses and governance policies, and allow participants to select configuration options that accommodate stakeholder needs.33 Figure 1 shows the process model we used to generate requirements for CER functionality. The CER process is divided into preparation and execution phases. The process begins with a research question, iterates through design and preparatory analyses, and a research plan that is submitted to appropriate parties for approval, to generate a final list of policies that must be enforced throughout the execution of the project. Later, in the execution phase, some studies require implementing interventions or new data collection modes, but in all cases, data must go through several stages of processing for both policy compliance and CER data quality validation. Using this process model, each endpoint in a CER workflow generates a policy compliance point—an opportunity to test for compliance with the stated policies of the resources in question. The boxes colored in black represent opportunities to gain efficiencies by codifying governance, data preparation, and policies.
Figure 1:
Comparative Effectiveness Study Process Model.
We conducted a series of interviews with institutional review board (IRB) and compliance officers and completed a regulatory content review34 to generate SCANNER requirements. This resulted in study-centric data sharing policy management requirements that resemble specifications that might be implemented in eIRB software—defining staff, roles, protocols, data definitions, and analysis methods. An important additional security requirement arising from this analysis is the ability to support menu-driven queries for computation of analytic models, to avoid distribution of opaque analysis programs that may introduce unknown security risks. In particular, these included methods that enable collaborative estimation of a single model without any need to share patient-level data. These requirements were used to design and develop the SCANNER system.
METHODS AND APPROACH
SCANNER software is intended to enable teams of investigators to initiate and manage studies that involve exposing data resources to a securely controlled network, with an emphasis on preserving the ability to conduct multivariate analysis without centralized pooling of data. SCANNER software is composed of a network portal, a set of web services, and virtual machines that host data and analysis programs that are controlled by the portal. Individual data contributors may join the SCANNER network by implementing one or more virtual machines and registering site authorities responsible for data stewardship. Alternatively, a private network may be created with an independent portal instance by downloading the SCANNER software. Joining as a member of the SCANNER network does not confer any privileges, each SCANNER study involves approval of data sharing policies—for example, a prep-to-research “cohort discovery” study commonly implemented in platforms such as SHRINE might enable all members of the study to query data from all other members of the study to retrieve counts without manual approval. Alternatively a “meta regression” study of congestive heart failure might enable investigators to run analyses on posted data sets adhering to a particular data dictionary, with some sites requiring “human in the loop” approval and others enabling synchronous access to the data set. Another option might be a single-site study where a principal investigator (PI) queries only data from his own site. The intent of SCANNER software is to clearly separate policy management from data management and queries so that role-based-access controls to data resources can be attributed at the level of a study rather than coupled tightly to software.
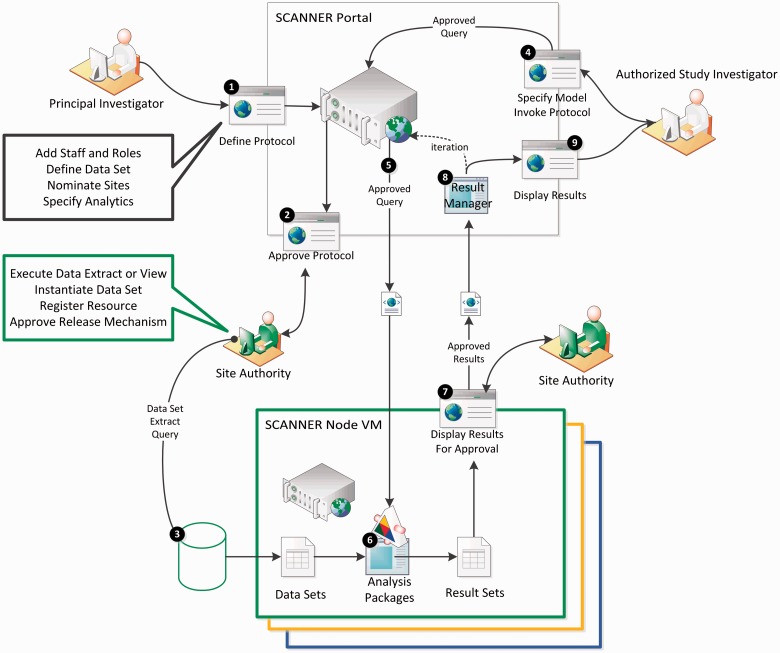
Figure 2 is a simplified view of the SCANNER architecture, representing the most important functional aspects of SCANNER. The SCANNER network consists of a web portal (top center) and a virtual machine server at each node in the network (bottom). The portal server integrates multiple components host the SCANNER website, which presents each user a view appropriate to their role in the network and the state of the different activities in which he participates. Screenshots from the portal are displayed in Figure 3 and in the Appendix.
Figure 2:
Simplified view of SCANNER architecture and functions: (1) Protocol definition; (2) protocol approval; (3) data location definition; (4) study member login and analysis request; (5) conversion into XML and distribution; (6) analysis; (7) results ready notification; (8) next parameter estimation iteration for map-reduce algorithms; and (9) results sent to requester.
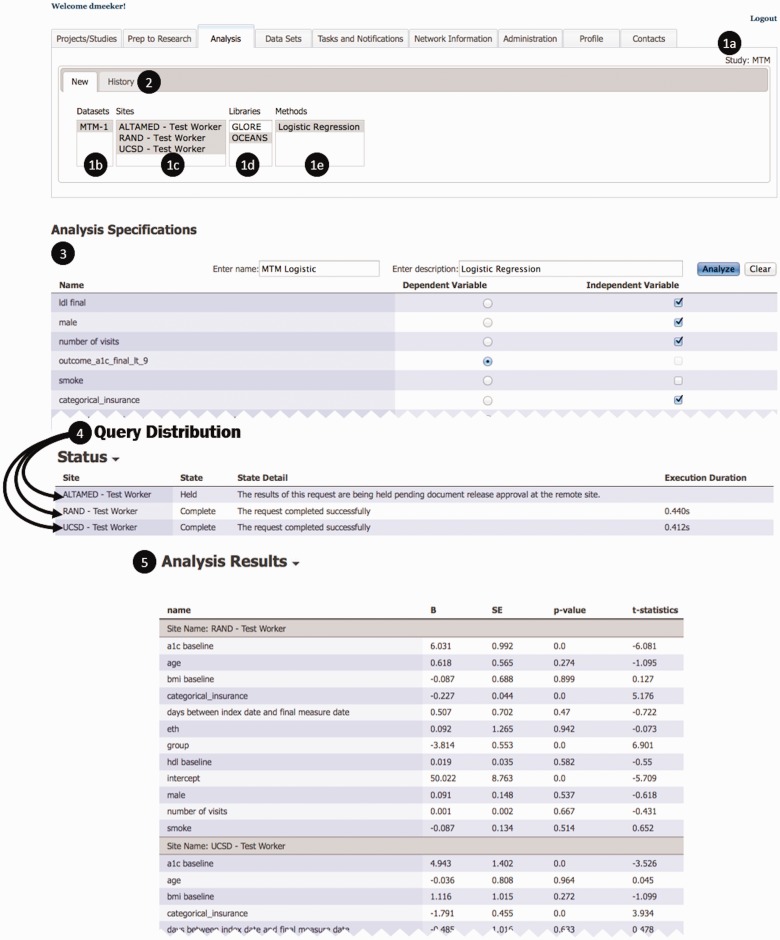
Figure 3:
Screenshots from a Distributed Multisite Logistic Regression Analysis.
In Figure 2, the first step in conducting a study is defining a computable study protocol (1). A designated PI may initiate a new study, assign staff to roles with varying levels of authorization, and define the detailed data elements in the analytic data set and methods that will be invoked after approval. The PI also nominates sites for participation. With these specifications in place, site authorities may then log into the portal’s approval forms (2). During approval, the site authorities (a) approve the set of study queries and data transfer modalities they will accept, (b) execute the extract queries needed to create the standardized analysis data set, and (c) post that data set to a network location, and add the resource address to the approval form.
With these pieces in place for each node participating in a study, an authorized investigator on the study team may log into the portal (4). At this point a graphical user interface (GUI) is presented where the user can specify the sites and analytic model parameters that have approved study protocols (e.g., selected variables within the approved data set, defining independent and dependent variables). This request is then converted into an XML-based query (5) that is distributed to the network nodes. The query invokes the analysis package hosted on the node and results are calculated (6). Results are retained at the site, if the protocol specified that approval was required, the site authority will receive an email message indicating that there is a pending result set (7). In the case of iterative map-reduce computations, the scheduler reformats the queries for the next iteration (8). Note that the “result manager” is the “master” where convergence algorithms are evaluated in each iteration of map-reduce algorithms, and only statistical aggregates are transferred. Approved results are returned to the portal. The result handler either schedules another iteration or returns final results to the user interface for display (9).
The main components of SCANNER are described below.
Study Manager
As described above, SCANNER data sharing policy management is based on a study, and resembles the types of specifications adopted by eIRB systems. A SCANNER study consists of computable protocols, staff, and their roles, the underlying data resources instantiating the protocol, and associated documentation. A protocol is instantiated with data by authorized individuals at each site. The SCANNER study manager includes GUIs for protocol specification (shown in the Appendix) that require a study designer to define 1) data set definitions, 2) data operations and analytics, and 3) options for mode of transfer of results, including an optional quarantine area so results can be inspected prior to transfer (i.e., the network can operate in synchronous as well as asynchronous modes). Protocol parameters are stored in the SCANNER study registry, so components (e.g., variable definitions and processing operations) can be reused.
A protocol must be approved and instantiated by participants. Each node on the network assigns an individual the role for study approval (this role might be labeled Site Principal Investigator). Protocols are proposed to these site authorities at the time of the study specification. At the time of agreement to participate, the site authority defines the local parameters in the protocol (e.g., the physical location of a data resource) and registers the study information in the SCANNER portal. Protocols are then made available in the portal. Distributed analytics can be executed with node operations and results are returned to the portal. A prep-to-research (cohort discovery) query informs researchers about the potential number of eligible subjects. In SCANNER these queries produce NIH targeted enrollment tables for each site (Appendix, Exhibit 4). This is a special case of distributed analytics. Cohort discovery typically limits analysis methods to count statistics.
Methods Library
SCANNER supports R, SAS, and other analytic engines that might be hosted on network nodes. In order to accommodate multiple possible analytic platforms for data operations, SCANNER created a “methods library” for plug-ins that enable analytic method developers to register new methods and code as SCANNER services. Methods plug-ins consist of three components—graphical user interfaces (GUIs) for users to invoke methods and see results at the portal, services to control the distribution and accumulation of results, and programs that are installed on the virtual machines (VMs) on each node to execute the data analysis. Different execution engines for the same analytic method can share the same templates for menu-driven GUIs for specifying parameters and displaying results of distributed analyses. For example, we have two libraries for logistic regression (LR) that have been fully integrated as SCANNER services. LR is one of the most frequently applied multivariate methods to adjust for confounders, develop propensity scores, and develop risk prediction tools.
To date, three methods libraries have been contributed to the SCANNER registry: a cohort discovery method, Observational Cohort Event Analysis and Notification System (OCEANS; http://sourceforge.net/projects/oceans/files/) and Grid LOgistic REgression (GLORE).44 We implemented web services for two open-source distributed analysis tools for LR. One of the LR tools performs meta-analyses using OCEANS, a statistical analysis and statistical process control tool. OCEANS produces independent parameter estimates for each participating site, and can be viewed as tool for meta-analysis.
The other service implements GLORE, which was initially conceived to address privacy-preserving data sharing through the NIH-funded iDASH national center for biomedical computing35,36 and applies principles of parallel distributed processing (PDP) and map-reduce algorithms,21 to achieve the same parameter estimates that might be achieved by centrally pooling data without requiring the release of patient-level data beyond institutional boundaries. We refer to this as “virtually pooled” regression. PDP is architecturally and algorithmically more complex than meta-analysis, and cannot be easily implemented in distributed networks with independently operating instances of programs like SAS because a centralized algorithm must manage estimation convergence across all sites.37 With this approach, regression parameter estimates are collected at every iteration and a new candidate vector of coefficients is generated for testing and correction at each site until estimations converge. These methods are not redundant, as GLORE and related parallel distributed methods can increase power if data are statistically homogeneous across all sites,37,38 while meta-regression methods like OCEANS compute independently estimated models and are most appropriate if data are not identically distributed at all sites. Calculation of propensity scores, for example, can involve several sites in GLORE but can only be calculated locally at each site in OCEANS. Other virtually pooled computing algorithms such as Cox Proportional Hazards are in development as web services for parallel distributed computation.
Software Architecture
Like many past approaches, SCANNER has adopted a Service-Oriented Architecture for managing network activities with web-based REST-style protocols over SSL-encrypted channels. An exception to REST conventions is necessary for iterative analytic methods like GLORE, which require preserving state within a network node across multiple requests. The SCANNER registry and service information model is based on PostgreSQL database hosted on the same server as the web portal. We used the Spring framework with Java to create the application interfaces that are based upon the registry.39 The two analytic libraries (OCEANS and GLORE) were both originally authored in Java, and modified for compatibility with the SCANNER Network Web Service API. The network API currently supports the HTTP verbs GET and POST. Both the portal and nodes share the same general REST-style functional semantics, with XML- and JSON-based representations of query and result specifications. The GUI is authored in HTML, CSS, and javascript, relying on the jQuery library for interface controls and menus. Implementing a SCANNER node involves installing a SCANNER virtual machine and opening ports to the master node hosting the SCANNER hub, which also hosts the portal. For any given analysis, data sets must conform to the data set definition registered at the time the study protocol was defined.
Any data model can be used to create data sets subject to queries. SCANNER offers a “data set authoring” interface for creating computable specifications for a data dictionary. We include a plug-in service that converts this computable dictionary into SQL queries for the Observational Medical Outcomes Partnership (OMOP) v4 common data model (CDM). While it is not required, partners that wish to leverage SCANNER data transformation resources for standardized multi-site data set preparation and quality assurance can invest in creating a data warehouse that employs terminology standards and a schema supported by a SCANNER plug-in (currently OMOP v4). While this initial investment is not required, it is most efficient for all sites that intend to participate in multiple studies, because it dramatically reduces data management burden for IT staff at the site.
Box 1 SCANNER Policy Management Features.
1. Data sharing policies are managed on the basis of a “study”—similar to some eIRB systems.
2. Any network participant may propose and lead a study (including persistent, real-time prep-to-research cohort discovery studies) through the network portal; participants may elect to participate as desired, allowing spontaneous formation of networks within the SCANNER network.
- 3. A study consists of
- a. Protocols
- b. Participating sites and data resources
- c. Staff and roles
4. Investigator initiated studies may propose protocols and associated data access control policies, participating sites may approve one or more protocol and policy.
5. Study-specific data sets that are generated and exposed to the network can be tagged with confidentiality settings.
6. Data set creation workflow and policies are separated from data analysis queries and workflow.
7. Both synchronous (pre-approved for query and response) and asynchronous (manually approved query and response) policies may be supported.
Box 2 SCANNER Analysis Libraries and Data Processing Features.
1. SCANNER includes an interface for authoring and specifying the rules for creating analysis data sets—A Computable Data Dictionary.
2. Data set rules can be parsed to generate executable programs for extracting and transforming source data into analytic data sets. SCANNER has created a web service that generates SQL for one particular data model, OMOP V4 CDM.
3. SCANNER was designed to support libraries of investigator-contributed methods for data analysis.
- 4. SCANNER Analysis Libraries are composed of three items:
- a. user interfaces that display the query and the response,
- b. the “master” query distribution and result aggregation methods
- c. the “worker” methods hosted on the VM
- 5. The three libraries that have been contributed include
- a. A native SCANNER library for cohort discovery for preparatory to research studies
- b. OCEANS, a library for meta-regression
- c. GLORE, a library for virtually pooled regression
Figure 3 shows screenshots and the steps in specifying and executing distributed LR analyses. In the analysis interface, a user is able to see all approved analyses and sites for a given study. The study (1a), the data set (1b), the sites that have agreed to participate (1c), the method library (1d) and the analytic model (1e). This query and its results are stored in a history (2) that can be accessed at a later time. After selecting the analysis model and data set, a template associated with the analysis model is populated with the variables in the data set, where the user may specify parameters (in this case, dependent and independent variables in a LR) (3). After the model has been specified, the query is distributed to sites where execution status can be tracked (4) and the fitted results are returned in a result template associated with the analytic model -- in this case, coefficients for the six selected variables and the intercept for the logistic regression (5).
RESULTS
We simulated two CER studies based on real projects focusing on 1) addition of a clinical pharmacist to the care team for medication therapy management,40 and 2) the effectiveness of providers’ public commitment to judicious antibiotic prescribing (Commitment).41 Data sets were simulated using model parameters from these studies and placed in three different institutions. The “Medication Therapy Management Simulated” data set included a binary outcome variable (HbA1c below 9.0%), a treatment indicator variable, 24 covariates, and 580 records on each node. The “Commitment Simulated” data sets included a binary outcome corresponding to prescribing practice, 5 covariates, and 10,000 records. Users with the role “Study PI” created two SCANNER studies, “MTM Study,” and “Commitment Study.” For each study, they proposed two analysis protocols to three sites hosting SCANNER nodes. For both studies the Study PI proposed protocols that requested Site PIs’ approval. The “Site PI” at each of the three sites approved protocols and created and registered data sets to the network test nodes. The Study PI for each of the two studies ran the protocols three times from the SCANNER portal and recorded execution times. Results are shown in Table 1.
Table 1:
Simulated Study Protocols
| Study-Protocol ID | Data set | Analysis Method Summary | Number of Sites | Number of coefficients estimated | Data Set size (KB) | Mean Response Time in Seconds (SD) | |
|---|---|---|---|---|---|---|---|
| 1.1 | MTM-Simulated | GLORE-Logit | Iterative convergence; single model for all sites | 3 | 25 | 580 records 126 KB | 0.015s (0.026) |
| 1.2 | MTM-Simulated | OCEANS-Logit | Non-iterative; 1 model per site | 3 | 75 | 580 records 126 KB | 0.6425 (0.148) |
| 2.1 | Commitment-Simulated | GLORE-Logit | Iterative convergence; single model for all sites | 3 | 5 | 10,000 records 147 KB | 27.02 (1.58) |
| 2.2 | Commitment-Simulated | OCEANS-Logit | Non-iterative; 1 model per site | 3 | 15 | 10,000 records 147 KB | 2.87 (0.54) |
MTM: Medication Therapy Management.
Simulations were conducted on three Virtual Machines with the following specifications:
CPU: Six-Core AMD Opteron(tm) Processor 2.4Ghz; RAM: 6.4GB; JVM: OpenJDK 1.7, 4GB Heap Memory
The differences between execution times can be explained in part by the different strategies for memory management used in the different methods. The virtual machines only had 6.4 GB of memory, and this limitation made GLORE slower for large data sets. When this protocol is preferred, sites may add additional nodes in order to speed up the parallel computations. The execution times were however, acceptable for CER. These results indicate that the SCANNER infrastructure has the capacity to handle complex study management rules and specifications of analytic protocols necessary for multi-site CER.
Discussion
Exhibit 1 displays some frequently asked questions about SCANNER.
Exhibit 1:Frequently Asked Questions.
How are SCANNER’s virtually pooled analysis methods different from meta-analysis?
SCANNER can produce meta-analyses to estimate one model for each data set shared on a node (using the OCEANS package). SCANNER’s virtually pooled regression methods (GLORE package) estimate a single model that is equivalent to what would be achieved by pooling patient-level data, except that it does not require the data to be transmitted to a central location. It does so by decomposing model estimation algorithms.42,43
Can virtually pooled analysis be implemented for all statistical models used for CER?
No, there are certain calculations that cannot be easily decomposed (e.g., XOR, clustering algorithms based on pairwise distances) and therefore SCANNER will not always produce results that are identical to those of a centralized resource. However, all algorithms that calculate sufficient statistics or gradients fit this model, covering a broad range of options. A wide variety of distributed algorithms and execution systems for hosting algorithms are available and may be incorporated into future versions of SCANNER.
Can SCANNER automatically map my data into a common model? Does SCANNER require a common data model?
SCANNER is neither a platform for easing data harmonization like the Reusable OMOP and SAFTINet Interface Adaptor (ROSITA) system,6 nor is it required that SCANNER networks harmonize all data domains to join the network. For purposes of scalability, many research networks require mapping the entirety of source data into a CDM in a materialized data warehouse that can generate a multiplicity of data sets. SCANNER does not include any tools for transforming or harmonizing data. In SCANNER, each analysis protocol includes a data set in a defined format that is committed to the data set registry. SCANNER’s data set authoring interface allows users to create data set and data processing specifications in an emerging standard format originally developed to represent data processing rules for clinical quality measures the Health Quality Measure Format (HQMF).44 These data processing specifications may be either manually interpreted at each site, or network nodes that share a common data model for their data warehouse may author a plug-in adapter that translates Health Quality Measure Format to executable programs. SCANNER developed such an adapter for the OMOP v4 data model. Thus, while SCANNER provides tools to encourage interoperability with a network information model, adherence to a particular standard is not required to participate in distributed analytics.
What is involved in implementing a SCANNER at my site?
At a minimum, sites that wish to set up a SCANNER node must install a SCANNER virtual machine and open appropriate ports to the master node hosting the SCANNER hub. For any given analysis, data sets must conform to the data set definition registered at the time the study protocol was defined. Sites that wish to leverage SCANNER resources for standardized multi-site data set preparation and quality assurance can invest in creating a data warehouse that employs terminology standards and a schema supported by SCANNER plug-ins (currently OMOP v4). While this investment is not required, it is most efficient for all sites that intend to participate in more than one study, because it dramatically reduces data management burden for IT staff at the site.
Is the SCANNER code open source? Where can I see it working?
Yes, the version of the SCANNER code described in this manuscript can be downloaded from the SCANNER website http://scanner.ucsd.edu/.
A SCANNER infrastructure demo is available at http://scanner.ucsd.edu/images/SCANNERdemo/SCANNERdemo.html using simulated data.
Limitations and Future Work
In deploying the network, some high-priority items for future work were identified by stakeholders as outstanding features. Some items will require further investigation, while others may be easily addressed with the addition of new analytic services (e.g., by adding R or SAS to the software stack available on each node). Future versions of SCANNER may take advantage of advances in computer science community for implementing platforms for distributed processing algorithms to dramatically enhance our methodological libraries to include a broad array of machine-learning and regression methods.32,45,46 Data quality and harmonization also represent considerable challenges,47,48 particularly in distributed networks.49,50 SCANNER’s service-oriented design allows method developers to register new methods such as these to the network with well-specified requirements for execution. Continuing to maintain and extend SCANNER interoperability with other networks has been prioritized—SCANNER was designed to support selection of data sets from a variety of source data, but the current automated translation service for data extraction and processing only includes a plug-in adapter for the OMOP V4 common data model.51 We have recently expanded SCANNER significantly by partnering with two other networks to develop a patient-centered system, patient-centered SCAlable National Network for Effectiveness Research (pSCANNER),52 funded by PCORI. Security hardening and scheduled testing protocols will be refined for future uses, particularly for cases that do not fall under HIPAA safe-harbor designation. Finally, features such as enabling granular patient control over data access may become of increasing importance in the near future.53
CONCLUSIONS
SCANNER has focused on systems for role-based study management and services for “in-node” data processing and analysis—these are services maintained centrally but executed at network nodes under full control of data partners. The OCEANS meta-regression and GLORE “virtually pooled” regression allow sites to perform computations at their nodes and participate in overall model development without transferring data. We encode local policies into the nodes, so that each can participate in different types of studies without the development of a completely new infrastructure every time a new study or cohort discovery collaboration is added. The infrastructure enables an “app store”–like scalability such that members of the community can author protocols, data operations, and other services and register them to the network as collaborative projects.
FUNDING
This work was supported by AHRQ grant R01HS019913, NIH grants U54HL108460 and UL1TR000100, and VA grant HSR&D CDA 08-020.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
All authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship. D.M. and L.O.M. contributed equally to the writing of this article and are responsible for the overall content as guarantors. The other authors (X.J., M.M., C.F., M.D., L.P., L.N., M.D., K.K., H.K., A.B., R.E., G.K., F.R., C.K.) are ranked according to their contributions. X.J. and M.M. developed analysis software; C.K. conducted oversight of software engineering; C.F., M.D., L.P. contributed to programming; K.K., G.K., R.E., A.B., L.N., and H.K. contributed to requirements generation and design specifications.
Acknowledgments
We thank several SCANNER colleagues and advisors for their present and past contributions: David Chang, Lola Ogunyemi, Paulina Paul, Seena Farzaneh, John Mattison, Deven McGraw, Serban Voinea, Shuang Wang, and Zia Agha. We thank Michael Kahn, Patrick Ryan, Christian Reich, and Lisa Schilling for collaborative efforts related to data modeling. We thank Gurvaneet Randhawa from AHRQ and Erin Holve from AcademyHealth for their support.
REFERENCES
- 1.Platt R, Carnahan R. The US Food and Drug Administration's Mini-Sentinel Program. Pharmacoepidemiol Drug Safety. 2012;21(S1):1–303. [DOI] [PubMed] [Google Scholar]
- 2.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). JAMIA. 2010;17(2):124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Eschenbach AC, Buetow K. Cancer informatics vision: caBIG™. Cancer Informatics. 2006;2:22. [PMC free article] [PubMed] [Google Scholar]
- 4.Ellisman M, Peltier S, eds. Biomedical Informatics Research Network at the Third International HealthGrid Conference; April 6, 2005; Oxford. [Google Scholar]
- 5.Helmer KG, Ambite JL, Ames J, et al. Enabling collaborative research using the biomedical informatics research network (BIRN). JAMIA. 2011;18(4):416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schilling LM, Kwan BM, Drolshagen CT, et al. Scalable Architecture for Federated Translational Inquiries Network (SAFTINet) Technology Infrastructure for a Distributed Data Network. eGEMs. 2013;1(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrest CB, Margolis P, Seid M, Colletti RB. PEDSnet: how a prototype pediatric learning health system is being expanded into a national network. Health Affairs. 2014;33(7):1171–1177. [DOI] [PubMed] [Google Scholar]
- 8.Kho AN, Hynes DM, Goel S, et al. CAPriCORN: Chicago Area Patient-Centered Outcomes Research Network. JAMIA. 2014;21:607–611. doi:10.1136/amiajnl-2014-002827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandl KD, Kohane IS, McFadden D, et al. Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS): architecture. JAMIA. 2014;21:615–620. doi:10.1136/amiajnl-2014-002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGlynn EA, Lieu TA, Durham ML, et al. Developing a data infrastructure for a learning health system: the PORTAL network. JAMIA. 2014;21:596–601. doi:10.1136/amiajnl-2014-002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeVoe JE, Gold R, Cottrell E, et al. The ADVANCE network: accelerating data value across a national community health center network. JAMIA. 2014;21:591–595. doi:10.1136/amiajnl-2014-002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaushal R, Hripcsak G, Ascheim DD, et al. Changing the research landscape: the New York City Clinical Data Research Network. JAMIA. 2014;21:587–590. doi:10.1136/amiajnl-2014-002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khurshid A, Nauman E, Carton T, et al. Louisiana Clinical Data Research Network: establishing an infrastructure for efficient conduct of clinical research. JAMIA. 2014;21:612–614. doi:10.1136/amiajnl-2014-002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin W, Tsui FR, Borromeo C, et al. PaTH: towards a learning health system in the Mid-Atlantic region. JAMIA. 2014;21:633–636. doi:10.1136/amiajnl-2014-002759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waitman LR, Aaronson LS, Nadkarni PM, et al. The Greater Plains Collaborative: a PCORnet Clinical Research Data Network. JAMIA. 2014;21:637–641. doi:10.1136/amiajnl-2014-002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleurence RL, Curtis LH, Califf RM, et al. Launching PCORnet, a national patient-centered clinical research network. JAMIA. 2014;21:578–582. doi:10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrne CM, Mercincavage LM, Pan EC, Vincent AG, Johnston DS, Middleton B. The value from investments in health information technology at the US Department of Veterans Affairs. Health Affairs. 2010;29(4):629–638. [DOI] [PubMed] [Google Scholar]
- 18.Ohno-Machado L, Agha Z, Bell DS, et al. pSCANNER: patient-centered Scalable National Network for Effectiveness Research. JAMIA. 2014;21(4):621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patient Centered Outcomes Research Institute. Improving Our National Infrastructure to Conduct Comparative Effectiveness Research. 2013 [cited 2014]. http://www.pcori.org/funding-opportunities/improving-our-national-infrastructure-to-conduct-comparative-effectiveness-research/.
- 20. Zaharia M, Chowdhury M, Franklin MJ, et al. Spark: cluster computing with working sets. In: 2nd USENIX Workshop on Hot Topics in Cloud Computing. Boston, MA: USENIX Association 2010.
- 21.Chu C, Kim SK, Lin Y-A, et al. Map-reduce for machine learning on multicore. Adv Neural Inf Process Syst. 2007;19:281. [Google Scholar]
- 22.Feldman V. A complete characterization of statistical query learning with applications to evolvability. J Comput Syst Sci. 2012;78(5):1444–1459. [Google Scholar]
- 23.Toh S, Baker MA, Brown JS, Kornegay C, Platt R. Rapid Assessment of Cardiovascular Risk Among Users of Smoking Cessation Drugs Within the US Food and Drug Administration's Mini-Sentinel Program. JAMA Int Med. 2013;173(9):817–819. [DOI] [PubMed] [Google Scholar]
- 24.McMurry AJ, Murphy SN, MacFadden D, et al. SHRINE: enabling nationally scalable multi-site disease studies. PLoS One. 2013;8(3):e55811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel AJ, Kamerick M, Berman D, Dahm L, University of California Research eXchange (UCReX): A Federated Cohort Discovery System. Healthcare Informatics, Imaging and Systems Biology. In: proceedings of the 2012 IEEE Second International Conference on Healthcare Informatics, Imaging and Systems Biology, p. 146. IEEE Computer Society; 2012. [Google Scholar]
- 26.Holmes JH, Elliott TE, Brown JS, et al. Clinical research data warehouse governance for distributed research networks in the USA: a systematic review of the literature. JAMIA. 2014;21:730–766. doi:10.1136/amiajnl-2013-002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ames MJ, Bondy MJ, Johnson MSC, Wade MTD, Davidson PA, Kahn MM. Analysis of Federated Data Sharing Platforms for a Regional Data Sharing Network. AMIA 2013 Summit on Clinical Research Informatics. San Francisco, CA. [Google Scholar]
- 28.Payne P, Ervin D, Dhaval R, Borlawsky T, Lai A. TRIAD: The Translational Research Informatics and Data Management Grid. Appl Clin Inform. 2011;2(3):331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown JS, Holmes JH, Shah K, Hall K, Lazarus R, Platt R. Distributed health data networks: a practical and preferred approach to multi-institutional evaluations of comparative effectiveness, safety, and quality of care. Med Care. 2010;48(6):S45–S51. [DOI] [PubMed] [Google Scholar]
- 30.Shvachko K, Kuang H, Radia S, Chansler R, eds. The hadoop distributed file system. Mass Storage Systems and Technologies (MSST), 2010 IEEE 26th Symposium on 2010. IEEE. [Google Scholar]
- 31.Orellana F, Niinimaki M, eds. Distributed Computing with RESTful Web Services. P2P, Parallel, Grid, Cloud and Internet Computing (3PGCIC), 2012 Seventh International Conference on 2012. IEEE. [Google Scholar]
- 32.Zaharia M, Chowdhury M, Franklin MJ, Shenker S, Stoica I, eds. Spark: cluster computing with working sets. In: Proceedings of the 2nd USENIX conference on Hot topics in cloud computing; 2010, Boston, MA. [Google Scholar]
- 33.Kim KK, Browe DK, Logan HC, et al. Data governance requirements for distributed clinical research networks: triangulating perspectives of diverse stakeholders. JAMIA. 2013;21:714–719. doi:10.1136/amiajnl-2013-002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim KK, McGraw D, Mamo L, Ohno-Machado L. Development of a privacy and security policy framework for a multistate comparative effectiveness research network. Med Care. 2013;51 (8 Suppl 3):S66–S72. [DOI] [PubMed] [Google Scholar]
- 35.Ohno-Machado L, Bafna V, Boxwala AA, et al. iDASH: integrating data for analysis, anonymization, and sharing. JAMIA. 2012;19(2):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno-Machado L. To share or not to share: that is not the question. Sci Transl Med. 2012;4(165):165cm15. [DOI] [PubMed] [Google Scholar]
- 37.Adams N, Kirby S, Harris P, Clegg D. A review of parallel processing for statistical computation. Stat Comput. 1996;6(1):37–49. [Google Scholar]
- 38.Xu M, Miller JJ, Wegman EJ, eds. Parallelizing multiple linear regression for speed and redundancy: an Empirical Study. Distributed Memory Computing Conference, 1990, Proceedings of the Fifth on 1990. IEEE. [Google Scholar]
- 39.Johnson R, Hoeller J, Donald K, et al. The spring framework, reference documentation. Interface. 21 (accessed 2004 07). 2004. [Google Scholar]
- 40.Johnson KA, Chen S, Cheng I-N, et al. The impact of clinical pharmacy services integrated into medical homes on diabetes-related clinical outcomes. Ann Pharmacother. 2010;44(12):1877–1886. [DOI] [PubMed] [Google Scholar]
- 41.Meeker D, Knight TK, Friedberg MW, et al. Nudging guideline-concordant antibiotic prescribing: a randomized clinical trial. JAMA Intern Med. 2014;174(3):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang W, Li P, Wang S, et al. WebGLORE: a Web service for Grid LOgistic REgression. Bioinformatics. 2013;29(24):3238–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, Jiang X, Kim J, Ohno-Machado L. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. JAMIA. 2012;19(5):758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behilng D, Davis D, Sherman G, et al. Quality Data Model Based Health Quality Measures Format (eMeasure) Implementation Guide, Release 1 (US Realm) Based on HL7 HQMF Release 2.0. [Google Scholar]
- 45.Panda B, Herbach JS, Basu S, et al. PLANET. Proc VLDB Endow 2009;2:1426–1437. doi:10.14778/1687553.1687569. [Google Scholar]
- 46.Oancea B, Dragoescu RM. Integrating R and Hadoop for Big Data Analysis. arXiv preprint arXiv:14074908. 2014. [Google Scholar]
- 47.Ohmann C, Kuchinke W. Future developments of medical informatics from the viewpoint of networked clinical research. Methods Inf Med. 2009;48(1):45–54. [PubMed] [Google Scholar]
- 48.Kahn MG, Batson D, Schilling LM. Data model considerations for clinical effectiveness researchers. Medical Care. 2012;50(Suppl):S60–S67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown JS, Kahn MG, Toh D. Data quality assessment for comparative effectiveness research in distributed data networks. Med Care. 2013;51(8 Suppl 3):S22–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avery TR, Kulldorff M, Vilk Y, et al. Near real-time adverse drug reaction surveillance within population-based health networks: methodology considerations for data accrual. Pharmacoepidemiol Drug Saf. 2013;22(5):488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Observational Medical Outcomes Partnership. OMOP Common Data Model V4.0. 2012. http://omop.org/CDM.
- 52.Ohno-Machado L, Agha Z, Bell DS, et al. pSCANNER: patient-centered SCAlable National Network for Effectiveness Research. JAMIA. 2014;21(4):621–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terry SF, Terry PF. Power to the people: participant ownership of clinical trial data. Sci Transl Med. 2011;3(69):69cm3. [DOI] [PubMed] [Google Scholar]