Abstract
Regular physical activity helps prevent heart disease, stroke, diabetes, and other chronic diseases, yet a broad range of conditions impair mobility at great personal and societal cost. Vast amounts of data characterizing human movement are available from research labs, clinics, and millions of smartphones and wearable sensors, but integration and analysis of this large quantity of mobility data are extremely challenging. The authors have established the Mobilize Center (http://mobilize.stanford.edu) to harness these data to improve human mobility and help lay the foundation for using data science methods in biomedicine. The Center is organized around 4 data science research cores: biomechanical modeling, statistical learning, behavioral and social modeling, and integrative modeling. Important biomedical applications, such as osteoarthritis and weight management, will focus the development of new data science methods. By developing these new approaches, sharing data and validated software tools, and training thousands of researchers, the Mobilize Center will transform human movement research.
Keywords: machine learning, biomechanics, wearable sensors, physical activity, obesity
MISSION
Movement is essential for human health. Regular physical activity helps prevent heart disease, stroke, and diabetes.1–3 It improves cognitive function among older adults, relieves symptoms of depression, and promotes weight loss.1,4 Unfortunately, many conditions limit mobility at a great cost to public health and personal well-being. Musculoskeletal disorders, such as osteoarthritis, affect one in two American adults during their lifetime at an estimated annual cost (direct plus indirect) of $870 billion.5 Obesity, which reduces mobility and contributes to conditions like cardiovascular disease, results in an estimated 20% of the US national health expenditures.6 Taken together, the consequences of limited mobility are substantial and severe.
The vast amount of movement data being collected today by clinical centers, biomechanics laboratories, and millions of wearable sensors and smartphone accelerometers have the potential to revolutionize the way human movement research is conducted. These large datasets allow us to investigate mobility limitations in novel ways and could lead to new paradigms in diagnosis and treatments, such as a system that predicts and improves the outcomes of surgeries for children with cerebral palsy or an application that tracks a person’s movement 24 hours a day to determine his risk of developing knee osteoarthritis.
However, the data describing human movement are underutilized. They are seldom analyzed in aggregate with data from other sources, and in many cases, have never been analyzed. The limit arises, in part, from the nature of the data: they are noisy, heterogeneous, incompatible, and incomplete. Much of the data are time series, representing coarse samples of a smoothly evolving physical process, and methods for optimal combination of statistical inference with physical, physiological, and behavioral knowledge have not been developed.
The Mobilize Center’s mission is to overcome the challenges of analyzing large data sets that describe human movement and to improve human movement across the wide range of conditions that limit mobility. The Center focuses on 4 data science areas: biomechanical modeling, statistical learning, behavioral and social modeling, and integrative modeling. To ensure applicability of our data science research to important health issues, the methods will be developed, tested, and validated within the context of three driving biomedical problems: cerebral palsy surgical planning, gait rehabilitation research, and weight management (Table 1). Through extensive training programs and the sharing of open-source software tools and unique data sets, the Center will also enable others to advance the integration of big data analytics into the field of mobility research and other areas of biomedicine.
Table 1:
Application of Data Science Core Research to Driving Biomedical Problems
| Core 1 | Core 2 | Core 3 | Core 4 | ||
|---|---|---|---|---|---|
| Driving Biomedical Problems | Biomechanical Modeling | Statistical Learning | Behavioral and Social Modeling | Integrative Modeling | |
| 1 | Cerebral Palsy Surgical Planning | ✓ | ✓ | ✓ | |
| 2 | Gait Rehabilitation Research | ✓ | ✓ | ✓ | |
| 3 | Weight Management | ✓ | ✓ | ✓ | ✓ |
The driving biomedical problems and the data science cores are closely coupled. The checkmarks indicate which cores apply to each driving biomedical problem. Note that each core is applied to at least 2 of the driving biomedical problems, enabling the assessment of the generalizability of the data science methods being developed.
IMPACTING MOBILITY RESEARCH AND HEALTH THROUGH DATA SCIENCE ADVANCES
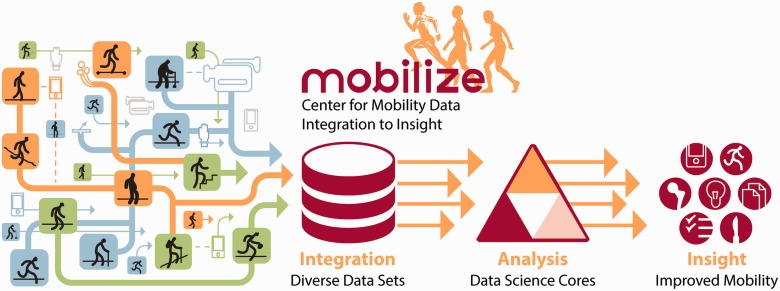
Big data analytics is poised to revolutionize the field of mobility research. Our 4 data science cores provide a set of generally applicable tools for analyzing biomedical big data. Our three driving biomedical problems apply methods developed within these data science cores, serving as examples of our general process for harnessing big data to improve the health of those with limited mobility (Figure 1). Each of the cores and driving biomedical problems are described below.
Figure 1:
Vision of the Mobilize Center. With our partner hospitals, collaborating biomechanics labs, and industry affiliates, we have assembled and continue to collect datasets describing human motions, including trajectories of markers placed on the body, ground forces, muscle electromyography, muscle strength, range-of-motion measurements, video, accelerometer and GPS recordings from wearable sensors and smartphones, treatment histories, clinical notes, food intake, sleep records, treatment outcome metrics, and published results. Tools developed within our data science cores will enable us to integrate and analyze these diverse data, leading to important new insights for surgical planning, gait modification, prosthesis design, and exercise prescriptions.
Driving Biomedical Problems
Cerebral Palsy Surgical Planning: Cerebral palsy affects approximately 3 of every 1000 births in the United States7 and leads to pathological movement patterns, such as crouch gait. Diagnosing and treating such pathologies is challenging and surgical outcomes variable. At one leading clinical center in the United States, only 50% of patients had significantly improved crouch gait despite receiving surgeries intended to improve walking.8 Our studies show that biomechanical modeling and statistical learning techniques hold great promise for improving surgical outcomes.9,10 We will develop a trained system—a data processing system combining statistical inference, domain knowledge, and data management techniques—to extend this research by analyzing large datasets to robustly predict the outcomes of surgeries intended to improve walking ability in children with cerebral palsy.
Gait Rehabilitation Research: Researchers currently do not understand movement dynamics well enough to identify individuals at risk for conditions such as osteoarthritis or running injuries. Barriers to achieving this understanding include the small number of subjects in past studies and the lack of tools for analyzing movement data. The Mobilize Center will address these problems by establishing a research network that integrates data from research labs and industry partners, and will apply big data analytical tools on the resulting database to identify individuals at risk for limited mobility due to osteoarthritis and running injuries. This framework will enable gait rehabilitation researchers to design effective interventions and answer long-standing questions utilizing mobility big data.
Weight Management: Over 35% of adults in the United States are obese, and another 35% are overweight according to recent estimates.11 The growing use of smartphones and wearable sensors, combined with our knowledge of biomechanics and behavior, provide a unique opportunity to understand how to encourage healthy physical activity and weight management. Our collaborators have designed biomechanically informed training programs to mitigate injury risk for individuals who are obese.12,13 Leveraging this work, we will design effective personalized interventions for weight management that integrate knowledge of biomechanics with the behavioral and social dynamic factors that influence individuals to engage in regular physical activity. As part of our investigation, we will examine different aspects of exercise, including duration and frequency, intensity, and type (e.g., running, walking, weight training), to identify interventions that successfully balance low joint loading with high energy consumption for an individual.
For each of our driving biomedical problems, we are initially focusing on the relationship between the driving biomedical problem, musculoskeletal factors like strength, and movement. In cases where other data, such as environmental, social, and dietary factors, are available, we will also incorporate that data into our analyses. The development of new methods in our data science cores is required to help elucidate these complex relationships. Together, the new methods and the new insights we gain from applying the methods to unique data resources will provide a foundation for a broad understanding of the driving biomedical problems, including the role of movement dynamics and other factors, such as genetics, that are likely involved.
Data Science Cores
Biomechanical Modeling: Biomechanical models, which represent the physics and physiology of human movement, can turn heterogeneous experimental data, from diverse labs and experimental protocols, into knowledge. Models also enable us to make predictions in domains where data are unavailable, such as predicting a patient’s response to a future surgery. However, analysis of existing data currently cannot identify new treatments or make robust predictions outside the measured domain of knowledge. Nor are there tools or standards to robustly integrate data from different research labs into subject-specific models and simulations for analysis and prediction.
Our biomechanical modeling core builds upon extensive past work using the OpenSim software to generate subject-specific models and simulations of able-bodied and pathological movement.14–18 By integrating advanced optimization tools,19,20 we will develop musculoskeletal model fitting methods that are robust to noisy data and capable of incorporating data from diverse sources (Figure 2). We continue to work on the field’s grand challenge: creating neural controllers that accurately represent the human response to perturbations like surgeries and pathology. These advancements will enable nearly automatic generation of subject-specific models and muscle-driven simulations, benefitting biomechanics researchers, as well as clinicians and scientists in neuroscience (e.g., aiding the design of neuroprosthetics) and ergonomics.
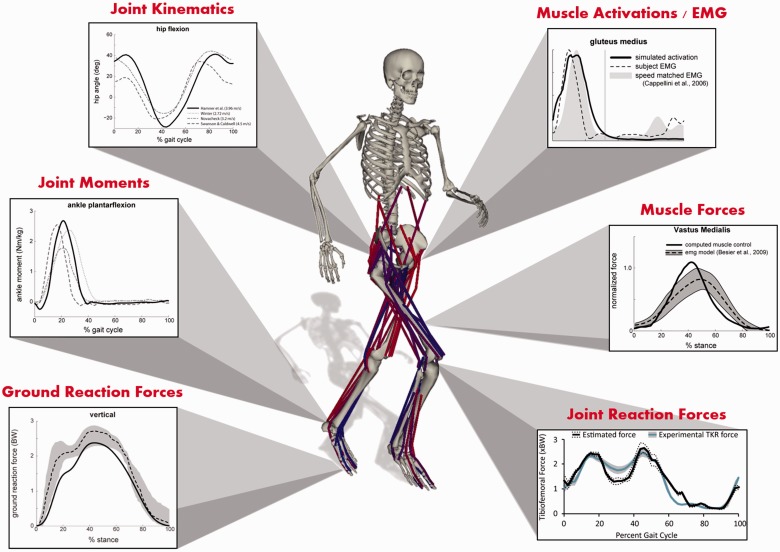
Figure 2:
Adapting Biomechanical Models and Simulations to Big Data. Biomechanical models and simulations fuse data about human movement, including joint kinematics, joint moments, and ground reaction forces, to produce estimates of muscle forces, muscle activations, and joint reaction forces. The data collection process and measurement accuracy for all these quantities vary from one research lab to another. Some quantities are not acquired in a given lab, while some datasets may include additional information, such as muscle strength or clinical notes. The development of predictive neural controllers and the integration of advanced optimization methods and statistical learning techniques will enable us to gain new insights from the growing data for human movement. Figure courtesy of Samuel Hamner.
Statistical Learning: Huge amounts of data about human movement are available from a range of sources. Although these data often have significant noise and measurement errors, they provide valuable information when analyzed in aggregate. However, we lack established scientific principles or methods to apply to such large volumes of noisy, sparse human activity data. Statistical learning approaches can aggregate and analyze these diverse data to identify risk factors for reduced mobility, correlations with improved mobility, and other data-driven insights to promote movement and health.
We will extend our work on large-scale, mixed-effects modeling and classification21–24 to apply to mobility data. These new models will address the sparse, noisy, time-series nature of mobility data, enabling the estimation of missing parameters so that datasets, even if they differ in some of their parameters, can still be combined. This will greatly expand the number of subjects and usable data available per subject to make predictions (e.g., treatment guidance for individuals with cerebral palsy) and classifications (e.g., identifying individuals with early stage osteoarthritis). The Mobilize Center will also develop a framework for extracting meaningful knowledge from large, raw sets of data that go beyond correlations between simple variables. Built upon our MetroMaps system, which produces structured summaries from large collections of documents,25 this framework would identify patterns within large datasets that are also plausible and previously unknown.
Behavioral and Social Modeling: Social and behavioral dynamics play powerful roles in motivating movement. We have begun investigating these dynamics using data from millions of users of iPhone applications from our industry partners, such as Azumio. We will model these dynamics and examine how to leverage social networks and motivational tools like badges to increase physical activity.
We know badges can influence and steer user behavior on a website (Figure 3),26,27 but their impact on behavior off-line is unclear. We will expand our current model for predicting user responses to badges to a multi-dimensional space, where multiple types of activity (rather than a single activity) are awarded and an individual’s actions are steered toward complex behavior (rather than narrow interaction with a single website). We are also developing network-based mathematical models to investigate the spread of peer effects and behavior change through social networks. These models will help us harness social networks to improve physical activity. The third thrust of this core is extending our current model of user evolution28 (e.g., progression through the stages of physical fitness) to develop personalized recommendations that account for the user’s expertise, knowledge, fitness, or individual abilities.
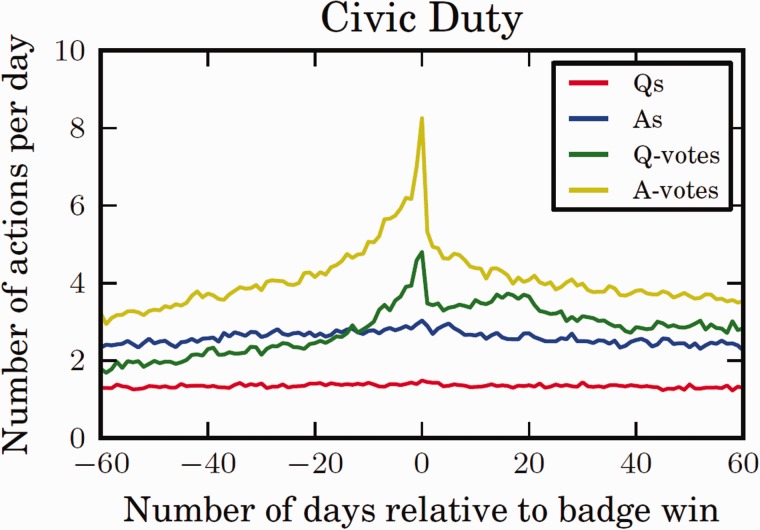
Figure 3:
Incentives to Motivate Physical Activity. We have demonstrated the effectiveness of badges to steer user activity on-line, as in this example, where a Civic Duty badge was introduced on the question-answering website Stack Overflow.26,27 The badge rewarded users for voting on the merit of questions and answers submitted to the site. The graph shows that an individual user’s voting activity (Q-votes and A-votes) increased as the time to achieving the badge decreased. Our social and behavioral modeling research will examine how social networks and motivational tools like badges, which influence behavior on-line, could be leveraged to change behavior off-line, e.g., to increase physical activity. Printed with permission from Jure Leskovec | Association for Computing Machinery 2013. The definitive Version of Record was published in The Proceedings for the 22nd International Conference on World Wide Web 2013.
Integrative Modeling: A key goal of the Center is generating robust and meaningful predictions to aid diagnosis and treatment planning for individuals with limited mobility. While many data sources (e.g., experimental research labs and wearable sensors) and models (e.g., biomechanical or statistical models) may aid this process, they are currently isolated and hard to integrate. We will dissolve these barriers and develop trained systems that integrate diverse models and data types using scalable statistical inference.
Specifically, we are utilizing the DeepDive framework,29–31 a trained computing system with the unique ability to ingest and process structured databases and unstructured information (e.g., clinical notes). Using DeepDive, we specify rules in a formal logic language that, for example, describe good indicators of a given outcome. These rules serve as features or signals to a statistical model, creating an inference and parameter estimation problem over large factor graphs. We will solve these problems in a computationally efficient and scalable manner by developing new methods that further improve our highly parallel statistical engine.32–34
TRAINING A NEW GENERATION OF INTERDISCIPLINARY RESEARCHERS
Training is an integral part of the Mobilize Center, helping to create the next generation of researchers who understand both data science and movement research. The Mobilize Center’s initial postdoctoral search revealed a dearth of these cross-trained individuals, and our Center is helping to fill this critical gap. We will build on our past successes in training 24 postdoctoral fellows and running 40+ workshops and 20+ webinars through the Simbios National Center for Biomedical Computing.35 Beyond our Distinguished Postdoctoral Fellows program, we will offer summer internships, massive open online courses, and a webinar and symposium series to train and engage tens of thousands of students around the world in data science methods for mobility research.
DEVELOPING RESOURCES FOR THE COMMUNITY
Software Tools: The Mobilize Center is committed to engaging others in biomedical big data efforts. We will enable individuals to investigate the potential of big data analytics by producing a set of open-source, extensible software tools:
OpenSim (http://opensim.stanford.edu)—biomechanical modeling and simulation software providing a common standard for integrating and analyzing mobility big data.
Optimization Software (http://stanford.edu/∼boyd/software.html)— tools to solve mathematical systems for modeling and analyzing big data.
R Statistical Learning Packages (http://web.stanford.edu/∼hastie/swData.htm)—tools to make predictions with time-series data and identify insightful connections from biomedical big data.
SNAP (http://snap.stanford.edu)—software for modeling and probing behavioral dynamics and decision making, as well as large social network dynamics.
DeepDive (http://deepdive.stanford.edu)—software for developing, testing, and understanding trained systems that integrate diverse data and model types through logic rules defined by domain researchers.
Data Resources: We will aggregate and share datasets describing human movement and lead an effort to develop the standards and metadata required for the effective sharing and mining of such data. Currently, movement datasets can be generated using different equipment, different measurement protocols, and different post-processing methods, to name just a few factors that differ from study to study. Standardization can facilitate bringing together these disparate datasets. The OpenSim software36 has already become a widely used standard for sharing a variety of movement data, and we will build upon this to enable the community to share and aggregate their datasets. A crucial component is the metadata associated with the data; information, such as the weight of the subject, identification of whether the data are pre- or post-treatment, the diagnoses and treatments, etc., facilitate data aggregation and analysis. We will help establish the minimal metadata needed to describe human movement to enable effective data integration. Even with a push for standardization, we expect many differences will remain; thus the data science tools we develop for statistical learning and integrative modeling will be designed to handle heterogeneous and noisy data.
Biomedical Computation Review: We will continue producing our Biomedical Computation Review magazine (http://bcr.org), which highlights research advances and tools from across the entire biocomputational community. It is written for a general audience, making it valuable for teaching and accessible to many.
VISION OF THE FUTURE
Through these diverse efforts, the Mobilize Center will improve mobility—and health—for the large portion of the population affected by musculoskeletal disorders and other factors that inhibit physical activity. We will transform the field of human movement research through our development and application of innovative data science methods and help ensure that the vast amounts of data continually being collected about movement today are routinely translated to insight and improved patient care.
FUNDING
The Mobilize Center and this work was supported by the National Institutes of Health Grant U54EB020405 awarded by the National Institute of Biomedical Imaging and Bioengineering through funds provided by the trans-National Institutes of Health Big Data to Knowledge (BD2K, http://www.bd2k.nih.gov) initiative.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
S.L.D. provides the vision for and leads the Mobilize Center. T.H. is the lead on statistical learning; J.L. leads the behavioral and social modeling effort; and C.R. heads the integrative modeling research. J.P.K., J.L.H., and S.L.D. manage the Center, including training efforts and integration of the data science cores with the driving biomedical problems. All authors wrote the manuscript, were responsible for the critical revision of the manuscript, and approved the final version.
REFERENCES
- 1.Promotion OoDPaH. 2008 Physical Activity Guidelines for Americans: U.S. Department of Health and Human Services, 2008. http://www.health.gov/paguidelines/guidelines/. Accessed February 16, 2015.
- 2.Helmrich SP, Ragland DR, Leung RW, et al. Physical activity and reduced occurrence of non-insulin-dependent diabetes mellitus. N Engl J Med. 1991;325(3):147–152. [DOI] [PubMed] [Google Scholar]
- 3.Blomstrand A, Blomstrand C, Ariai N, et al. Stroke incidence and association with risk factors in women: a 32-year follow-up of the Prospective Population Study of Women in Gothenburg. BMJ Open 2014; 4(10):e005173. doi:10.1136/bmjopen-2014-005173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langa KM, Levine DA. The diagnosis and management of mild cognitive impairment: a clinical review. JAMA 2014;312(23):2551–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.United States Bone and Joint Initiative: The Burden of Musculoskeletal Diseases in the United States. 3rd ed. Rosemont, IL: United States Bone and Joint Initiative; 2014. http://www.boneandjointburden.org Accessed February 15, 2015.
- 6.Cawley J, Meyerhoefer C. The medical care costs of obesity: an instrumental variables approach. J Health Econ. 2012;31(1):219–230. [DOI] [PubMed] [Google Scholar]
- 7.Honeycutt A, Dunlap L, Chen H, et al. Economic Costs Associated with Mental Retardation, Cerebral Palsy, Hearing Loss, and Vision Impairment - United States, 2003: Centers for Disease Control and Prevention; 2004:57–59. [PubMed]
- 8.Hicks JL, Delp S, Andriacchi TP, et al. Biomechanical guidelines for the treatment of crouch gait in children with cerebral palsy. Stanford, CA: Stanford University; 2010. [Google Scholar]
- 9.Hicks JL, Delp SL, Schwartz MH. Can biomechanical variables predict improvement in crouch gait? Gait Posture. 2011;34(2):197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinbolt JA, Fox MD, Schwartz MH, et al. Predicting outcomes of rectus femoris transfer surgery. Gait Posture. 2009;30(1):100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flegal KM, Carroll MD, Kit BK, et al. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA. 2012;307(5):491–497. [DOI] [PubMed] [Google Scholar]
- 12.Ehlen KA, Reiser RF, 2nd, Browning RC. Energetics and biomechanics of inclined treadmill walking in obese adults. Med Sci Sports Exerc. 2011;43(7):1251–1259. [DOI] [PubMed] [Google Scholar]
- 13.Haight DJ, Lerner ZF, Board WJ, et al. A comparison of slow, uphill and fast, level walking on lower extremity biomechanics and tibiofemoral joint loading in obese and nonobese adults. J Orthop Res. 2013;32(2):324–330. [DOI] [PubMed] [Google Scholar]
- 14.Damiano DL, Arnold AS, Steele KM, et al. Can strength training predictably improve gait kinematics? A pilot study on the effects of hip and knee extensor strengthening on lower-extremity alignment in cerebral palsy. Phys Ther. 2010;90(2):269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerus P, Sartori M, Besier TF, et al. Subject-specific knee joint geometry improves predictions of medial tibiofemoral contact forces. J Biomech. 2013;46(16):2778–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steele KM, van der Krogt MM, Schwartz MH, et al. How much muscle strength is required to walk in a crouch gait? J Biomech. 2012;45(15):2564–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knarr BA, Reisman DS, Binder-Macleod SA, et al. Understanding compensatory strategies for muscle weakness during gait by simulating activation deficits seen post-stroke. Gait Posture. 2013;38(2):270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards C, Higginson JS. Knee contact force in subjects with symmetrical OA grades: differences between OA severities. J Biomech. 2010;43(13):2595–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd SP, Vandenberghe L. Convex Optimization. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- 20.Do CB, Batzoglou S. What is the expectation maximization algorithm? Nat Biotechnol. 2008;26(8):897–899. [DOI] [PubMed] [Google Scholar]
- 21.James GM, Hastie TJ. Functional linear discriminant analysis for irregularly sampled curves. J R Stat Soc. 2001;63(3):533–550. [Google Scholar]
- 22.James GM, Hastie TJ, Sugar CA. Principal component models for sparse functional data. Biometrika. 2000;87(3):587–602. [Google Scholar]
- 23.James GM, Sugar CA. Clustering for sparsely sampled functional data. J Am Stat Assoc. 2003;98(462):397–408. [Google Scholar]
- 24.Bachrach LK, Hastie T, Wang MC, et al. Bone mineral acquisition in healthy Asian, Hispanic, black, and Caucasian youth: a longitudinal study. J Clin Endocrinol Metab. 1999;84(12):4702–4712. [DOI] [PubMed] [Google Scholar]
- 25.Shahaf D, Yang J, Suen C, et al. Information cartography: creating zoomable, large-scale maps of information. Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA; 2013:1097–1105. [Google Scholar]
- 26.Anderson A, Huttenlocher D, Kleinberg J, et al. Steering user behavior with badges. 22nd International Conference on World Wide Web, Rio de Janeiro, Brazil; 2013:95–106. [Google Scholar]
- 27.Anderson A, Huttenlocher D, Kleinberg J, et al. Engaging with massive online courses. 23rd International Conference on World Wide Web, Seoul, Korea; 2014:687–698. [Google Scholar]
- 28.McAuley JJ, Leskovec J. From amateurs to connoisseurs: modeling the evolution of user expertise through online reviews. Proceedings of the 22nd International Conference on World Wide Web, Rio de Janeiro, Brazil; 2013:897–908. [Google Scholar]
- 29.Kumar A, Ré C. Probabilistic management of OCR data using an RDBMS. Proc VLDB Endowment. 2011;5(4):322–333. [Google Scholar]
- 30.Niu F, Zhang C, Ré C, et al. DeepDive: web-scale knowledge-base construction using statistical learning and inference. VLDS, Istanbul, Turkey. 2012;25–28. [Google Scholar]
- 31.Zhang C, Govindaraju V, Borchardt J, et al. GeoDeepDive: statistical inference using familiar data-processing languages. ACM SIGMOD International Conference on Management of Data, SIGMOD, New York, NY, USA. 2013;993–996. [Google Scholar]
- 32.Liu J, Wright S, Ré C, et al. An asynchronous parallel stochastic coordinate descent algorithm. 31th International Conference on Machine Learning (ICML), 2014, Beijing, China. 2014;469–477. [Google Scholar]
- 33.Niu F, Recht B, Ré C, et al. Hogwild!: a lock-free approach to parallelizing stochastic gradient descent. Advances in Neural Information Processing Systems 24: 25th Annual Conference on Neural Information Processing Systems, Granada, Spain; 2011:693–701. [Google Scholar]
- 34.Zhang C, Ré C. Towards high-throughput Gibbs sampling at scale: a study across storage managers. ACM SIGMOD International Conference on Management of Data, SIGMOD, New York, NY, USA; 2013:397–408. [Google Scholar]
- 35.Delp SL, Ku JP, Pande VS, et al. Simbios: an NIH national center for physics-based simulation of biological structures. JAMIA. 2012;19(2):186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delp SL, Anderson FC, Arnold AS, et al. OpenSim: open-source software to create and analyze dynamic simulations of movement. IEEE Trans Biomed Eng. 2007;54(11):1940–1950. [DOI] [PubMed] [Google Scholar]