Abstract
Background Electronic health records (EHRs) are increasingly used for clinical and translational research through the creation of phenotype algorithms. Currently, phenotype algorithms are most commonly represented as noncomputable descriptive documents and knowledge artifacts that detail the protocols for querying diagnoses, symptoms, procedures, medications, and/or text-driven medical concepts, and are primarily meant for human comprehension. We present desiderata for developing a computable phenotype representation model (PheRM).
Methods A team of clinicians and informaticians reviewed common features for multisite phenotype algorithms published in PheKB.org and existing phenotype representation platforms. We also evaluated well-known diagnostic criteria and clinical decision-making guidelines to encompass a broader category of algorithms.
Results We propose 10 desired characteristics for a flexible, computable PheRM: (1) structure clinical data into queryable forms; (2) recommend use of a common data model, but also support customization for the variability and availability of EHR data among sites; (3) support both human-readable and computable representations of phenotype algorithms; (4) implement set operations and relational algebra for modeling phenotype algorithms; (5) represent phenotype criteria with structured rules; (6) support defining temporal relations between events; (7) use standardized terminologies and ontologies, and facilitate reuse of value sets; (8) define representations for text searching and natural language processing; (9) provide interfaces for external software algorithms; and (10) maintain backward compatibility.
Conclusion A computable PheRM is needed for true phenotype portability and reliability across different EHR products and healthcare systems. These desiderata are a guide to inform the establishment and evolution of EHR phenotype algorithm authoring platforms and languages.
Keywords: electronic health records, phenotype algorithms, computable representation, phenotype standardization, data models
INTRODUCTION
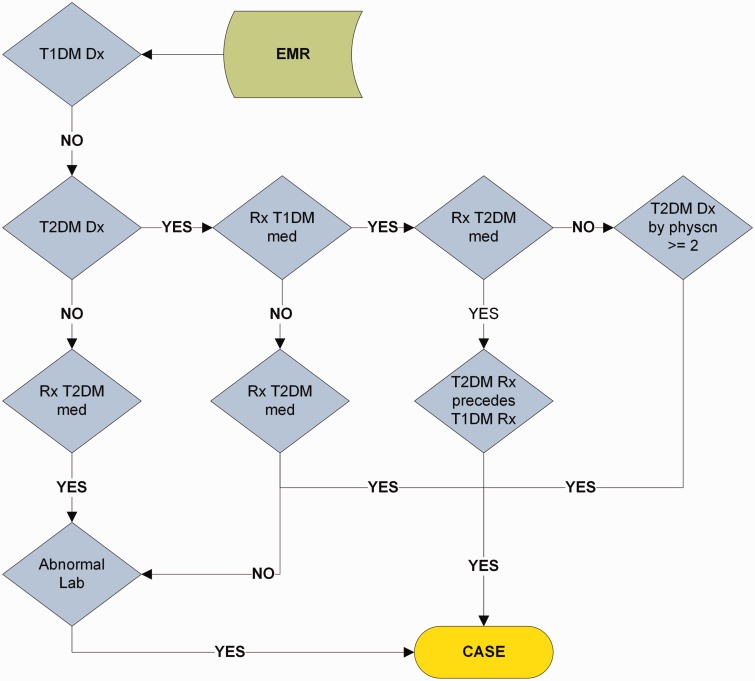
Electronic health records (EHRs) are increasingly used for clinical and translational research through the creation of phenotype algorithms, consisting of structured selection criteria designed to produce research-quality phenotypes.1–7 These algorithms operate on diverse classes of EHR data to select individuals with given traits (e.g., identifying records for continuous trait analyses or marking records as a case, a control, or neither for given conditions).8,9 Examples include identifying patients with hypothyroidism matched to hypothyroidism-free controls,5 evaluating cardiac conduction duration in electrocardiograms of “heart-healthy” individuals,10 and determining medication responses.11–18 Typically, these algorithms define the workflow for querying clinical data regarding diagnoses, procedures, medications, laboratory or radiology reports, and other EHR data, and can require natural language processing (NLP) or text mining. Multi-site studies have shown that these algorithms often are portable between sites.5,19,20 Currently, most phenotype algorithms are recorded as human-readable descriptive text documents that can be shared via knowledge bases such as the Phenotype KnowledgeBase (PheKB, http://phekb.org) and PhenotypePortal (http://phenotypeportal.org). Algorithms described via text and flowcharts (such as the type 2 diabetes mellitus [T2DM] algorithm shown in Figure 1 and the Desiderata section) require human translation to computable formats and are often ambiguous. Implementation across different institutions requires human experts to interpret the algorithm and translate it into executable operations and queries. This situation has hampered cross-institutional collaboration.21
Figure 1:
Phenotype algorithm for identifying type 2 diabetes mellitus (T2DM) from electronic medical records (EMR or EHR). T1DM: type 1 diabetes mellitus; Dx: diagnoses, defined as recorded using International Classification of Diseases, 9th Revision (ICD-9) codes; med: medication; physcn: physicians; Rx: prescriptions. More details can be found in the appendix and on PheKB.org.
To enable cross-site phenotype execution, we suggest two needed initiatives: (1) creation of a common phenotype representation model (PheRM) as a computable representation of phenotype algorithms and (2) development of infrastructure to allow standards-based authoring and execution of PheRM-based algorithms for a variety of EHR systems. In this paper, we leveraged our experiences with the Electronic Medical Records and Genomics (eMERGE) Network,22 Pharmacogenomics Research Network (PGRN),23 Strategic Health IT Advanced Research Project (SHARP),24 and the National Patient-Centered Clinical Research Network (PCORnet)25 to propose desiderata for PheRM (Table 1).
Table 1:
A list of desiderata
|
BACKGROUND
With the implementation of Meaningful Use (MU),26 EHRs have been increasing in ubiquity, functionality, and comprehensiveness. One recent advance has been the coupling of DNA bio-repositories to EHR data27–30 to enable genomic discoveries.31 In particular, the eMERGE network, a large scale, multi-site network of research organizations of 11 academic medical centers, has been at the forefront of mining biobank resources (both EHRs and associated DNA samples) for genomic medicine. Identification of research subjects from patient populations using phenotype algorithms is the starting point for these projects.
Data components in phenotyping may include the full range of clinical data stored in the EHR, such as demographics, vital signs, laboratory tests, medication, diagnoses, procedures, and other documentation.32 However, each EHR can have a different data model. One approach to facilitate research interoperability among different sites has been the Observational Health Data Sciences and Informatics (OHDSI) program, which has built on the Observational Medical Outcomes Partnership (OMOP) common data model (CDM).33–37 This CDM provides a standardized data interface for a vibrant ecosystem of healthcare big-data analyses (http://omop.org/OSCAR), including tools, web applications, and application program interfaces. Similarly, PCORnet25 and the Informatics for Integrating Biology and the Bedside (i2b2) based Shared Health Research Information Network38,39 are advancing common data models among their groups. These CDMs typically cover more focused, common data elements to enable a broad range of queries.
Phenotype algorithms are typically developed in an iterative fashion with expert review for validation21 to rule-based models, but can also utilize machine learning methods.40–42 The efficacy of a phenotype algorithm is usually measured with information retrieval metrics, such as sensitivity, specificity, positive predictive value, and F-measure.
At present, most existing phenotype algorithms are expressed in pseudo-code and not directly executable, because there is no widely adopted standards and underlying data structures. Thus, implementation requires human experts to translate descriptive algorithms from documents to ad hoc queries in local EHR research repositories, a process which is prone to inconsistencies or errors.43 One of the major efforts in establishing a standard language for a related task is the Quality Data Model (QDM) from National Quality Forum, which has been designed to represent electronic clinical quality measures.44 QDM has been shown to be capable of representing many phenotype algorithms from PheKB.45,46 Systems such as i2b2 system,47 SHARP,24 and Eureka! Clinical Analytics48 all have internal data and algorithm representations, some of which may be shared across sites. In general, these systems provide graphical interfaces that can standardize queries, but complex scoring metrics, counting rules, and nested temporal references or sequencing of events—such as found in many eMERGE algorithms49—often exceed their capability.
Phenotype algorithms have adopted a variety of logical and computational modalities.9,50 Modalities (e.g., scoring rules, counting rules) adopted in clinical diagnostic criteria51–53 have potential application in phenotype algorithms. In addition, machine learning and statistical model-based phenotype algorithms have been increasingly reported.54–57
Most current phenotype algorithms (CPT) use both structured and unstructured EHR elements.9 Structured EHR data usually include demographic information (e.g., age, sex, race, death), billing codes (i.e., International Classification of Disease version 9 (ICD-9), Current Procedural Terminology), most laboratory tests, vital signs, medications, and more. Unstructured EHR elements usually include clinical notes (e.g., history and physical examinations, progress notes, discharge summaries, nursing notes), some non-billing medical problems and most family history elements, some medications records and refills, diagnostic reports (e.g., radiology, microbiology, pathology), and more.
METHODS
A group of clinicians and informaticians reviewed 21 eMERGE phenotype algorithms (Table 2) and several authoring tools (Measure Authoring Tool [www.emeasuretool.cms.gov], i2b2, Eureka!, PhenotypePortal, the Vanderbilt Synthetic Derivative,27 and the Marshfield Personalized Medicine Research Project interface58) for common features. These phenotyping algorithms were of different complexity and included both disease and drug response phenotypes using algorithms from the eMERGE22 and Pharmacogenomics of Very Large Populations (PGPop) networks. We also evaluated the ability to represent selected well-known diagnostic criteria (e.g., Duke criteria for infective endocarditis,52 CHADS2 criteria for anticoagulation therapy in atrial fibrillation (AF)51) as potential phenotypes (see Supplementary Appendix Part 2). After proposal by a smaller team of investigators, the desiderata were evaluated and refined by all authors, which included investigators from eMERGE, PGRN, PGPop, SHARPn, PCORNet, and HMO Research Network.
Table 2:
Features of selected algorithms available on PheKB
| Algorithms | Data elements | Challenges informing desiderataa |
|---|---|---|
| Atrial fibrillation | CPT, ICD-9, ECG reports |
|
| Cardiac conduction10,94,95 |
|
|
| Cataract96,97 |
|
|
| Clopidogrel poor metabolizers11 |
|
|
| Crohn’s disease |
|
|
| Dementia | ICD-9, medication | Code counts (D5) |
| Diabetic retinopathy |
|
|
| Drug-induced liver injury14,98 | ICD-9, medications, laboratories |
|
| Height | ICD-9, laboratories, medications, height, age |
|
| HDL99,100 | ICD-9, laboratories, medications |
|
| Hypothyroidism5 | CPT, ICD-9, laboratories, medications, clinical documents |
|
| Lipids | ICD-9, laboratories, medications | Event selection (D4, D6) |
| Multiple sclerosis | ICD-9, medications, PL, H&P, discharge summaries, other notes |
|
| Peripheral arterial disease | CPT, ICD-9, laboratories, medications, clinical notes, radiological reports |
|
| RBC indices101 | CPT, ICD-9, laboratories, medications | Event selection and exclusion (D4, 6) |
| Rheumatoid arthritis |
|
|
| Severe early childhood obesity | ICD-9, medications, vital signs, age |
|
| Type 2 diabetes mellitus19,102 | ICD-9, laboratories, medications | Complex nested Boolean logic (D5) |
| Warfarin dose and response103 | Medications, laboratories, notes from anticoagulation clinics |
|
| WBC indices104 | CPT, ICD-9, laboratories, medications | Complex selection and exclusion of events (D4–6) |
aD1–D10 in parentheses represent the desiderata elements corresponding to each challenge. All phenotype algorithms benefit from D1, D2, and D7.
BMI: body mass index; CPT: current procedural terminology; ECG: electrocardiogram; HDL: high-density lipoprotein; H&P: history and physical examination (notes); ICD-9: International Classification of Diseases, 9th Revision; NLP: natural language processing; PL: problem list; PMH: past medical history; QRS: the QRS complex which indicates ventricular depolarization in ECG; RA: rheumatoid arthritis; RBC: red blood cells; WBC: white blood cells.
DESIDERATA
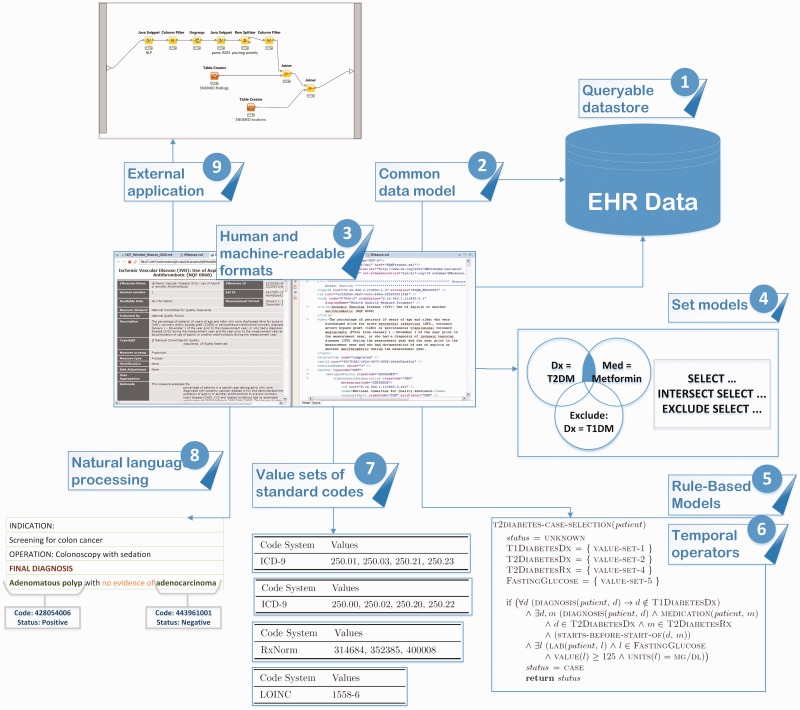
Based on our review, we propose the following desiderata for PheRM and its software implementation (see Figure 2 and Table 1). We acknowledge that phenotyping is not a standalone practice, and, instead, is closely coupled with EHR infrastructure and clinical practice. Therefore, we have included recommendations (representing the phenotyping community) to the EHR development community (Desiderata 1 and 2) as well as those regarding PheRM itself (Desiderata 3–10).
Figure 2:
Schematic of desiderata for computable phenotype electronic health record-driven phenotyping. Numerals 1–9 in the figure correspond to Desiderata 1–9 (Desideratum 10 is not depicted in this Figure).
Recommendations for clinical data representation to support phenotype
1. Structure clinical data into queryable forms
Clinical data are practically structured to promote efficient queries of all clinical information for an individual patient. On the other hand, phenotyping requires population-wide searching of individuals with similar characteristics (e.g., elevated LDL for a hyperlipidemia phenotype). Relational databases have been widely used for data storage as parts of enterprise data warehouse solutions. To further facilitate querying, where possible, clinical data stored in such data warehouses should be atomized (as first normal form59), such as storing a blood pressure into a systolic reading and a diastolic reading. Precalculating commonly derived observations (e.g., periods of drug exposure, as implemented in the OMOP drug era model33–37) also facilitate more efficient querying. Currently available documents are mostly poorly structured, and require information extraction or indexing60 to make them queryable.
2. Recommend a common data model, but also support customization for the variability and availability of EHR data among sites
To achieve a common PheRM, a common EHR data representation should be implemented where possible. Huser and Cimino analyzed three public integrated data repositories (IDRs) and proposed desiderata for their common design patterns.61 Potential candidates for CDM include Clinical Information Modeling Initiative,62 Mini-Sentinel Common Data Model (recommended by US Food and Drug Administration, www.mini-sentinel.org), i2b2 Star Schema,63 and OMOP CDM.33,35,36,64 Additionally, the Institute of Medicine has recently initiated an effort to standardize structured capture of social and behavioral domains in the EHR.65
EHR implementations and systems are heterogeneous, and CDMs must have the flexibility to adapt to a variety of institutional IDRs. One challenge in standardization is labeling and referencing of specific document types, and many EHR sites may have specific but nonstandard documents that address a particular question.66 Custom approaches can generically circumnavigate this limitation. For example, the colon polyp phenotype in the eMERGE network67 used colonoscopy surgical and pathology reports, which are not yet labeled in a standard manner or mapped to CDMs in most of the IDR systems in the network. This algorithm separates the implementation into transportable tasks (e.g., concept extraction through NLP, grouping, extraction of covariates) implemented as a fully executable Konstanz Information Miner (KNIME) package with institutional adaptation tasks (i.e., database querying for the proper document types). Creating a portable infrastructure that implements the algorithmic rules and thus only requires the user to build the “last mile” of the solution can accelerate algorithm implementation across other sites.
Recommendations for phenotype representation models
3. Support both human-readable and computable representations
The investigators and initiators for most phenotype projects are clinical experts, epidemiologists, geneticists, and other subject matter experts. As important communication tools among researchers of different expertise, the phenotype representations should support a human-readable format or transformation for clinical experts to ensure medical accuracy. Additionally, phenotype algorithms should include clear scientific and clinical definitions to enable creation of gold standards for evaluation and to facilitate reuse. For example, one algorithm may allow any cause of hypothyroidism when evaluating treatment efficacy while another may focus on only on primary autoimmune hypothyroidism when evaluating genetic causes. It is strongly preferable that the human-readable format and computable formats be computationally coupled such that one can be automatically generated from the other; otherwise it would risk inconsistency between them. For example, the QDM provides a transformation from machine-readable XML to human-readable HTML via automated Extensible Stylesheet Language Transformations.
4. Implement set operations and relational algebra
Phenotyping is a population level process, which includes intersection (e.g., patients billed with ICD-9 codes for T2DM and patients treated with oral hypoglycemic medications), union (e.g., patients treated with angiotensin converting enzyme inhibitors or patients treated with angiotensin receptor blockers), or exclusion (e.g., patients who have diabetes but have never had retinopathy diagnosed). Relational algebra in database theory is a typical set model. The capability to handle set operations and seamless connections to rule-based models (see Desideratum 5) will directly affect the usability of phenotype algorithms.
Virtually all phenotype algorithms explicitly exclude certain other conditions, exposures, or laboratory results operating on either the patient-level or on particular episode(s) of care. Such exclusions are commonly present in control algorithms but also present in many case algorithms. For example, the methotrexate toxicity algorithm68 excludes patients with known organic liver disease, and for cases, excludes episodes of liver function test elevation while the patient is taking leflunomide (another common rheumatoid arthritis medication with liver toxicity as a side effect).
5. Represent phenotype criteria with structured rules
PheRM should support structured and rule-based logical representations, which has been successfully adopted in QDM, OMOP Health Outcomes of Interest (HOI http://omop.org/HOI),34,37,69 and JBOSS® Drools based phenotyping.45,70
Nested logical structure
Phenotyping algorithms can involve multiple complex logical steps, integrating various operations (e.g., Boolean, comparative, aggregative, temporal). A complex, nested logical structure is supported by QDM.46 On the other hand, while interface tools such as i2b2 may limit the number of nested levels, some allow users to reference prior patient sets to support more complex workflows.
Boolean
Boolean values can be generated with comparative or temporal operations (see below), set projection (i.e., nonempty set as TRUE, empty set as FALSE). Many of the phenotyping rules are Boolean operations.50 Common Boolean operators include AND, OR, and NOT (similar to intersection and union). For example, in the T2DM algorithm19 (Figure 1), every step generates a Boolean value for each patient to follow a decision tree to determine if the patient is a case or control.
Comparative operations
In phenotyping, comparative operators can be used to threshold a variable (e.g., the numeric result of a laboratory test, such as a white blood cell count), or to compare a numeric variable to another numeric variable (e.g., comparing the LDL value after statin treatment to LDL value before treatment). In addition, important raw data are not always ready to be used directly from an EHR. For example, body mass index (BMI) often needs to be calculated from weight and height. Thus, supporting basic arithmetic functions will broaden the application. Rules to exclude nonbiologic values may also be needed, such as a BMI of 1000 kg/m2.
Aggregative operations
Aggregative functions (e.g., COUNT, FIRST) bridge across different levels of clinical information (e.g., from events to patients). In addition, more complex counting and scoring rules should be implemented.46 In fact, these rules are extremely popular in clinical diagnostic criteria (see Supplementary Appendix Part 2), including the Modified Duke Criteria for diagnosis of infective endocarditis,52 the CHADS2 score for antithrombotic therapy in AF,51 or the 2013 guidelines for cholesterol management.71 In addition, most regression-based predictive models in phenotyping can be represented as a scoring system, such as an algorithm to find rheumatoid arthritis.54
Negation
In phenotyping, negation has two meanings. It can be a negative assertion (e.g., “patient denies headache”), which can be extracted with NLP,72–74 or an empty set from aggregation in many computer languages (e.g., Perl, Python), similar to exclusion (see Desideratum 5). These two interpretations can be conflicting, and need to be distinguished. Care must be taken to not imply negation from missing values that are not available due to the variability of the EHR systems.
6. Support defining temporal relations between events
Temporal relationships are widely used in phenotype algorithms,75 especially for studying response and side effects of medications.11,14 The first type is sequential clinical events, such as an algorithm to identify patients that have subsequent cardiovascular events while still on clopidogrel,11 which requires ordered and appropriately spaced sequences of ischemic and medication events computed from the timestamps of records. On the other hand, temporality can also be captured through narrative text, requiring advanced NLP to parse grammatical features (past tense of verbs) and relative temporal expressions (“five years ago,” “1980s,” or location within a “past medical history” section). This strategy has been tested in the 2012 i2b2 challenge,75,76 and applied in a prior analysis of colorectal cancer screening77 and in an identification of methotrexate-induced liver toxicity.68 Frequently in an EHR, the true incident date for a disease is not defined even when using NLP, since it may precede the patient’s enrollment in the given clinic or hospital system.
7. Use standardized terminologies, ontologies, and facilitate reuse of value sets
To allow phenotype algorithms in PheRM to be supported in different EHR systems, accommodating non-standardized terminologies is important. Many EHR systems employ local ad hoc terminologies, but the use of local terminology should be limited in PheRM, because it will hinder the portability of algorithms. Both HL7 and OMOP CDM recommend standardized coding systems for clinical terminology,78 such as ICD-9/10 for billed diagnoses, RxNorm for medication, Logical Observation Identifiers Names and Codes (LOINC) for laboratory tests, and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) for describing medical conditions. Therefore, EHR databases should provide mapping between standardized terminology systems and their local systems.61
Phenotype algorithms and quality measures often enumerate lists of concepts to define a medical condition, and these lists have been conventionally called value sets, such as all the ICD-9 codes to define T2DM. Authoring these value sets requires expertise and manual curation, and such sets should be available for reuse by other investigators. To facilitate authoring, i2b2 uses the intrinsic hierarchical structure of medical ontology79 to allow a user to select all concepts under the same semantic nodes. Local ontologies are supported in i2b2 for the convenience that it offers for their research domain. Broad pathophysiological groupings of ICD-9 codes have been developed for genetic and clinical research, including codes designed to enable phenome-wide association studies,80–82 and groupings designed for the Agency for Healthcare Research and Quality Clinical Classifications Software.83
The same value set sometimes can be reused in a variety of projects. For instance, the value set of all the angiotensin-converting-enzyme inhibitors can be used in research projects on diabetic nephropathy, congestive heart failure, or adverse drug reaction. Such information can be stored and managed in the Value Set Authority Center (provided by the National Library of Medicine, https://vsac.nlm.nih.gov/), and the Common Terminology Services 2 (an Object Management Group standard, http://www.omg.org/spec/CTS2/).
8. Define representations for text searching and NLP
Documentation of a detailed description of a patient’s clinical presentation and management in free text is indispensable in clinical care and in validating that a patient has a given disease. Clinical documents are commonly used for phenotype research.6 Text searching and NLP are major strategies to validate coded data or define more granular phenotypes than what is possible via structured data, such as subtypes of multiple sclerosis,84 physical exam findings,85,86 or the collection of all blood pressure measures.87 NLP-derived features have been widely applied for machine learning-based phenotype algorithms.88
PheRM should include NLP and text searching. Patterns of NLP recurring in phenotype algorithms have included: identifying targeted document types (e.g., colonoscopy reports), section location,89 concept identification,90–92 and negation and context filtering.72–74 Here, we propose the PheRM should allow for specification of inclusion or exclusions of elements based on: document type, section location, concept instances (with removal of non-patient and negated concepts), and keywords.
In addition to NLP, keyword and regular-expression text searches have been applied widely in phenotype algorithms. For example, an AF algorithm includes a keyword search from electrocardiogram reports for different variances in phrasing AF, such as “A-fib”, “Atr. Fibrillation.”6,10 With assistance from section separators and negation masks, text searching can achieve a higher accuracy and faster execution (than comprehensive de novo NLP) for many phenotypes.
9. Provide interfaces for external software algorithms
Development of phenotype algorithms is a rapidly evolving field, as are complementary computational algorithms and tools, such as NLP and statistical models. For example, the severe childhood obesity algorithm2 requires age appropriate percentiles for BMI, which may require an external calculator and/or additional percentile data. These dynamic tasks are difficult to represent or program with static languages (such as XML). Likely the optimal method to “interface” with external software packages would be to allow inclusion of new specifications of data elements that could be calculated external to the phenotype algorithm. As a related endeavor, the eMERGE colon polyps algorithm67 was delivered as a standard executable KNIME workflow, with a simple Java Snippet unit connecting to a customized NLP package to parse the colonoscopy reports. The T2DM algorithm has a KNIME workflow implementation available on PheKB.
10. Maintain backward compatibility
A PheRM must be developed according to current existing EHR data, but robust enough to evolve to make use of new clinical data and standards. In addition, unlike a quality measure, which only focuses on records of a limited and recent period, phenotype algorithms frequently use information dated back to as early as the first day of utilization of the EHR to obtain enough data for statistical significance. The information usually comes from records across multiple distinct historical eras of EHR development, and from multiple generations of EHR client software and templates. An obvious example is the need to support both ICD9 and ICD10, as well as different historical versions of ICD9 (e.g., allergic bronchopulmonary aspergillosis was billed as “518.89,” but has been billed as “518.6” since 199793). Since phenotype algorithms often examine historical data, such capabilities are still required even after the United States formally adopts ICD10.
Acknowledging that robust data normalization across EHRs (especially for historical data) is also a difficult and yet unachieved task, we recommend prioritizing the development of functionality and support of data elements for PheRM. For example, data elements that have been widely used in previous phenotype algorithms should be standardized first: billing codes, RxNorm codes for medications, Logical Observation Identifiers Names and Codes for laboratory tests, and diagnoses on problem lists. Progressive normalization of EHR data with CDMs may simplify backward compatibility.
An example: the desiderata applied to T2DM phenotype algorithm
The T2DM algorithm19 first ascertains T2DM diagnosis with grouped T2DM ICD-9 codes, use of oral hypoglycemic medications represented in grouped RxNorm codes (as Desideratum 7), or multiple mentions of T2DM in clinical narratives (Desiderata 5 [a counting rule] and 8); then it differentiates T2DM from type 1 diabetes mellitus (T1DM) patients by excluding patients with T1DM ICD-9 codes (as Desideratum 4 [exclusion]), enforcing absence of insulin use or oral medications should preceded insulin use (as Desiderata 5 [aggregation function of first appearance] and 6); for some cases, it confirms diabetes diagnoses with laboratory values. Its implementation and inter-institutional operation requires supports of other listed desiderata (with details in Supplementary Appendix A Part 1).
DISCUSSION
To develop these desiderata for a standardized PheRM, we have investigated phenotyping modalities adopted in algorithms from eMERGE, PGRN, SHARPn, and PGPop networks (Table 2), and evaluated popular clinical diagnostic and decision-making algorithms. We have also investigated currently available phenotyping tools, and find that these tools are evolving along with our proposed desiderata and are able to perform increasingly complex phenotype queries. As tests for the feasibility and sufficiency of these modalities, algorithms, and tools, the ongoing Phenotype Execution Modeling Architecture (PhEMA) (http://projectphema.org) collaboration has been actively implementing these desiderata and delivering phenotype workflows (Supplementary Appendix Part 3).
Since phenotyping is a knowledge-intensive process based on a global evaluation of each patient,105 missing only a few features in a phenotyping platform or standard language will result in difficulty representing elementary algorithms. It is challenging to list all the technical requirements and details in one paper. Thus, ongoing collaboration between developers of phenotype languages and tools, and user communities (including both geneticists and clinicians) will be imperative.
The desiderata (D1–10) we proposed cover multiple domains:
Partnership with evolutions of EHR repositories (D1, D2);
A balance between human-readable and computational representations (D3);
Common computational elements in phenotype algorithms (D4–D8);
Extensibility with external tools and modules (D9);
Flexibility in accommodating to different institutions and states of the art (D2, D10).
While there are similarities between phenotype algorithms and healthcare-focused algorithms like quality measures,106 eligibility criteria for clinical trials, and clinical decision support rules, the implementation for each has differences. For example, quality measures often are more focused on sensitivity while phenotype algorithms for research studies, including EHR-based genomics studies, are typically more focused on positive predictive value.21 In addition, many phenotype algorithms use NLP49,50 and corroboration with different data elements, whereas quality measures and clinical decision support utilize predominantly structured data. For the purpose of this paper, “phenotype algorithm” typically refers to the application of decision logic applied for EHR-based biomedical research purposes. Nevertheless, we anticipate most desiderata for phenotyping algorithms may apply to other healthcare applications. For example, we have successfully translated the “last mile” solution in phenotyping (described in Desideratum 2) to electronic clinical quality measures.107 However a formal evaluation across all categories of algorithms is outside the scope of this paper.
Strategies of phenotyping are evolving with new informatics and data representation methods, new EHR data elements, and new medical knowledge. A PheRM will also need to be able to evolve continually. A persistent trend, however, has been the need to access detailed information and the context of information from a variety of sources. For example, “glaucoma” diagnosed or mentioned by an ophthalmologist (a matched specialist) provides much higher confidence than when mentioned as self-report or by non-ophthalmologist clinicians.
In addition, a diagnosis is typically developed and confirmed by a clinician over time. Thus, a standalone assertion in the medical records can be misleading. Computational reconstruction of the clinical timelines and connecting diverse clinical elements using medical knowledge may provide a more accurate capture of phenotypes. For example, elevated liver function tests in a patient with rheumatoid arthritis can be a side effect of a medication but may also result from a primary viral infection, heart failure, sepsis, or other causes. Designing computational medical knowledge maps to interrelate different information sources may improve phenotyping. Examples include historical expert systems such as INTERNIST-1108 and DXplain.109 More recent, data-driven approaches include resources such as Side Effect Resource -2110 and MEDication-Indication.111
Limitations caution interpretation of this work. First, these desiderata are based on the experiences of the authors and the algorithms and systems explored to date. A robust community-based phenotyping ecosystem will sustain the continuous evolution of these desiderata with ever expanding knowledge and experience. Second, these desiderata mainly focus on knowledge-driven phenotype approaches, and have not yet addressed data-driven approaches, such as unsupervised or deep learning.112 Third, these desiderata are written during a period of rapid EHR evolution and adoption due to MU incentives. Availability of CDMs, standards, and data types available will evolve similarly as the field continues maturing. Fourth, it is unclear the degree to which these desiderata will apply to international experiences with computable phenotypes. Finally, these desiderata may not be addressable by any one single system, but represent an overarching series of goals for such work.
Our mission is to create research quality information from data gathered in a non-research enterprise. Clinically-derived data comes with the advantages of larger scale, reduced cost, repeated observations, and the ability to observe rare events. It is important to understand that this is not just about technology; efforts by clinicians to record quality data and robustly use EHRs enable greater secondary use potential. We are optimistic that this new endeavor will lead us to align and expand the “research quality prospective data” from direct clinical trials.
FUNDING
This work was funded primarily by R01 GM105688 from the National Institute of General Medical Sciences. Additional contribution came from the eMERGE Network sites funded by the National Human Genome Research Institute through the following grants: U01 HG006828 (Cincinnati Children’s Hospital Medical Center); U01-HG004610 and U01-HG006375 (Group Health Cooperative/University of Washington); U01-HG004608 (Marshfield Clinic); U01-HG04599 and U01-HG06379 (Mayo Clinic); U01-HG004609 and U01-HG006388 (Northwestern University); U01-HG006389 (Essentia Institute of Rural Health); U01-HG04603 and U01-HG006378 (Vanderbilt University); and U01-HG006385 (Vanderbilt University serving as the Coordinating Center). Additional support came from R01-LM010685 and R01 GM103859.
COMPETING INTEREST
None.
CONTRIBUTORS
J.C.D., J.P., and W.K.T. provided leadership for the project; H.M., J.C.D, J.P., W.K.T., J.A.P., L.V.R., C.G.C., G.T. drafted desiderata elements; H.M and J.C.D. drafted the manuscript; W.K.T., J.A.P., L.V.R., R.K., and H.M. led executability and adaptability studies; G.J and R.K. lead standardization and library development; J.X. and E.M. led environmental scan and usability studies; Q.Z., J.A.P., and H.M. led algorithm modeling studies; L.V.R. and P.S. led authoring environment and modularization studies; F.W., A.N.K., and G.J. led clinical data modeling; W.K.T. and C.A.B. led NLP studies; all authors contributed expertise and edits.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Mosley JD, Van Driest SL, Larkin EK, et al. Mechanistic phenotypes: an aggregative phenotyping strategy to identify disease mechanisms using GWAS data. PLoS ONE. 2013;8:e81503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Namjou B, Keddache M, Marsolo K, et al. EMR-linked GWAS study: investigation of variation landscape of loci for body mass index in children. Front Genet. 2013;4:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pathak J, Kiefer RC, Bielinski SJ, et al. Mining the human phenome using semantic web technologies: a case study for Type 2 Diabetes. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2012;2012:699–708. [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Ruau D, Chen R, et al. Systematic identification of risk factors for Alzheimer’s disease through shared genetic architecture and electronic medical records. Pac Symp Biocomput Pac Symp Biocomput. 2013;2013:224–235. [PubMed] [Google Scholar]
- 5.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. JAMIA. 2010;17:568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. JAMIA. 2013;20:e206–e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. JAMIA. 2014;21:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie MD, Denny JC, Zuvich RL, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaney JT, Ramirez AH, Bowton E, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther. 2012;91:257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam JR, Schneider JL, Zhao W, et al. PRoton pump inhibitor and histamine 2 receptor antagonist use and vitamin b12 deficiency. JAMA. 2013;310:2435–2442. [DOI] [PubMed] [Google Scholar]
- 13.Wei W-Q, Feng Q, Jiang L, et al. Characterization of statin dose response in electronic medical records. Clin Pharmacol Ther. 2014;95:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overby CL, Pathak J, Gottesman O, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. JAMIA. 2013;20:e243–e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Salmasian H, Vilar S, et al. A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. JAMIA. 2014;21:308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel VN, Kaelber DC. Using aggregated, de-identified electronic health record data for multivariate pharmacosurveillance: a case study of azathioprine. J Biomed Inform. 2013;52:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein RH, St Jacques P, Stockin M, et al. Automated identification of drug and food allergies entered using non-standard terminology. JAMIA. 2013;20:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6:234cm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. JAMIA. 2012;19:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. JAMIA. 2013;20:e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giacomini KM, Brett CM, Altman RB, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chute CG, Pathak J, Savova GK, et al. The SHARPn project on secondary use of Electronic Medical Record data: progress, plans, and possibilities. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2011;2011:248–256. [PMC free article] [PubMed] [Google Scholar]
- 25.Collins FS, Hudson KL, Briggs JP, et al. PCORnet: turning a dream into reality. JAMIA. 2014;21:576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blumenthal D, Tavenner M. The “Meaningful Use” regulation for electronic health records. N Engl J Med. 2010;363:501–504. [DOI] [PubMed] [Google Scholar]
- 27.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarty CA, Chapman-Stone D, Derfus T, et al. Community consultation and communication for a population-based DNA biobank: the Marshfield clinic personalized medicine research project. Am J Med Genet A. 2008;146A:3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott CT, Caulfield T, Borgelt E, et al. Personal medicine—the new banking crisis. Nat Biotechnol. 2012;30:141–147. [DOI] [PubMed] [Google Scholar]
- 30.Bielinski SJ, Chai HS, Pathak J, et al. Mayo Genome Consortia: a genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clin Proc. 2011;86:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–428. [DOI] [PubMed] [Google Scholar]
- 32.Denny JC. Chapter 13: mining Electronic Health Records in the Genomics Era. PLoS Comput Biol. 2012;8:e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisinger SJ, Ryan PB, O’Hara DJ, et al. Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases. JAMIA. 2010;17:652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stang PE, Ryan PB, Dusetzina SB, et al. Health outcomes of interest in observational data: issues in identifying definitions in the literature. Health Outcomes Res Med. 2012;3:e37–e44. [Google Scholar]
- 35.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. JAMIA. 2012;19:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reich C, Ryan PB, Stang PE, et al. Evaluation of alternative standardized terminologies for medical conditions within a network of observational healthcare databases. J Biomed Inform. 2012;45:689–696. [DOI] [PubMed] [Google Scholar]
- 37.Reich CG, Ryan PB, Schuemie MJ. Alternative outcome definitions and their effect on the performance of methods for observational outcome studies. Drug Saf. 2013;36 (Suppl 1):S181–S193. [DOI] [PubMed] [Google Scholar]
- 38.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): A Prototype Federated Query Tool for Clinical Data Repositories. JAMIA. 2009;16:624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurry AJ, Murphy SN, MacFadden D, et al. SHRINE: Enabling Nationally Scalable Multi-Site Disease Studies. PLoS ONE. 2013;8(3):e55811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Carroll RJ, Hinz ERM, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. JAMIA. 2013;20:e253–e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll RJ, Eyler AE, Denny JC. Naïve Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 42.Peissig PL, Santos Costa V, Caldwell MD, et al. Relational machine learning for electronic health record-driven phenotyping. J Biomed Inform. 2014;52:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pathak J, Bailey KR, Beebe CE, et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. JAMIA. 2013;20:e341–e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amster A, Jentzsch J, Pasupuleti H, et al. Completeness, accuracy, and computability of National Quality Forum-specified eMeasures. J Am Med Inform Assoc. 2015;22(2):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li D, Endle CM, Murthy S, et al. Modeling and executing electronic health records driven phenotyping algorithms using the NQF Quality Data Model and JBoss® Drools Engine. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2012;2012:532–541. [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2012;2012:911–920. [PMC free article] [PubMed] [Google Scholar]
- 47.Payne PRO, Johnson SB, Starren JB, et al. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Investig Med Off Publ Am Fed Clin Res. 2005;53:192–200. [DOI] [PubMed] [Google Scholar]
- 48.Post AR, Krc T, Rathod H, et al. Semantic ETL into i2b2 with Eureka! AMIA Summits Transl Sci Proc. 2013;2013:203–207. [PMC free article] [PubMed] [Google Scholar]
- 49.Rasmussen LV, Thompson WK, Pacheco JA, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J Biomed Inform. 2014;51:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2011;2011:274–283. [PMC free article] [PubMed] [Google Scholar]
- 51.Fuster V, Rydén LE, Cannom DS, et al. ACC/AHA/ESC 2006 Guidelines for the Management of Patients With Atrial Fibrillation—Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation) Developed in Collaboration With the European Heart Rhythm Association and the Heart Rhythm Society. J Am Coll Cardiol. 2006;48:854–906. [DOI] [PubMed] [Google Scholar]
- 52.Durack DT, Lukes AS, Bright DK. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am J Med. 1994;96:200–209. [DOI] [PubMed] [Google Scholar]
- 53.Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. [DOI] [PubMed] [Google Scholar]
- 54.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. JAMIA. 2012;19:e162–e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawaler E, Cobian A, Peissig P, et al. Learning to Predict Post-Hospitalization VTE Risk from EHR Data. AMIA Annu Symp Proc. 2012;2012:436–445. [PMC free article] [PubMed] [Google Scholar]
- 56.Mani S, Chen Y, Elasy T, et al. Type 2 diabetes risk forecasting from EMR data using machine learning. AMIA Annu Symp Proc. 2012;2012:606–615. [PMC free article] [PubMed] [Google Scholar]
- 57.Fine AM, Reis BY, Nigrovic LE, et al. Use of population health data to refine diagnostic decision-making for pertussis. JAMIA. 2010;17:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarty CA, Peissig P, Caldwell MD, et al. The Marshfield Clinic Personalized Medicine Research Project: 2008 scientific update and lessons learned in the first 6 years. Pers Med. 2008;5:529–542. [DOI] [PubMed] [Google Scholar]
- 59.Date CJ. An Introduction to Database Systems. Boston: Pearson/Addison Wesley; 2004.. [Google Scholar]
- 60.Jon Duke, Charity A., Hilton, Chris Beesley, et al. Linking Structured and Unstructured Clinical Phenotypes through the OMOP Common Data Model. 2015. http://www.slideshare.net/jduke1/amia-cri-2015-structured-vs-unstructured-duke Accessed 31 March 2015. [Google Scholar]
- 61.Huser V, Cimino JJ. Desiderata for healthcare integrated data repositories based on architectural comparison of three public repositories. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2013;2013:648–656. [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang G, Evans J, Oniki TA, et al. Harmonization of detailed clinical models with clinical study data standards. Methods Inf Med. 2015;54(1):65–74. [DOI] [PubMed] [Google Scholar]
- 63.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. JAMIA. 2012;19:181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matcho A, Ryan P, Fife D, et al. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug Saf. 2014;37:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Committee on the Recommended Social and Behavioral Domains and Measures for Electronic Health Records, Board on Population Health and Public Health Practice, Institute of Medicine. “Front Matter.” Capturing Social and Behavioral Domains and Measures in Electronic Health Records: Phase 2. Washington, DC: The National Academies Press, 2014. [PubMed] [Google Scholar]
- 66.Hyun S, Shapiro JS, Melton G, et al. Iterative evaluation of the Health Level 7–Logical Observation Identifiers Names and Codes Clinical Document Ontology for representing clinical document names: a case report. JAMIA. 2009;16:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gawron AJ, Thompson WK, Keswani RN, et al. Anatomic and advanced adenoma detection rates as quality metrics determined via natural language processing. Am J Gastroenterol. 2014;109:1844–1849. [DOI] [PubMed] [Google Scholar]
- 68.Lin C, Karlson EW, Dligach D, et al. Automatic identification of methotrexate-induced liver toxicity in patients with rheumatoid arthritis from the electronic medical record. JAMIA. Published Online First: 25 October 2014, doi:10.1136/amiajnl-2014-002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyce RD, Ryan PB, Norén GN, et al. Bridging islands of information to establish an integrated knowledge base of drugs and health outcomes of interest. Drug Saf. 2014;37:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peterson K, Pathak J. Scalable and high-throughput execution of clinical quality measures from electronic health records using MapReduce and the JBoss(R) Drools Engine. In: AMIA Annu Symp Proc. 2014:1864–1873. [PMC free article] [PubMed] [Google Scholar]
- 71.Stone NJ, Robinson J, Lichtenstein AH, et al. 2013 ACC/AHA Guideline on the Treatment of Blood Cholesterol to Reduce Atherosclerotic Cardiovascular Risk in Adults A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129(25 Suppl 2):S1–S45. [DOI] [PubMed] [Google Scholar]
- 72.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. [DOI] [PubMed] [Google Scholar]
- 73.South BR, Phansalkar S, Swaminathan AD, et al. Adaptation of the NegEx algorithm to Veterans Affairs electronic text notes for detection of influenza-like illness (ILI). AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2007;2007:1118. [PubMed] [Google Scholar]
- 74.Chapman BE, Lee S, Kang HP, et al. Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform. 2011;44:728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. JAMIA. 2013;20:806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikfarjam A, Emadzadeh E, Gonzalez G. Towards generating a patient’s timeline: extracting temporal relationships from clinical notes. J Biomed Inform. 2013;46 (Suppl):S40–S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denny JC, Peterson JF, Choma NN, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. JAMIA. 2010;17:383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helleman J, Goossen WTF. Modeling nursing care in health level 7 reference information model. Comput Inform Nurs. 2003;21:37–45. [DOI] [PubMed] [Google Scholar]
- 79.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 80.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinforma Oxf Engl. 2014;30:2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinforma Oxf Engl. 2010;26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cowen ME, Dusseau DJ, Toth BG, et al. Casemix adjustment of managed care claims data using the clinical classification for health policy research method. Med Care. 1998;36:1108–1113. [DOI] [PubMed] [Google Scholar]
- 84.Davis MF, Sriram S, Bush WS, et al. Automated extraction of clinical traits of multiple sclerosis in electronic medical records. JAMIA. 2013;20:e334–e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Denny JC, Arndt FV, Dupont WD, et al. Increased hospital mortality in patients with bedside hippus. Am J Med. 2008;121:239–245. [DOI] [PubMed] [Google Scholar]
- 86.Meystre SM, Deshmukh VG, Mitchell J. A clinical use case to evaluate the i2b2 Hive: predicting asthma exacerbations. AMIA Annu Symp Proc. 2009;2009:442–446. [PMC free article] [PubMed] [Google Scholar]
- 87.Turchin A, Kolatkar NS, Grant RW, et al. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. JAMIA. 2006;13:691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bejan CA, Xia F, Vanderwende L, et al. Pneumonia identification using statistical feature selection. JAMIA. 2012;19:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Denny JC, Spickard A, 3rd, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. JAMIA. 2009;16:806–815. 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Denny JC, Smithers JD, Armstrong B, et al. “Where do we teach what?” Finding broad concepts in the medical school curriculum. J Gen Intern Med. 2005;20:943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. JAMIA. 2010;17:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Annu Symp AMIA Symp. 2001;2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 93.The National Center for Health Statistics (NCHS) and the Centers for Medicare & Medicaid Services (CMS). Conversion Table of New ICD-9-CM Codes, October 2013. 2013. http://www.cdc.gov/nchs/data/icd/ICD-9-CM_FY14_CNVTBL_Final.pdf. Accessed July 30, 2015.
- 94.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramirez AH, Schildcrout JS, Blakemore DL, et al. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm Off J Heart Rhythm Soc. 2011;8:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peissig PL, Rasmussen LV, Berg RL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. JAMIA. 2012;19:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waudby CJ, Berg RL, Linneman JG, et al. Cataract research using electronic health records. BMC Ophthalmol. 2011;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Overby CL, Weng C, Haerian K, et al. Evaluation considerations for EHR-based phenotyping algorithms: A case study for drug-induced liver injury. AMIA Summits Transl Sci Proc. 2013;2013:130–134. [PMC free article] [PubMed] [Google Scholar]
- 99.Feng Q, Jiang L, Berg RL, et al. A common CNR1 (cannabinoid receptor 1) haplotype attenuates the decrease in HDL cholesterol that typically accompanies weight gain. PloS One. 2010;5:e15779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turner SD, Berg RL, Linneman JG, et al. Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PLoS ONE. 2011;6:e19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kullo IJ, Ding K, Jouni H, et al. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE. 2010;5:e13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei W-Q, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. JAMIA. 2012;19:219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ramirez AH, Shi Y, Schildcrout JS, et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Crosslin DR, McDavid A, Weston N, et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum Genet. 2012;131:639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. JAMIA. 2013;20:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kizer KW. Establishing health care performance standards in an era of consumerism. JAMA. 2001;286:1213–1217. [DOI] [PubMed] [Google Scholar]
- 107.Mo H, Pacheco J, Rasmussen L, et al. A prototype for executable and portable electronic clinical quality measures using the KNIME analytics platform. AMIA Jt Summits Transl Sci Proc. 2015 Mar 25;2015:127–31. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26306254. [PMC free article] [PubMed] [Google Scholar]
- 108.Miller RA, Pople HE, Myers JD. Internist-1, an experimental computer-based diagnostic consultant for general internal medicine. N Engl J Med. 1982;307:468–476. [DOI] [PubMed] [Google Scholar]
- 109.Barnett GO, Cimino JJ, Hupp JA, et al. DXplain. An evolving diagnostic decision-support system. JAMA. 1987;258:67–74. [DOI] [PubMed] [Google Scholar]
- 110.Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei W-Q, Cronin RM, Xu H, et al. Development of an ensemble resource linking MEDications to their Indications (MEDI). AMIA Summits Transl Sci Proc. 2013;2013:172. [PubMed] [Google Scholar]
- 112.Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PloS One. 2013;8:e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]