AMIGO2 is a novel scaffold protein that regulates PDK1 membrane localization and Akt activation in endothelial cells, and inhibition of the interaction between PDK1–AMIGO2 results in impaired neovascularization, pathological angiogenesis, and tumor angiogenesis.
Abstract
The phosphoinositide 3-kinase–Akt signaling pathway is essential to many biological processes, including cell proliferation, survival, metabolism, and angiogenesis, under pathophysiological conditions. Although 3-phosphoinositide–dependent kinase 1 (PDK1) is a primary activator of Akt at the plasma membrane, the optimal activation mechanism remains unclear. We report that adhesion molecule with IgG-like domain 2 (AMIGO2) is a novel scaffold protein that regulates PDK1 membrane localization and Akt activation. Loss of AMIGO2 in endothelial cells (ECs) led to apoptosis and inhibition of angiogenesis with Akt inactivation. Amino acid residues 465–474 in AMIGO2 directly bind to the PDK1 pleckstrin homology domain. A synthetic peptide containing the AMIGO2 465–474 residues abrogated the AMIGO2–PDK1 interaction and Akt activation. Moreover, it effectively suppressed pathological angiogenesis in murine tumor and oxygen-induced retinopathy models. These results demonstrate that AMIGO2 is an important regulator of the PDK1–Akt pathway in ECs and suggest that interference of the PDK1–AMIGO2 interaction might be a novel pharmaceutical target for designing an Akt pathway inhibitor.
Introduction
The phosphoinositide 3-kinase (PI3K)–3-phosphoinositide–dependent kinase 1 (PDK1)–protein kinase B (Akt) signaling pathway plays vital roles in the transduction of extracellular cues that control multiple aspects of biological processes, including cell growth, survival, protein translation, metabolism, and angiogenesis. Dysregulation of this pathway is also thought to be correlated with the pathogenesis of many human diseases including cancer, as well as metabolic, cardiovascular, and neurological disorders (Raff, 1992; Thompson, 1995; Toker and Newton, 2000; Dimmeler and Zeiher, 2000a; Chang et al., 2010; Portt et al., 2011). Numerous studies illustrate that abnormal activation of the Akt pathway is one of the principal causative factors for the onset and progression of human cancers (Vivanco and Sawyers, 2002). Oncogenic mutations of Akt pathway regulators such as PI3K, PTEN, and PDK1 were commonly detected in many types of cancers in the breast, endometrium, prostate, liver, lung, brain, and skin (Raimondi and Falasca, 2011; Sheppard et al., 2012). The Akt pathway is involved in tumor angiogenesis and the epithelial to mesenchymal transition process, which play essential roles in cancer metastasis and the generation of cancer stem cells (Sheppard et al., 2012; Chang et al., 2013). Moreover, Akt serves as a crucial downstream mediator of angiogenic ligands in endothelial cells (ECs), including VEGF, and coordinates diverse aspects of vascular functionality, including EC survival, proliferation, migration, permeability, vascular tone, and angiogenesis (Liu et al., 2000; Dimmeler and Zeiher, 2000b; Vicent et al., 2003). Thus, the regulators of the PI3K–Akt pathway have become attractive targets for cancer prevention and chemotherapy. Currently, diverse classes of PI3K–Akt pathway inhibitors are being assessed for cancer-related clinical trials.
In general, the PI3K–Akt pathway is triggered by multiple stimuli such as growth factors, cytokines, cell to cell junctions, and the ECM (Bischoff, 1995; Strömblad and Cheresh, 1996; Dimmeler and Zeiher, 2000b; Lamalice et al., 2007). Once PI3K signaling is activated by a stimulus, phosphatidylinositol-(3,4,5)-triphosphate (PIP3), a product of PI3K, recruits the pleckstrin homology (PH) domain of PDK1 to the plasma membrane, which results in activation of membrane-associated Akt at threonine 308 (Datta et al., 1999; Lim et al., 2003; Mora et al., 2004; Primo et al., 2007; Pearce et al., 2010). Alternatively, when PIP3-induced Akt conformation changes occur before threonine 308 phosphorylation by PDK1, conformational changes that permit serine 473 phosphorylation by mammalian target of rapamycin complex 2 can likewise occur. However, Akt phosphorylation at serine 473 also occurred by mammalian target of rapamycin complex 2 independently of PIP3 (King et al., 1997; Huang et al., 2011). In addition, PIP3 binding activates PDK1 by promoting serine 241 autophosphorylation (Gao and Harris, 2006). The mutation of PDK1 at the serine 241 residue causes a significant reduction in PDK1 activity toward Akt (Casamayor et al., 1999). However, the additional mechanisms of PDK1 localization to the plasma membrane after Akt activation require further clarification.
The adhesion molecule with IgG-like domain (AMIGO) family was identified by ordered differential display, which revealed a novel sequence induced by amphoterin, the neurite outgrowth-promoting factor in neurons (Kuja-Panula et al., 2003). AMIGO2 promotes the depolarization-dependent survival of cerebellar granule neurons (Ono et al., 2003). A recent study suggested that AMIGO2 may be potentially involved in vascular development and angiogenesis; the down-regulation of AMIGO2 appears to cause EC death, and overexpression of AMIGO2 seems to protect ECs from death in vitro (Hossain et al., 2011). Furthermore, the stable expression of antisense AMIGO2 in gastric adenocarcinoma cell lines leads to abrogated tumorigenicity (Rabenau et al., 2004). AMIGO2 contains six leucine-rich repeats (LRRs), a single IgG-like domain, a transmembrane domain, and a cytosolic domain (CD; Kuja-Panula et al., 2003; Chen et al., 2006). An algorithm analysis suggested that the CD of AMIGO2 might provide a docking site for signaling pathway intermediates, thereby making it an important participant in signal transduction (Rabenau et al., 2004). AMIGO2 plays important roles in neuronal development and survival as well as tumorigenicity, but the function and precise mechanism of AMIGO2 in ECs have not been determined.
Here, we show that AMIGO2 directly interacts with PDK1 to activate the Akt pathway. Also, using a synthetic peptide, we elucidate the role of AMIGO2 in PDK1–Akt activity–dependent tumor growth and angiogenesis.
Results
AMIGO2 regulates EC survival, migration, and capillary-like network formation
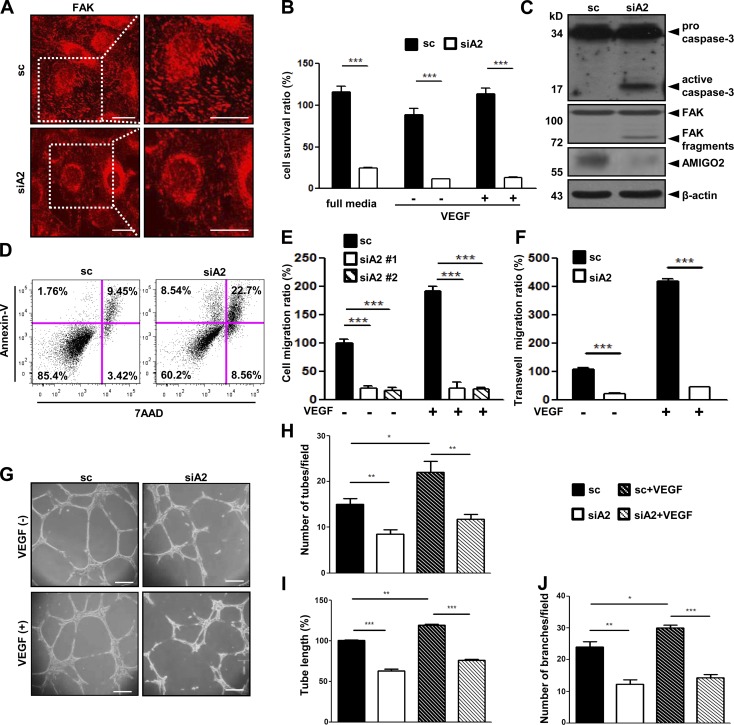
Affymetrix gene chip analysis (available in GEO under accession no. GSE12891; Maeng et al., 2009) revealed that AMIGO2 is highly expressed in ECs. We also found that the up-regulation of AMIGO2 differed in ECs and hemopoietic monocytes (Fig. S1 A) and was specifically expressed in human umbilical vein ECs (HUVECs) among the AMIGO family (Fig. S1 B). Because AMIGO2 affects cell adhesion and survival in other cell types, we examined these functions in ECs. AMIGO2 siRNA–transfected HUVECs (Fig. S1, C and D) displayed reduced adhesion on all three ECM substrates (gelatin, fibronectin, and collagen type I) compared with scrambled siRNA–transfected (control) cells (Fig. S1 E). Endothelial focal adhesions in ECs were examined by immunostaining for FAK. AMIGO2 siRNA–transfected ECs revealed the loss of FAK, which suggests defective cell–ECM adhesion (Fig. 1 A and Fig. S1 F). In the presence or absence of VEGF, the viability of AMIGO2 siRNA–transfected ECs was decreased (Fig. 1 B). Over time, AMIGO2 siRNA–transfected ECs exhibited F-actin depolymerization and an apoptotic morphology, which are morphological markers of apoptosis, whereas control siRNA–treated ECs displayed a cobblestone shape (Fig. S1 G). Active caspase-3 and FAK fragments, which are markers of cell apoptosis, were detected in AMIGO2 knockdown cells (Fig. 1 C). Cell death was identified by TUNEL staining, which revealed that TUNEL-positive nuclei were merged with AMIGO2 knockdown ECs (Fig. S1 I). Furthermore, flow cytometry analysis after annexin V staining revealed increased levels of apoptosis in AMIGO2 knockdown ECs (Fig. 1 D). These results indicated that AMIGO2 regulates EC–ECM interactions and cell viability. Because cell–ECM adhesion is crucial for cell migration and the formation of tube-like structures (Ridley et al., 2003; Mammoto et al., 2009), we further examined the angiogenic function of AMIGO2 in ECs. Both wound healing and chemotactic motility were blocked in AMIGO2 siRNA–treated ECs regardless of VEGF induction (Fig. 1, E and F; and Fig. S1 H). Furthermore, AMIGO2 siRNA–transfected ECs displayed severely impaired tube formation and appeared to undergo anoikis (Fig. 1, G–J; Fig. S1, J–L; and Video 1). Conversely, overexpression of AMIGO2 in ECs revealed increased branching and robust lumen structures (Fig. S1, M–O). These results indicate that AMIGO2 could modulate the angiogenic function of ECs.
Figure 1.
AMIGO2 regulates EC viability and angiogenesis. (A–J) HUVECs were pretransfected with scrambled siRNA and AMIGO2-specific siRNA and harvested for analysis 48 h after transfection. (A) FAK immunostaining was performed in parallel. Images on the right are enlargements of the boxed regions on the left. Bars, 20 µm. (B) Cell viability of AMIGO2 siRNA–transfected HUVECs in complete media and under conditions of serum-free starvation with or without VEGF (n = 6). Data were collected from independent experiments and analyzed using a two-tailed unpaired t test. Data are means ± SD. (C) Expression of active caspase-3 and FAK fragments in AMIGO2 siRNA–transfected ECs. (D) Different stages of apoptosis were detected by phycoerythrin-conjugated annexin V flow cytometry and PerCP–7-AAD staining. Quadrant gates are based on unstained, annexin V, or 7-AAD alone. Quadrant regions show the percentage of living cells (7-AAD−/annexin V−), early apoptotic cells (7-AAD−/annexin V+), late apoptotic cells (7-AAD+/annexin V+), and necrotic cells (7-AAD+/annexin V−). The data are representative of three experiments conducted by using different samples. (E and F) Wound healing migration and gelatin-coated Transwell migrations with or without VEGF were performed. Two types of siA2 were evaluated for wound healing migration. n = 3 and n = 6. (G) Matrigel-induced tube formation by AMIGO2 siRNA–transfected HUVECs was assessed. (H and J) Mean numbers of tubule branch points and of tubes per field in randomly selected images were quantified. (I) Tube length is presented as the percentage of total tube length per field versus that of untreated control cells. (H–J) Data were collected from independent experiments and analyzed using a two-tailed unpaired t test. Data are means ± SD. n = 4. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. sc, scrambled siRNA; siA2, AMIGO2 siRNA.
Reduction of AMIGO2 exhibits angiogenic defects in vivo
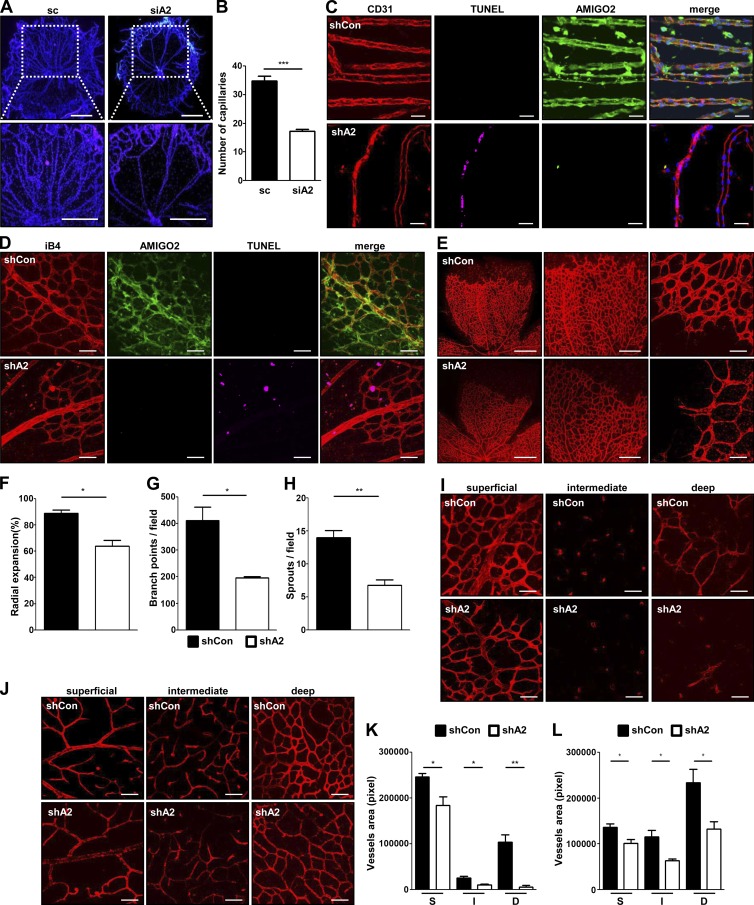
To determine the physiological relevance of AMIGO2 in vivo, we examined expression levels of Amigo family members in the adult mouse. Amigo2 was mainly expressed in the eye, retina, brain, lung, and lymph node (Fig. S2 A). Because the level of Amigo2 expression in the eye was elevated, we investigated the expression of Amigo2 in the hyaloid and retinal vessel systems. In hyaloid vessels, AMIGO2 was detectable from postnatal day 3.5 (P3.5), with the highest expression at P5.5, after which expression decreased (Fig. S2, B and C). In retinal vessels, AMIGO2 was detected at P5.5 and consistently increased in retinal vessels over time (Fig. 2 D and Fig. S2, D, H, and J). To define the function of AMIGO2 in vivo, P3.5 mice were injected with Amigo2-specific or control siRNA and shRNA, and experiments were performed at P5.5, P8.5, and P15.5. Western blotting indicated that Amigo2 siRNA– and shRNA-injected mice exhibited significant reductions in the expression of AMIGO2 in hyaloid vessels and retinas at P5.5 (Fig. 2 D; Fig. 3, C and D; and Fig. S2, F–K). At P5.5, Amigo2 knockdown mice exhibited significant changes in hyaloid vessel structure (Fig. 2, A–C) and reductions in retinal vessel growth, branch points, and vascular sprouts (Fig. 2, E–H). Furthermore, hyaloid and retinal vessels from Amigo2 knockdown mice exhibited increased CD31-positive cells merged with TUNEL-positive cells compared with those from control shRNA mice (Fig. 2, C and D). Additionally, stable Amigo2 knockdown mice at P8.5 and 15.5 revealed reductions in superficial, intermediate, and deep layers of vessels compared with those from control shRNA mice (Fig. 2, I–L; and Fig. S2, H and J). These data demonstrate that AMIGO2 regulates survival, growth, and development of hyaloid and retinal vasculature in vivo.
Figure 2.
AMIGO2 is required for angiogenesis in vivo. P3.5 mice were injected with Amigo2-specific or control siRNA and shRNA, and experiments were performed at P5.5, P8.5, and P15.5. (A) Impairment of hyaloid vessel structures in Amigo2 siRNA–injected mice. Bars, 200 µm. (B) Capillary quantification. n = 5. (C) Hyaloid vessel staining with CD31, AMIGO2, and TUNEL was performed in Amigo2 shRNA–injected P5.5 mice. (D and E) Visualization of blood vessels by iB4 staining with AMIGO2 and TUNEL staining of Amigo2-depleted retinas from P5.5 mice. Bars: (D) 50 µm; (E, left to right) 500 µm, 200 µm, and 50 µm. (E) Middle and right images are enlargements of the left image. (F–H) Radial expansions, retinal vascular branch points, and sprouts per field were quantified. n = 5. (I–L) Immunostaining of blood vessels by iB4-staining of Amigo2 shRNA–injected P8.5 retinas (I and K) and P15.5 retinas (J and L). Superficial (S), intermediate (I), and deep (D) layers of retinal vessels were taken and quantified. Bars, 50 µm. n = 5. Data were collected from mice (n = 5–6) and analyzed using a two-tailed unpaired t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. Data are means ± SD. shCon, control shRNA; shA2, Amigo2 shRNA.
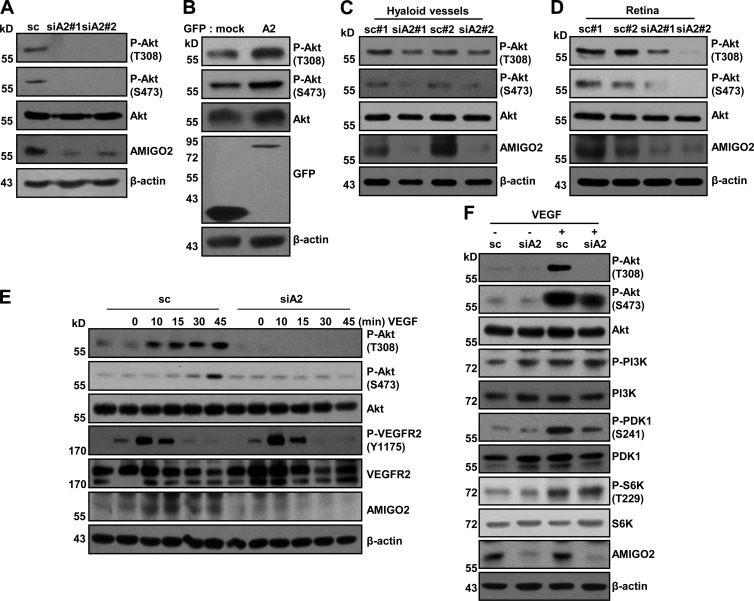
Figure 3.
AMIGO2 modulates the PDK1–Akt pathway. (A) The endogenous effect of AMIGO2 knockdown on Akt signaling in HUVECs. Two types of siA2 were evaluated. (B) Endogenous effects of AMIGO overexpression on Akt signaling activation in HUVECs. (C and D) Akt phosphorylation in hyaloid vessels (C) and retinas (D) from Amigo2-depleted mice was evaluated. The data are representative of six mice. (E) Temporal effects of AMIGO2 knockdown on VEGF-induced signaling pathways (20 ng/ml) in HUVECs. (F) VEGF-induced PI3K–PDK1–Akt signaling was evaluated in AMIGO2 siRNA–transfected HUVECs.
AMIGO2 controls Akt activation in vitro and in vivo
EC survival is mainly affected by Akt signaling, which is regulated by VEGF and anchorage-dependent signaling (Fujio and Walsh, 1999). We examined whether Akt phosphorylation was affected in ECs that were AMIGO2 deficient or that overexpressed AMIGO2. Akt phosphorylation (threonine 308 and serine 473) was reduced in AMIGO2 siRNA–transfected ECs incubated in normal growth media (Fig. 3 A), whereas overexpression of AMIGO2 induced Akt activation (Fig. 3 B). It is recognized that microvascular patterning is controlled by Akt signaling (Sun et al., 2005; Alvarez et al., 2009), and thus we examined Akt activation in mice. Interestingly, depletion of Amigo2 in mice using in vivo siRNA revealed decreased Akt phosphorylation in both hyaloid vessels and retinas (Fig. 3, C and D). To investigate whether VEGF-induced survival signaling was affected in AMIGO2-deficient ECs, AMIGO2 knockdown ECs were treated with VEGF at different time points (Fig. 3 E). Phosphorylation of VEGF receptor 2 (VEGFR2) at tyrosine 1175 was unaffected, but Akt activation (threonine 308 and serine 473) was inhibited in AMIGO2 knockdown ECs treated with VEGF (Fig. 3 E). To narrow down the effect of AMIGO2 in the PI3K–Akt signaling pathway, we further examined the activation of PI3K and PDK1, which are upstream molecules of Akt, after VEGF stimulation. AMIGO2 knockdown in ECs did not affect the phosphorylation of PI3K, but phosphorylation of PDK1 was inhibited (Fig. 3 F). Interestingly, the activation of p70 ribosomal S6 kinase (S6K), which is a PDK1 substrate, was not affected by AMIGO2 knockdown in ECs (Fig. 3 F). These data suggest that AMIGO2 modulates EC survival and angiogenesis by regulating the PDK1–Akt signaling pathway.
AMIGO2 interacts with PDK1
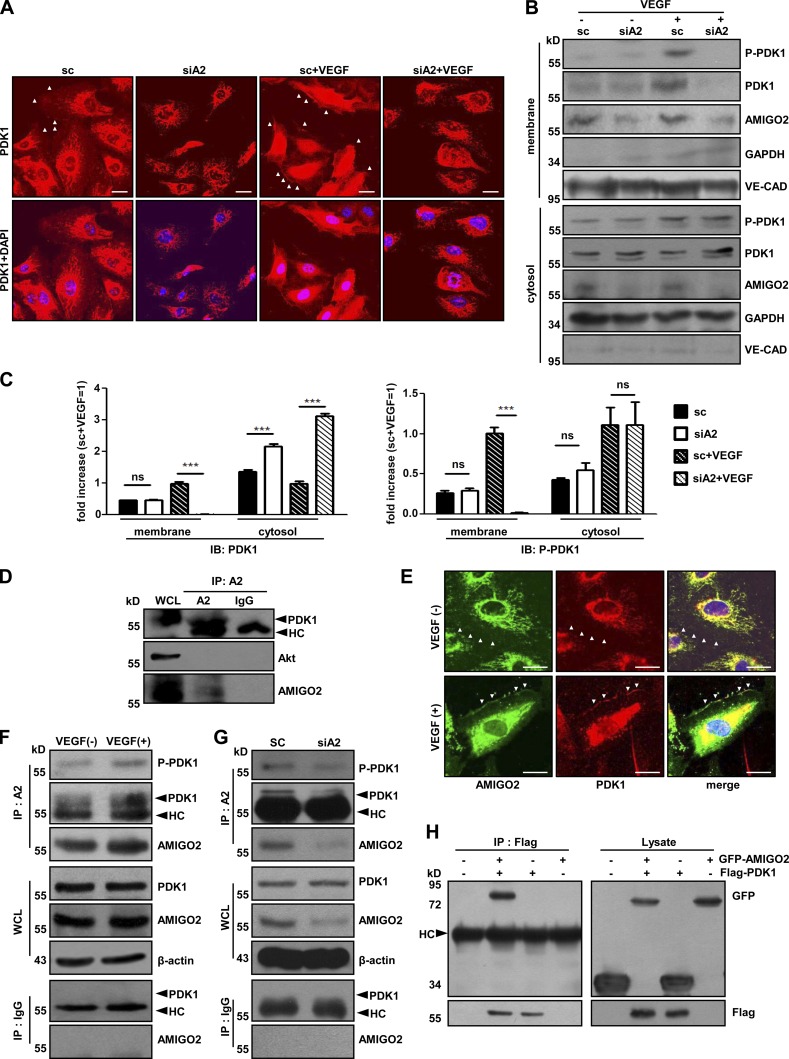
To determine how AMIGO2 regulates the PDK1-mediated Akt pathway, we examined the cellular localization of PDK1 in AMIGO2 knockdown ECs. To examine the translocation of PDK1 in ECs, control and AMIGO2-knockdown ECs were treated with or without VEGF and immunolabeled with anti-PDK1. Plasma membrane translocation of PDK1 was reduced in AMIGO2 knockdown ECs compared to control ECs upon VEGF stimulation (Fig. 4, A–C; and Fig. S3 A). Furthermore, subcellular localization analysis revealed that plasma membrane translocation of PDK1 in AMIGO2 knockdown ECs was impaired, predominantly remaining in the cytosol upon VEGF stimulation (Fig. 4, B and C). Plasma membrane localization of phosphorylated PDK1 (at serine 241) in AMIGO2-depleted ECs was much lower than in control ECs (Fig. 4, B and C). It is well known that PDK1 activates Akt, S6K, and serum/glucocorticoid-regulated kinase (SGK). Phosphorylation of S6K (at threonine 229) and SGK (at threonine 256) occurred in the cytoplasm. Interestingly, phosphorylation of S6K and SGK was unaffected in VEGF-stimulated AMIGO2 knockdown ECs (Fig. S3 B). These data suggest that AMIGO2 may control PDK1 localization to the plasma membrane, thus specifically mediating Akt activation.
Figure 4.
AMIGO2 regulates subcellular localization and interacts with PDK1. (A) PDK1 immunostaining was performed in AMIGO2-inhibited HUVECs in the presence or absence of VEGF. Cells were starved for 6 h and treated with VEGF. White arrowheads indicate membrane localization of PDK1. Bars, 20 µm. (B) Subcellular localization analysis was performed by cell fractionation in VEGF-treated, AMIGO2-deficient HUVECs. (C) Quantification of Western blots. Subcellular localization analysis was performed by cell fractionation with or without VEGF in control siRNA and AMIGO2-deficient HUVECs. ***, P < 0.0001. Data are means ± SD. (D) Endogenous AMIGO2 interacts with PDK1 in HUVECs. AMIGO2 was immunoprecipitated and blotted with anti-PDK1 and -Akt antibodies. (E) Colocalization of PDK1 and AMIGO2 in the plasma membrane and cytosol of HUVECs in the presence of VEGF. HUVECs were starved for 6 h and stimulated with or without VEGF. White arrowheads indicate membrane edge areas of HUVECs. Bars, 20 µm. (F) HUVECs were starved for 6 h, stimulated with VEGF for 4 h, and immunoprecipitated with an AMIGO2 antibody and blotted with endogenous PDK1. (G) AMIGO2-deficient HUVECs were immunoprecipitated with an AMIGO2 antibody and blotted with an anti-PDK1 antibody. (H) Flag-PDK1 was immunoprecipitated and blotted with an anti-GFP antibody tagged to AMIGO2. A2, AMIGO2; HC, heavy chain; IB, immunoblotting; IgG, normal IgG; IP, immunoprecipitation; ns, not significant; WCL, whole cell lysate.
We next investigated whether PDK1 localization is involved in the physical interaction of AMIGO2 and PDK1. PDK1, but not Akt, was detected in endogenous AMIGO2 immunoprecipitates from HUVECs (Fig. 4 D). Colocalization of AMIGO2 and PDK1 was observed at the plasma membrane and the cytoplasm in the presence of VEGF (Fig. 4 E). Furthermore, PDK1–AMIGO2 association was increased in VEGF-stimulated HUVECs (Fig. 4 F). PDK1 was almost undetectable in AMIGO2 immunoprecipitates from AMIGO2 knockdown ECs (Fig. 4 G). To confirm the interactions between AMIGO2 and PDK1, GFP-AMIGO2 and Flag-PDK1 were cotransfected into HEK293T cells. GFP-AMIGO2 was coimmunoprecipitated with Flag-PDK1, but not from cells expressing either AMIGO2 or PDK1 alone (Fig. 4 H). Collectively, these results indicate that AMIGO2 associates with PDK1.
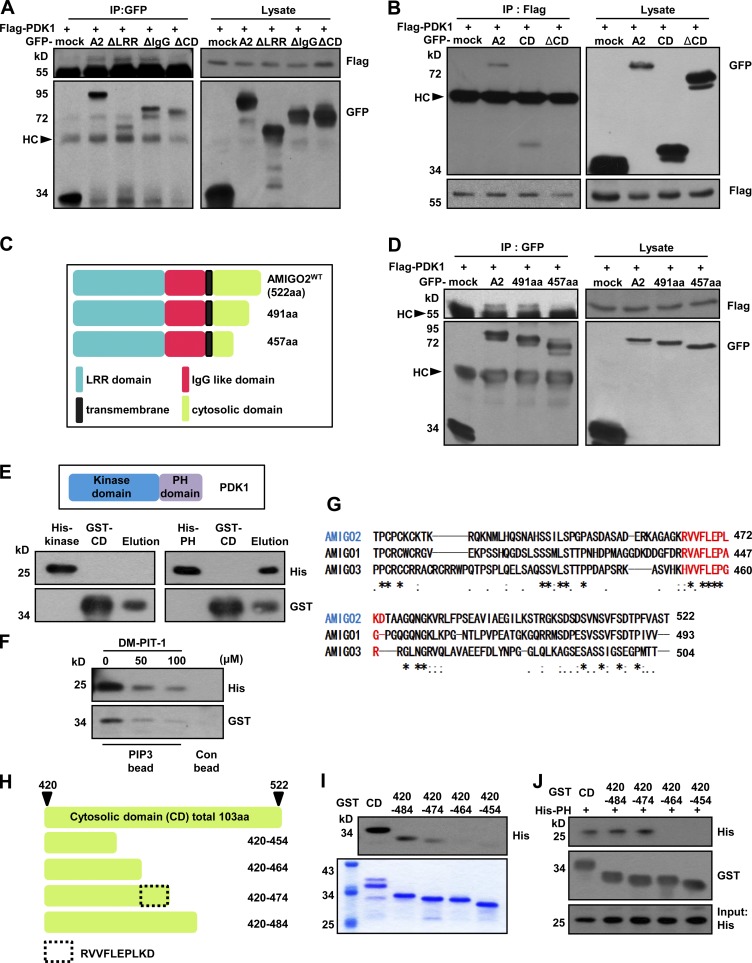
The AMIGO2 CD interacts with the PDK1 PH domain
To explore the binding domain of AMIGO2 and PDK1 further, HEK293T cells were cotransfected with Flag-PDK1 and one of the following AMIGO2 deletion constructs: AMIGO2WT, AMIGO2ΔLRR, AMIGO2ΔIgG, or AMIGO2ΔCD (Fig. S3 C). Interestingly, GFP-AMIGO2ΔCD did not coimmunoprecipitate with Flag-PDK1 (Fig. 5 A). To verify the interaction of the CD and PDK1, HEK293T cells were cotransfected with Flag-PDK1 and AMIGO2WT, AMIGO2CD (which only contains the CD domain), or AMIGO2ΔCD. AMIGO2CD and AMIGO2WT were detected in Flag-PDK1 immunoprecipitated fractions, but not AMIGO2ΔCD (Fig. 5 B). Next, we investigated the segment of the CD of AMIGO2 that interacts with PDK1. HEK293T cells were cotransfected with Flag-PDK1 and one of the AMIGO2 and truncated CD constructs (Fig. 5 C). GFP-AMIGO2WT and AMIGO2491aa coimmunoprecipitated with Flag-PDK1, but not AMIGO2457aa (Fig. 5 D). Because the PH domain of PDK1 binds to PIP3 and activates Akt signaling (Bayascas et al., 2008), we examined the direct binding of the AMIGO2 CD (AMIGO2CD) and the PDK1 PH domain (PDK1PH). As anticipated, His-PDK1PH proteins, but not His-PDK1kinase, could bind to GST-AMIGO2CD under cell-free conditions (Fig. 5 E and Fig. S3 D). The negative control did not bind to GST-AMIGO2CD (Fig. S3 E). To confirm that the PIP3-mediated interaction between AMIGO2CD and PDK1PH could be blocked by a PIP3 antagonist, we examined the binding of AMIGO2CD and PDK1PH with PIP3-coated beads and DM-PIT-1, which is a PIP3 antagonist that competes with PIP3 for binding to PH domains of PDK1 and effectively blocks the PI3K–PDK1–Akt signaling pathway. The binding of PDK1PH and AMIGO2CD with DM-PIT-1 was observed to decrease in a dose-dependent manner (Fig. 5 F). Next, we examined the possible interaction between the Akt PH domain and AMIGO2CD. The results showed that the Akt PH domain did not bind to AMIGO2CD (Fig. S3 F). Analysis of the AMIGO family CD revealed highly conserved sequences between 457 and 491 aa (Fig. 5 G). To clarify the regions that directly interact with the PH domain of PDK1, we generated GST-tagged truncated CD mutants (Fig. 5 H and Fig. S3 G). The purified GST-tagged AMIGO2CD 420–484 and 420–474 aa directly interacted with purified His-PDK1PH, but not GST-tagged 420–464 and 420–454 aa (Fig. 5, I and J). Interestingly, the peptide sequences between 464 and 475 are RVVFLEPLKD, which is highly conserved among the AMIGO family (Fig. 5 G). Thus, we determined that RVVFLEPLKD, the C-terminal amino acids of AMIGO2, directly bind to the PH domain of PDK1 to activate the PDK1–Akt pathway.
Figure 5.
The PH domain of PDK1 binds to the CD of AMIGO2. (A and B) The CD of AMIGO2 interacts with PDK1. HEK293 cells were transfected with Flag-PDK1 and GFP-tagged AMIGO2 or domain deletion mutants. Lysates were immunoprecipitated with GFP (A) or Flag (B) antibodies and blotted with the GFP or Flag antibody. PDK1 was immunoprecipitated with the Flag antibody and detected with the anti-GFP antibody. A2, AMIGO2WT; ΔLRR, AMIGO2ΔLRR; ΔIgG, AMIGO2ΔIgG; ΔCD, AMIGO2ΔCD; CD, CD domain of AMIGO2. (C) Schematic of 491 aa and 457 aa of the AMIGO2 constructs. (D) PDK1 associates with the region between 491 aa and 457 aa of AMIGO2. HEK293 cells were transfected with Flag-PDK1– and GFP-tagged AMIGO2 or domain deletion mutants. Lysates were immunoprecipitated with GFP antibody and blotted with the Flag and GFP antibodies. (E) Each His-tagged PDK1kinase and His-tagged PDK1PH was incubated with purified GST-AMIGO2CD. Mixtures of His-tagged PDK1kinase or PDK1PH proteins and GST-AMIGO2CD proteins were pulled down with GST resin and analyzed by Western blotting using anti-His and -GST antibodies. His-kinase, His-PDK1kinase; His-PH, His-PDK1PH; GST-CD, GST-AMIGO2CD. (F) Mixtures of His-PDK1PH and GST-CD proteins were incubated with PIP3-coated or control beads and treated with DM-PIT-1. The PIP3-bound proteins were analyzed with His and GST antibodies. (G) Multiple sequence alignments of the AMIGO family CD domains were performed. An asterisk indicates that the alignment contains identical amino acid residues in all sequences (or identical bases if DNA sequences are aligned); a colon indicates that the alignment contains different but highly conserved (very similar) amino acids; a period indicates that the alignment contains different amino acids that are somewhat similar; and a blank space indicates that the alignment contains dissimilar amino acids or gaps (or different bases if DNA sequences are aligned). (H) Diagram showing the protein sequences of cytosolic-truncated mutants. The total length of the cytosolic tail is 103 aa. (I) Far-western analysis was performed for each truncated mutant. Coomassie blue staining revealed purified GST-tagged truncated mutants that were previously loaded for far-western analysis. (J) Mixtures of purified GST-tagged truncated mutants and His-tagged PH domain proteins were pulled down with GST beads and eluted, and the Western blot was performed using His and GST antibodies. HC, heavy chain; IP, immunoprecipitation.
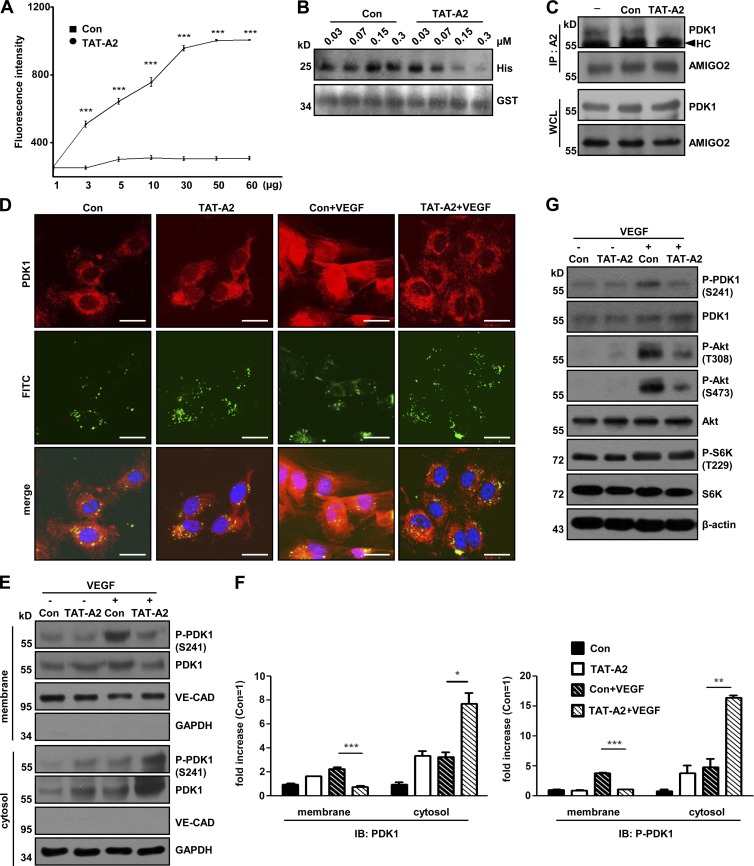
Protein transduction domain (PTD)–A2, a small peptide from the CD of AMIGO2, controls EC survival, Akt activation, and angiogenesis
To evaluate whether the AMIGO2 CD regulates cell viability, HUVECs were transfected with AMIGO2CD, and cell apoptosis was examined. CD-transfected ECs revealed higher apoptotic rates than control cells in both the presence and absence of VEGF (Fig. S3, H and I). To provide further evidence that the PH domain binding site of AMIGO2 regulates cell viability, we generated a small peptide containing RVVFLEPLKD conjugated with FITC and cell-penetrating peptide (PTD-A2; TAT sequence for general cell penetration and NGR sequence for vessel-specific penetration). PTD-A2 bound to the PDK1 PH domain (Fig. 6 A and Fig. S4 A) and inhibited the interaction of the CD and PH domains (Fig. 6 B and Fig. S4 B). Endogenous PDK1 was detected in immunoprecipitated fractions from control-treated HUVECs, but PDK1 was barely detected in PTD-A2–treated HUVECs (Fig. 6 C and Fig. S4 C), which indicated that PTD-A2 might compete with endogenous AMIGO2 for PDK1 binding. Interestingly, PTD-A2–treated HUVECs exhibited a dose- and time-dependent inhibitory effect on cell viability compared with control peptides (Fig. S4, D and E). PTD-A2 penetrated to HUVECs within 40 min and colocalized with PDK1 in the cytoplasm under our experimental conditions (Fig. S4 F). To test the hypothesis that PTD-A2 blocks PDK1 localization to the plasma membrane, we verified the alteration of PDK1 localization after VEGF treatment by using PDK1 staining and cell fractionation assays. PDK1 was unable to localize to the plasma membrane after VEGF treatment in PTD-A2–treated HUVECs compared with that in Con-treated ECs; moreover, the phosphorylation of PDK1 was located in the cytoplasm (Fig. 6, D–F). Also, PTD-A2–treated HUVECs reduced the activity of both Akt and PDK1, but not the phosphorylation of S6K (Fig. 6 G).
Figure 6.
PTD-A2, an AMIGO2 competitive peptide, blocks PDK1 translocation and Akt activation. (A) The TAT-A2 binding assay was performed. PH domain proteins (0–60 µg) were incubated with 1-µM FITC–TAT-A2 and detected by fluorescence. Data were collected from independent experiments and analyzed using a two-tailed unpaired t test. n = 4. (B) Peptide competition assay with Con or TAT-A2 and CD to PH domains. His-tagged PDK1PH was incubated with purified GST-AMIGO2CD and Con or TAT-A2 in a dose-dependent manner. His-tagged PH domain proteins were pulled down with GST resins, eluted, and analyzed by Western blotting. (C) Con- and TAT-A2– treated HUVECs were immunoprecipitated with an AMIGO2 antibody and blotted with anti-PDK1 and AMIGO2 antibodies. (D) Localization of PDK1 in the plasma membrane and cytosol of HUVECs in the presence of VEGF after Con or TAT-A2 treatment. Bars, 20 µm. (E) Subcellular localization analysis was performed by cell fractionation in VEGF-treated, TAT-A2–treated HUVECs. (F) Quantification of Western blots. Subcellular localization analysis was performed by cell fractionation with or without VEGF in control and TAT-A2 HUVECs. HUVECs were affected by both PTD-A2 peptides. (G) Effects of TAT-A2 on VEGF-induced Akt signaling in HUVECs. Con, control peptide; TAT-A2, TAT-RVVFLEPLKD peptide. PTD-A2 means both TAT-A2 and NGR-A2. Data are means ± SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. HC, heavy chain; IB, immunoblotting; IP, immunoprecipitation; WCL, whole cell lysate.
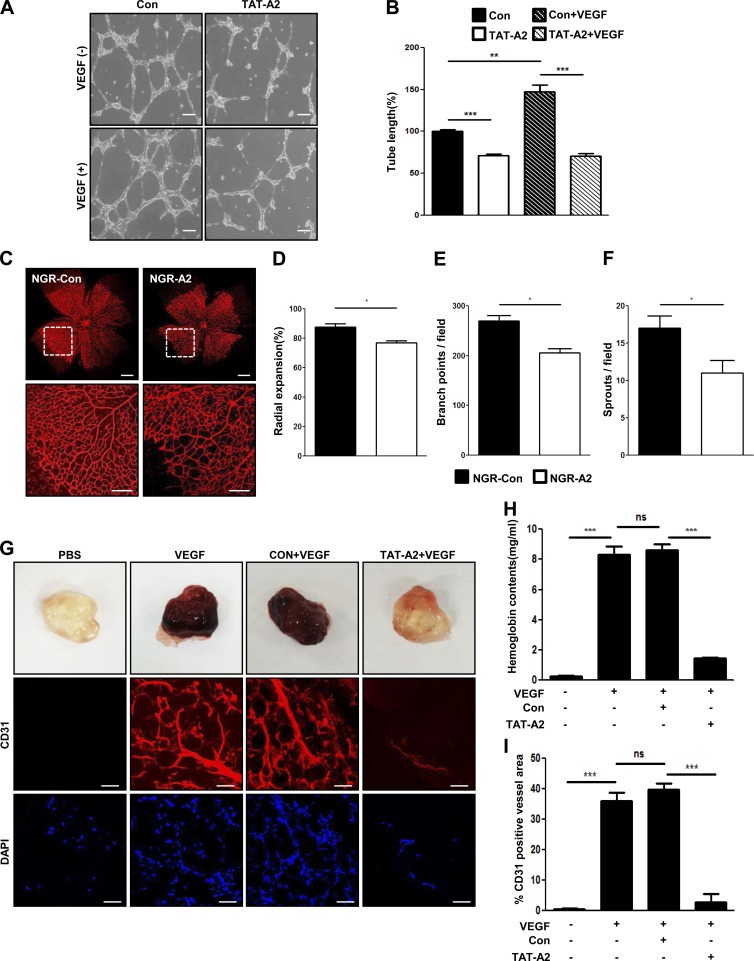
Because PTD-A2–treated ECs regulate cell viability, we speculated that PTD-A2 might play an important role in angiogenesis. As expected, PTD-A2–treated ECs revealed severely impaired tube-like structures in both the presence and absence of VEGF compared with Con-treated ECs (Fig. 7, A and B). To evaluate the angiogenic activity of PTD-A2 in vivo, P3.5 mice were injected with either Con or PTD-A2. PTD-A2–injected retinas at P5.5 exhibited decreased retinal vessel outgrowth and branch points (Fig. 7, C–F). We further examined the angiogenic activity of PTD-A2 in vivo using mouse Matrigel plug assays. Compared with Matrigel plugs containing VEGF or control peptide with VEGF, Matrigel plugs containing PTD-A2 with VEGF revealed a dramatic inhibition of neovascularization, which was confirmed by hemoglobin contents and histological staining with CD31 (Fig. 7, G–I; and Fig. S4 G). Moreover, CD31-positive cells containing PTD-A2 with VEGF merged with TUNEL-positive cells (Fig. S4, H and I). Collectively, the results indicate that PTD-A2 affects cell viability and angiogenesis by blocking the localization and activation of PDK1.
Figure 7.
PTD-A2 blocks retinal and Matrigel-induced angiogenesis. (A and B) TAT-A2 Matrigel-induced tube formation was assessed in HUVECs. Bars, 200 µm. Tube lengths are presented as the percentage of total tube length per field versus control cells. n = 5. (C) Visualization of blood vessels by iB4 staining of NGR-A2–injected retinas from P5.5 mice. Bars: (top) 500 µm; (bottom) 200 µm. Bottom images are enlargements of the boxed regions in the top panel. (D–F) Radial expansions (D), retinal vascular branch points (E), and sprouts per field (F) were quantified. n = 4–5. (G) Male C57BL/6 mice (n = 6 per group) received subcutaneous injections of Matrigel containing PBS, 200 ng VEGF, 25-µM Con with VEGF, or 25-µM TAT-A2 with VEGF. Matrigel plugs were removed 5 d after implantation, fixed, sectioned, and stained for immunohistochemistry with anti-CD31 antibody (red) for the identification of endothelial vessels. DAPI (blue) was used for nuclei labeling, and visualization was performed using confocal microscopy. Bars, 50 µm. (H) Quantification of neovessel formation by measuring hemoglobin in the Matrigel. (I) Quantitative assessment of CD31-positive ECs. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ns, not significant. Data are means ± SD.
PTD-A2 reveals an antiangiogenic activity in pathological angiogenesis and tumor growth
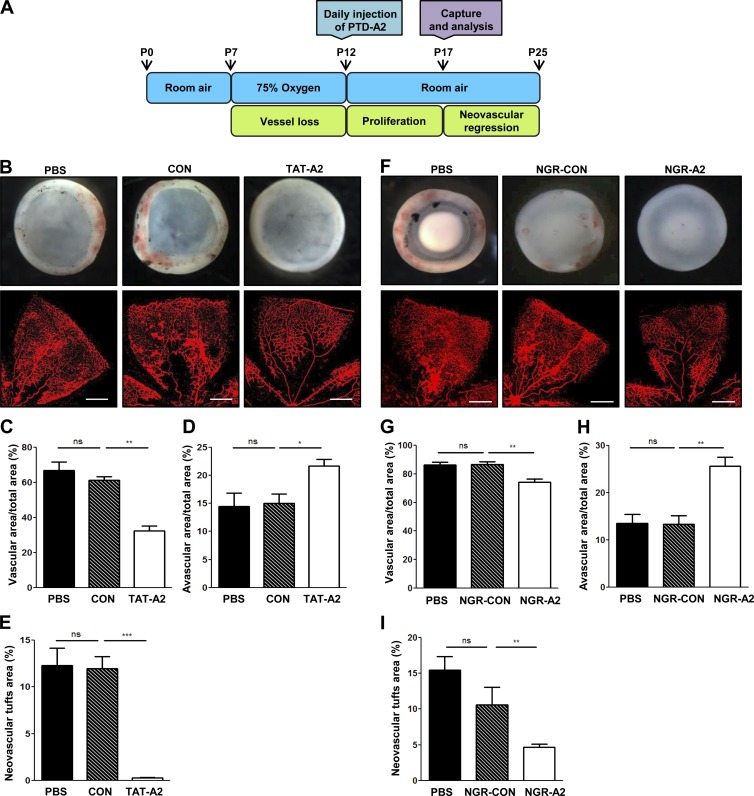
The model of oxygen-induced retinopathy (OIR) has been used extensively in retinopathy of prematurity, proliferative diabetic retinopathy, and for evaluating the efficacy of antiangiogenic molecules (Connor et al., 2009). To determine the antiangiogenic activity of PTD-A2 in pathological conditions, P7 mouse pups were subjected to hypoxia for 5 d. P12 mice were returned to normal oxygen levels and were administered an intraperitoneal injection of PBS, Con, or PTD-A2 (Fig. 8 A). Interestingly, compared with PBS- and Con-injected mice, mice injected with PTD-A2 (both TAT-A2 and NGR-A2) exhibited increased avascular areas and significantly reduced retinal hemorrhage, vascular areas, and tuft formation (Fig. 8, B–I).
Figure 8.
PTD-A2 contributes to retinal neovascularization in an OIR model. (A) Scheme and schedule of PTD-A2 treatment in the mouse OIR model. (B and F) Stereomicroscopy and iB4 staining of whole-mount retinas from Con- and TAT-A2– or NGR-Con– and NGR-A2–injected mice (n = 7–8 mice per group; 8 mg/kg of peptide administered every day during P12–P16) with ischemic retinopathy at P17, and quantification of vascular (C and G) and avascular areas (D and H) and the neovascular tufts (E and I). (B and F) Top images are whole-mount retinas and indicate retinal hemorrhage. The bottom images are iB4-staining of flat-mount retinas and indicate the avascular area and neovascular tuft formation. Bars, 200 µm. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ns, not significant. Data are means ± SD.
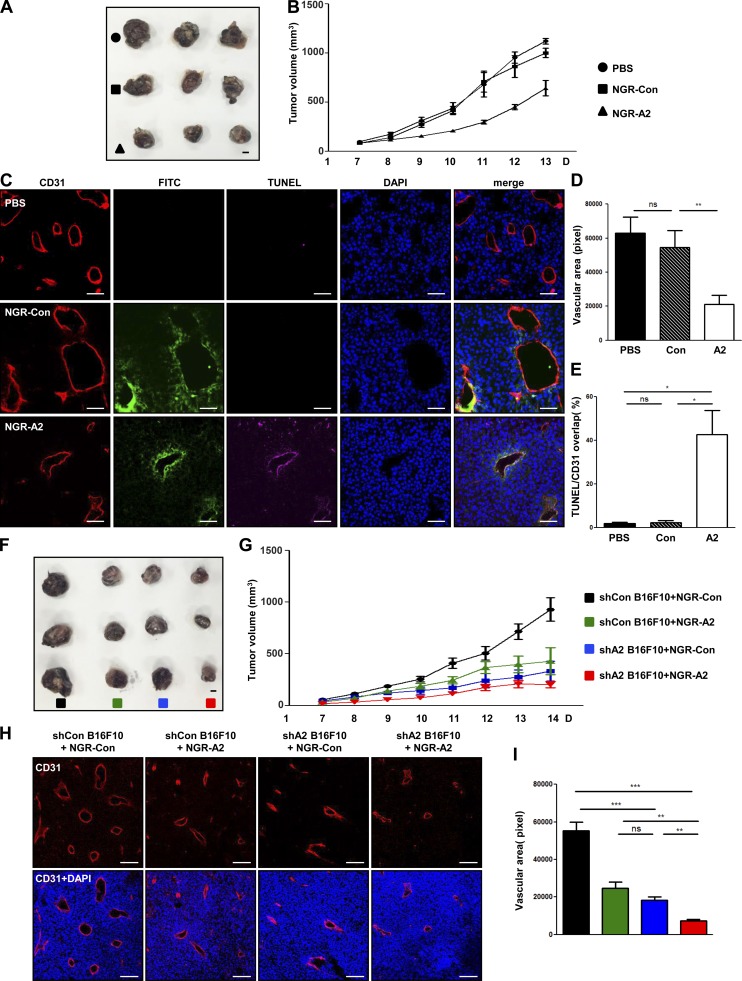
To examine the antitumor angiogenic activity of PTD-A2, we first evaluated the expression of AMIGO2 in B16F10 melanoma tumors and verified that AMIGO2 was expressed in B16F10 melanoma tumor vessels and cells (Fig. S5 A). Furthermore, B16F10 melanoma cell viability was decreased in PTD-A2–treated B16F10 melanoma cells (Fig. S5 B). Next, we administered intratumoral injections of PBS, Con, or PTD-A2 to B16F10 melanoma tumor–bearing mice every other day when tumors reached 100 mm3. Intriguingly, compared with PBS- or Con-injected mice, both TAT-A2 and NGR-A2 peptide–injected mice exhibited decreased tumor growth (Fig. 9, A and B; and Fig. S5, C and D) and vessel density (Fig. 9, C and D; and Fig. S5, E and F) and increased apoptotic regions in CD31-positive areas (Fig. 9, C and E). To further verify the effect of AMIGO2 in B16F10 melanoma cells with PTD-A2, we first stably transfected Amigo2-specific shRNA into these cells (Fig. S5 G). Next, control shRNA and Amigo2-depleted B16F10 melanoma cells were injected into mice, and Con or PTD-A2 was administrated by intratumoral injections every other day to both control shRNA and Amigo2-depleted B16F10 melanoma tumor–bearing mice. Amigo2-depleted B16F10 melanoma tumors exhibited decreased tumor volume and vessel density compared with control shRNA melanoma tumors (Fig. 9, F–I). Intriguingly, Amigo2-depleted B16F10 melanoma tumors with PTD-A2 showed a dramatic reduction in tumor volume and vessel density (Fig. 9, F–I). These results demonstrate that treatment with PTD-A2 might be essential for blocking tumor growth and pathological angiogenesis.
Figure 9.
Treatment with PTD-A2 inhibits tumor growth and tumor angiogenesis. (A–E) Treatment with FITC–NGR-A2 inhibits tumor growth, neovessel formation, and tumor cell death in B16F10 tumors. (A) Photograph of B16F10 melanoma tumors resected after 13 d. Bar, 5 mm. (B) Comparison of tumor volume (n = 7 mice per group; 4 mg/kg NGR-Con or NGR-A2 was administered every other day). (C–E) Images (C) and quantification (D) of CD31-positive blood vessels (red) and TUNEL-positive cells (pink) over CD31-positive blood vessels (E) in B16F10 melanoma tumors. FITC, green; DAPI, blue. Bars, 50 µm. (F–I) Amigo2 shRNA–transfected B16F10 tumors with NGR-A2 display dramatically inhibited tumor growth and tumor vessel formation. (F) Image of Amigo2 shRNA–transfected B16F10 melanoma tumors resected after 14 d. Bar, 5 mm. (G) Comparison of tumor volumes (n = 7–8 mice per group; 4 mg/kg NGR-Con or NGR-A2 was administered every other day). (H and I) Images (H) and quantification (I) of CD31-positive blood vessels (red). (B and G) Data were analyzed using repeated measures of two-way analysis of variance. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. ns, not significant. Data are means ± SD.
Discussion
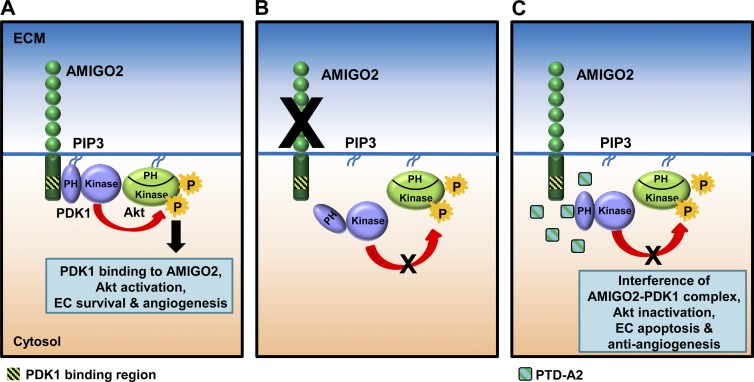
In this study, we demonstrate that AMIGO2 is an important regulator of the PDK1–Akt signaling pathway, which in turn modulates cell survival, adhesion, migration, and angiogenesis (Fig. 10). We provide three lines of evidence, including indications that the loss of AMIGO2 attenuated the phosphorylation of PDK1 and Akt as well as EC survival and adhesion ability. We also demonstrated that the direct binding of AMIGO2 to the PH domain of PDK1 enhanced the activation of PDK1 and Akt in the plasma membrane and that PTD-A2, a specific peptide in the C-terminal region of AMIGO2, effectively abrogated phosphorylation of PDK1 and Akt, cell survival, OIR-induced angiogenesis, tumor angiogenesis, and tumor growth.
Figure 10.
The AMIGO2–PDK1–Akt signaling pathway. (A) This model proposes that the direct binding of AMIGO2 to the PDK1 PH domain enhanced the activation of PDK1 and Akt in the plasma membrane, which resulted in increased EC viability, adhesion, migration, and angiogenesis. (B) Loss of AMIGO2 attenuated the phosphorylation of PDK1 and Akt and EC survival and adhesion. (C) PTD-A2, a specific peptide in the C-terminal region of AMIGO2, effectively abrogated the phosphorylation of PDK1 and Akt, cell survival, OIR-induced angiogenesis, tumor angiogenesis, and tumor growth. P, phosphorylation.
PDK1 possesses a PH domain and a catalytic domain, which contains the PDK1-interacting fragment binding pocket and activates a group of kinases including Akt, S6K, and SGK (Pullen et al., 1998; Pearce et al., 2010). PDK1 is constitutively active, owing to the autophosphorylation ability of the serine 241 residue (Casamayor et al., 1999; Gao and Harris, 2006; Pearce et al., 2010). Given that constitutive PDK1 activity was detected in cellular compartments including the nucleus, cytoplasm, and the plasma membrane (Gao and Harris, 2006), PDK1 activity appears to be controlled by an independent mechanism that activates its substrate in response to extracellular signaling and the environment. In insulin signaling, growth factor receptor–bound protein 14 (Grb14) directly interacts with PDK1 (King and Newton, 2004; Goenaga et al., 2009). The mutation of Grb14 in the PDK1-binding motif revealed a significant reduction in insulin-dependent Akt activation, which suggested the role of Grb14 as an adapter molecule for the modulation of the membrane association of PDK1 in response to insulin (King and Newton, 2004). Freud1/Aki1 interacts with both the catalytic and PH domains of PDK1 and Akt and activates Akt signaling in EGF receptor–mediated signaling (Nakamura et al., 2008; Yamada et al., 2013). Given Grb14 and Freud/Aki1 are scaffolding components that facilitate the localization of PDK1 to the plasma membrane to activate Akt, the context-dependent optimal activation mechanism of PDK1 and Akt was apparently required. Our immunoprecipitation and pull-down assay results indicated that AMIGO2 selectively interacted with the PH domain of PDK1 and subsequently facilitated Akt activation at the plasma membrane in response to VEGF signaling. The specificity of AMIGO2 to PDK1 was also confirmed by the application of PTD-A2, a small peptide from the C terminus of AMIGO2. PTD-A2 inhibited PDK1 and Akt phosphorylation, but did not affect S6K phosphorylation. S6K is activated by the PDK1-interacting fragment binding pocket of PDK1 in a PIP3-independent manner (Pullen et al., 1998; Pearce et al., 2010), indicating that S6K is activated in the cytoplasm without recruitment to the plasma membrane. The role of AMIGO2 was also supported by the observation that knockdown of AMIGO2 in ECs abrogated the phosphorylation of PDK1 and Akt, but not the phosphorylation of S6K, which was located in the cytoplasm. We therefore suggest that AMIGO2 is a novel scaffold molecule that ensures the proper localization of PDK1 to the edge of the plasma membrane to activate PIP3-dependent Akt signaling.
During Akt signaling, PDK1 is recruited to the membrane-bound PIP3 second messenger through a direct interaction with the PH domain of PDK1 and is activated by autophosphorylation at serine 241 within the catalytic domain and at threonine 513 within the PH domain (Gao and Harris, 2006). Autophosphorylation of PDK1 at serine 241 can be easily autoinhibited by the PH domain without binding to PIP3 (Gao and Harris, 2006). The binding of the PDK1 PH domain to PIP3 leads to the trans-phosphorylation of PDK1 at threonine 513 (Gao and Harris, 2006), which results in destabilization of the autoinhibition conformation and maintenance of the catalytic activity of PDK1 to finally activate Akt. The interaction of AMIGO2 and the PDK1 PH domain might be related to PDK1 conformational changes that promote the autophosphorylation of PDK1 at serine 241 after recruitment to the plasma membrane; otherwise, PDK1 activity is autoinhibited. Indeed, our studies showed that AMIGO2 deficiency in ECs attenuated PDK1 phosphorylation by the loss of AMGIO2 binding to PDK1 and reduced the recruitment of PDK1 to the plasma membrane to achieve Akt phosphorylation. More interestingly, soluble PTD-A2 also inhibited PDK1 phosphorylation, which provided the plausible explanation that AMIGO2 seems to protect phospho-PDK1 (serine 241) from its autoinhibition by facilitating conformational changes and is likely to bring PIP3-mediated PDK1 into proximity with PIP3-mediated Akt. However, a more detailed PDK1 and PTD-A2 mechanism requires further investigation.
Several studies have shown the importance of PDK1 in regulating Akt activation and cell viability in a vascular system. Endothelial-specific PDK1 knockout mice (embryonic day [E] 10.5) display apoptotic ECs (Feng et al., 2010). Platelet-specific PDK1-deficient mice exhibit thrombocytopenia and inhibition of platelet aggregation and Akt activation (Chen et al., 2013). Moreover, the disruption of the PH domain in PDK1 knockin embryoid bodies resulted in impairment of Akt activation, vessel formation, and EC migration (Primo et al., 2007; Bayascas et al., 2008). Likewise, depletion of AMIGO2 using siRNA resulted in inhibition of PDK1 localization to the plasma membrane as well as activation and caused defective EC survival, migration, Akt activation, and angiogenesis in vitro and in vivo. Furthermore, treatment with PTD-A2 revealed antiangiogenic activities as well as the reduction of pathological retinal angiogenesis and tumor vessel formation.
It is well established that the PI3K-mediated PDK1–Akt signaling pathway plays a crucial role in regulating tumorigenesis and angiogenesis through VEGF and several other growth factors (Jiang and Liu, 2008; Meuillet et al., 2010). The ablation of Akt interfered with mammary tumor growth in MMTV-PyMT and MMTV-ErbB2/Neu transgenic mice (Maroulakou et al., 2007). Inhibition of Akt by siRNA revealed decreased ovarian tumor growth, angiogenesis, EC tube formation, and morphogenesis (Xia et al., 2006; Kitamura et al., 2008). However, recent studies show that inhibition of Akt did not affect capillary-like network formation, and expression of constitutively active Akt led to nonfunctional vasculature formation in ECs after co-culture with vascular smooth muscle cells (Hellesøy et al., 2014; Hellesøy and Lorens, 2015). In addition, partial inhibition of PI3K signaling activates the Erk1/2 (MAPK) pathway by suppressing Akt1 activity and promotes arteriogenesis, though expression of constitutively active Akt promotes venous specification (Hong et al., 2006; Ren et al., 2010). Therefore, the balance of PI3K–Akt and MAPK signaling could be determined by various endothelial cellular contexts, thus regulating vasculogenesis and angiogenesis in different conditions. It has also been reported that the copy number of PDK1 is often increased in human breast cancer, and increased levels of PDK1 have been reported in 45% of patients with acute myeloid leukemia (Pearn et al., 2007; Raimondi and Falasca, 2011). Therefore, PDK1 is an attractive target for potential anticancer drugs. Most kinase inhibitors have been focused on ATP-binding pockets because of the similarity between serine/threonine kinases, which caused difficulty in achieving the target specificity (Garuti et al., 2010). MK2206, an allosteric Akt inhibitor that binds outside the PH domain of Akt and provides selectivity, is an alternative approach under clinical trial (Hirai et al., 2010; Meuillet et al., 2010). The Akt inhibitor perifosine, which effectively inhibits tumor growth by targeting the lipid-binding PH domain to block the translocation of Akt to the plasma membrane, is also under clinical trials (Martelli et al., 2003). PHT-427 is an inhibitor of PDK1 and Akt that binds to the PH domain of PDK1 and Akt, which was demonstrated by the structure-based design of small molecules that resulted in inhibition of their activities (Meuillet et al., 2010). PHT-427, when administered to non–small cell lung cancer, breast cancer, and pancreatic cancer xenografts, exhibited antitumor effects, and the inhibition of PDK1 was more closely correlated with antitumor activity than Akt inhibition (Meuillet et al., 2010). It is interesting that the PH domains of PDK1 and Akt are required in order for several signaling proteins in the plasma membrane to cause the allosteric activation and the intimate interaction between effectors and substrates that leads to the activation of the PDK1–Akt signaling pathway. Along with a previous study that revealed AMIGO2 is highly expressed in the peripheral zone of pancreatic tumors (Nakamura et al., 2007), we also detected robust expression of AMIGO2 in both melanoma tumor vessels and tumor cells. Administration of PTD-A2 significantly reduced tumor growth and vessel density in a xenograft B16F10 melanoma tumor model. This suggests that PTD-A2 attenuated angiogenesis and tumor growth by alleviating PDK1 and Akt activities through competition with AMIGO2. It is tempting to suggest that the peptide PTD-A2 can effectively attenuate PDK1–Akt activation in both tumor cells and ECs that are expressing AMIGO2.
The amino acid alignment among the AMIGO family revealed that the binding of the C-terminal region of AMIGO2 to the PH domain of PDK1 is highly conserved, although the amino acid homologies between AMIGO2 and AMIGO1 and between AMIGO2 and AMIGO3 are both 48% (Kuja-Panula et al., 2003). The similarity of the binding sequence to the PH domain indicates that PDK1 is also likely to interact with AMIGO1 and AMIGO3 expressed in a variety of cell types. Thus, PTD-A2, a small peptide that selectively binds to the PH domain of PDK1 and ultimately leads to the reduction of PDK1 and Akt activities, might provide novel therapeutic implications for tumor growth and pathological angiogenesis.
In summary, the work presented here supports the new mechanistic insight that PDK1 localization is regulated by AMIGO2 to activate Akt signaling, which regulates EC adhesion, migration, survival, and angiogenesis. We also demonstrate that the CD of AMIGO2 directly binds to the PH domain of PDK1. Through these findings, we generated PTD-A2, which inhibited the interaction between AMIGO2 and PDK1 and resulted in inhibition of neovascularization, pathological angiogenesis, and tumor angiogenesis. Collectively, the data suggest that PTD-A2 could potentially serve as a new therapeutic target for tumorigenesis and angiogenesis-related diseases.
Materials and methods
Isolation and culture of endothelial progenitor cells and ECs
Endothelial progenitor cells were isolated from human umbilical cord blood as previously described (Maeng et al., 2009). In brief, human umbilical cord blood samples (∼50 ml each) were collected from fresh placentas with attached umbilical cords by gravity flow. This protocol was approved by the local ethics committee. HUVECs were isolated from human umbilical cord veins that were cannulated, perfused with PBS to remove blood, and then incubated with 250 U/ml collagenase type 2 in PBS for 10 min at 37°C as described previously (Marin et al., 2001). Collagenase (Worthington Biochemical Corporation) solution was collected and centrifuged at 1,200 rpm for 5 min. The pellet was resuspended in 3 ml of complete M199 media (HyClone) containing 20% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 3 ng/ml basic FGF (EMD Millipore), and 5 U/µl heparin. HUVECs were cultured on 2% gelatin-coated dishes at 37°C in a 5% CO2-humidified atmosphere using M199 medium and used at passages 3–8.
Transfection of siRNAs and plasmids into HUVECs
HUVECs were transfected with scrambled (control) and human AMIGO2 siRNAs using Lipofectamine (Invitrogen) for 3 h, and cells were assayed 48 h after transfection. Human AMIGO2 siRNA was designed by Invitrogen using the following sequences: 5′-UUAGGAUGCCCUCAGCUAUCACUGC-3′ and 5′-AUUGUUGUAAAGCAGAAGCACUUCC-3′; and the control siRNA sequence is 5′-AUGUAUUGGCCUGUAUUAG-3′. HUVECs were transfected with EGFP-tagged plasmids using Lipofectamine LTX and PLUS reagent (Invitrogen) for 2 h, and cells were assayed 24 h after transfection.
Plasmids and recombinant protein production
Full-length human AMIGO2 was purchased from GeneCopoeia (EX-E1271-M02) and subcloned into the EGFP-tagged vector using HindIII (5′-CCCAAGCTTGGCGACCATAATGTCGTTACGTGTACACACT-3′) and BamHI (5′-CGGGATCCCGTTAAGTGGACGCCACAAAAG-3′). AMIGO2ΔLRR (40–282 aa) was cloned using HindIII (5′-GATGATGCTGTCAGACCCAGAGGCACCAGG-3′ [megaprimer of signal peptide to IgG domain] and 5′-CCTGGTGCCTCTGGGTCTGACAGCATCATC-3′ [megaprimer of signal peptide to IgG domain]) and BamHI. AMIGO2ΔIgG (295–381 aa) was cloned using HindIII (5′-GAAATTGCTCACATTGCAATTCATAAAGCT-3′ [megaprimer of LRR to transmembrane domain] and 5′-AGCTTTATGAATTGCAATGTGAGCAATTTC-3′ [megaprimer of LRR to transmembrane domain]) and BamHI. AMIGO2ΔCD (Δ420–522 aa) was cloned using HindIII (5′-CCCAAGCTTGGCGACCATAATGTCGTTACGTGTACACACT-3′) and BamHI with a stop codon (5′-CGGGATCCCGTTACAGATAGAGGTACAAAA-3′). AMIGO2CD (420–522 aa) was cloned using HindIII (5′-CCCAAGCTTGATGACTCCATGCCCCTGCAA-3′) and BamHI (5′-CGGGATCCCGTTAAGTGGACGCCACAAAAG-3′). The 491-aa and the 457-aa constructs were subcloned with BamHI (5′-CGGGATCCCGTTACACTGCCTCGCTGGGAAAGA-3′ and 5′-CGGGATCCCGTTATTCATCAGCGGAGGCATCAC-3′, respectively). For protein production, the plasmids were constructed with pGEX, 4T1, and pET32a. The GST-CD, 65, 55, 45, and 35 used BamHI (5′-CGCGGATCCGCGGAAAACCTGTATTTTCAGGGCACTCCATGCCCCTGCAAG-3′) and XhoI (5′-CCGCTCGAGCGGTCATGATTAAGTGGACGCCACAAA-3′ for CD; 5′-CCGCTCGAGCGGTCATGATTACCTGACTTTCCCGTT-3′ for 65; 5′-CCGCTCGAGCGGTCATGATTAATCCTTCAGGGGTTC-3′ for 55; 5′-CCGCTCGAGCGGTCATGATTATTTACCTGCACCTGC-3′ for 45; and 5′-CCGCTCGAGCGGTCATGATTAGGAGGCATCACTAGC-3′ for 35). For His-PDK1PH, BamHI (5′-CGCGGATCCGGCAGCAACATAGAGCAG-3′) and XhoI (5′-CCGCTCGAGCGGTCACTGCACAGCGGCGTCGGG-3′) were used. For the His-Akt PH domain, BamHI (5′-CGGGATCCATGAGCGACGTGGCTATTGTGA-3′) and HindIII (5′-CCCAAGCTTCGAAGTCCATCTCCTCCTCCT-3′) were used. Thioredoxin fusion containing a hexa-His tag and GST-fusion proteins were expressed in Escherichia coli BL21 and purified with glutathione Sepharose 4B resin and Ni2+–nitrilotriacetic acid (NTA) agarose.
EC adhesion assay
HUVECs transfected with siRNA were trypsinized and washed twice in phenol-free M199. Before seeding, a 96-well plate was coated with 2% gelatin, 3 µg/ml fibronectin in PBS, and 10 µg/ml collagen type I in PBS and incubated at 37°C for 1 h in a humidified incubator. Before the initiation of the experiments, the wells were rinsed twice with PBS. HUVECs were labeled with acetomethoxy derivative of calcein, and 1.5 × 104 cells were seeded into each well of the 96-well plates. Cells were incubated for 1 h at 37°C, and nonadherent, calcein-labeled cells were removed by four washes. M199 containing 20% FBS was added to the cells in each well, and the fluorescence was measured (maximum absorbance at 494 nm and maximum emission at 517 nm) using a fluorescein filter set by FLUOstar Omega (BGM LABTECH).
Flow cytometry analysis of apoptosis
Flow cytometry was performed using a flow cytometer (FACSCanto II; BD). Annexin V–phycoerythrin and 7–amino actinomycin D (7-AAD) staining with an annexin V–phycoerythrin apoptosis detection kit (BD) were used to identify cells in various stages of apoptosis. HUVECs were transfected and detached using cell dissociation buffer (StemPro Accutase; Life Technologies), and annexin V and 7-AAD staining was performed. The annexin V–phycoerythrin apoptosis detection kit was used to evaluate apoptosis.
EC migration assay
The chemotactic motility of HUVECs was assayed in Transwell chambers (Corning) using polycarbonate filters (8-µm pore size; 6.5-mm diameter). In brief, the lower surface of the filter was coated with 0.1% gelatin. Fresh M199 medium containing 1% FBS and 20 ng VEGF was added to the lower wells. HUVECs were trypsinized and resuspended in M199 containing 1% FBS to a final concentration of 1 × 106 cells/ml. A 100-µl aliquot of the cell suspension was added to each of the upper wells and incubated at 37°C for 4 h. Cells were then fixed and stained with hematoxylin and eosin. Nonmigrating cells on the upper surface of the filter were removed by wiping with a cotton swab, and chemotaxis was quantified by counting the cells that migrated to the lower side of the filter using optical microscopy (200× magnification). 10 fields were counted for each assay. Each sample was assayed in triplicate, and the assays were repeated three times.
In vitro tube formation assay
The tube formation was assayed as previously described (Choi et al., 2009). In brief, 250 µl of growth factor–reduced Matrigel (BD) was added to a 16-mm-diameter tissue culture well and allowed to polymerize for 30 min at 37°C. Matrigel cultures were incubated at 37°C and photographed at various time points (200× magnification). The area covered by the tube network was determined using an optical imaging technique. The images of the tubes were scanned into Photoshop (Adobe) and quantified using ImageJ software (National Institutes of Health).
RT-PCR and real-time PCR primers
The human AMIGO2 RT-PCR forward and reverse primers were 5′-GATACTGCAGCAGGGCAGAA-3′ and 5′-GACGCCACAAAAGGTGTGTC-3′, respectively. The forward and reverse murine AMIGO2 primers were 5′-GGCACTTTAGCTCCGTGATG-3′ and 5′-GTCTCGTTTAACAGCCGCTG-3′, respectively, as previously described (Kuja-Panula et al., 2003). For the mouse GAPDH control, the forward and reverse primers were 5′-CAACGACCCCTTCATTGACC-3′ and 5′-AGTGATGGCATGGACTGTGG-3′, respectively. Quantitative real-time PCR was performed in a real-time PCR system (PikoReal; Thermo Fisher Scientific).
In vivo siRNA, shRNA, and peptide injection and analysis
In vivo siRNA injections were performed as previously described (Behlke, 2006; Mammoto et al., 2009; Bonifazi et al., 2010; Inaba et al., 2012). The mouse AMIGO2 siRNA was designed by GE Healthcare using the sequence 5′-GUACAAAAGAUCUCGCUGA-3′. Each mouse received, in its dorsal neck, a subcutaneous injection with AMIGO2-specific siRNA (5 µg/P3.5 mouse) or an equivalent dose of a nonspecific control siRNA duplex in 50 µl of delivery solution from the TransIT-QR starter kit (Mirus) carefully using a 1-ml insulin syringe. After 2 d, siRNA-injected mice were sacrificed, and eyes were quickly enucleated to collect the hyaloid vessels and retinas. shRNA lentiviral transduction particles were purchased from Sigma-Aldrich (mouse AMIGO2 MISSION shRNA lentiviral transduction particles). Each P3.5 mouse received, in its dorsal neck, a subcutaneous injection with shRNA lentiviral transduction particles (2.5 × 106 transduction units/50 µl) or an equivalent dose of nonspecific control shRNA particles. For peptide injection, each P3.5 mouse received, in its dorsal neck, a subcutaneous injection with 4 mg/kg PTD-A2 or Con. After 2 d, PTD-A2 injected mice were sacrificed. Moreover, topical (direct) shRNA viral and peptide delivery to the eye was assessed by using a microsyringe pump controller (Micro4; WPI Inc.). Each eye of each P3.5 mouse was topically applied with 1 µl of shRNA lentiviral particles (5 × 104 transduction units) or 25 µg/µl peptide. siRNA-, shRNA lentiviral particle–, and peptide-injected mice were reared until P5.5, P8.5, or P15.5 and then sacrificed under institutional guidelines established for the Animal Core Facility at the Yonsei University College of Medicine. Hyaloid vessels and retinas were prepared as previously described (Lobov et al., 2005). TUNEL labeling of apoptotic cells was performed using an in situ cell death detection kit (Roche). Mouse anti-AMIGO2 antibody (Santa Cruz Biotechnology, Inc.) was used for mouse tissue staining, and Alexa Fluor 594–conjugated isolectin B4 (iB4) was used to label the vessels.
Immunofluorescence
HUVECs were fixed in 3.7% formaldehyde for 10 min and permeabilized with 0.1% Triton X-100 in PBS. Cells were labeled with rabbit anti-FAK antibody (Santa Cruz Biotechnology, Inc.), rabbit anti-PDK1 antibody (Cell Signaling Technology), and goat anti-AMIGO2 antibody (Santa Cruz Biotechnology, Inc.) for 2 h at room temperature or overnight at 4°C. Afterward, the cells were rinsed in PBS and incubated with anti–goat Alexa Fluor 488 and anti–rabbit Alexa Fluor 546 secondary antibodies (Invitrogen) for 60 min at room temperature. Samples were then examined under a fluorescence microscope (LSM 700 META; Carl Zeiss).
Immunoprecipitation and Western blot analysis
For the immunoprecipitation of endogenous proteins, HUVECs were cultured on 2% gelatin-coated dishes by using endothelial basal medium 2 (Lonza). The medium was supplemented with 10% FBS, human VEGF-A, human ΦΓFGF-B, human EGF, insulin-like growth factor 1, and ascorbic acid in appropriate amounts. HUVECs were lysed in 1 ml NP-40 lysis buffer (50-mM Tris/HCl, pH 8.0, 150-mM NaCl, 1% NP-40, and protease inhibitor cocktail). For coimmunoprecipitation, HEK293T cells were lysed in 1 ml lysis buffer (20 mmol/liter Tris/HCl, pH 8.0, 2 mmol/liter EDTA, 137 mmol/liter NaCl, 1 mmol/liter NaVO4, 1 mmol/liter PMSF, 10% glycerol, and 1% Triton X-100). Cell lysates were centrifuged at 14,000 g for 15 min. The supernatants were immunoprecipitated with antibodies against mouse anti-FLAG (Sigma-Aldrich) and mouse anti-GFP (Santa Cruz Biotechnology, Inc.) at 4°C overnight, followed by the addition of protein A agarose beads (EMD Millipore) at 4°C for 2 h. Immunoprecipitates were washed three times with lysis buffer, resuspended in SDS-PAGE sample buffer containing β-mercaptoethanol, and further analyzed by Western blotting. For Western blot analysis, antibodies against rabbit anti-phosphorylated VEGFR2, rabbit anti-VEGFR2, rabbit anti-phosphorylated Akt (P-Akt at threonine 308 and serine 473), rabbit anti-Akt, rabbit anti-phosphorylated PDK1, rabbit anti-PDK1, rabbit anti-phosphorylated PI3K, rabbit anti-PI3K, and rabbit anti-S6K were purchased from Cell Signaling Technology. Rabbit anti-phosphorylated S6K was purchased from Abcam. Rabbit anti–caspase 3 and goat anti–vascular endothelial cadherin antibodies were purchased from Santa Cruz Biotechnology, Inc. Mouse and goat anti-AMIGO2 antibodies for immunoprecipitation and Western blotting were purchased from R&D Systems and Santa Cruz Biotechnology, Inc., respectively. Mouse anti-GAPDH antibody was purchased from Thermo Fisher Scientific, and the ProteoExtract Subcellular Proteome Extraction kit was purchased from EMD Millipore.
Pull-down assays
For pull-down assays using His- and GST-fused proteins, equal amounts of GST- and His-fused proteins (∼1.5:1.5 mg/ml) were incubated for 2 h at 4°C for in vitro binding. Ni2+-NTA acid resins or glutathione Sepharose 4B beads were equilibrated in sodium phosphate buffer, pH 7.5. The mixture of GST- and His-fused proteins were then immobilized on Ni2+-NTA resins or glutathione Sepharose 4B beads and incubated for 30 min at 4°C with gentle rotation. After a series of wash steps with sodium phosphate buffer, proteins were eluted with elution buffer (25-mM NaPi, 300-mM NaCl, 5-mM β-mercaptoethanol, and 500-mM imidazole, pH 7.5) and analyzed by Western blotting with rabbit anti-GST and -His antibodies.
Far-western analysis
Far-western analysis was performed as previously described (Wu et al., 2007). In brief, 1 µg of purified GST-tagged proteins was loaded and separated by SDS-PAGE. The prey proteins were transferred to polyvinylidene fluoride membranes, denatured, and renatured on the membrane, and then the proteins were blocked. The membranes were incubated with 10 µg of purified bait protein (His-tagged PH domain), and the bait proteins were detected with anti-His antibody.
Peptide binding and competition assays
For the peptide binding assay, the purified PH domain of proteins were incubated with Ni2+-NTA resins for 1 h, incubated with the FITC–PTD-A2 or Con for 1 h, washed five times, and eluted with elution buffer, and the fluorescence was measured using a FLUOstar Omega FITC filter set. For the peptide competition assay, the purified protein PH domains were incubated with Ni2+-NTA resins for 1 h, incubated with the CD of AMIGO2 proteins and FITC–PTD-A2 or Con for 1 h, washed five times, and eluted with elution buffer, and then the fluorescence was measured.
Peptide preparation and treatment
The FITC linker YGRKKRRQRRR-GKRVVFLEPLKDTA (TAT-A2), FITC linker YGRKKRRQRRR-GRVKTDFLAVPEKL (the control peptide), FITC linker GNGRGG-GKRVVFLEPLKDTA (NGR-A2 for vascular-specific penetration), and FITC linker GNGRGG-GRVKTDFLAVPEK (for control peptide) were obtained from Peptron. The 5-µM peptide concentration was optimized using a dose-dependent cell survival assay. Peptide-treated HUVECs were incubated with serum-free M199 medium at 37°C.
Matrigel plug angiogenesis assay
Angiogenesis was evaluated in 5–6-wk-old male C57BL/6 mice using the Matrigel plug assay as previously described (Choi et al., 2009). 500 µl of growth factor–reduced Matrigel containing 200 ng VEGF and 10 U heparin was mixed with the indicated amount of either PTD-A2 or 25-µM control peptide. After injection, the Matrigel rapidly formed a single, solid gel plug. After 5 d, the skin of the mouse was pulled back to expose the Matrigel plug, which remained intact. To quantify blood vessel formation, hemoglobin was measured using the Drabkin method and the Drabkin reagent kit 525 (Sigma-Aldrich). The concentration of hemoglobin was calculated by comparison to a known amount of hemoglobin assayed in parallel. To identify infiltrating ECs, immunohistochemistry was performed using rat anti-CD31 antibody (BD).
Mouse model of OIR
OIR was induced in C57BL/6J mice as previously described (Connor et al., 2009). In brief, litters of P7 C57BL/6J pups and their mothers were placed in a 75 ± 2% oxygen atmosphere (hyperoxia) for 5 d and then returned to room air at age P12. After returning to room air, the pups received an intraperitoneal injection of PBS, Con, or PTD-A2 every day for 5 d. After the injections, the avascular retinal areas of the mice became hypoxic, and intraretinal physiological revascularization of avascular areas and preretinal pathological neovascularization developed simultaneously. Pathological neovascularization reached its maximum at P17. Mice were euthanized by cervical dislocation, and the eyes were enucleated. The quantification of OIR model was previously described (Connor et al., 2009). In brief, a screenshot of one retinal quadrant with the total vascular area is traced. The tools used to obtain the total retinal area are the polygonal lasso tool and the add to selection function in Photoshop CS4. To obtain a screenshot of one retinal quadrant with quantification of the avascular zone, the functions and settings used are the lasso tool, subtract from selection, histogram function with refresh key, and pixel record. Once the desired area is outlined, click the refresh key and record the number of pixels in the area. To obtain a screenshot of a retinal quadrant with the neovascular tufts highlighted, the necessary tools and settings are magic wand and add to selection. The tolerance should be set to 50, and the anti-alias and contiguous boxes should be checked.
Tumor model, treatment of peptide, immunofluorescence staining, and TUNEL assay
To generate the tumor implantation model, suspensions of B16F10 melanoma cells (5 × 105 cells in 100 µl) were subcutaneously injected into the dorsal flank of 6–7-wk-old mice, and tumor volumes were measured at given time points. Tumor volume was calculated according to the formula 0.52 × A × B, where A is the largest diameter of a tumor and B is the perpendicular diameter. PTD-Con or PTD-A2 peptide (4 mg/kg every other day) was administered by intratumoral injection after the tumor volume exceeded 100 mm3. Tumor tissues were fixed in 4% PFA-PBS, pH 7.4, overnight at 4°C and rinsed with PBS at room temperature. Tissues were incubated in 15% sucrose solution overnight at 4°C and then transferred to 30% sucrose at 4°C until the tissues sank. The tissues were infiltrated in optimal cutting temperature (OCT) embedding medium (Tissue Tek) for 30 min at room temperature before freezing. Tissues were then transferred to an embedding mold filled with OCT embedding medium, and the mold was cooled with dry ice. After the material had frozen, the tissues were stored at –70°C. Sections (10–50-µm thick) were cut at –20°C, prefixed in acetone for 30 min at –70°C, and dried briefly until the acetone was removed. The OCT embedding medium was removed with water, and sections were incubated in blocking solution for 2 h at room temperature, followed by overnight incubation with primary antibody at 4°C. After five washes in 0.1% Triton X-100 in PBS for 15 min each, the sections were incubated with secondary antibody overnight at 4°C. Before washing, the sections were treated with 1 µg/ml DAPI and washed five more times with 0.1% Triton X-100 in PBS for 30 min each. All antibodies were dissolved in antibody diluent (Dako). For the TUNEL assay, an in situ cell death detection kit (Roche) was used for detecting cell death in tumors. Samples were mounted in fluorescent mounting medium (Dako). Confocal images were captured at RT using ZEN software on an upright confocal microscope (LSM 700; Carl Zeiss; Apochromat 40×/1.2 NA, W Korr UV-visible infrared objective and Plan Apochromat 20×/0.8 NA) using the predefined ZEN software configurations for Alexa Fluor 546, Alexa Fluor 488, and DAPI.
Animal studies
All animal experiments were conducted under the institutional guidelines established for the Animal Core Facility at Yonsei University College of Medicine and were approved by the Laboratory Animal Research Center Institutional Animal Care and Use Committee at Yonsei University
Statistical analysis
Data are presented as means ± SD. All experiments were performed in triplicate, and representative results are shown.
Online supplemental material
Fig. S1 shows the expression level and angiogenic roles of AMIGO2 in ECs. Fig. S2 shows that AMIGO2 is expressed in retinal and hyaloid vessels and the effect of AMIGO2 inhibition in retinal and hyaloid vessels during eye development. Fig. S3 shows the binding of the PH domain of PDK1 and the CD of AMIGO2. Fig. S4 shows that a synthetic peptide, PTD-A2, binds to the PH domain of PDK1 and inhibits AMIGO2 and PDK1 interaction. Fig. S5 shows that AMIGO2 is expressed in B16F10 melanoma tumor vessels and PTD-A2 has an anti-angiogenic activity in tumorigenesis. Video 1 shows the effect of AMIGO2 knockdown in EC tube formation on Matrigel. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503113/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201503113.dv.
Supplementary Material
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (MEST; grants NRF-2015R1A2A1A05001859 and NRF-2013M3A9B6046563) and by the Bio and Medical Technology Development Program of the NRF funded by MEST (grant NRF-2011-0019267). This work was also supported by the Mid-career Researcher Program (grant NRF-2013R1A2A2A01068963) through an NRF grant funded by MEST.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AMIGO
- adhesion molecule with IgG-like domain
- CD
- cytosolic domain
- EC
- endothelial cell
- HUVEC
- human umbilical vein EC
- LRR
- leucine-rich repeat
- NTA
- nitrilotriacetic acid
- OCT
- optimal cutting temperature
- OIR
- oxygen-induced retinopathy
- PH
- pleckstrin homology
- SGK
- serum/glucocorticoid-regulated kinase
References
- Alvarez Y., Astudillo O., Jensen L., Reynolds A.L., Waghorne N., Brazil D.P., Cao Y., O’Connor J.J., and Kennedy B.N.. 2009. Selective inhibition of retinal angiogenesis by targeting PI3 kinase. PLoS One. 4:e7867 10.1371/journal.pone.0007867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayascas J.R., Wullschleger S., Sakamoto K., García-Martínez J.M., Clacher C., Komander D., van Aalten D.M.F., Boini K.M., Lang F., Lipina C., et al. 2008. Mutation of the PDK1 PH domain inhibits protein kinase B/Akt, leading to small size and insulin resistance. Mol. Cell. Biol. 28:3258–3272. 10.1128/MCB.02032-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke M.A. 2006. Progress towards in vivo use of siRNAs. Mol. Ther. 13:644–670. 10.1016/j.ymthe.2006.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J. 1995. Approaches to studying cell adhesion molecules in angiogenesis. Trends Cell Biol. 5:69–74. 10.1016/S0962-8924(00)88949-7 [DOI] [PubMed] [Google Scholar]
- Bonifazi P., D’Angelo C., Zagarella S., Zelante T., Bozza S., De Luca A., Giovannini G., Moretti S., Iannitti R.G., Fallarino F., et al. 2010. Intranasally delivered siRNA targeting PI3K/Akt/mTOR inflammatory pathways protects from aspergillosis. Mucosal Immunol. 3:193–205. 10.1038/mi.2009.130 [DOI] [PubMed] [Google Scholar]
- Casamayor A., Morrice N.A., and Alessi D.R.. 1999. Phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo. Biochem. J. 342:287–292. 10.1042/bj3420287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Graham P.H., Hao J., Ni J., Bucci J., Cozzi P.J., Kearsley J.H., and Li Y.. 2013. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 4:e875 10.1038/cddis.2013.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Zhang Q., Feng Q., Xu J., Teng T., Luan Q., Shan C., Hu Y., Hemmings B.A., Gao X., and Yang Z.. 2010. Deletion of Akt1 causes heart defects and abnormal cardiomyocyte proliferation. Dev. Biol. 347:384–391. 10.1016/j.ydbio.2010.08.033 [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang Y., Wang Y., Li D., Zhang L., Wang K., Luo X., Yang Z., Wu Y., and Liu J.. 2013. PDK1 regulates platelet activation and arterial thrombosis. Blood. 121:3718–3726. 10.1182/blood-2012-10-461897 [DOI] [PubMed] [Google Scholar]
- Chen Y., Aulia S., Li L., and Tang B.L.. 2006. AMIGO and friends: An emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res. Brain Res. Rev. 51:265–274. 10.1016/j.brainresrev.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Choi Y.S., Choi H.J., Min J.K., Pyun B.J., Maeng Y.S., Park H., Kim J., Kim Y.M., and Kwon Y.G.. 2009. Interleukin-33 induces angiogenesis and vascular permeability through ST2/TRAF6-mediated endothelial nitric oxide production. Blood. 114:3117–3126. 10.1182/blood-2009-02-203372 [DOI] [PubMed] [Google Scholar]
- Connor K.M., Krah N.M., Dennison R.J., Aderman C.M., Chen J., Guerin K.I., Sapieha P., Stahl A., Willett K.L., and Smith L.E.H.. 2009. Quantification of oxygen-induced retinopathy in the mouse: A model of vessel loss, vessel regrowth and pathological angiogenesis. Nat. Protoc. 4:1565–1573. 10.1038/nprot.2009.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S.R., Brunet A., and Greenberg M.E.. 1999. Cellular survival: A play in three Akts. Genes Dev. 13:2905–2927. 10.1101/gad.13.22.2905 [DOI] [PubMed] [Google Scholar]
- Dimmeler S., and Zeiher A.M.. 2000a Akt takes center stage in angiogenesis signaling. Circ. Res. 86:4–5. 10.1161/01.RES.86.1.4 [DOI] [PubMed] [Google Scholar]
- Dimmeler S., and Zeiher A.M.. 2000b Endothelial cell apoptosis in angiogenesis and vessel regression. Circ. Res. 87:434–439. 10.1161/01.RES.87.6.434 [DOI] [PubMed] [Google Scholar]
- Feng Q., Di R., Tao F., Chang Z., Lu S., Fan W., Shan C., Li X., and Yang Z.. 2010. PDK1 regulates vascular remodeling and promotes epithelial-mesenchymal transition in cardiac development. Mol. Cell. Biol. 30:3711–3721. 10.1128/MCB.00420-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujio Y., and Walsh K.. 1999. Akt mediates cytoprotection of endothelial cells by vascular endothelial growth factor in an anchorage-dependent manner. J. Biol. Chem. 274:16349–16354. 10.1074/jbc.274.23.16349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., and Harris T.K.. 2006. Role of the PH domain in regulating in vitro autophosphorylation events required for reconstitution of PDK1 catalytic activity. Bioorg. Chem. 34:200–223. 10.1016/j.bioorg.2006.05.002 [DOI] [PubMed] [Google Scholar]
- Garuti L., Roberti M., and Bottegoni G.. 2010. Non-ATP competitive protein kinase inhibitors. Curr. Med. Chem. 17:2804–2821. 10.2174/092986710791859333 [DOI] [PubMed] [Google Scholar]
- Goenaga D., Hampe C., Carré N., Cailliau K., Browaeys-Poly E., Perdereau D., Holt L.J., Daly R.J., Girard J., Broutin I., et al. 2009. Molecular determinants of Grb14-mediated inhibition of insulin signaling. Mol. Endocrinol. 23:1043–1051. 10.1210/me.2008-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellesøy M., and Lorens J.B.. 2015. Cellular context-mediated Akt dynamics regulates MAP kinase signaling thresholds during angiogenesis. Mol. Biol. Cell. 26:2698–2711. 10.1091/mbc.E14-09-1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellesøy M., Blois A.L., Tiron C.E., Mannelqvist M., Akslen L.A., and Lorens J.B.. 2014. Akt1 activity regulates vessel maturation in a tissue engineering model of angiogenesis. Tissue Eng. Part A. 20:2590–2603. 10.1089/ten.tea.2013.0399 [DOI] [PubMed] [Google Scholar]
- Hirai H., Sootome H., Nakatsuru Y., Miyama K., Taguchi S., Tsujioka K., Ueno Y., Hatch H., Majumder P.K., Pan B.S., and Kotani H.. 2010. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Ther. 9:1956–1967. 10.1158/1535-7163.MCT-09-1012 [DOI] [PubMed] [Google Scholar]
- Hong C.C., Peterson Q.P., Hong J.Y., and Peterson R.T.. 2006. Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr. Biol. 16:1366–1372. 10.1016/j.cub.2006.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S., Ahmed M.U., Alam S., Watanabe A., Harashima A., Yonekura H., and Yamamoto H.. 2011. Expression and roles of AMIGO gene family in vascular endothelial cells. Int. J. Mol. Med. Adv. Sci. 7:5–11. 10.3923/ijmmas.2011.5.11 [DOI] [Google Scholar]
- Huang B.X., Akbar M., Kevala K., and Kim H.Y.. 2011. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 192:979–992. 10.1083/jcb.201005100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba S., Nagahara S., Makita N., Tarumi Y., Ishimoto T., Matsuo S., Kadomatsu K., and Takei Y.. 2012. Atelocollagen-mediated systemic delivery prevents immunostimulatory adverse effects of siRNA in mammals. Mol. Ther. 20:356–366. 10.1038/mt.2011.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.H., and Liu L.Z.. 2008. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim. Biophys. Acta. 1784:150–158. 10.1016/j.bbapap.2007.09.008 [DOI] [PubMed] [Google Scholar]
- King C.C., and Newton A.C.. 2004. The adaptor protein Grb14 regulates the localization of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 279:37518–37527. 10.1074/jbc.M405340200 [DOI] [PubMed] [Google Scholar]
- King W.G., Mattaliano M.D., Chan T.O., Tsichlis P.N., and Brugge J.S.. 1997. Phosphatidylinositol 3-kinase is required for integrin-stimulated AKT and Raf-1/mitogen-activated protein kinase pathway activation. Mol. Cell. Biol. 17:4406–4418. 10.1128/MCB.17.8.4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Asai N., Enomoto A., Maeda K., Kato T., Ishida M., Jiang P., Watanabe T., Usukura J., Kondo T., et al. 2008. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat. Cell Biol. 10:329–337. 10.1038/ncb1695 [DOI] [PubMed] [Google Scholar]
- Kuja-Panula J., Kiiltomäki M., Yamashiro T., Rouhiainen A., and Rauvala H.. 2003. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J. Cell Biol. 160:963–973. 10.1083/jcb.200209074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamalice L., Le Boeuf F., and Huot J.. 2007. Endothelial cell migration during angiogenesis. Circ. Res. 100:782–794. 10.1161/01.RES.0000259593.07661.1e [DOI] [PubMed] [Google Scholar]
- Lim M.A., Kikani C.K., Wick M.J., and Dong L.Q.. 2003. Nuclear translocation of 3′-phosphoinositide-dependent protein kinase 1 (PDK-1): a potential regulatory mechanism for PDK-1 function. Proc. Natl. Acad. Sci. USA. 100:14006–14011. 10.1073/pnas.2335486100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Ahmad S.A., Reinmuth N., Shaheen R.M., Jung Y.D., Fan F., and Ellis L.M.. 2000. Endothelial cell survival and apoptosis in the tumor vasculature. Apoptosis. 5:323–328. 10.1023/A:1009679307513 [DOI] [PubMed] [Google Scholar]
- Lobov I.B., Rao S., Carroll T.J., Vallance J.E., Ito M., Ondr J.K., Kurup S., Glass D.A., Patel M.S., Shu W., et al. 2005. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 437:417–421. 10.1038/nature03928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng Y.S., Choi H.J., Kwon J.Y., Park Y.W., Choi K.S., Min J.K., Kim Y.H., Suh P.G., Kang K.S., Won M.H., et al. 2009. Endothelial progenitor cell homing: Prominent role of the IGF2-IGF2R-PLCβ2 axis. Blood. 113:233–243. 10.1182/blood-2008-06-162891 [DOI] [PubMed] [Google Scholar]
- Mammoto A., Connor K.M., Mammoto T., Yung C.W., Huh D., Aderman C.M., Mostoslavsky G., Smith L.E.H., and Ingber D.E.. 2009. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 457:1103–1108. 10.1038/nature07765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin V., Kaplanski G., Grès S., Farnarier C., and Bongrand P.. 2001. Endothelial cell culture: Protocol to obtain and cultivate human umbilical endothelial cells. J. Immunol. Methods. 254:183–190. 10.1016/S0022-1759(01)00408-2 [DOI] [PubMed] [Google Scholar]
- Maroulakou I.G., Oemler W., Naber S.P., and Tsichlis P.N.. 2007. Akt1 ablation inhibits, whereas Akt2 ablation accelerates, the development of mammary adenocarcinomas in mouse mammary tumor virus (MMTV)-ErbB2/neu and MMTV-polyoma middle T transgenic mice. Cancer Res. 67:167–177. 10.1158/0008-5472.CAN-06-3782 [DOI] [PubMed] [Google Scholar]
- Martelli A.M., Tazzari P.L., Tabellini G., Bortul R., Billi A.M., Manzoli L., Ruggeri A., Conte R., and Cocco L.. 2003. A new selective AKT pharmacological inhibitor reduces resistance to chemotherapeutic drugs, TRAIL, all-trans-retinoic acid, and ionizing radiation of human leukemia cells. Leukemia. 17:1794–1805. 10.1038/sj.leu.2403044 [DOI] [PubMed] [Google Scholar]
- Meuillet E.J., Zuohe S., Lemos R., Ihle N., Kingston J., Watkins R., Moses S.A., Zhang S., Du-Cuny L., Herbst R., et al. 2010. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol. Cancer Ther. 9:706–717. 10.1158/1535-7163.MCT-09-0985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A., Komander D., van Aalten D.M.F., and Alessi D.R.. 2004. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. 15:161–170. 10.1016/j.semcdb.2003.12.022 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Naito M., Tsuruo T., and Fujita N.. 2008. Freud-1/Aki1, a novel PDK1-interacting protein, functions as a scaffold to activate the PDK1/Akt pathway in epidermal growth factor signaling. Mol. Cell. Biol. 28:5996–6009. 10.1128/MCB.00114-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Kuwai T., Kitadai Y., Sasaki T., Fan D., Coombes K.R., Kim S.J., and Fidler I.J.. 2007. Zonal heterogeneity for gene expression in human pancreatic carcinoma. Cancer Res. 67:7597–7604. 10.1158/0008-5472.CAN-07-0874 [DOI] [PubMed] [Google Scholar]
- Ono T., Sekino-Suzuki N., Kikkawa Y., Yonekawa H., and Kawashima S.. 2003. Alivin 1, a novel neuronal activity-dependent gene, inhibits apoptosis and promotes survival of cerebellar granule neurons. J. Neurosci. 23:5887–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L.R., Komander D., and Alessi D.R.. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22. 10.1038/nrm2822 [DOI] [PubMed] [Google Scholar]
- Pearn L., Fisher J., Burnett A.K., and Darley R.L.. 2007. The role of PKC and PDK1 in monocyte lineage specification by Ras. Blood. 109:4461–4469. 10.1182/blood-2006-09-047217 [DOI] [PubMed] [Google Scholar]
- Portt L., Norman G., Clapp C., Greenwood M., and Greenwood M.T.. 2011. Anti-apoptosis and cell survival: A review. Biochim. Biophys. Acta. 1813:238–259. 10.1016/j.bbamcr.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Primo L., di Blasio L., Roca C., Droetto S., Piva R., Schaffhausen B., and Bussolino F.. 2007. Essential role of PDK1 in regulating endothelial cell migration. J. Cell Biol. 176:1035–1047. 10.1083/jcb.200607053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen N., Dennis P.B., Andjelkovic M., Dufner A., Kozma S.C., Hemmings B.A., and Thomas G.. 1998. Phosphorylation and activation of p70s6k by PDK1. Science. 279:707–710. 10.1126/science.279.5351.707 [DOI] [PubMed] [Google Scholar]
- Rabenau K.E., O’Toole J.M., Bassi R., Kotanides H., Witte L., Ludwig D.L., and Pereira D.S.. 2004. DEGA/AMIGO-2, a leucine-rich repeat family member, differentially expressed in human gastric adenocarcinoma: Effects on ploidy, chromosomal stability, cell adhesion/migration and tumorigenicity. Oncogene. 23:5056–5067. 10.1038/sj.onc.1207681 [DOI] [PubMed] [Google Scholar]
- Raff M.C. 1992. Social controls on cell survival and cell death. Nature. 356:397–400. 10.1038/356397a0 [DOI] [PubMed] [Google Scholar]
- Raimondi C., and Falasca M.. 2011. Targeting PDK1 in cancer. Curr. Med. Chem. 18:2763–2769. 10.2174/092986711796011238 [DOI] [PubMed] [Google Scholar]
- Ren B., Deng Y., Mukhopadhyay A., Lanahan A.A., Zhuang Z.W., Moodie K.L., Mulligan-Kehoe M.J., Byzova T.V., Peterson R.T., and Simons M.. 2010. ERK1/2-Akt1 crosstalk regulates arteriogenesis in mice and zebrafish. J. Clin. Invest. 120:1217–1228. 10.1172/JCI39837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: Integrating signals from front to back. Science. 302:1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Sheppard K., Kinross K.M., Solomon B., Pearson R.B., and Phillips W.A.. 2012. Targeting PI3 kinase/AKT/mTOR signaling in cancer. Crit. Rev. Oncog. 17:69–95. 10.1615/CritRevOncog.v17.i1.60 [DOI] [PubMed] [Google Scholar]
- Strömblad S., and Cheresh D.A.. 1996. Cell adhesion and angiogenesis. Trends Cell Biol. 6:462–468. 10.1016/0962-8924(96)84942-7 [DOI] [PubMed] [Google Scholar]
- Sun J.F., Phung T., Shiojima I., Felske T., Upalakalin J.N., Feng D., Kornaga T., Dor T., Dvorak A.M., Walsh K., and Benjamin L.E.. 2005. Microvascular patterning is controlled by fine-tuning the Akt signal. Proc. Natl. Acad. Sci. USA. 102:128–133. 10.1073/pnas.0403198102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.B. 1995. Apoptosis in the pathogenesis and treatment of disease. Science. 267:1456–1462. 10.1126/science.7878464 [DOI] [PubMed] [Google Scholar]
- Toker A., and Newton A.C.. 2000. Cellular signaling: Pivoting around PDK-1. Cell. 103:185–188. 10.1016/S0092-8674(00)00110-0 [DOI] [PubMed] [Google Scholar]
- Vicent D., Ilany J., Kondo T., Naruse K., Fisher S.J., Kisanuki Y.Y., Bursell S., Yanagisawa M., King G.L., and Kahn C.R.. 2003. The role of endothelial insulin signaling in the regulation of vascular tone and insulin resistance. J. Clin. Invest. 111:1373–1380. 10.1172/JCI15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I., and Sawyers C.L.. 2002. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2:489–501. 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- Wu Y., Li Q., and Chen X.Z.. 2007. Detecting protein–protein interactions by far western blotting. Nat. Protoc. 2:3278–3284. 10.1038/nprot.2007.459 [DOI] [PubMed] [Google Scholar]
- Xia C., Meng Q., Cao Z., Shi X., and Jiang B.H.. 2006. Regulation of angiogenesis and tumor growth by p110 alpha and AKT1 via VEGF expression. J. Cell. Physiol. 209:56–66. 10.1002/jcp.20707 [DOI] [PubMed] [Google Scholar]
- Yamada T., Takeuchi S., Fujita N., Nakamura A., Wang W., Li Q., Oda M., Mitsudomi T., Yatabe Y., Sekido Y., et al. 2013. Akt kinase-interacting protein1, a novel therapeutic target for lung cancer with EGFR-activating and gatekeeper mutations. Oncogene. 32:4427–4435. 10.1038/onc.2012.446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.