Abstract
Arsenic (As) contamination of water is a global concern and rice consumption is the biggest dietary exposure to human posing carcinogenic risks, predominantly in Asia. Sulfur (S) is involved in di-sulfide linkage in many proteins and plays crucial role in As detoxification. Present study explores role of variable S supply on rice leaf proteome, its inclination towards amino acids (AA) profile and non protein thiols under arsenite exposure. Analysis of 282 detected proteins on 2-DE gel revealed 113 differentially expressed proteins, out of which 80 were identified by MALDI-TOF-TOF. The identified proteins were mostly involved in glycolysis, TCA cycle, AA biosynthesis, photosynthesis, protein metabolism, stress and energy metabolism. Among these, glycolytic enzymes play a major role in AA biosynthesis that leads to change in AAs profiling. Proteins of glycolytic pathway, photosynthesis and energy metabolism were also validated by western blot analysis. Conclusively S supplementation reduced the As accumulation in shoot positively skewed thiol metabolism and glycolysis towards AA accumulation under AsIII stress.
Arsenic (As) exposure is a serious threat to human beings and its key source of exposure is the As tainted water. Unfortunately, in many parts of the world rice is grown in highly As contaminated areas which leads to contamination of food chain1. Rice is the staple crop of 3 billion people worldwide and Asian countries account for 90% of its production as well as consumption2. Unlike other cereal crops, rice is particularly efficient AsIII accumulator that leads to impaired cellular functions through strapping of sulfhydryl groups of enzymes and proteins3. In addition, inhibition of SH containing enzymes by As, alters cellular redox state and finally leads to cytotoxicity4.
Sulfur, an essential element, plays vital role in the plant growth and defense response by regulating the expression of genes involved in the uptake and assimilation of sulfate5,6. Plant deficient in sulfur status have lower non protein thiol content, metal(loid) complexing ligands such as PCs7, thus they have higher As translocation from root to shoot8. In plants, inorganic sulfate is reduced to sulfide and further assimilated to form Cys9. Cysteine serves as a precursor for a range of S-containing defense compounds, such as methionine, S-adenosylmethionine (SAM), glucosinolates, and GSH10. Sulfur moiety of Cys is responsible for di-sulfide bonding in proteins for their proper function and also important for the formation of the Fe–S cluster in the photosynthetic apparatus and electron transport chain11. A set of Cys containing redox-sensitive proteins e.g., glyceraldehyde-3-P-dehydrogenase (GAPDH), malate dehydrogenase (MDH) and elongation factor-thermo unstable (EF-TU) are well known for their roles in oxidative thiol modifications12.
Rice grain contains 3–7% protein, comprising of different essential and non essential amino acids (EAAs and NEAAs), in which glutamic acid (Glu), proline (Pro) and lysine (Lys) are the major components. Various studies13,14 indicate variable responses of AAs metabolism including EAAs and NEAAs during heavy metal stresses. Dwivedi et al. (2010a)15 demonstrated a positive correlation between total AAs content and As accumulation. Further, the stress responsive AAs like Pro, glycine (Gly), Cys, Glu and methionine (Met) showed higher accumulation in high As accumulating rice genotype in comparison to low As accumulating rice genotype16.
Genome-wide gene expression profiling experiments have revealed that a wide array of genes (stress-responsive gene, heat-shock proteins, metallothioniens, sulfate-metabolizing proteins and assimilation pathway) are differentially expressed during As stress in rice plant17. Over the years, the proteomic approach has been used extensively to analyze the proteins involved in various stress responses in plants18,19. Arsenate (AsV) induced NADP-dependent malic enzyme (NADP-ME), aspartate aminotransferase, NAD-dependent formate dehydrogenase (FDH), GAPDH and ATP-dependent protease, proteolytic subunit ClpP-like protein have been reported in rice leaves19. In another study, AsV stress altered the chloroplast 23 kDa polypeptide of photosystem II, Chloroplastic aldolase and Photosystem II stability/assembly factor HCF136 that leads to reduced photosynthetic rate and distorted chloroplast structure in rice leaves20. Studies on nutritional aspects vis-à-vis As have been targeted only to nitrogen21 and phosphorus supplementations22. As AsIII is more toxic than AsV in fields and also limits uptake of macro and micro nutrients23, it is important to study AsIII and S interaction in rice. In order to gain an insight into AsIII induced stress under different S regimes in rice leaves, hydroponic studies were conducted in rice (IR-36) with respect to combined proteome, AA, thiol metabolism and western blot analysis. Besides deprivation, higher S dose was also used during experiments to observe role of S amelioration of As toxicity as S has potential to mitigate As toxicity through chelation. The study emphasizes role of SH under AsIII toxicity as S containing enzymes are prime target during AsIII exposure. It is hypothesized that S supplementation would alleviate the AsIII toxicity in rice plants.
Results
Impact of Sulfur and Arsenite on Rice Leaf Morphology, Photosynthetic Pigments and Arsenic Accumulation
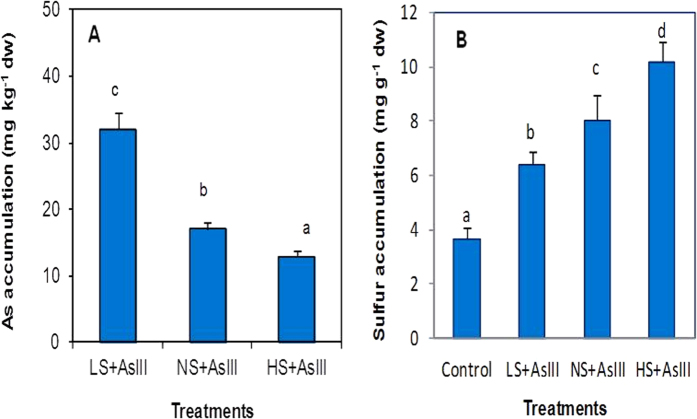
The S conditions were abbreviated as follows: LS for the low sulfur conditions, NS for standard sulfur conditions, and HS for the high sulfur concentration. All the treatments were compared to NS control unless otherwise mentioned in the text. AsIII hampered plant growth through reduction of shoot length and weight. LS + AsIII showed considerable reduction (20%) in shoot length; however, HS + AsIII ameliorated this effect by increasing shoot length by 17%. Similarly LS + AsIII reduced shoot weight by 47% and HS + AsIII ameliorated it by 68% (Supplemental Information 3 Fig. S1 and S2 A, B). Chl a, b and total Chl contents were substantially reduced (31, 43 and 31%) in LS + AsIII while S supplementation elevated Chl levels (19, 44 and 21%) in NS + AsIII and (41, 91 and 32%) in HS + AsIII, respectively. Carotenoid contents were enhanced in LS + AsIII (108%) and decreased in NS + AsIII (19%) and (53%) in HS + AsIII, respectively as compared to LS + AsIII (Supplemental Information 3 Fig. S2 C, D, E, F). Arsenic accumulation was analyzed in twenty days old hydroponically grown rice seedling leaves (Fig. 1A). Results indicated that LS + AsIII increased As accumulation (32.12 mg kg−1 dw) and reduced in HS + AsIII (12.92 mg kg−1 dw) treatments in comparison to NS + AsIII (17.03 mg kg−1 dw). Arsenic exposure enhanced the S accumulation in shoot than the plants not exposed to As. Sulfur accumulation in shoot was allied to S concentration in the nutrient solution (Fig. 1B).
Figure 1. Effect of different sulfur doses on arsenic (A) sulfur (B) accumulation under arsenite stress in leaves of rice (Oryza sativa L.) plant.
All the values are means of triplicate ± S.D. ANOVA significant at p ≤ 0.01. Different letters indicate significantly different values at a particular treatment (DMRT, p ≤ 0.05).
Rice leaf proteome modulation during S and AsIII interaction
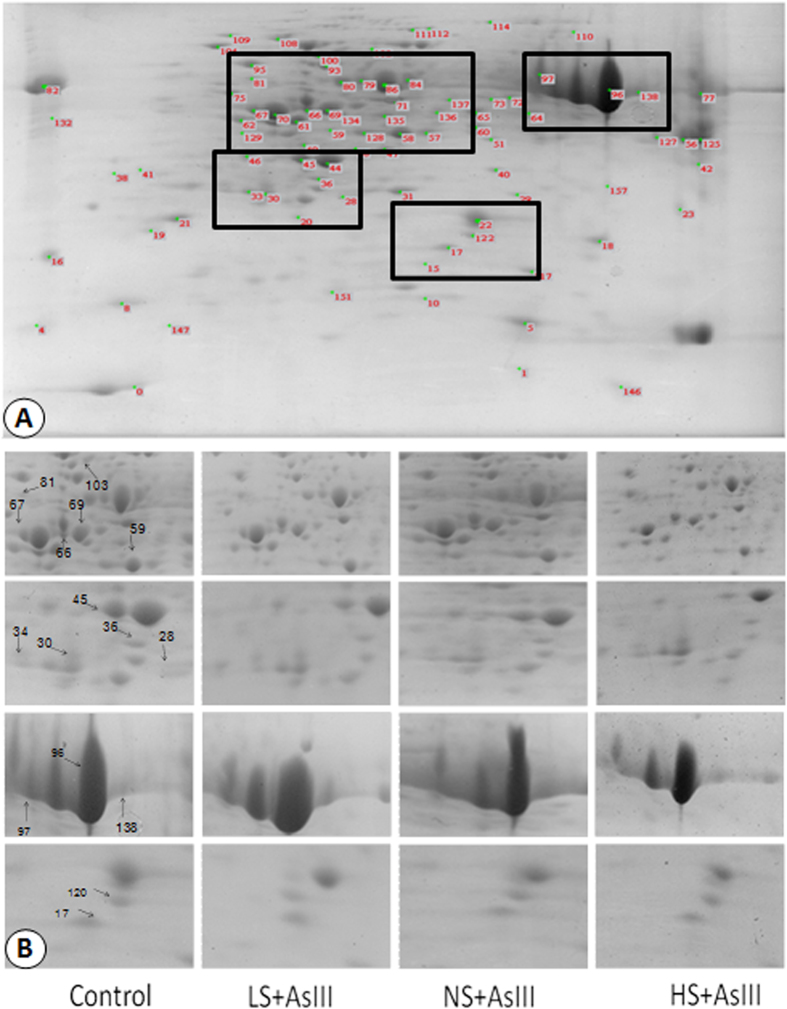
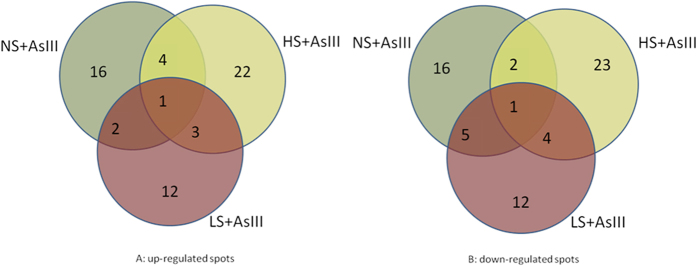
A total 282 spots were detected using CBB staining; reference gel depicting all spots and four typical enlarged regions are shown in Fig. 2A,B. Out of 282 spots, 113 spots were differentially expressed in comparison to control. 60 spots were up regulated while 53 spots were down regulated. All the spots showed at least 1.5 fold changes in abundance. Out of these 113 (60 + 53), 80 spots showed more than 2 fold changes in abundance (Table 1). Venn diagrams of differentially expressed proteins indicated that out of 60 up regulated spots, one common spot was present in all the treatments, 9 spots showed the enhanced abundance in any of two different treatments (Fig. 3A). Out of 53 down regulated spots, one spot was down regulated under all three treatments and 11 spots diminished in abundance in any of two different treatments (Fig. 3B).
Figure 2.
(A) Reference 2-DE image of rice leaves proteome. Proteins were stained with CBB G-250. (B) Close-up of some up and down regulated proteins detected by 2-DE (gel image with identified proteins) and their expression profile patterns.
Table 1. Differentially expressed proteins identified by MALDI-TOF-TOF (negative sign represents down regulation of protein).
| Functional category & Match ID | NCBI accession no. | Protein name | Average fold change |
Score | Mass | % coverage | ||
|---|---|---|---|---|---|---|---|---|
| LS + AsIII | NS + AsIII | HS + AsIII | ||||||
| Amino acid biosynthesis | ||||||||
| 64 | gi|29468084 | aminotransferase, classes I and II, domain containing protein | −2.599 | −2.269 | nd | 51 | 46016 | 13% |
| 66 | gi|19387272 | glutamine synthetase, catalytic domain containing protein | 8.316 | nd | nd | 336 | 49770 | 17% |
| 69 | gi|19387272 | glutamine synthetase, catalytic domain containing protein | 1.015 | 1.105 | −2.004 | 626 | 49770 | 30% |
| 72 | gi|127046 | S-adenosylmethionine synthetase, putative | −1.082 | 1.248 | 2.178 | 46 | 43618 | 3% |
| 77 | gi|29569153 | aminotransferase, classes I and II, domain containing protein | −1.09 | −1.648 | −3.419 | 467 | 53947 | 27% |
| 136 | gi|255571784 | aminotransferase, putative | nd | 1.049 | −2.63 | 57 | 50836 | 5% |
| Carbohydrate metabolism | ||||||||
| 57 | gi|115471157 | NAD dependent epimerase/dehydratase family protein, putative | 1.501 | 1.065 | 1.202 | 267 | 41268 | 14% |
| 73 | gi|110289082 | NAD dependent epimerase/dehydratase family protein, putative | 1.462 | −2.479 | nd | 64 | 41235 | 11% |
| Cell wall polysaccharide metabolism | ||||||||
| 40 | gi|297604125 | glycosyl hydrolase, putative | 1.674 | 1.319 | 1.436 | 277 | 32757 | 28% |
| Cytosketon | ||||||||
| 81 | gi|493725 | tubulin/FtsZ domain containing protein, putative | 1.903 | 1.376 | 2.575 | 199 | 50703 | 18% |
| Energy metabolism | ||||||||
| 23 | gi|3345477 | carbonic anhydrase, chloroplast precursor, putative | −2.651 | 2.695 | −1.202 | 220 | 29498 | 22% |
| 67 | gi|8918361 | AAA-type ATPase family protein, putative | −1.141 | −1.053 | −1.575 | 335 | 48128 | 19% |
| 74 | gi|8918361 | AAA-type ATPase family protein, putative | −0.684 | nd | −2.52341 | 276 | 48128 | 22% |
| 75 | gi|8918361 | AAA-type ATPase family protein, putative | −1.685 | −2.492 | −2.556 | 586 | 48128 | 30% |
| 79 | gi|11466794 | ATP synthase subunit beta, putative | −1.265 | 1.712 | 1.174 | 846 | 54037 | 45% |
| 85 | gi|3676294 | ATP synthase, putative | −1.183 | −4.003 | 2.670 | 708 | 60045 | 24% |
| 87 | gi|11466794 | ATP synthase subunit beta, putative | −1.577 | −1.173 | −1.127 | 1680 | 54037 | 40% |
| 97 | gi|11466784 | ATP synthase subunit alpha, mitochondrial, putative | 1.965 | 1.937 | 2.704 | 1323 | 55687 | 37% |
| 114 | gi|11583 | ATP synthase subunit beta, putative | 1.077 | −3.633 | −1.042 | 285 | 53899 | 18% |
| 127 | gi|285014508 | ATP synthase gamma chain, putative | nd | nd | 2.359 | 144 | 40012 | 12% |
| Glycolysis | ||||||||
| 28 | gi|553107 | triosephosphate isomerase, cytosolic, putative | −1.268 | 1.574 | −2.254 | 413 | 27816 | 26% |
| 32 | gi|553107 | triosephosphate isomerase, cytosolic, putative | −2.040 | nd | −1.197 | 474 | 27816 | 31% |
| 58 | gi|108864048 | fructose-bisphospate aldolase isozyme, putative | 1.132 | 1.043 | −1.657 | 41808 | 527 | 27% |
| 53 | gi|118175929 | fructose-1,6-bisphosphatase, putative | −3.535 | −0.949 | −2.043 | 79 | 42825 | 12% |
| 39 | gi|115450493 | glyceraldehyde-3-phosphate dehydrogenase, putative | 2.612 | 2.568 | nd | 259 | 47537 | 16% |
| 84 | gi|780372 | enolase, putative | 1.15 | 1.594 | 1.318 | 222 | 48299 | 10% |
| 125 | gi|968996 | glyceraldehyde-3-phosphate dehydrogenase, putative | nd | −1.659 | 1.244 | 545 | 36641 | 32% |
| 128 | gi|108864048 | fructose-bisphospate aldolase isozyme, putative | nd | 1.688 | −2.57 | 685 | 41808 | 45% |
| 131 | gi|114386664 | phosphoglycerate kinase protein, putative | nd | 1.568 | 0.934 | 71 | 42224 | 12% |
| Hydrolase | ||||||||
| 36 | gi|115453797 | cbbY, putative | −5.075 | nd | −1.487 | 225 | 34138 | 23% |
| Pentose phosphate pathway | ||||||||
| 56 | gi|125580 | phosphoribulokinase/Uridine kinase family protein | −2.784 | nd | nd | 400 | 45512 | 21% |
| Protein metabolism | ||||||||
| 10 | gi|18103931 | RNA recognition motif containing protein | 1.15 | −1.525 | nd | 207 | 19579 | 30% |
| 38 | gi|125559266 | RNA recognition motif containing protein, putative | 1.128 | −1.597 | nd | 59 | 27820 | 9% |
| 41 | gi|56784713 | nascent polypeptide-associated complex subunit alpha, putative | −1.201 | 1.599 | nd | 47 | 57717 | 3% |
| 48 | gi|115444057 | peptidase, T1 family, putative | 2.012 | nd | nd | 284 | 29897 | 22% |
| 71 | gi|6525065 | chloroplast translational elongation factor Tu [Oryza sativa Japonica Group] | 1.185 | 1.035 | −1.451 | 390 | 50555 | 12% |
| 92 | gi|15231255 | T-complex protein, putative | −3.235 | nd | −1.721 | 347 | 63702 | 14% |
| 90 | gi|115488160 | T-complex protein, putative | 1.129 | 1.514 | 1.115 | 323 | 61150 | 17% |
| 101 | gi|75114857 | OsFtsH2 FtsH protease, homologue of AtFtsH2/8 | 1.2003 | 2.495 | 2.507 | 120 | 72607 | 6% |
| 110 | gi|18423214 | ATP-dependent Clp protease ATP-binding subunit clpA homolog CD4B,chloroplast precursor, putative | 1.442 | nd | 2.636 | 455 | 102241 | 18% |
| 118 | gi|108711192 | eukaryotic translation initiation factor 5A, putative | nd | 0.679 | 1.553 | 188 | 17930 | 25% |
| 154 | gi|108706511 | peptidase, T1 family, putative | nd | 2.2917 | nd | 99 | 32472 | 12% |
| PS calvin cycle | ||||||||
| 4 | gi|671740 | ribulose bisphosphate carboxylase small chain, chloroplast precursor, putative | −1.275 | −1.303 | −14.731 | 168 | 15111 | 35% |
| 8 | gi|56966763 | ribulose bisphosphate carboxylase small chain, chloroplast precursor, putative | −1.164 | 1.199 | −1.521 | 352 | 15091 | 47% |
| 29 | gi|4105561 | ribulose-phosphate 3-epimerase, chloroplast precursor, putative | −1.371 | −1.575 | 2.835 | 416 | 29234 | 34% |
| 42 | gi|2961307 | ribulose bisphosphate carboxylase large chain precursor, putative | −2.962 | −1.52 | nd | 165 | 53618 | 6% |
| 82 | gi|11466795 | ribulose bisphosphate carboxylase large chain precursor, putative | −2.429 | −1.812 | nd | 1020 | 53418 | 27% |
| 96 | gi|11466795 | ribulose bisphosphate carboxylase large chain precursor, putative | 1.395 | 1.107 | 1.564 | 1025 | 53418 | 30% |
| 113 | gi|28190676 | transketolase, chloroplast precursor, putative | 1.064 | nd | 4.902 | 104 | 80549 | 8% |
| 112 | gi|28190676 | transketolase, chloroplast precursor, putative | 1.255 | −2.032 | −2.085 | 388 | 80549 | 23% |
| 132 | gi|328682245 | ribulose bisphosphate carboxylase large chain precursor, putative | nd | −6.512 | 11.681 | 77 | 50092 | 7% |
| 148 | gi|146741370 | dehydrogenase, putative | nd | 2.399 | nd | 169 | 48012 | 6% |
| 138 | gi|11466795 | ribulose bisphosphate carboxylase large chain precursor, putative | nd | 1.717 | 1.657 | 1018 | 53418 | 40% |
| PS light reaction | ||||||||
| 0 | gi|115465862 | proteinplastocyanin, chloroplast precursor, putative | 3.045 | −2.879 | nd | 72 | 15624 | 15% |
| 2 | gi|1835731 | photosystem II 10 kDa polypeptide, chloroplast precursor, putative | −1.663 | nd | nd | 329 | 12885 | 24% |
| 17 | gi|34394725 | photosystem I reaction center subunit IV A, chloroplast precursor, putative | −1.132 | 1.107 | −1.601 | 182 | 15537 | 31% |
| 20 | gi|115467828 | chlorophyll A-B binding protein, putative | 1.76 | 1.226 | 1.242 | 70 | 26397 | 4% |
| 24 | gi|115470529 | PsbP, putative | 2.184 | nd | 4.424 | 694 | 27094 | 42% |
| 30 | gi|62733870 | chlorophyll A-B binding protein, putative | 1.634 | nd | −1.505 | 79 | 24317 | 17% |
| 37 | gi|19184 | chlorophyll A-B binding protein, putative | −3.581 | −1.069 | nd | 58 | 30575 | 6% |
| 34 | gi|108864186 | chlorophyll A-B binding protein, putative | 2.859 | 2.712 | 1.374 | 240 | 24038 | 27% |
| 45 | gi|739292 | oxygen-evolving enhancer protein 1, chloroplast precursor, putative | −1.355 | −1.031 | −1.937 | 92 | 26603 | 19% |
| 49 | gi|27261025 | FAD dependent oxidoreductase domain containing protein | −3.744 | −1.163 | nd | 239 | 37156 | 19% |
| 50 | gi|41052915 | ferredoxin–NADP reductase, chloroplast precursor, putative | 1.031 | 1.943 | 1.956 | 199 | 41095 | 19% |
| 144 | gi|11466848 | photosystem I iron-S center, putative | nd | nd | −1.55 | 252 | 9406 | 54% |
| Nuclic acid metaboism | ||||||||
| 129 | gi|186463816 | phosphoribosylformylglycinamidine synthase, putative | nd | nd | 3.345 | 53 | 11324 | 27% |
| 134 | gi|78708842 | IAP100, putative | nd | nd | 1.621 | 125 | 47305 | 14% |
| 147 | gi|115466468 | profilin domain containing protein | nd | 2.63 | nd | 77 | 14352 | 24% |
| Stress | ||||||||
| 13 | gi|42408425 | copper/zinc superoxide dismutase, putative | 1.216 | 2.087 | nd | 341 | 20633 | 46% |
| 16 | gi|18698985 | 2Fe-2S iron-S cluster binding domain containing protein | −1.534 | −1.679 | 5.052 | 123 | 15082 | 33% |
| 18 | gi|15667623 | abscisic stress-ripening, putative | −1.717 | 1.429 | nd | 398 | 15923 | 24% |
| 19 | gi|6002472 | 2-Cys peroxiredoxin BAS1, chloroplast precursor, putative | −1.351 | 1.357 | 2.051 | 223 | 29490 | 37% |
| 21 | gi|115446541 | peroxiredoxin, putative | 1.008 | 1.318 | −1.595 | 233 | 28307 | 37% |
| 47 | gi|162461576 | glyoxalase family protein, putative | −1.925 | 1.407 | 1.626 | 80 | 32450 | 10% |
| 102 | gi|115448989 | DnaK family protein, putative | 1.995 | nd | nd | 130 | 73081 | 10% |
| 103 | gi|6746592 | DnaK family protein, putative | −1.601 | nd | 1.637 | 151 | 77230 | 6% |
| 108 | gi|39104468 | heat shock protein, putative | −1.384 | 1.287 | 1.877 | 315 | 80449 | 11% |
| 116 | gi|115477014 | heat shock protein, putative | nd | 1.5903 | 0.088 | 286 | 88749 | 14% |
| 151 | gi|42408425 | copper/zinc superoxide dismutase, putative | nd | −1.52 | nd | 116 | 20633 | 18% |
| Seed storage non enzymatic proteins | ||||||||
| 120 | gi|4239821 | Cupin domain containing protein | nd | −1.532 | −1.075 | 155 | 22017 | 10% |
| TCA Cycle | ||||||||
| 60 | gi|15982948 | lactate/malate dehydrogenase, putative | 1.079 | −1.323 | 1.643 | 285 | 35817 | 27% |
nd: not detected.
Figure 3. Venn diagram analysis of the differentially expressed protein spots in rice (Oryza sativa L.) leaves.
The numbers of differentially expressed protein spots with up- or down-regulation under different concentration of sulfur and equimolar concentration of arsenite are shown in the different segments. (A) The up regulated protein spots. (B) The down regulated protein spots.
Identification and functional categorization of the differentially expressed proteins
A total 113 differentially expressed protein spots were analyzed by MALDI-TOF-TOF. Of these, 80 spots were successfully identified by MS/MS as listed in Table 1. Proteins were identified, and their functions were determined using the classifications by Bevan et al. (1998)24. On the basis of functionality, proteins were classified into 14 major categories (Fig. 4).
Figure 4. Functional classification and distribution of all identified proteins of rice (Oryza sativa L.) leaves proteome as classified by Bevan et al. (1998) on non-redundant basis.
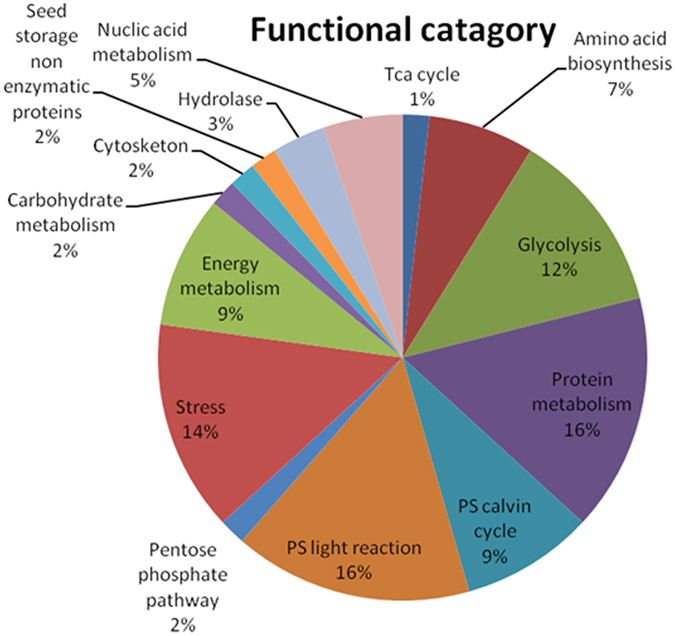
Functional categorization of proteins was done on non-redundant basis. Taken together, the 80 identified proteins represented 57 unique proteins; rest may be their corresponding isoforms. The proteins involved in photosynthesis and protein metabolism were represented more (16% each) of total identified proteins. Notably LS + AsIII had a positive influence on photosynthetic light reaction proteins (Spots no0, 20, 24, 30, 34 and 50). Proteins influencing AA metabolism included glycolysis (12%), AA biosynthesis (7%) and TCA cycle (1%) proteins, comprising 20% of whole proteome. Energy metabolism related proteins (9%) primarily involved divergent subunits of ATP synthase. ATP synthase (Spots no79, 85) and iron-sulfur protein (Spot no16) levels were enhanced considerably by higher S supply while Ferredoxin NADP reductase (Spot no50) level was increased at all the S supplies + AsIII. Many proteins with a potential role in protection against As induced oxidative damage changed in abundance in rice leaves during As stress e.g. Cu, Zn SOD, 2-cys peroxiredoxin, glyoxalase, heat shock proteins though their expression levels varied at different S supplies (Table 1). Among stress proteins (14%), iron-sulfur protein (Spot no16) was up regulated under HS + AsIII, while LS + AsIII conditions had negative effect on its abundance. Though nucleic acid metabolic proteins (5%) were not detected in LS + AsIII, however, HS + AsIII treatment up regulated these proteins (Spots no129 and 134). Cytoskeleton (2%) and carbohydrate metabolism (2%) proteins were enhanced during AsIII stress throughout all the S regimes except HS + AsIII where NAD dependent epimerase was not detectable. Hydrolase and seed storage non enzymatic proteins were down regulated in all the NS + AsIII exposure conditions.
Biochemical changes during AsIII Stress under different S conditions
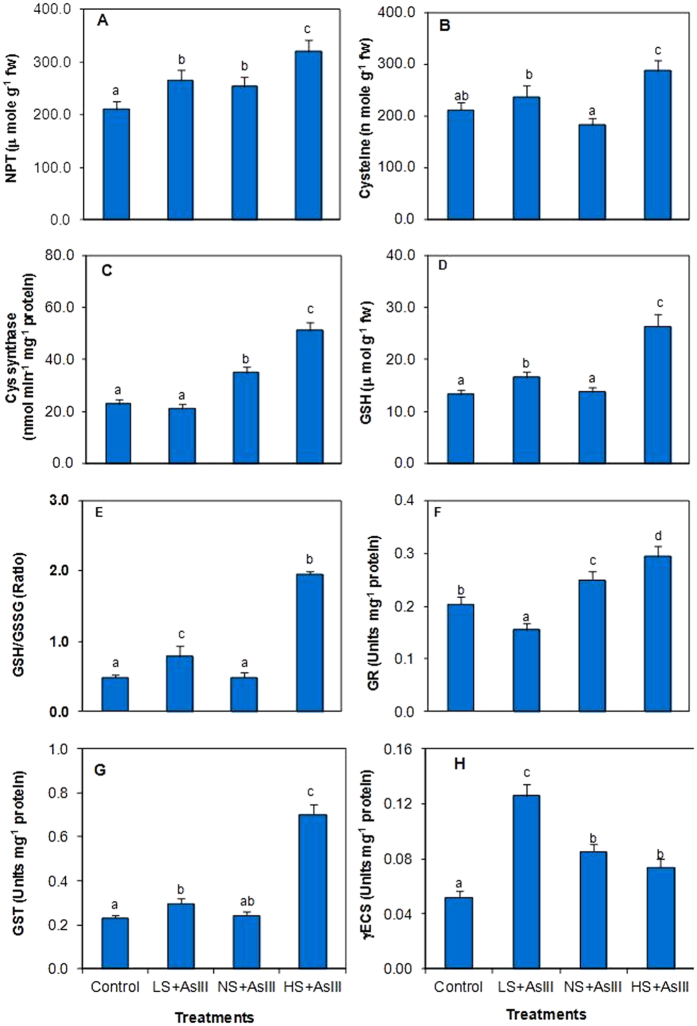
Different S treatments illustrated statistically significant variation in thiolic ligands. Plants exposed to HS + AsIII had elevated levels of thiolic compounds. Total NPTs, Cys and GSH levels were enhanced up to 52%, 46% and 99%, respectively in HS + AsIII plants. LS + AsIII exposed plants also had enhanced thiolic compounds as NPTs (26%) and GSH (24%), while Cys level declined by 9%. Activities of enzymes related to thiol metabolism (CS, GR, GST and γ-ECS) modulated considerably in various S treatments. Cysteine synthase was enhanced (139%) in NS + AsIII treatment, whereas 9% reduction was observed in LS + AsIII plants. GR activity was substantially different under different S treatments, while LS + AsIII exposure resulted in 24% decline, HS + AsIII plants demonstrated higher increase (204%). Similarly activity of GST was higher (189%) in HS + AsIII than NS + AsIII and activity of γ-ECS was increased by 142% LS + AsIII exposure than NS + AsIII (Fig. 5A–H).
Figure 5. Effect of different S doses on the level of (A) non protein thiols (NPTs), (B) cysteine, (C) reduced glutathione (GSH), (D) ratio of reduced to oxidized glutathione (E), cysteine synthase (CS), (F) glutathione reeducates (GR), (G) glutathione-S-transferase (GST), (H) γ-glutamylcysteine synthetase (γ-ECS) under arsenite stress in rice (Oryza sativa L.) leaves.
All the values are means of triplicate ± S.D. ANOVA significant at p ≤ 0.01. Different letters indicate significantly different values at a particular treatment (DMRT, p ≤ 0.05).
Gene expression analysis and western blot analysis
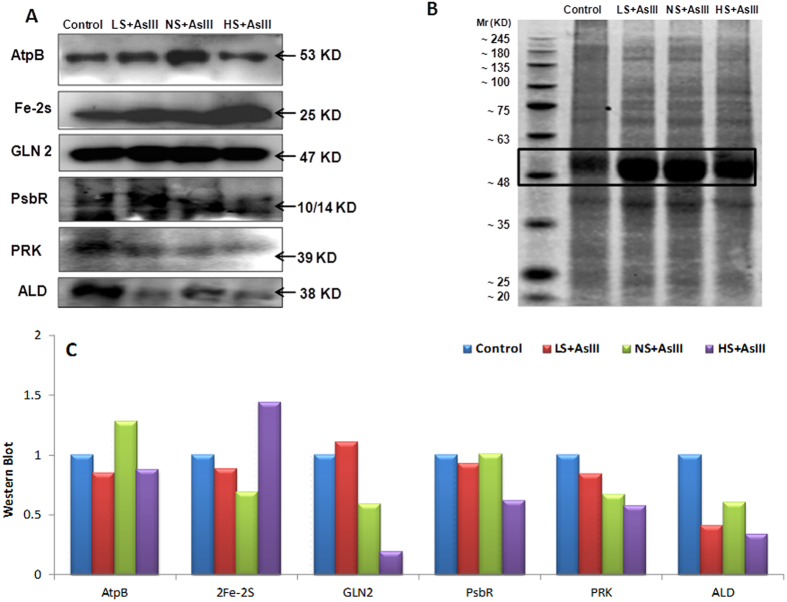
To investigate the changes in gene expression at the mRNA level, qPCR analysis was performed (Supplemental Information 3, Fig. S3). The genes encoding 15 proteins (PKR, 2Fe-2S, TPI, GAPDH, OEE, FNR, PsbR, Fru-bisP aldolase, GLN1, GLN2, AtpB, RuP3-E, aminotransferase) in O. sativa (Table 1) were analyzed. Transcript level analysis revealed induction at mRNA level in most of the proteins under LS + AsIII. However, in seedlings exposed to HS + AsIII and NS + AsIII decrease in expression level was observed.
To validate the results of 2DE gel analysis, western blot of selected candidate proteins were performed (Fig. 6A–C). Based on our matching rationales from the literatures, supporting their roles on the cellular physiology of sulfur and arsenic interactions, we selected AtpB, 2Fe-2S, GLN2, PsbR, PRK, and ALD proteins to verify their expression in treated conditions by western blotting. The densitometry analysis of these proteins on blot showed a similar trends expression by the effect of treatment as it observed by 2DE analysis.
Figure 6. Western blot analysis of selected candidate proteins (AtpB, 2Fe-2S, GLN2, Psb R, PRK, ALD) with their corresponding molecular weight in rice leaves during As stress under various S regimes (A); RuBisCo large subunit a band in coomassie blue staining (CBB) SDS gel served as a loading control (B), which used for normalization of detected proteins in densitometry studies (C) fold change indicated with respect of control samples.
Amino acid profiling
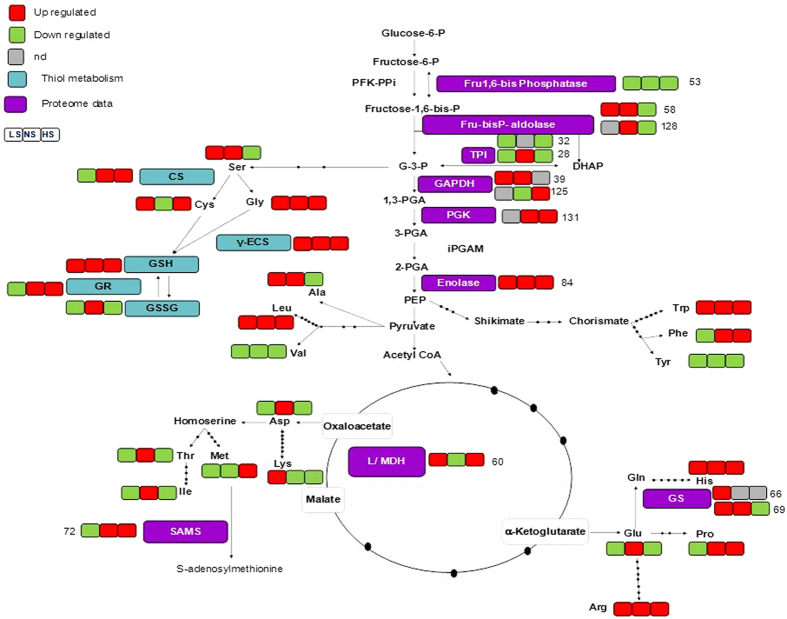
The fueling reactions of central metabolism provide precursor metabolites for synthesis of the 20 AAs which are incorporated into proteins. The diversions of AA biosynthetic pathways from the glycolytic pathway and the TCA of central metabolism are shown in (Fig. 7). Free AAs were examined viz., aspartic acid (Asp), threonine (Thr), serine (Ser), glutamic acid (Glu), proline (Pro), glycine (Gly), alanine (Ala), valine (Val), methionine (Met), leucine (Leu), tyrosine (Tyr), phenylalanine (Phe), lysine (Lys), histidine (His), arginine (Arg), isoleucine (Ile) and cysteine (Cys) in rice shoot during As stress with different S doses. Histidine was induced maximally (333%) among all the AAs followed by gly (274%) during HS + AsIII. Valine showed highest reduction of 97% under LS + AsIII followed by Lys (94%) HS + AsIII. In all the treatments, levels of His, Trp, Gly, Arg and Leu enhanced, however, content of Val and Tyr were declined (Table 2).
Figure 7. Pathways involved in the biosynthesis of amino acids.
All data were extracted from Tables 1; Purple boxes represent protein change from proteome analysis. Metabolite abbreviations are as follows: nd, not detected; LS, low sulfur; S, optimum sulfur; HS, high sulfur; PFK-PPi, PPi-Fru-6-P 1-phosphotransferase; fructose 1,6-bisphosphatase, FBPase-1; fructose bisphosphate aldolase, Fru-bisP aldolase; DHAP, dihydroxyacetone-phosphate; TPI, triosephosphate isomerase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; G-3P, glyceraldehyde-3-phosphate; 1,3PGA, 1,3-bisphosphoglycerate; 3PGA, 3-phosphoglycerate; 2PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate; L/MDH, lactate/malate dehydrogenase; SAMS, S-adenosine methionine synthetase; GS, glutamine synthetase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; CS, cysteine synthase; γECS, glutamylcysteine synthetase; GSH, reduced glutathione; GSSG, oxidized glutathione; GR, glutathione reductase; Aspartic Acid(Asp), Threonine (Thr), Serine (Ser), Glutamic Acid (Glu), Proline (Pro), Glycine (Gly), Alanine (Ala), Valine (Val), Methionine (Met), Leucine (Leu), Tyrosine (Tyr), Phenylalanine (Phe), Lysine (Lys), Histidine (His), Arginine (Arg), Isoleucine (Ile), Cysteine (Cys).
Table 2. Amino acid content was expressed in mg kg−1 fw.
| Amino acid | Control | LS + AsIII | NS + AsIII | HS + AsIII |
|---|---|---|---|---|
| SER | 4.72b ± 0.31 | 16.13d ± 1.06 | 12.70c ± 0.84 | 1.73a ± 0.11 |
| THR | 5.75c ± 0.38 | 1.49a ± 0.10 | 13.56d ± 0.91 | 3.73b ± 0.25 |
| HIS | 2.23a ± 0.17 | 3.94b ± 0.30 | 5.23c ± 0.39 | 9.64d ± 0.72 |
| ALA | 4.95b ± 0.39 | 5.73b ± 0.45 | 8.71c ± 0.69 | 3.12a ± 0.25 |
| MET | 4.83b ± 0.35 | 3.37a ± 0.24 | 3.65a ± 0.26 | 5.65c ± 0.41 |
| TRP | 2.36a ± 0.16 | 3.80b ± 0.25 | 2.65a ± 0.18 | 5.08c ± 0.34 |
| GLY | 7.22a ± 0.64 | 18.86c ± 1.68 | 10.41b ± 0.93 | 26.98d ± 2.41 |
| ARG | 3.20a ± 0.19 | 8.63c ± 0.51 | 5.68b ± 0.33 | 10.77d ± 0.63 |
| ILE | 8.05b ± 0.61 | 4.18a ± 0.32 | 12.50c ± 0.95 | 3.75a ± 0.29 |
| PRO | 11.26b ± 0.89 | 1.85a ± 0.15 | 15.32c ± 1.21 | 15.08c ± 1.19 |
| LYS | 3.78c ± 0.27 | 6.94d ± 0.49 | 1.40b ± 0.10 | 0.21a ± 0.01 |
| TYR | 1.34d ± 0.08 | 0.69b ± 0.05 | 0.39a ± 0.03 | 1.13c ± 0.08 |
| LEU | 5.58a ± 0.49 | 11.96b ± 1.06 | 15.51c ± 1.37 | 10.78b ± 0.95 |
| CYS | 3.10b ± 0.25 | 1.13a ± 0.09 | 7.99d ± 0.63 | 5.83c ± 0.46 |
| ASP | 8.96b ± 0.55 | 5.25a ± 0.32 | 14.01c ± 0.87 | 4.64a ± 0.29 |
| VAL | 8.23a ± 0.69 | 0.27b ± 0.02 | 3.78b ± 0.32 | 3.11c ± 0.26 |
| PHE | 3.92b ± 0.32 | 2.00a ± 0.16 | 4.58b ± 0.37 | 6.08c ± 0.49 |
| GLU | 26.97b ± 2.31 | 3.89a ± 0.33 | 60.75c ± 5.21 | 3.19a ± 0.27 |
Chromatograms were kintegrated using Em power 2 HPLC software v 6.0. All values are mean of triplicates ± S.D. ANOVA significant at p ≤ 0.01.
Discussion
The aim of the present study was to validate the hypothesis that high and adequate S may mitigate As stress through various metabolic pathways. In this study results obtained provide insight on the influence of different S conditions on proteomic changes in rice leaves during As stress. Results indicated that As accumulation is considerably affected by different S regimes. Limiting S enhanced As accumulation in aerial part of the plants, perhaps due to lesser chelation of As by thiols in root25. Contrastingly high S supplementation results in lesser As accumulation in leaves, as high As-phytochelatin complexation in root restricts entry of As in shoot. High PC-As chelation has been found to immobilize As in the root and minimizing its level in shoot and grain26 in transgenic rice. In HS + AsIII leaves, the excess thiolic metabolites such as glutathione may serve as antioxidant leading to reduction in toxicity. Limiting S reduced shoot length as well as weight, while HS + AsIII reverted this effect. Similar study in Arabidopsis plants supported the finding27. Decline in growth under stress has been regarded as an effect of reduced photosynthetic content28 (Supplemental Information 3 Fig. S1, S2 C–F).
In the present proteomic study, a number of proteins were differentially expressed among different S treatments (Table 1). Functional classification of differently expressed proteins reveled that major proteins were dedicated to central metabolic pathways (glycolysis, TCA cycle and AA biosynthesis) followed by photosynthesis (electron transport chain and Calvin cycle), energy, stress, nucleic acid and protein metabolism (Fig. 4).
Effect of sulfur and arsenic interaction on glycolysis and TCA cycle
In the glycolysis, preparatory phase is energy consuming, starting from glucose and ending with splitting of hexose to triose. During the present study different enzymes of preparatory and payoff phase of glycolysis behaved differentially during low and high S regimes in combination with As. Results indicated that enzymes of preparatory phase specifically fructose 1,6-bisphosphatase (FBPase-1), fructose bisphosphate aldolase (Fru-bisP aldolase) and triose phosphate isomerase (TPI) were down regulated in LS + AsIII and HS + AsIII combinations except upregulatory response of Fru-bisP aldolase under LS + AsIII. To abate the As toxicity, plant required more reducing equivalents so that FBPase-1 might be inhibited to compel glycolysis to move in forward direction29. Though LS + AsIII enhanced the Fru-bisP aldolase while decreased during HS + AsIII exposure, this may be due to the excess GSH formation that inhibits enzyme activity30. Higher level of GSSG and GSH may reduce TPI level in LS + AsIII and HS + AsIII treatments respectively and it is also evident by study of Ito et al. (2003)30.
In contrast to preparatory phase, enzymes of payoff phase were up regulated under low and high S regimes along with As. However, similar to preparatory phase, enzymes of payoff phase (PGK and enolase) also showed up-regulation under As exposure alone except GAPDH, which showed differential response, might be due to the different isoforms (Table 1, spots no39 and 125) of this enzyme. Differential expression of GAPDH has also been observed in several other proteomic studies in response to metal and metalloid stress19,31. In the present study enzymes of payoff phase of glycolysis viz., GAPDH, phosphoglycerate kinase (PGK) and enolase played active role to fulfill the energy requirement during stress conditions probably by using photo assimilates directly from chloroplasts for mitochondrial respiration19. The enzymes of preparatory phase were down regulated in high sulfur combined with AsIII, and enzymes of pay off phase were up regulated under same circumstances, then there must be any alternative source of substrate. Pentose phosphate pathway may provide substrate to pay off phase, as glyceraldehyde 3-phosphate and fructose 6-phosphate are the byproduct of pentose phosphate pathway32.
Lactate/malate dehydrogenase (L/MDH) is chloroplast localized enzyme of TCA cycle and catalyses malate to oxaloacetate or vice-versa was positively regulated in HS + AsIII treatment. Higher requirement of oxaloacetate occurs during stress conditions in cells to synthesize various stress related AAs (Met, Lys, Thr) to maintain the homeostasis. While malate counterbalance the uptake of anions and cations in plant cells in order to maintain the anion-cation charge balance and the cytoplasmic pH, oxaloacetate is required for synthesis of Met involving various steps. S-adenosylmethionine synthetase (SAMS) catalyzes the biosynthesis of S-adenosyl-L-methionine (SAM) from Met and ATP, which is a universal methyl group donor in several transmethylation reactions33. SAMS were elevated at HS + AsIII in the present study, similarly34 concluded that different isogenes of SAMS play important role during hexavalent Cr stress.
Sulfur deficiency is considered as S shortage relative to nitrogen (N) nutrition, depicting symptoms of excess nitrogen35. Enhanced levels of glutamine synthetase (GS) as observed during LS + AsIII, suggest that low S nutrition mimics excess N nutrition. Glutamine synthetase functions as the major assimilatory enzyme for ammonia produced from N fixation or nitrate reduction pathway36. Additionally GS is involved in the synthesis of GSH through Glu biosynthesis pathway29. The enhanced expression of GS leads to more GSH formation which would enhance cellular defense against oxidative stress by participating in the ascorbate/GSH cycle37. At HS + AsIII, glutamine synthetase activity was down regulated imitating deficiency of N in comparison to S nutrition. This result is further corroborated by the downregulation of aminotrasferases at almost all the S supply. Aminotransferases are involved in number of cellular processes including glycolysis, AA metabolism, photorespiration and N use efficiency38. Therefore down regulation of this enzyme would have resulted in lesser nitrogen in rice leaves.
Effect of arsenic sulfur interaction on amino acid level and glutathione metabolism
Protein and AA composition are the vital component of rice nutrient quality, however, literature indicated As induced changes in various AAs due their role for chelation of metal(iod)s and cellular homeostasis16. During the present study As and S interaction differentially affected the AAs content, e.g. NS + AsIII enhanced the Thr, Met, Ile, Asp except Lys. HS + AsIII exposure resulted in high Met level and also the downstream enzyme S-adenosyl methionine synthatase (SAMS). Similarly HS + AsIII treatment resulted in higher Cys and Met levels and associated enzymes in submerged macrophyte Hydrilla verticillata39. Induced Met level also indicates its role in AdoMet-dependent reactions, the nitrogen–carbon skeleton and its S derivatives recycling40. These cycles could be of importance in situations of low S availability for Met synthesis as a consequence of a high demand of Cys for GSH synthesis under stress conditions39. Glutamic acid, Cys and Gly are the main components of GSH, induction of these AAs in all the treatments except Glu and Cys at LS + AsIII may be due to the limiting S in the medium which is essential for the formation of GSH39. In addition, higher level of GSH is maintained in HS + AsIII due to higher activity of GR41,42,43. Glutathione is involved in detoxification processes, and plays a role as an enzyme cofactor, and a storage and transport form of Cys44. GSTs are involved in conjugation of GSH to metal or metalloid and metabolites induced due to oxidative stress39,45 and its enhanced activity would be helpful in preventing oxidative stress16. At LS + AsIII higher activity of γ-ECS that catalyses the ATP-dependent ligation of Cys and Glu to form γ-EC may occur to fulfill demand of GSH. The low levels of Glu and Cys were obtained in the present study suggesting rationale for higher rate of enzymatic activity of γ-EC. In LS + AsIII, As accumulation enhanced NPTs to chelate endogenous AsIII using most of the available GSH pool. However, HS + AsIII induced NPTs because of higher exogenous supply of S resulting into higher GSH pool37. Which may have played crucial role in detoxification of As during high S supply. Ratio of GSH:GSSG is crucial to maintain cellular homeostasis, HS + AsIII treatment has been found to achieve this aim through maintaining this homeostasis when plant was suffering from AsIII toxicity. This is in accordance with study demonstrating protection of Arabidopsis plants from heavy metal toxicity by maintaining GSH homeostasis through recycling Glu46.
Effect of S and As interaction on proteins of photosynthesis, energy and stress
Various proteins of photosynthesis machinery have been shown to be differentially affected by metal stress19.Proteomic analysis revealed oxygen evolving enhancer (OEE) protein was down regulated under AsIII stress with various S regimes. Leaf proteome of Agrostis tenuis has shown drastic reduction of OEE protein in response to 134 μM As(V)47,48. Considerable reduction in photo system I reaction center subunit IV (Spot no17) was observed under HS + AsIII condition, which is in agreement with earlier study showing similar trend45 irrespective of As stress. This contrast may be due to the varietal difference among the rice genotypes, behaving differentially under As stress15,23.
Recently, D’Hooghe et al. (2013)49 concluded that S limitation results in reduction of plastocyanin and Ferredoxin–NADP reductase (FNR) in Brassica napus. However, current study on rice demonstrating up regulation of plastocyanin may be due to additional As stress. Sulfate restriction reduces S assimilation into cysteine27, also reduces carbon assimilation and photosynthetic activity, resulting into distortion of glycolytic flux that leads to reduction of AA accumulation.
In the present study, NS + AsIII and HS + AsIII up regulated FNR level, that catalyses the production of NADPH + H+ required for CO2 assimilation and energy production. Srivastava et al. (2011)4 reported that plant requires more energy during As stress to maintain cellular homeostasis. Recycling of NADPH serves to strengthen antioxidant system under salt stress in olive plants50.Thus it can be inferred that to full-fill energy requirement, S enhanced energy production through enhancing NADPH + H+ availability to cope up As stress, as lower S limits the availability of NADPH + H+ 50.
Many proteins which showed different pIs and/or Mws were identified as ribulose-1,5-biphosphate carboxylase/oxygenase (RuBisCo) subunits (Table 1). Interestingly ribulose bisphosphate carboxylase (RuBisCo) small chain precursor (Spots no4 and 8) was down regulated during all the AsIII treatments irrespective of S condition, whereas RuBisCo large chain precursor was both up regulated (Spots no96, 132 and 138) and down regulated (Spot no42 and 82). Small subunit being a limiting factor, may down regulate overall RuBisCo mediated reaction leading to reduced carbon fixation (Calvin cycle) that lead to reduced photosynthesis rate19,49.
Reduction in ATP synthase along with its subunits during LS + AsIII exposure could be drawn in, for S remobilization processes through the maintenance of an efficient sulfate efflux from the vacuole. Similar results were reported in leaves of Brassica involving tonoplast sulfate transporters (BnSult4;1 and BnSult4;2)51. At higher S, increased abundance of ATP synthase (alpha and beta subunits) and Ferredoxin-NADP reductase suggest their likely role in As tolerance mechanism. In general, plants exposed to abiotic stresses show downregulation of linear electron flow (LEF) and activation of cyclic electron transport (CET) when LEF becomes saturated52. It could be possible that under high S the LEF is partially replaced by up regulated CET which provides energy to Calvin cycle53, thereby meeting energy demand.
Iron-sulfur clusters is essential and versatile cofactors of proteins involved in catalysis, electron transport, vitamin B1 synthesis and sensing of ambient conditions54,55. During current study this protein was found up regulated with HS + AsIII while LS + AsIII reduced this protein, suggesting an imbalance of this protein under deprived S condition, and this modulation by S concentration is first time reported during current study.
IAP100 is a member of ‘inhibitor of apoptosis’ (IAP) gene family which was discovered less than a decade ago, serves as anti-apoptotic protein56. This protein, reported for the first time in rice, was found up regulated in HS + ASIII condition tentatively suggesting role of S in apoptosis inhibition, though this requires further investigation.
Materials and Methods
Plant Material and Stress Treatment
Rice seeds (Oryza sativa L.), cultivars IR-36, collected from RRS, Chinsurah India, were surface sterilized using 10% H2O2 for 30 s, followed by thorough washing with de-ionized water, and then were soaked in distilled water for 24 h. Seeds were germinated in the dark for 4 d at 37 ± 1 °C. Uniform germinated seedlings were selected and transplanted to trays containing fixed PVC cups (4 cm diameter and 5 cm height, 10 plants per cup) and grown in modified Hewitt’s media57,58 supplemented with low sulfur (0.5 mM), normal sulfur (3.5 mM) as used in standard Hewitt media, or high sulfur (5.0 mM)39 for 10 d. Then As was added as AsIII (NaAsO2; 25 μM)56 for 7 d in a controlled growth environment at 28/21 °C at light intensity of 210 μ mol cm−2s−1 (16-h light/8-h dark) with relative humidity of 70%. The S conditions were abbreviated as follows: LS for the low sulfur conditions (0.5 mM), NS for standard sulfur conditions (3.5 mM), and HS for the high sulfur concentration (5.0 mM). All the experiments were conducted with three replicates (biological replicates) for each treatment combination. Plants were harvested, washed three times with milli-Q water, and the plant material was divided up into different aliquots for various analyses. In all the analyses only plant leaves were used, except for the determination of total S and As in which both roots and leaves were used.
Protein Extraction, 2-DE, Gel Staining, and Image Analysis
Leaf proteins from three biological replicates were extracted using a trichloroacetic acid/acetone precipitation method followed by phenol extraction as per method described by Deeba et al. (2012)59. Protein were collected from three biological replicates and pooled to make technical replicates to normalize the effect of variation in the biological replicates. Total protein was estimated by Bradford reagent (Bradford 1976)60 using BSA as standard. For First-dimensional electrophoresis an equal amount (100 μg) of protein was loaded on each immobilized pH gradient (IPG) strips (7 cm, pH 4–7 linear) diluted with an iso electric focusing (IEF) rehydration solution (7 M urea, 2 M Thiourea, 2% CHAPS, 20 mM DTT, 0.5% v/v IPG buffers) in a re-swelling tray (GE Healthcare, USA) at room temperature for overnight. Then focusing was performed on the IPGphore-3 (GE Healthcare, USA) under the following conditions: ramping to 200 V for 1 h, ramping to 500 V for 1 h, 3000 V for 2 h, and 8000 V for 2 h for a total of 12 kVh.
Before the SDS-PAGE, the strips were in 10 ml of reducing equilibration buffer [6 M urea, 50 mM Tris-HCl (pH 8.8), 30% (v/v) glycerol, 2% (w/v) SDS, a trace of bromophenol blue, and 1% (w/v) DTT] for 15 min, and another 15 min in alkylating equilibration buffer that contained 2.5% (w/v) iodoacetamide instead of 1% DTT. The strips were placed on the top of 10% SDS-polyacrylamide gels and sealed with 0.5% agarose solution. The electrophoresis was carried out at a constant voltage of 200 V at 25 °C (Bio-Rad mini-gel apparatus, BioRad) with a standard Tris Glycine running buffer. Protein spots in 2-DE gels were detected by CBB G-250 staining. The 2-DE gels were scanned using scanner (HP precision scan pro 3.02, USA). 2-DE gels were analyzed using the Image MasterTM 2-D platinum software (version 7.0; GE Healthcare, USA). Spot detection parameters were as follows: smoothness 2; saliency 1; minimum area 5. Quantification was carried out using the percent volume criterion. The match analysis was performed in an automatic mode, and further manual editing was performed to correct the mismatched and unmatched spots (Supplemental Information 1, Table S1 A–B). The relative volume of each spot was represented as expression level. Total spot intensity per gel was used to normalize spot intensities (% of individual spot intensity/% spot intensity of each gel) to compensate for variations between gel replicates. A criterion of p < 0.001 was used to define the significant difference when analyzing the parallel spots between groups with analysis of one-way variance (ANOVA). Protein spots with an abundance ratio of at least 1.5 folds were selected as differentially expressed.
In-Gel Digestion, MS Analysis, and Database Searching
Protein spots showing significant changes in abundance between different treatments were selected and excised manually with scalpel. Excised gel pieces were destained with 50% MetOH and 50 mM ammonium bicarbonate (ABC) followed by a washing with 25 mM ABC. Selected gel pieces were dehydrated with 50% acetonitrile (ACN) and 50 mM ABC mixed in 2:1 ratio. The cycle of dehydration followed by rehydration by 25 mM ABC was repeated 3 times. Destained gel pieces were dried in speed vac (Labconco, USA) and rehydrated in trypsin solution (20 μg/ml) at 1:20 ratio of protein. Gel particles were immersed in 25 mM ammonium bicarbonate (ABC) and samples were digested overnight at 37 oC (about 16–18 h digestion). Peptides were extracted twice with 50% ACN/1% tri flouro acetic acid (TFA). Extracted peptides were mixed with matrix solution (5 mg/ml a-Cyano-4-hydroxycinnamic acid in 50% ACN containing 0.1% TFA) in 1:1 ratio. Samples were spotted and air dried at room temperature on stainless steel 384 wells target plate. External calibration of mass spectrometer was performed with a mixture of angiotensin I, Glufibrino-peptide B, ACTH (1–17), and ACTH (18–39) while for MS/MS with fragment of Glufibrino-peptide B.
All samples were analyzed using a MALDI-TOF-TOF (Model 4800, Applied Biosystems, USA). The monoisotopic peptide masses obtained from MALDI-TOF-TOF were analyzed by the 4000 Series Explorer software (version 3.5, Applied Biosystems, USA). On the basis of mass signals, protein identification was performed with the Mascot software (http://www.matrixscience.com) to search proteins against SwissProt, NCBInr and MSDB databases (Supplemental Information 1, Table S1 C). The following parameters were used for database searches: monoisotopic mass accuracy, <100 ppm; missed cleavages, 1; carbamidomethylation of Cys as fixed modification and oxidation of Met; keratin, known contaminant was excluded. Additionally the theoretical values of molecular weight (Mr) and pI of identified proteins were calculated by using the Peptide Mass program (ExPASy). The score threshold to achieve p < 0.05 was set by the Mascot algorithm and was based on the size of the database used in the search. Peptides were again searched against Rice Genome Database Project to identify the locus ID and encoded proteins names are assigned (Supplemental Information 1, Table S1 D). Peptide view of Mascot search results is mentioned in Supplemental Information 2, Table S2. Hierarchical clustering of S responsive proteins was conducted using MeV software (Supplemental Information 3, Figure S4).
Expression Analysis using Quantitative RT-PCR
Approximately, 5 μg RNase free DNase-treated total RNA (5 μg) isolated from leaves of rice plants exposed to various treatments and control was reverse-transcribed using SuperScriptII (Fermentas, USA), following the manufacturer’s recommendation. The synthesized cDNA was diluted 1:5 in DEPC water and subjected to quantitative RT-PCR (qRT-PCR) analysis. The qRT-PCR was performed using an ABI 7500 instrument (ABI Biosystems, USA) using primers (Supplemental Information 3, Table S1). Each qPCR reaction contained 5 μl of SYBR Green Supermix (ABI Biosystems, USA), 1 μl of the diluted cDNA reaction mixture (corresponding to 5 ng of starting amount of RNA) and 10 pM of each primer in a total reaction volume of 10 μl. qPCR reactions were performed under the following conditions: 10 min at 95 °C and 40 cycles of the one step thermal cycling of 3 s at 95 °C and 30 s at 60 °C in a 96-well reaction plate. Actin gene was used as an internal control to estimate the relative transcript levels of the target gene. Specificity of amplicons generated in qPCR reactions was verified by melt curve analysis. Each qPCR reaction was performed in triplicate (technical replicates) for each biological replicate (three for each treatment). Relative gene expression was calculated using ∆∆CT method61.
Crude extracts of plant tissues and western blot analysis
Protein extraction was carried out by Tanou et al. (2009)62 with slight modifications. Leaf tissues were ground in liquid N2 using a mortar and pestle. Soluble proteins were extracted from the powdered tissue at 4 °C in 1 ml of buffer (pH = 7.5) containing 50 mM HEPES-KOH, 1 mM EDTA, 5 mM DTT, 10% (v/v) glycerol, 2 mM benzamidine, 2 mM aminocuprioc acid, protease inhibitor cocktail mini from Roche Diagnostic. The extract was stirred for 20 min at 4 °C then centrifuged (14000 g for 10 min at 4 °C). Supernatant was used for further analysis. Protein estimation in supernatant was carried out by Bradford method (1976)60.
Western blotting is carried out by method of Mitani et al. (2009)63. Briefly, 50 μg of proteins of control and treated samples were resolved on 12% 1D-PAGE and blotted onto PVDF membrane. Further, the blocking of blotting membrane was done for 1 h using 1x blocking buffer (Sigma-Aldrich). Further, these membranes were probed by polyclonal primary antibodies against AtpB (AS05 085), GLN2 (AS08 296), 2Fe-2S (AS12 1852), PsbR (AS 05–059), PRK (AS09 464), ALD (AS08 294) proteins at recommended dilution for overnight. All antibodies were procured from Agrisera, Sweden. In the next step, HRP conjugated cross-reactive secondary antibodies was incubated and their bands were visualized by chemiluminescence system. For the analysis of their expression equal amount of proteins was loaded on another 12% 1D-PAGE for normalization of protein expression of the corresponding western blot64. The intensities of each blot were quantified by using UN-SCAN IT software (Orem, UT, USA). Data were analyzed by comparing relative pixel density of the protein bands normalized to with bands of single lane RuBisCo large subunit of commassie stained gel.
Arsenic and Sulfur Estimation
For analysis of total As the analytical procedure was performed according to Dwivedi et al. (2010b)23 using Inductively Coupled Plasma-Mass Spectrometry (ICP-MS) (7500 cx; Agilent, Tokyo, Japan). Total S concentration was estimated by Chesnin and Yien (1951)65. Detailed methodology is provided in Supplemental Information 3 Text S1 A.
Plant growth parameters
Shoot length was measured on a metric scale and samples were oven dried for biomass measurements. Photosynthetic pigments measurement was carried out after extraction in 80% chilled acetone66 and carotenoids by Duxbury and Yenstch (1956)67 method.
Amino Acid Profiling
Amino acid analysis was performed by HPLC (Waters model 2475, column-C18) using the pico tag method14,68. Detailed methodology is provided in Supplemental information (Detailed methodology is provided in Supplemental Information 3 Text S1 B).
Estimation of Thiol Compounds and Enzymes
The level of nonprotein thiols (NPTs) was measured using Ellman’s et al. (1959)69 reagent. Estimation of Cys was performed using acid ninhydrin reagent70. The levels of reduced (GSH) and oxidized (GSSG) glutathione were determined fluorometrically using o-phthaldialdehyde (OPT) as fluorophore on fluorescence spectrophotometer (Hitachi F 7000, Japan)71. For the assay of cysteine synthase (CS; EC 2.5.1.47) and γ-glutamylcysteine synthetase (γECS; EC 6.3.2.2) activities, homogenization and assay were performed following Saito et al. (1994)72 and Seelig and Meister (1984)73 respectively, with slight modifications. Glutathione S-transferase (GST; EC 2.5.1.18) activity was assayed following Habig and Jacoby (1981)74. The GR activity was assayed by following Smith et al. (1988)75. Detailed methodology is provided in Supplemental Information 3 Text S1 C. The concentration of total phytochelatins (PCs) was calculated as PCs = NPTs − total GSH (Hartley‐Whitaker et al. 2001)76.
Additional Information
How to cite this article: Dixit, G. et al. Sulfur alleviates arsenic toxicity by reducing its accumulation and modulating proteome, amino acids and thiol metabolism in rice leaves. Sci. Rep. 5, 16205; doi: 10.1038/srep16205 (2015).
Supplementary Material
Acknowledgments
The authors are thankful to Director, CSIR-National Botanical Research Institute (CSIR-NBRI), Lucknow for the facilities and for the financial support from the network projects (CSIR-INDEPTH), New Delhi, India. The authors are grateful to the Joint Director, Rice Research Station (RRS), Chinsurah to provide rice germplasm. GD and APS are thankful to Council of Scientific and Industrial Research and University Grant Commission (UGC) New Delhi, India respectively, for the award of Junior/Senior Research Fellowship and Academy of Scientific and Innovative Research (AcSIR) for their Ph.D. registration. We are also thankful to Mr. Dilip Chakraborty for technical assistance and special thanks to Mr. P.K. Singh for editing of manuscript. Award of Emeritus Scientist (CSIR) project to RDT is gratefully acknowledged.
Footnotes
Author Contributions R.D.T., P.K.T. and V.P. designed experiments and reviewed manuscript. A.P.S. and G.D. performed experimental work and prepared figures. A.K. performed statistical analysis. F.D. Operated MALDI-TOF-TOF. S.S. operated Western blot analysis. S.K., S.D., B.A. and Y.S. reviewed manuscript. All authors have read and approved the manuscript.
References
- Williams P. N. et al. Variation in arsenic speciation and concentration in paddy rice related to dietary exposure. Environ. Sci. Technol. 39, 5531–5540 (2005). [DOI] [PubMed] [Google Scholar]
- Stone R. Arsenic and paddy rice: a neglected cancer risk? Science 321, 184–185 (2008). [DOI] [PubMed] [Google Scholar]
- Tripathi R. D. et al. Arsenic hazards: strategies for tolerance and remediation by plants. Trends Biotechnol 25, 158–165 (2007). [DOI] [PubMed] [Google Scholar]
- Srivastava S. et al. Redox state and energetic equilibrium determine the magnitude of stress in Hydrilla verticillata upon exposure to arsenate. Protoplasma 248, 805–815 (2011). [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct Integr Genomics 11, 259–273 (2011). [DOI] [PubMed] [Google Scholar]
- Duan G. L. et al. Evidence for a role of phytochelatins in regulating arsenic accumulation in rice grain. Environ Exp Bot 71, 416–421 (2011). [Google Scholar]
- Reid R. et al. Arsenite elicits anomalous sulfur starvation responses in barley. Plant physiol, 162, 401–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al., Influence of sulphur on arsenic accumulation and metabolism in rice seedlings. Environ Exp Bot 72, 34–40 (2011). [Google Scholar]
- Smith F. W. et al. Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92, 9373–9377 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T. & Wachter A. Sulfur metabolism: a versatile platform for launching defence operations. Trends Plant Sci 10, 503–509 (2005). [DOI] [PubMed] [Google Scholar]
- Rochaix J. D. Assembly of the photosynthetic apparatus. Plant physiol 155, 1493–1500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert L. I. et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA 105, 8197–8202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P. et al. Arsenite tolerance in rice (Oryza sativa L.) involves coordinated role of metabolic pathways of thiols and amino acids. Environ. Sci. Pollut. Res. 20, 884–896 (2013). [DOI] [PubMed] [Google Scholar]
- Kumar A. et al. Evaluation of amino acid profile in contrasting arsenic accumulating rice genotypes under arsenic stress. Biol. Plant. 58, 733–742 (2014). [Google Scholar]
- Dwivedi S. et al. Arsenate exposure affects amino acids, mineral nutrient status and antioxidants in rice (Oryza sativa L.) genotypes. Environ. Sci. & technol. 44, 9542–9549 (2010). [DOI] [PubMed] [Google Scholar]
- Dave R. et al. Arsenate and arsenite exposure modulate antioxidants and amino acids in contrasting arsenic accumulating rice (Oryza sativa L.) genotypes. J. Hazard. Mater. 262, 1123–1131 (2013). [DOI] [PubMed] [Google Scholar]
- Norton G. J. et al. Arsenic influence on genetic variation in grain trace-element nutrient content in Bengal Delta grown rice. Environ. Sci. Technol. 44, 8284–8288 (2010). [DOI] [PubMed] [Google Scholar]
- Ahsan N. et al. Comparative proteomic study of arsenic‐induced differentially expressed proteins in rice roots reveals glutathione plays a central role during As stress. Proteomics 8, 3561–3576 (2008). [DOI] [PubMed] [Google Scholar]
- Ahsan N. et al. Analysis of arsenic stress-induced differentially expressed proteins in rice leaves by two-dimensional gel electrophoresis coupled with mass spectrometry. Chemosphere 78, 224–231 (2010). [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. Comparative proteomic analysis of rice shoots exposed to high arsenate. J Integr Plant Biol 55, 965–978 (2013). [DOI] [PubMed] [Google Scholar]
- Shibato J. et al. Gel-based proteomics approach for detecting low nitrogen-responsive proteins in cultivated rice species. Physiol. and Mol. Biol. Plants 15, 31–41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S. et al. A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 9, 159–170 (2009). [DOI] [PubMed] [Google Scholar]
- Dwivedi S. et al. Arsenic affects mineral nutrients in grains of various Indian rice (Oryza sativa L.) genotypes grown on arsenic-contaminated soils of West Bengal. Protoplasma 245, 113–124 (2010). [DOI] [PubMed] [Google Scholar]
- Bevan M. et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488 (1998). [DOI] [PubMed] [Google Scholar]
- Dixit G. et al. Sulfur mediated reduction of arsenic toxicity involves efficient thiol metabolism and the antioxidant defense system in rice. J. Hazard. Mater. 298, 241–251 (2015). [DOI] [PubMed] [Google Scholar]
- Shri M. et al. Heterologous expression of Ceratophyllum demersum phytochelatin synthase, CdPCS1, in rice leads to lower arsenic accumulation in grain. Sci. Rep. 4, 5784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V. J. et al. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant physiol. 138, 304–318 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. P. Salicylic acid modulates arsenic toxicity by reducing its root to shoot translocation in rice (Oryza sativa L.). Front plant sci 6, 340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z. & Komatsu S. Contribution of proteomic studies towards understanding plant heavy metal stress response. Front Plant Sci 3, 310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Iwabuchi M. & Ogawa K. I. The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant and Cell Physiol 44, 655–660 (2003). [DOI] [PubMed] [Google Scholar]
- Kieffer P. et al. Combining proteomics and metabolite analyses to unravel cadmium stress-response in poplar leaves. J Proteome Res 8, 400–417 (2008). [DOI] [PubMed] [Google Scholar]
- Wamelink M. M. C., Struys E. A. & Jakobs C. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J Inherit Metab Dis 31, 703–717 (2008). [DOI] [PubMed] [Google Scholar]
- Bitrián M. et al. Polyamines under abiotic stress: metabolic crossroads and hormonal cross talks in plants. Metabolites 2, 516–528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F. et al. Physiological and proteomic alterations in rice (Oryza sativa L.) seedlings under hexavalent chromium stress. Planta, 2, 291–308 (2014). [DOI] [PubMed] [Google Scholar]
- Hirai M. Y. & Saito K. Post-genomics approaches for the elucidation of plant adaptive mechanisms to sulphur deficiency. J. Exp. Bot. 55, 1871–1879 (2004). [DOI] [PubMed] [Google Scholar]
- Miflin B. J. & Habash D. Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987 (2002). [DOI] [PubMed] [Google Scholar]
- Verbruggen N., Hermans C. & Schat H. Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant. Biol. 12, 364–372 (2009). [DOI] [PubMed] [Google Scholar]
- Good A. G. et al., Engineering nitrogen use efficiency with alanine aminotransferase. Botany 85, 252–262 (2007). [Google Scholar]
- Srivastava S. & D’souza S. F. Increasing sulfur supply enhances tolerance to arsenic and its accumulation in Hydrilla verticillata (Lf) Royle. Environ. Sci. Technol. 43, 6308–6313 (2009). [DOI] [PubMed] [Google Scholar]
- Rébeillé F. et al. Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103, 15687–15692 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M. Sulfur assimilation and the role of sulfur in plant metabolism: a survey. Photosynth. Res. 79, 331–348 (2004). [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Serrano M. A. R. Í. A. et al. Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 29, 1532–1544 (2006). [DOI] [PubMed] [Google Scholar]
- Srivastava S. et al. Phytochelatins and antioxidant systems respond differentially during arsenite and arsenate stress in Hydrilla verticillata (Lf) Royle. Environ. Sci. Technol. 41, 2930–2936 (2007). [DOI] [PubMed] [Google Scholar]
- Noctor G. et al. Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J. Exp. Bot. 53, 1283–1304 (2002). [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. Omics and biotechnology of arsenic stress and detoxification in plants: Current updates and prospective. Environ. Int. 74, 221–230 (2015). [DOI] [PubMed] [Google Scholar]
- Paulose B. et al. A γ-Glutamyl Cyclotransferase Protects Arabidopsis Plants from Heavy Metal Toxicity by Recycling Glutamate to Maintain Glutathione Homeostasis. Plant Cell 25, 4580–4595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. S. et al. Evaluation of metal-ion stress in sunflower (Helianthus annuus L.) leaves throug proteomic changes. Metallomics 1, 107–113 (2009). [Google Scholar]
- Duquesnoy I. et al. Identification of Agrostis tenuis leaf proteins in response to As (V) and As (III) induced stress using a proteomics approach. Plant Science 176, 206–213 (2009). [Google Scholar]
- D’Hooghe P. et al. Sulphur limitation provokes physiological and leaf proteome changes in oilseed rape that lead to perturbation of sulphur, carbon and oxidative metabolisms. BMC Plant Biol 13, 23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama R. et al. The dehydrogenase‐mediated recycling of NADPH is a key antioxidant system against salt‐induced oxidative stress in olive plants. Plant Cell Environ. 29, 1449–1459 (2006). [DOI] [PubMed] [Google Scholar]
- Krebs M. et al. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107, 3251–3256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir B., Sharmila P. & Saradhi P. P. Photosynthetic performance of Salvinia natans exposed to chromium and zinc rich wastewater. Brazil J Plant Physiol. 20, 61–70 (2008). [Google Scholar]
- Hurley J. K. et al. Structure–function relationships in Anabaena ferredoxin/ferredoxin: NADP+ reductase electron transfer: insights from site-directed mutagenesis, transient absorption spectroscopy and X-ray crystallography. Biochim. Biophys. Acta -Bioenergetics 1554, 5–21 (2002). [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron–sulphur proteins. Nature 460, 831–838 (2009). [DOI] [PubMed] [Google Scholar]
- Raschke M. et al. Vitamin B1 biosynthesis in plants requires the essential iron–sulfur cluster protein, THIC. Proc Natl Acad Sci USA 104, 19637–19642 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen G. S. & Duckett C. S. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Bio 3, 401–410 (2002). [DOI] [PubMed] [Google Scholar]
- Liu W. J. et al. Do phosphorus nutrition and iron plaque alter arsenate (As) uptake by rice seedlings in hydroponic culture? New Phytol. 162, 481–488 (2004). [Google Scholar]
- Dave R. et al. Arsenite tolerance is related to proportional thiolic metabolite synthesis in rice (Oryza sativa L.). Arch. Environ. Contam. Toxicol. 64, 235–242 (2013). [DOI] [PubMed] [Google Scholar]
- Deeba F. et al. Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol. Biochem. 53, 6–18 (2012). [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Tanou G. et al. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J 60, 795–804 (2009). [DOI] [PubMed] [Google Scholar]
- Mitani N. et al. Identification and characterization of maize and barley Lsi2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. Plant Cell 21, 2133–2142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. et al. Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol, 161, 108–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnin L. & Yien C. H. Turbidimeteric determination of available sulphates. Proc. Soil Sci. Soc. Amer. 15, 149–151 (1951). [Google Scholar]
- Arnon D. I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury A. C. & Yenstch C. S. Plankton pigment nomographs. J. Marine Res. 15, 91- 101 (1956). [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A. & Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr B Biomed Sci App 336, 93–104 (1984). [DOI] [PubMed] [Google Scholar]
- Ellman G. L. Tissue sulfhydryl groups. Arch. Biochem. Biophys 82, 70–77 (1959). [DOI] [PubMed] [Google Scholar]
- Gaitonde M. K. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J 104, 627–633 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hissin P. J. & Hilf R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal. Biochem. 74, 214–226 (1976). [DOI] [PubMed] [Google Scholar]
- Saito K. et al. Modulation of cysteine biosynthesis in chloroplasts of transgenic tobacco over expressing cysteine synthase [O-acetylserine (thiol)-Iyase]. Plant Physiol. 106, 887–895 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig G. F. & Meister A. Gamma-glutamylcysteine synthetase. Interactions of an essential sulfhydryl group. J. Biol. Chem. 259, 3534–3538 (1984). [PubMed] [Google Scholar]
- Habig W. H. & Jakoby W. B. Assays for differentiation of glutathione S-Transferases. Methods Enzymol. 77, 398–405 (1981). [DOI] [PubMed] [Google Scholar]
- Smith I. K., Vierheller T. L. & Thorne C. A. Properties and functions of glutathione reductase in plants. Physiol. Plant. 77, 449–456 (1989). [Google Scholar]
- Hartley-Whitaker J. et al. Phytochelatins Are Involved in Differential Arsenate Tolerance in Holcus lanatus. Plant Physiol. 126, 299–306 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.