Crispr/Cas9-mediated mutation of the transcription factor GRHL2 or either of its predicted downstream targets ZNF750 and SMAGP in primary human bronchial epithelial basal cells leads to defects in ciliogenesis and/or barrier function.
Abstract
Pseudostratified airway epithelium of the lung is composed of polarized ciliated and secretory cells maintained by basal stem/progenitor cells. An important question is how lineage choice and differentiation are coordinated with apical–basal polarity and epithelial morphogenesis. Our previous studies indicated a key integrative role for the transcription factor Grainyhead-like 2 (Grhl2). In this study, we present further evidence for this model using conditional gene deletion during the regeneration of airway epithelium and clonal organoid culture. We also use CRISPR/Cas9 genome editing in primary human basal cells differentiating into organoids and mucociliary epithelium in vitro. Loss of Grhl2 inhibits organoid morphogenesis and the differentiation of ciliated cells and reduces the expression of both notch and ciliogenesis genes (Mcidas, Rfx2, and Myb) with distinct Grhl2 regulatory sites. The genome editing of other putative target genes reveals roles for zinc finger transcription factor Znf750 and small membrane adhesion glycoprotein in promoting ciliogenesis and barrier function as part of a network of genes coordinately regulated by Grhl2.
Introduction
The structural organization and cellular composition of the pseudostratified mucociliary epithelium lining the conducting airways of the mammalian lung is essential to its many physiological functions. For example, apical–junctional complexes between polarized luminal cells provide the first line of defense against external pathogens and allergens, while allowing selective paracellular transport of ions and macromolecules. Different kinds of secretory cells produce antimicrobial agents and mucus, and the multiciliated cells drive secretions out of the lung (Knight and Holgate, 2003; Proud and Leigh, 2011). Executing these functions comes at a price, however, because the specialized luminal cells are vulnerable to damage by agents such as viruses, inhaled toxicants, and aspirated stomach acid. Fortunately, the loss of luminal cells is normally compensated for by the mobilization of undifferentiated stem/progenitor cells localized close to the basal lamina. At steady state, these basal cells (BCs) are relatively quiescent, but after a loss of luminal cells they proliferate and differentiate into ciliated and secretory cells (Borthwick et al., 2001; Hogan et al., 2014). Considerable progress is being made in understanding the transcriptional circuits and signaling pathways that regulate the self-renewal of BCs and the lineage choices of their descendants (Rock et al., 2011b; Tata et al., 2013; Paul et al., 2014; Tadokoro et al., 2014; Zhao et al., 2014; Danahay et al., 2015; Mori et al., 2015; Pardo-Saganta et al., 2015; Watson et al., 2015). However, less is known about the cellular mechanisms that drive morphological changes in the daughters of the BCs as they acquire apical–basal polarity and integrate seamlessly into the epithelium. Indeed, from a broader perspective and considering other tissue systems, we need to better understand how genes regulating epithelial cell shape, cytoskeletal organization, and intercellular adhesion are linked to networks controlling lineage choice and differentiation (Connelly et al., 2010; Buske et al., 2012; Folgueras et al., 2013; Ader and Tanaka, 2014; Castanieto et al., 2014; Lancaster and Knoblich, 2014).
Previous studies identified the evolutionarily conserved grainyhead and grainyhead-like transcription factors as master regulators of epithelial morphogenesis (Wang and Samakovlis, 2012). Processes regulated by these factors include apical–basal polarity, intercellular adhesion, barrier function, and motility, as well as lineage choice and differentiation of progenitor populations (Wilanowski et al., 2002; Paré et al., 2012; Senga et al., 2012; Gao et al., 2013; Li et al., 2013). Epithelial tissues in which vertebrate grainyhead-like genes are known to be important include skin, developing neural tube, placental trophoblast, hepatic biliary cells, inner ear, and embryonic kidney (nephric duct; Ting et al., 2005; Chalmers et al., 2006; Werth et al., 2010; Pyrgaki et al., 2011; Tanimizu and Mitaka, 2013; Petrof et al., 2014; Aue et al., 2015; Walentin et al., 2015). In humans, a recent study has shown that some individuals homozygous for missense mutations in GRHL2 not only have conditions like ectodermal dysplasia, deafness, and hypodontia, but also asthma (Petrof et al., 2014). This condition is associated with excess mucus production, subepithelial inflammation and fibrosis, and potential defects in epithelial barrier function (Holgate, 2011; Rezaee and Georas, 2014).
Previously, we addressed the role of Grhl2 in human airway epithelium using Krt5+ Trp63+ primary BCs in culture (Gao et al., 2013). We found that expression of dominant-negative Grhl2 protein inhibited both the ability of daughter cells to form a polarized epithelium with barrier function and their differentiation. By combining transcriptomics and genome-wide chromatin immunoprecipitation sequencing (ChIP-Seq), we identified several hundred putative Grhl2 target genes with binding sites near the promoter region. Although many of these genes have been implicated in the adhesion, polarity, motility, and differentiation of cell lines, much less is known about their role in the morphogenesis and physiological function of specialized epithelial tissues.
Here, we use conditional deletion of a new Grhl2flx allele in mouse tracheal BCs to further define the role of this transcription factor during the regeneration of the mucociliary epithelium from basal progenitors in vivo and in 3D organoid cultures. We also use CRISPR/Cas9 genome editing in primary human BCs to screen multiple putative Grhl2 target genes for functions in airway epithelium using air–liquid interface (ALI) and organoid cultures. Together, these experiments establish that Grhl2 coordinately regulates airway cell polarity, barrier function, and lineage differentiation through multiple downstream effectors. These include the Notch signaling pathway and known ciliogenesis genes, as well as the transcription factor zinc finger protein 750 (ZNF750 in humans and Znp750 in mice) and small membrane adhesion glycoprotein (SMAGP), genes that had not previously been implicated in airway epithelial cell biology.
Results
Grhl2 expression and changes in BC phenotype during airway repair after exposure to SO2
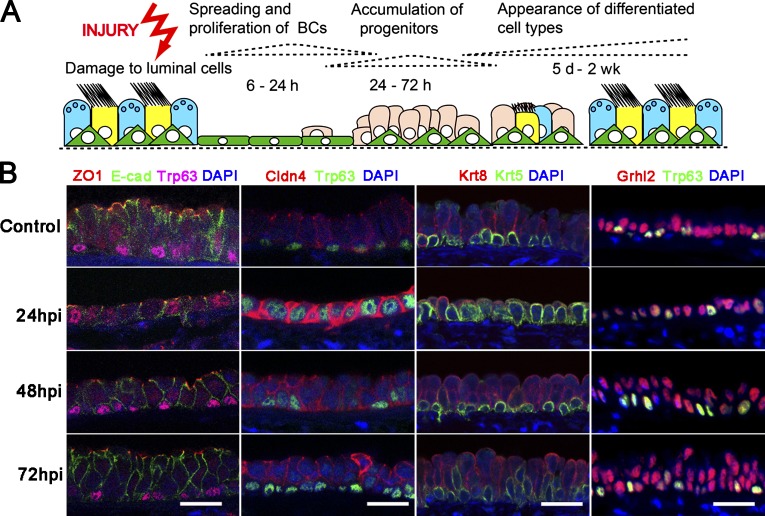
The mucociliary epithelium of the mouse trachea is composed of approximately equal proportions of Foxj1+ multiciliated cells, Scgb1a1+ secretory cells, and Krt5+ Trp63+ BCs. If luminal cells are destroyed by brief exposure to SO2, they are replaced by the proliferation and differentiation of surviving BCs (Fig. 1 A; Rock et al., 2009; Tadokoro et al., 2014; Pardo-Saganta et al., 2015). In the normal epithelium, ZO1 (Tjp1) and Claudin 4 (Cldn4) proteins are localized to apical–junctional complexes between the polarized luminal cells. In contrast, the BCs do not have distinct apical domains delimited by localized ZO1 expression (Fig. 1 B). One of the earliest responses of the BCs to injury, within 24 h, is spreading to cover the denuded basal lamina and then proliferating to self-renew and generate undifferentiated progenitors. Significantly, all of the squamous epithelial cells present at 24 h postinhalation (hpi), including those that express Trp63, have distinct ZO1+ apical junctions. They also show elevated levels of Cldn4 expression that is not restricted to apical junctions (Fig. 1 B). The localization of ZO1 between all cells at this time was confirmed by whole-mount visualization of tracheas from Tjp1tm1lch mice expressing a ZO1:GFP fusion protein from the endogenous Tjp1 allele (Fig. S1; Huebner et al., 2014). At 48 and 72 hpi, when the Krt8+ progenitor cells have become more columnar in shape, the Krt5+ Trp63+ cells no longer express localized ZO1, and the level of Cldn4 is down-regulated (Fig. 1 B).
Figure 1.
Changes in BC shape and protein expression during regeneration of airway mucociliary epithelium. (A) Schematic for repair of mouse tracheal epithelium from BCs after SO2 injury. (B) Confocal images of epithelium at steady state and 24, 48, and 72 hpi to show distribution of Trp63 and Krt5 (BC markers), ZO1 (Tjp1) and Cldn4 (components of apical tight junctions), E-cadherin (E-cad; component of adhesion junctions), Krt8 (luminal cell marker), and transcription factor Grhl2. Note that Grhl2 is expressed in both Trp63+ BCs and luminal cells. Bars, 20 µm.
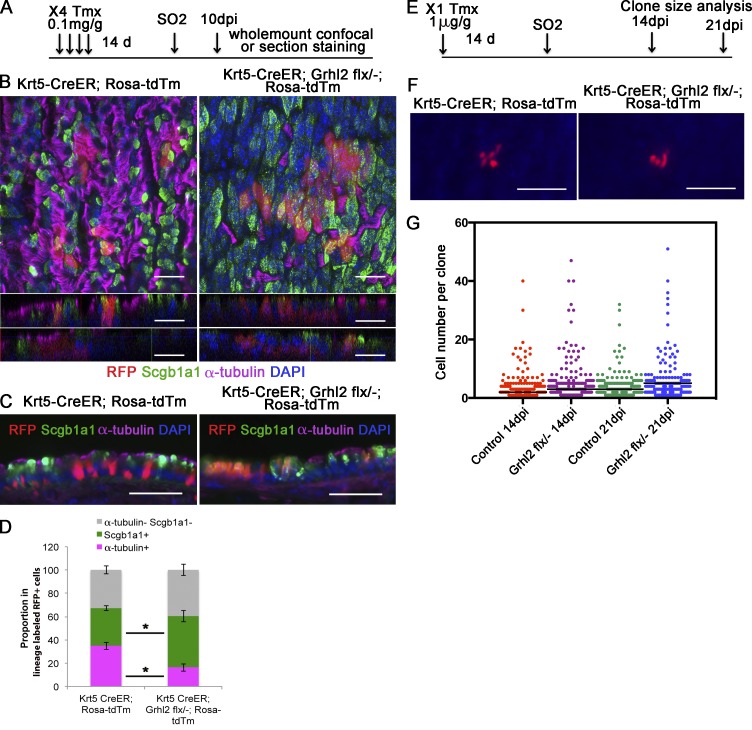
Grhl2 is expressed in all tracheal epithelial cells both before and during the repair process, including the Krt5+ Trp63+ BCs (Fig. 1 B). To test the function of Grhl2 in Krt5+ cells during repair in vivo, we generated a Grhl2flx allele in which recombination deletes exon 3 (see Materials and methods section Mice). Adult male Krt5-CreER;Grhl2flx/null;Rosa-tdTm experimental mice and Krt5-CreER;Grhl2+/+;Rosa-tdTm controls were treated with tamoxifen (Tmx) 2 wk before exposure to SO2 according to two different regimens. In one cohort (Fig. 2 A), a relatively high dose (four doses of 0.1 mg/g body weight through gavage) was used to delete Grhl2 in ∼32% of the Krt5+ cells. In the second cohort, a single low dose (1 µg/g) was given to label only a few cells so that their clonal expansion could be assayed (Fig. 2 E). In both cases, tracheas were examined at times when repair is normally complete (10, 14, and 21 d postinhalation [dpi]).
Figure 2.
Conditional deletion of Grhl2 in tracheal BCs inhibits ciliated cell differentiation but not clonal expansion. (A) Schematic for lineage labeling and deleting Grhl2 in BCs before injury, and analysis of regenerated epithelium. Four injections of 0.1 mg/g Tmx were given every other day. (B) Whole-mount staining of tracheal epithelium of Krt5-CreER;Rosa-tdTm (left) and Krt5-CreER;Grhl2flx/−;Rosa-tdTm (right) mouse for RFP (red), Scgb1a1 (green), α-tubulin (magenta), and DAPI (blue) 10 dpi. Top panels are maximum intensity projections generated from z-stack confocal images. Bottom panels are z-direction images from two different regions. (C) IHC of tracheal sections stained for α-tubulin, Scgb1a1, RFP, and DAPI. (D) Quantification of the proportion of lineage labels scored as ciliated (magenta) and secretory (green) cells or neither (gray). Data are from tracheas of three mice of each genotype, and quantification is described in text (Results, third paragraph) and Materials and methods (section Quantification of cell types). Error bars are SEM. *, P < 0.05 by Student’s t test. (E) Schematic for clonal analysis during repair. Before SO2 exposure, mice were given a low dose of Tmx that was shown to label single BCs. (F) Whole-mount fluorescence image of the surface of the tracheas of Krt5-CreER;Rosa-tdTm and Krt5-CreER;Grhl2flx/−;Rosa-tdTm mice 14 d after injury. (G) Clone size of lineage-labeled cells at 14 and 21 dpi. Control mice were Ghrl2flx/+ or Grhl2+/+. Numbers of mice and clones analyzed were as follows: control 14 dpi (5, 202); Grhl2flx/− 14 dpi (4, 313); control 21 dpi (5, 214); Grhl2flx/− 21 dpi (3, 237). P > 0.05 by unpaired Student’s t test. Horizontal bars represent mean. Bars: (B) 20 µm; (C and F) 50 µm.
The effect of deleting Grhl2 in many Krt5+ cells is shown in Fig. 2 (B–D). Whole-mount and section immunohistochemistry (IHC) coupled with confocal analysis and quantification showed that deleting Grhl2 resulted in a reduction in the proportion of Krt5+ cells that become multiciliated cells at 10 dpi (32.6 ± 4.7% for wild type vs. 16.3 ± 3.2% for mutant; n = 3 mice, P = 0.004). At the same time, there was a significant increase in the proportion of cells that express Scgb1a1, a marker for the Club cell secretory lineage (29.9 ± 3.6% vs. 44.0 ± 5.0%; P = 0.02). There was, however, no change in the proportion of cells that express neither acetylated tubulin nor Scgb1a1, presumably BCs and undifferentiated progenitors (37.5 ± 7.2% vs. 39.7 ± 4.7%; Fig. 2 D).
To study the effect of deleting Grhl2 on the clonal expansion of Krt5+ stem and progenitor cells, we performed a whole-mount analysis using tracheas from mice exposed to only a single low dose of Tmx (Fig. 2 E). Clones were examined in whole mount at several different positions along the dorsal–ventral and anterior–posterior axis, at both 14 and 21 dpi. As shown in Fig. 2 (F and G), there was no statistically significant difference in the mean size of mutant versus wild-type or heterozygous clones. Collectively, these results indicate that loss of Grhl2 from Krt5+ BCs does not affect their ability to proliferate in the context of in vivo epithelial repair. Rather, it preferentially reduces the differentiation of mutant cells along the ciliated lineage, and more daughters become secretory cells.
Loss of Grhl2 in mouse BCs inhibits epithelial morphogenesis in clonal organoid culture and differentiation into ciliated cells
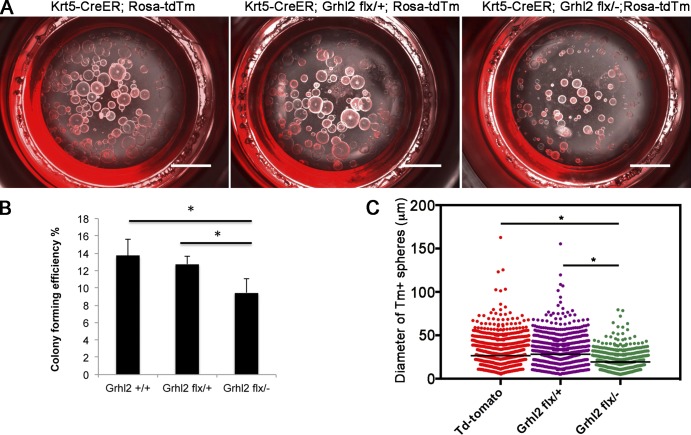
To further explore the mechanism by which Grhl2 regulates the behavior of basal progenitors, we exploited the ability of single BCs to give rise in 3D culture to organoids (tracheospheres) containing basal, secretory, and ciliated lineages around a central lumen (Rock et al., 2009; Tadokoro et al., 2014). Because spheres are clonal, mutant cells are associated only with other mutant cells and are not intermingled with wild-type or heterozygous cells, as in tracheal repair in vivo. The factors affecting the efficiency with which single cells generate clonal tracheospheres have not been completely defined but include the ability to survive after dissociation and the probability of BC self-renewal versus differentiation. In addition, the diameter of a sphere will depend not only on cell proliferation but also on lumen formation (Jaffe et al., 2008), polarized fluid transport, and barrier function. For organoid culture, BCs that express both the Tm+ lineage label and the surface marker Ngfr were isolated by FACS (Fig. S2) from tracheas of Krt5-CreER;Rosa-tdTm mice that were either Grhl2+/+ or Grhl2flx/+ (controls) or Grhl2flx/−. Cells were embedded in extracellular matrix and cultured for up to 21 d. As shown in Fig. 3 (A and B), there was a significant decrease in the colony forming efficiency (CFE) of cells lacking Grhl2 compared with controls (CFE = 13.71 for wild-type cells and 9.45 for mutant; n = 3, P = 0.025). In addition, the mutant spheres were on average ∼36% smaller in diameter than controls (292.5 ± 4.6 µm for wild type, 308.7 ± 5.9 µm for heterozygous cells, and 193.1 ± 3.8 µm for mutants; P < 0.0001; Fig. 3 C).
Figure 3.
Deletion of Grhl2 in BCs reduces CFE in 3D tracheosphere cultures and sphere size. (A) Representative differential interference contrast (DIC) and fluorescence microscopy images at culture day 14 of spheres formed in Matrigel from lineage-labeled Ngfr+ BCs isolated by FACS (red). (B) CFE of the lineage-labeled BCs. Data are reported as mean ± SEM. (C) The diameters of spheres in triplicate wells from four biological replicates. *, P < 0.05. Bars, 2 mm.
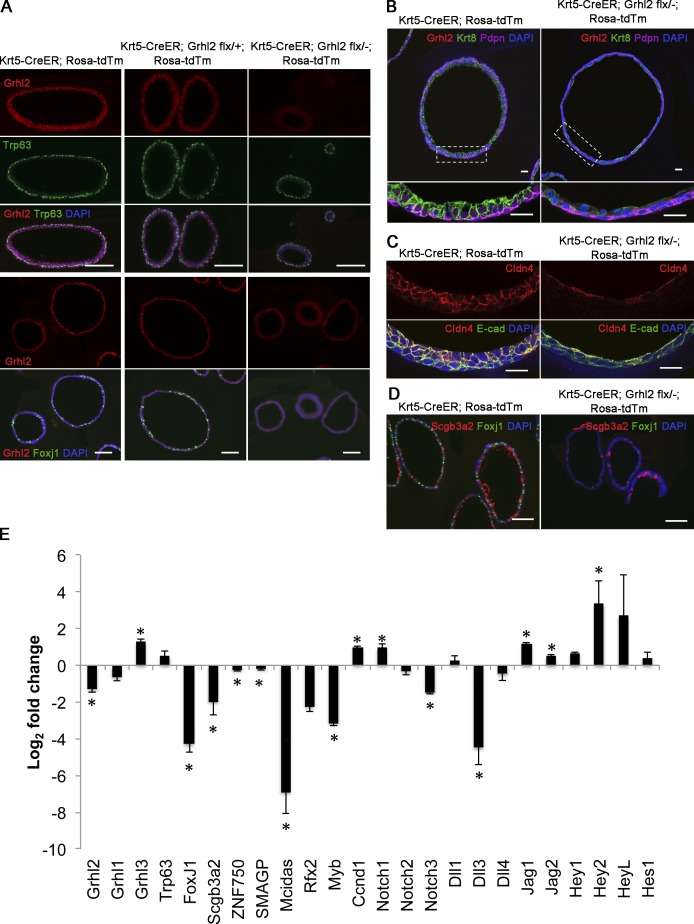
To follow the fate of lineage-labeled cells in individual spheres, organoid cultures were analyzed by IHC for markers of basal (Trp63+ Pdpn+), luminal (Krt8), secretory (Scgb3a2), and ciliated (Foxj1) cells. Because some Tm+ BCs isolated from Krt5-CreER;Rosa-Tm;Ghl2flx/− mice may have recombined the reporter allele but not Grhl2flx, spheres were confirmed as being derived from mutant cells by an absence of Grhl2 protein. As shown in Fig. 4 (A and B), mutant spheres contained both Trp63+ Pdpn+ BCs and Krt8+ luminal cells. However, the mutant luminal cells were flatter and more squamous than the columnar Krt8+ control cells and had shorter basolateral membrane domains (Fig. 4 B). They also appeared to express less Cldn4 than controls (Fig. 4 C). Luminal cells positive for the secretory marker, Scgb3a2, were present, but there were very few Foxj1+ ciliated cells (Fig. 4, A and D). This deficiency was still seen after 21-d culture, indicating that it is not just caused by a delay in differentiation. To extend these findings, we performed a quantitative RT-PCR (qRT-PCR) analysis of genes expressed in mutant and control spheres at 14 d. As shown in Fig. 4 E, the expression of genes encoding transcription factors that are known to promote ciliogenesis (Multicilin [Mcidas], Rfx2, and Myb) are all significantly down-regulated in Grhl2 mutant spheres. This is consistent with our previous ChIP-Seq evidence that these genes are direct targets for Grhl2 (Gao et al., 2013). Transcripts of the ciliated cell–specific gene, Foxj1, are also reduced, although this gene does not have associated Grhl2-binding sites. In contrast, expression of Grhl3 and Ccnd1 (cyclin D1), two other potential direct Grhl2 targets, is up-regulated. Although Club cells are present in mutant spheres after 14 d of culture (Fig. 4 D), levels for the secretory cell–specific gene, Scgb3a2 (which is not a potential target), were down-regulated as judged by qRT-PCR, whereas there was no change in the levels of Scgb1a1 RNA.
Figure 4.
Deletion of Grhl2 in BCs impairs apical–basal polarity of daughter cells in clonal spheres and their differentiation into ciliated cells. (A) At day 14, spheres were fixed, sectioned, and stained for Grhl2, Trp63 or Foxj1, and DAPI. Note the presence of Trp63+ BCs and absence of Foxj1 cells in spheres that do not stain for Grhl2 (right). (B) Sections of spheres stained for Grhl2, Krt8, Pdpn (BC marker), and DAPI. Bottom panels are higher magnifications of boxed regions. Note the low cuboidal shape of mutant Krt8+ luminal cells. (C) Sections of similar spheres stained for E-cadherin (E-cad) and Cldn4. (D) Sections stained for Scgb3a2, Foxj1, and DAPI. Note the absence of ciliated cells and the presence of secretory cells in mutant spheres. (E) qRT-PCR analysis of grainyhead-like genes and genes involved in cell differentiation, ciliogenesis, and Notch signaling in spheres cultured for 14 d. Data (±SEM) are from triplicates of two biological replicates. *, P < 0.05. Bars: (A and D) 100 µm; (B and C) 20 µm.
Previous studies have shown a key role for Notch signaling in the self-renewal and differentiation of adult airway BCs, with high levels of Notch promoting the secretory lineage. Specifically, recent studies have implicated Notch3 in promoting the differentiation of adult basal stem cells into parabasal progenitors and Notch1 and 2 in fate selection between secretory and ciliated linages (Rock et al., 2011b; Danahay et al., 2015; Mori et al., 2015; Pardo-Saganta et al., 2015). ChIP-Seq has shown Grhl2 occupancy of binding sites near NOTCH1, NOTCH3, JAG1, and JAG2 (Gao et al., 2013). We therefore assayed the level of expression of various Notch pathway components in organoid cultures and found that in mutant spheres Notch1, Jag1, and Jag2 were significantly up-regulated, whereas Notch3 and Dll3 were down-regulated compared with wild type. These data suggest that Grhl2 is a direct regulator of the Notch pathway in airway progenitors.
Use of CRISPR/Cas9 genome editing to test the function of GRHL2 and putative downstream targets in primary human basal airway cells
In previous studies, we used a combination of ChIP-Seq and transcriptomic analysis to identify several hundred genes as likely direct targets of Grhl2 in primary human bronchial epithelial (HBE) cells. These genes are therefore potentially part of a network that regulates and integrates airway epithelial morphogenesis and differentiation from basal progenitors (Gao et al., 2013). However, the functions of most of these genes in airway epithelial cell biology are not known. To address this challenge, we used CRISPR/Cas9 genome editing technology to generate mutations in a subset of target genes in HBE cells and assayed their effect on cell proliferation, polarity, barrier function, and lineage differentiation. We used two different assay conditions. The first involves ALI culture in which the starting population of Krt5+ Trp63+ BCs differentiates to form a mucociliary epithelium with barrier function. The second assay uses clonal organoid (bronchosphere) culture in which single BCs both self-renew and give rise to ciliated and secretory cells.
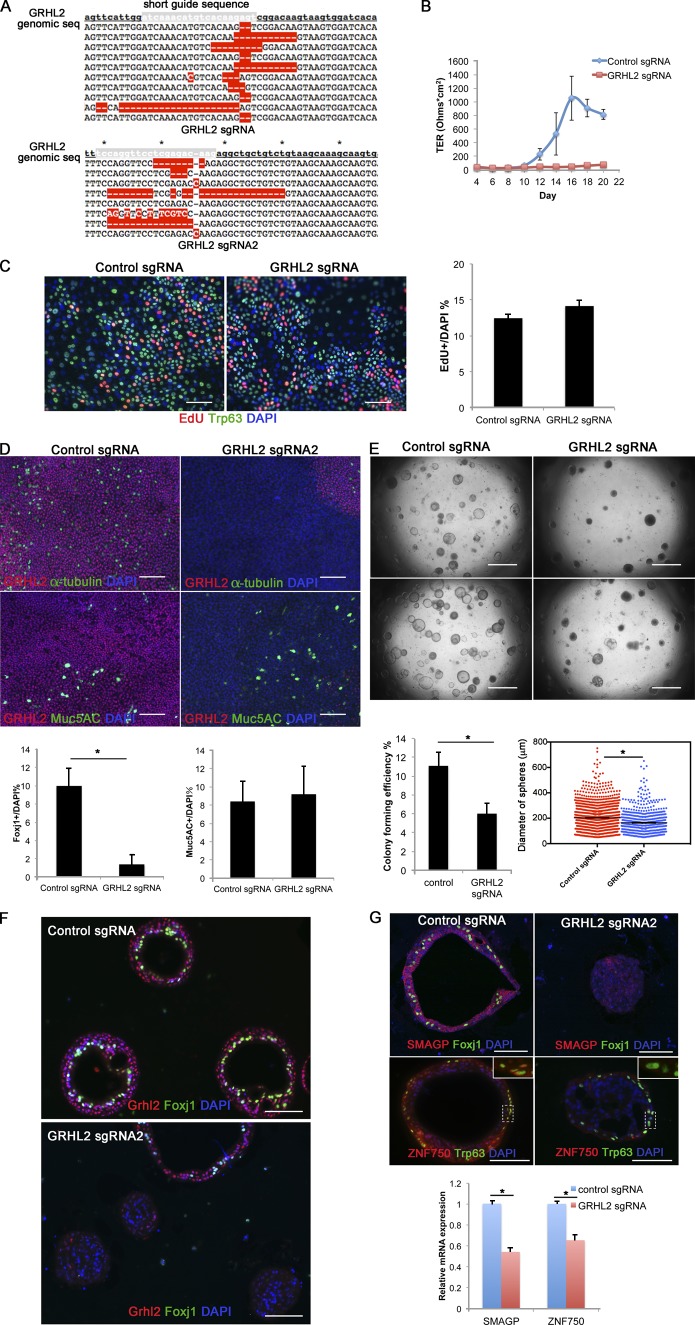
As proof of principle, we first generated small deletions in GRHL2 using a puromycin-inducible lentivirus expressing short guide (sg) RNAs complementary to sequences in the trans-activating domain of the protein (Shalem et al., 2014). We used two independent sgRNAs to be sure that results were reproducible, and both gave similar results. As shown in Fig. 5 A, the two lentiCRISPR-sgGRHL2 vectors induced insertions or deletions (indels) in most of the sequenced genomic DNAs assayed from a population of puromycin-resistant cells. 80% of the mutant alleles were predicted to encode mutant or truncated protein. Control experiments using cells from three different donors showed that after selection, there was a decrease in wild-type RNA transcripts from the GRHL2 locus and Grhl2 protein levels in cells transfected with lentiCRISPR-sgGRHL2 virus (Fig. S5).
Figure 5.
Mutation of GRHL2 in HBE cells using CRISPR/Cas9 inhibits the development of TER and differentiation of ciliated cells. (A) Sequences of 18 clones amplified from the target region of genomic DNA of selected cells transfected with any lentiCRISPR-GRHL2 sgRNA viruses. Target regions amplified from control cells showed no mutations. The red highlights are mismatched, missing, or inserted nucleotides. Asterisks indicate every 10 nucleotides of the genomic sequence. (B) Selected cells were cultured in ALI, and TER was measured every other day. Values are mean and SEM of triplicate wells, and data are from one of n = 4 biological replicates. Other replicates gave similar results. (C) At ALI day 3, cell proliferation was assayed using EdU incorporation over 2 h, and EdU and the BC marker Trp63 were analyzed by IHC. The right panel is quantification of the proportion of EdU+ cells. P = 0.077 > 0.05. (D) Whole-mount IHC of control and GRHL2 mutant ALI cultures at day 21 for α-tubulin (ciliated cell marker), Muc5AC (secretory cell marker), and Grhl2. Note the presence of a small patch of cells that still express Grhl2 protein in the top right panel. The bottom panel is the quantification of the proportion of Foxj1+ ciliated and Muc5AC+ secretory cells. Values are mean and SEM of triplicate wells from one donor. Other donors gave similar results. *, P < 0.05. (E) Representative DIC microscopy images of spheres at day 21 from BCs from two different donors. (Bottom) CFE of GRHL2-sgRNA mutant cells is decreased compared with control, and the diameters of mutant spheres are smaller. *, P < 0.05. Means are from triplicate wells from four donors. Horizontal bars represent mean. (F) At day 21, spheres were fixed, sectioned, and stained for GRHL2, Foxj1, and DAPI. (Top) Spheres from control culture. (Bottom) Spheres from GRHL2 sgRNA–transfected cultures. One sphere contains Grhl2+ and Foxj1+ cells. All other spheres are Grhl2 and Foxj1–. (G) Reduced expression of Smagp and Znf750 in GRHL2 mutant cells. (Top) IHC for Smagp and Znf750 protein in sections of wild-type (left) and GRHL2 mutant (right) spheres. Insets show higher magnification of Znf750 staining in wild-type, but not mutant, Trp63+ cells. (Bottom) Reduced expression of SMAGP and ZNF750 RNA in GRHL2 mutant cells grown for 21 d in ALI culture. Bars: (C, D, F, and G) 100 µm; (E) 2 mm. Data are reported as mean ± SEM.
When sgGRHL2 mutated cells were cultured in ALI, the epithelial monolayer did not establish electrical resistance (Fig. 5 B), as previously observed using cells expressing a dominant-negative Grhl2 protein (Gao et al., 2013). In contrast, control cells developed electrical resistance beginning at ∼10 d. Control experiments based on 5-ethynl-2′-deoxyuridine (EdU) incorporation over 2 h showed that mutant cells proliferated at the same rate as controls (Fig. 5 C). After 21 d in ALI culture, monolayers were stained in whole mount using antibodies to Foxj1, Muc5AC, and Grhl2. Quantification of the results showed that mutant cells had greatly reduced the ability to differentiate into multiciliated cells, whereas differentiation into secretory cells was not affected (Fig. 5 D).
In a second set of experiments, single cells infected with either of the two lentiCRISPR-sgGRHL2s or control vector were seeded into extracellular matrix and grown in organoid (bronchosphere) culture. As shown in Fig. 5 E, the CFE of mutant cells was reduced compared with controls, and spheres were smaller on average. IHC showed that control spheres have a single lumen lined by an epithelium with multiciliated cells (Fig. 5 F). This was also true for the few spheres in cultures of resistant cells that were still Ghrl2+, presumably because they were derived from single cells in which the indel did not generate a mutant protein or were not GRHL2 homozygous mutant. In contrast, none of the small mutant spheres lacking Grhl2 protein contained Foxj1+ cells (Fig. 5 F).
We then used lentiCRISPR vectors to induce mutations in nine genes predicted from a previous ChIP-Seq and transcriptomic study (Gao et al., 2013) to be direct positive targets of Grhl2 and for which no function is known in the lung. These were ANKRD22, MPZL2, PVRL4, SCRIB, SMAGP, RHBDL2, VGLL1, WDR5B, and ZNF750. The sgRNA sequences were selected to generate indels near the 5′ end of the gene (Shalem et al., 2014). As shown in Fig. S3, most of the lentiCRISPR vectors induced indels in ∼60–100% of the alleles sequenced from puromycin-resistant populations. However, efficiency of indel induction was only 33% for WDR5B. We then tested the ability of mutant cells to differentiate in organoid cultures. As shown in Fig. S4, there was a significant reduction in CFE for cells mutant in SMAGP, ZNF750, PVRL4, and ANKRD22, but not the other genes tested.
For further studies, we focused on SMAGP (encoding a small transmembrane and glycosylated protein; Tarbé et al., 2004) and ZNF750 (also known as zinc finger protein 750, or Zfp750; encoding a member of the C2H2 subclass of zinc finger transcription factors; Birnbaum et al., 2006; Cohen et al., 2012; Sen et al., 2012; Boxer et al., 2014). As summarized in Fig. S4, our ChIP-Seq analysis of primary HBE cells had shown that these two genes have occupied Grhl2-binding sites near the 5′ end (Gao et al., 2013). Two independent lentiCRISPR-sgSMAGPs and -sgZNF750s reduced levels of the corresponding RNA and protein in donor cells after selection (Fig. S5). In addition, transcripts for the two genes were reduced in mouse BCs lacking Grhl2 (Fig. 4 E), and both proteins and RNAs were down-regulated in HBE cells mutant for GRHL2 (Fig. 5 G).
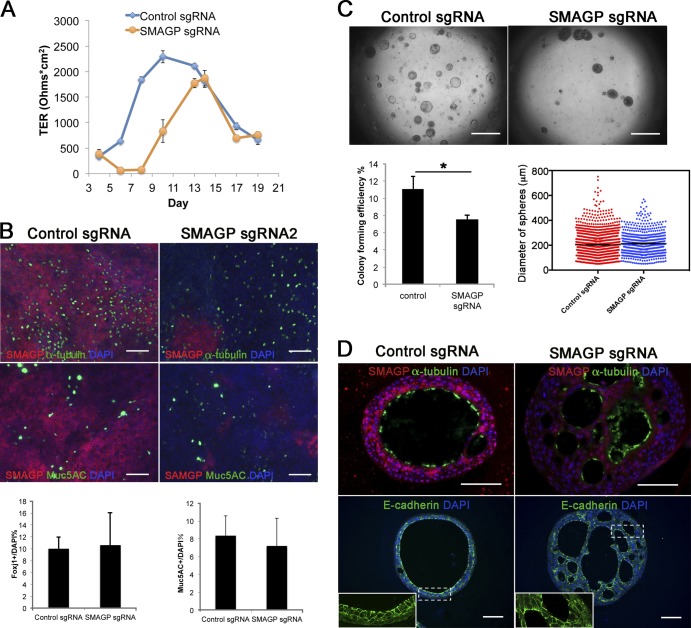
In the case of SMAGP, when cells treated with the two different lentiCRISPR-sgRNAs were assayed in ALI culture, there was an ∼2-d delay in establishing electrical resistance but no significant reduction in the differentiation of the epithelial cells into ciliated and secretory cells after 21 d (Fig. 6, A and B). When cells were placed in organoid culture, there was a 31.4% reduction of CFE, but the spheres that did form had, on average, about the same diameter as controls, although they had a denser morphology (Fig. 6 C). Histological analysis showed that mutant spheres did contain multiciliated cells (Fig. 6 D). However, many spheres had multiple lumens, and the E-cadherin+ cells lining them were not as uniform in shape as those in control spheres (Fig. 6 D, inset). Smagp encodes a conserved transmembrane protein and has been localized to the lateral membrane of well-polarized epithelial cells in several tissues (Tarbé et al., 2004). However, its expression in the pseudostratified epithelium of the normal human lung has not been reported. We therefore stained sections of normal lung with antibodies to Smagp and observed strong expression in the basolateral membrane of the columnar luminal cells in bronchial epithelium (see Fig. 8 B).
Figure 6.
Effect of mutation of SMAGP on phenotype of HBE cells. (A) ALI cultures were tested for TER as described in Materials and methods (section HBE cell culture). Values are mean and SEM of triplicate wells, and data are from one of n = 3 biological replicates. (B) Whole-mount IHC of control and SMAGP mutant cultures at ALI day 21 for Smagp, Foxj1, and Muc5AC. A few small patches of Smagp+ cells are present in the cultures of mutant cells. Bottom panel is quantification of the proportion of ciliated and secretory cells. Values are mean and SEM of duplicate/triplicate wells from one of three donor samples. P > 0.05. (C, top) Representative DIC microscopy images of spheres at day 21. (Bottom) The CFE of SMAGP sgRNA mutant cells is decreased compared with control. However, the diameters of control and mutant spheres are similar. *, P > 0.05. Spheres were analyzed in triplicate wells from three biological replicates. Horizontal bars represent mean. Data are reported as mean ± SEM. (D) At day 21, spheres were fixed, sectioned, and stained for SMAGP, α-tubulin or E-cadherin, and DAPI (blue). Insets are higher magnification of the boxed regions. Bars: (B and D) 100 µm; (C) 2 mm.
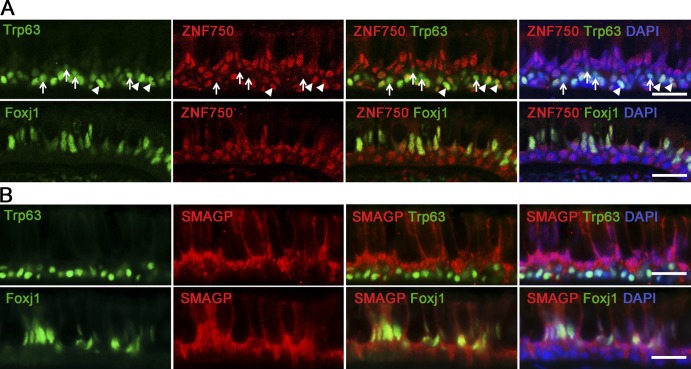
Figure 8.
Localization of ZNF750 and SMAGP proteins in human airway epithelial cells. (A) IHC of section through human lung (6-mm-diameter airway) for ZNF750, Trp63 (top), Foxj1 (bottom), and DAPI. Note that some Trp63+ BCs express ZNF750 (arrowheads), whereas others do not (arrows). (B) IHC for SMAGP, Trp63 (top), Foxj1 (bottom), and DAPI. Single images from confocal stacks. Bars, 25 µm.
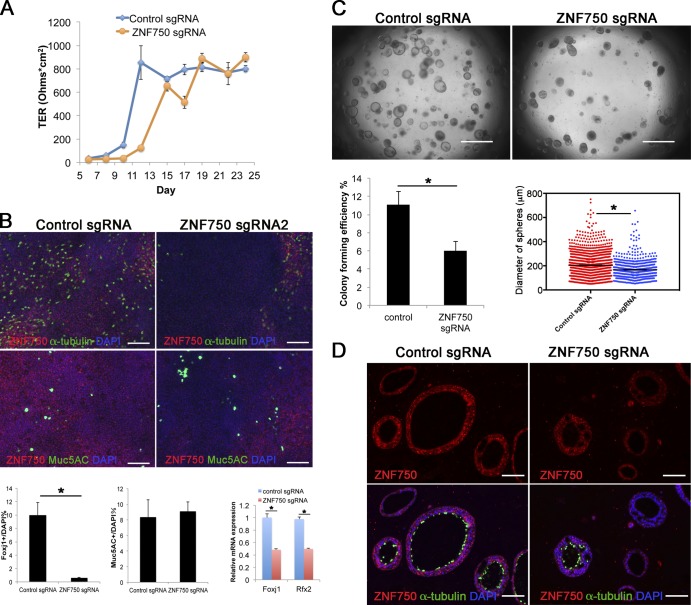
In the case of ZNF750, when cells treated with lentiCRISP-sgRNAs were assayed in ALI culture, there was also a delay of ∼2 d in establishing electrical resistance compared with controls (Fig. 7 A). Whole-mount IHC of the monolayers after 21 d showed a reduction in the number of acetylated tubulin+ ciliated cells, but there was no change in the proportion of secretory cells. This result was confirmed by qRT-PCR showing a reduction in both FoxJ1 and Rfx2 transcripts in mutant cells (Fig. 7 B). Organoid culture demonstrated a 45.5% reduction in both CFE and sphere size in ZNF750 mutant cells comparable with that seen with cells treated with lentiCRISPR-sgGRHL2 (Fig. 7 C). Likewise, there was reduced differentiation of cells lacking ZNF750 into ciliated cells (Fig. 7 D). Expression of ZNF750 protein has been reported in several epithelial tissues including the suprabasal (but not basal) cells of the epidermis (Boxer et al., 2014). However, there is no study of expression in airway mucociliary epithelium. We therefore examined expression in sections of normal human lung. ZNF750 is present in most of the nuclei of luminal cells, including Foxj1+ ciliated cells, and in ∼63% of the Trp63+ BCs (Fig. 8 A). Znf750 staining was also seen in ∼73% of Trp63+ HBE cells in organoid culture (Fig. 5 G). As discussed below (Discussion section Grhl2 and epithelial differentiation), this expression in a subpopulation of BCs has implications for models of Znf750 function in airway differentiation.
Figure 7.
Effect of mutation of ZNF750 on phenotype of HBE cells. (A) ALI cultures were tested for TER as described in Materials and methods (section HBE cell culture). Values are mean and SEM of triplicate wells, and data are from one of n = 3 donor samples. (B, top) Whole-mount IHC of control and ZNF750 mutant cultures at day 21 ALI stained for α-tubulin, Muc5AC, and Znf750. (Bottom) Proportion of ciliated and secretory cells in ALI culture. (Bottom right) qRT-PCR analysis of FoxJ1 and Rfx2 RNA in control and ZNF750 mutant cells cultured in ALI for 21 d. Values are mean and SEM of triplicate wells from one of three donor samples. *, P < 0.05. (C, top) Representative DIC microscopy images of spheres at day 21. (Bottom) CFE of ZNF750 sgRNA mutant cells is decreased compared with control, and the diameters of mutant spheres are smaller. *, P < 0.05. Spheres were analyzed in triplicate wells from three donor samples. Horizontal bars represent mean. Data are reported as mean ± SEM. (D) At day 21, spheres were fixed, sectioned, and stained for ZNF750 and α-tubulin. Note that the spheres that do not express ZNF750 do not have ciliated cells. Bars: (B and D) 100 µm; (C) 2 mm.
Discussion
In this paper, we use a combination of conditional gene deletion and CRISPR/Cas9 gene editing technology to elucidate the role of Grhl2 in coordinating the morphogenesis and differentiation of airway epithelium from unpolarized basal progenitors. Our results and recent studies by others with kidney and placenta (Aue et al., 2015; Walentin et al., 2015) clearly establish that grainyhead-like proteins coordinately regulate processes such as cell polarity, adhesion, and barrier function. Significantly, we also show that Grhl2 positively regulates genes associated with progenitor cell differentiation and lineage choice. These include multiple Notch pathway and ciliogenesis genes and the transcription factor Znf750, which has recently been implicated in driving epidermal keratinocyte differentiation (Sen et al., 2012; Boxer et al., 2014). Collectively, our findings establish a role for Grhl2 as a key integrator of epithelial morphogenesis and differentiation in multiple vertebrate organ systems.
Grhl2 and airway epithelial architecture
Previously, we used expression of a dominant-negative Grhl2 protein to inhibit Grhl2 function in primary HBE cells (Gao et al., 2013). This prevented the cells from developing electrical resistance in ALI culture, even though they formed a confluent monolayer. At the same time, there was transcriptional down-regulation of many potential Grhl2 target genes (Gao et al., 2013) implicated in cell–cell interactions and selective permeability. Here, we elaborate on these findings to show defects in both barrier function (development of electrical resistance) and epithelial morphogenesis in mouse and human BCs in which Grhl2 has been conditionally deleted or GRHL2 mutated using CRISPR/Cas9. These defects are observed when mutant cells are either isolated by FACS or selected in culture and subsequently grown in monolayer or clonal organoid culture assays. These findings are consistent with recent studies of small and collapsed lumens in the nephric ducts of Grhl2-null mutant mouse embryos (which die before birth; Aue et al., 2015). Collectively, it appears that the absence of Grhl2 in all or the majority of epithelial cells in a tissue or in culture results in defects in columnar morphology and permeability that may ultimately lead to leakiness, reduced fluid transport, and small lumen size.
Given these observations, the phenotype of the regenerated tracheal epithelium of mice, in which Grhl2 was conditionally deleted in ∼30% of the BCs, was somewhat surprising. Although there was a reduction in the proportion of multiciliated cells (discussed in the next section), the overall gross morphology of the epithelium was relatively normal. In particular, there was no evidence for subepithelial inflammation or edema, as might be expected if the epithelium was leaky or if barrier function was locally disrupted. It therefore appears that in vivo, the presence of adjacent wild-type cells can compensate for some of the phenotypic defects of mutant cells seen in vitro.
Grhl2 and epithelial differentiation
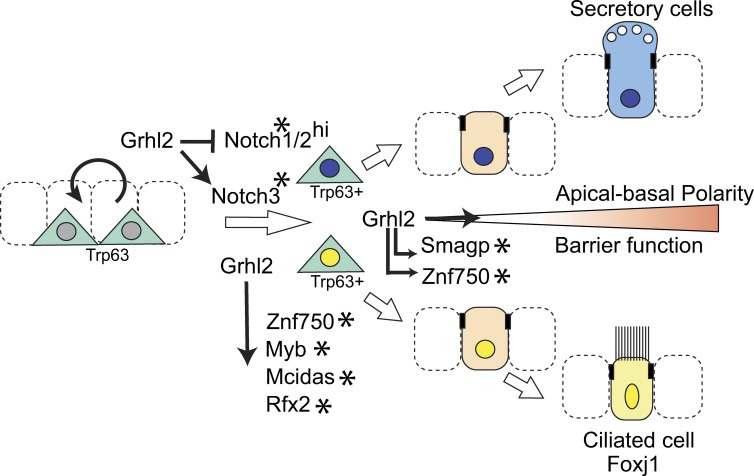
A consistent finding in this study is that conditional deletion or mutation of Grhl2 in mouse or human BCs leads to their reduced differentiation into Foxj1+ multiciliated luminal cells. This inhibition does not appear to be merely secondary to changes in cell morphogenesis because BCs in which SMAGP is mutated do differentiate into ciliated and secretory cells as normal, even though barrier function is delayed and mutant spheres do not form a single lumen (Fig. 6). Rather, we favor the explanation that Grhl2 regulates ciliogenesis by acting directly through transcriptional activation of target genes known to regulate ciliogenesis such as Mcidas (Multicilin), Myb, and Rfx2 (Fig. 9). Two lines of evidence indicate that these genes are direct targets of Grhl2. First, each is associated with robust binding sites that are occupied in HBE cells, as shown by ChIP-Seq (Gao et al., 2013), and second, their transcript levels decrease in spheres derived from Grhl2 mutant BCs (Fig. 4). In addition, Grhl2 likely regulates ciliogenesis by promoting transcription of Notch3 and inhibiting Notch1, both of which have binding sites. Evidence from other studies suggests that Notch3 promotes transition of basal stem cells into differentiating progenitors, whereas Notch1 and 2 promote differentiation into secretory cells (Mori et al., 2015).
Figure 9.
Model for Grhl2 function in coordinating differentiation and morphogenesis of airway epithelium. Krt5+ Trp63+ BCs of the pseudostratified mucociliary epithelium both self-renew and give rise to both Foxj1+ ciliated and secretory cells. This differentiation involves notch signaling and is thought to proceed via Trp63+ progenitors committed to either lineage pathway (yellow or blue nuclei; Mori et al., 2015; Pardo-Saganta et al., 2015; Watson et al., 2015). As progenitors and their descendants integrate into the epithelium, they acquire apical–basal polarity and apical–junctional complexes mediating barrier function. We propose that Grhl2 coordinates these changes by acting through multiple downstream effectors, including direct Grhl2 target genes indicated by asterisks.
Another significant finding from this work is that mutation of ZNF750 (Zfp750 in mice), a putative Grhl2 target gene encoding a zinc finger transcription factor, leads to a delay in barrier formation and inhibition of ciliogenesis (Fig. 7). This raises the possibility that Znf750 is both a novel ciliogenesis gene and a regulator of luminal cell–cell interactions and selective permeability. Previous studies have shown that Znf750 is expressed in suprabasal epidermal keratinocytes in vivo and plays an important role in both inhibiting the progenitor phenotype of BCs and promoting their terminal differentiation and barrier function (Cohen et al., 2012; Sen et al., 2012; Boxer et al., 2014). This is achieved by Znf750 acting as a gene repressor in conjunction with chromatin-modifying proteins and as a transcriptional activator in conjunction with Klf4. In the epidermis, Znf750 is not expressed in BCs. In contrast, we found that in the bronchial epithelium of the adult human lung, ZNF750 is expressed in all luminal cells and in ∼63% of BCs (Fig. 8). This pattern of expression is consistent with a role in promoting the differentiation of ciliated cells and in suppressing the multipotency of ∼37% of BCs. Significantly, recent mathematical modeling of BC lineage–tracing experiments in the uninjured mouse trachea suggests that about half of the Krt5+ BCs are already committed to luminal differentiation (Watson et al., 2015). By analogy, based on studies in the epidermis (Cohen et al., 2012; Sen et al., 2012; Boxer et al., 2014), we speculate that in the pseudostratified mucociliary epithelium, Znf750 marks the BC population that has a high probability of differentiation. Moreover, we suggest that it functions to both repress multipotency and promote expression of genes related to establishment of barrier function and ciliogenesis (Fig. 9). In the future, this model can be tested by lineage tracing Znf750+ cells during repair as well as with ChIP-Seq and transcriptomic studies.
In conclusion, we have used a combination of in vivo conditional gene deletion and in vitro CRISPR/Cas9 gene editing techniques to demonstrate that Grhl2 functions in basal stem cells as a central coordinator of both epithelial morphogenesis and barrier function as well as lineage commitment and differentiation. Moreover, we show that CRISPR/Cas9 technology in combination with morphogenetic assays is a powerful method to rapidly screen for genes regulating epithelial cell morphogenesis and differentiation and identify two new downstream effectors of Grhl2 action in mucociliary epithelium, namely Smagp and Znf750.
Materials and methods
Mice
The Grhl2flox targeting construct was obtained from the KOMP Repository (project CSD30984; clone PRPGS00075_B_H01) and confirmed by DNA sequencing. The linearized vector was electroporated into C57BL/6N-PRX-B6N #1 mouse embryonic stem cells (The Jackson Laboratory). Long-range PCR was used to verify homologous recombination in the 5′ and 3′ homology arms and DNA sequencing to verify the LoxP site. Three correctly targeted ES clones were used to generate chimeras by blastocyst injection. Grhl2flox mice were crossed with ACTB:FLPeB6J mice (The Jackson Laboratory) to remove the Neo and LacZ cassette. Mice carrying the null Grhl2tm1.1Jane (provided by S. Jane, Monash University, Melbourne, Victoria, Australia; Rifat et al., 2010), Krt5-CreER (Van Keymeulen et al., 2011), and Tjp1tm1lch alleles (provided by T. Lechler, Duke University School of Medicine, Durham, NC; Foote et al., 2013; Huebner et al., 2014) and ROSA-CAG-tdTomato (Rock et al., 2011a) were maintained on a C57BL/6 background. Adult mice were 8–12 wk old. Between two and five animals were used in each experiment as indicated.
To induce recombination in BCs, Tmx (Sigma-Aldrich) in corn oil was administered through gavage. To label many BCs, 0.1 mg/g of body weight Tmx was given to adult mice every other day for a total of four times. For clonal analysis, a single dose of 1 µg/g of body weight Tmx was given. For SO2 exposure experiments, the final Tmx dose was given 2 wk before injury. Adult male mice were placed in individual compartments within a chamber and exposed to 500 parts per million SO2 in air for 4 h.
IHC
Normal human lungs, which were obtained under Institutional Review Board–approved protocols from donors without preexisting lung disease, and mouse tracheas were fixed in 4% (wt/vol) PFA at 4°C for 3 h. ALI cultured cells and spheres were fixed in 4% PFA for 10 min and 30 min at RT, respectively. Samples were embedded in paraffin, sectioned at 7 µm, and stained as described previously (Rock et al., 2011b). For whole-mount staining of ALI culture, after fixation and washing with PBS, the membrane was cut from the insert and stained by standard protocols. Antibodies were rabbit GRHL2 (1:450; human protein atlas number HPA004820; Sigma-Aldrich), rat E-cadherin (1:500; 13-1900; Invitrogen), mouse monoclonal Trp63 (1:50; model sc-8431; Santa Cruz Biotechnology, Inc.), mouse monoclonal MUC5A (1:200; MS145-PO; Lab Vision), mouse Foxj1 (1:1,000; 14-9965; eBioscience), mouse acetylated tubulin (1:1,000; T-7451; Sigma-Aldrich), rabbit Scgb3a2 (1:5,000, a gift from S. Kimura, National Cancer Institute, Bethesda, MD; mature 70 amino acids used as antigen), Hamster Pdpn (1:1,000; Developmental Studies Hybridoma Bank; 8.1.1), rabbit Scgb1a1 (1:10,000, a gift from B. Stripp, Cedars-Sinai Medical Center, Los Angeles, CA; 4× His-tagged full-length recombinant rat Scgb1a1 used as antigen), rabbit RFP (1:250; 600-401379; Rockland) rabbit Cldn4 (1:150; 36-4800; Invitrogen), rabbit SMAGP (1:100; human protein atlas number HPA039711; Sigma-Aldrich), and rabbit ZNF750 (1:50; human protein atlas number HPA021573; Sigma-Aldrich). Antigen retrieval on paraffin sections used 10-mM sodium citrate butter, pH 6, in a pressure cooker–like device (2100 Retriever; PickCell Laboratories).
Image acquisition
Images were recorded on the following microscopes: Axiovert 200 (Carl Zeiss) with 1.25×/0.035 and 2.5×/0.075 Plan-Neofluar objective lenses and Axio Imager D10 (Carl Zeiss) with 40×/0.75 EC Plan-Neofluar and 20×/0.8 Plan-Apochromat objective lenses. AxioCam MRm and HRc cameras (Carl Zeiss) were used to capture the images. Confocal images were recorded on an inverted confocal microscope (710; Carl Zeiss) with 20×/0.8 Plan-Apochromat, 40×/1.30 EC Plan-Neofluar, and 63×/1.4 Plan-Apochromat objective lenses. All images were taken at RT. The acquisitions were performed with Zen software (Carl Zeiss), and subsequent images were cropped and contrast adjusted using Photoshop (Adobe).
Quantification of cell types
For quantification of lineage-traced cells in tracheas, the following methods were used. For whole-mount staining, tracheas were fixed in 4% PFA at 4°C for 3 h, bisected longitudinally, and stained in whole mount with antibody to RFP, acetylated tubulin, and Scgb1a1. At least five random areas along the proximal–distal axis were imaged in an inverted confocal microscope (Carl Zeiss) and Tm+ lineage-labeled cells were scored as either ciliated (α-tubulin+), secretory (Scgb1a1+), or neither (basal or undifferentiated progenitors). For IHC of sections, tracheas were embedded in an optimal cutting temperature compound, and 8-µm sections were cut longitudinally in the midline. After IHC, several areas from three sections were imaged using Axio Imager D10, and cells were scored as in whole mounts. A total of 681 lineage-labeled cells were scored from control (Krt5-CreEr;Rosa-tdTomato) and 668 cells from experimental (Krt5-CreEr;Rosa-tdTomato;Grhl2flx/−) tracheas (from n = 3 different mice).
For ALI cultures, after fixation in 4% PFA at room temperature for 15 min and washing with PBS, membranes were cut from triplicate inserts from each of three to five donors and stained in whole mount by standard protocols. Two to four areas near the middle of each insert were imaged using Axio Imager D10, and Foxj1+, Muc5AC+, and DAPI+ cells were counted. Values are means and SEM of triplicate samples. Because of variability between donors in the differentiation of HBE cells, only quantification from one donor is shown in each figure, but all results were similar.
FACS experiments
To isolate mouse BCs, tracheas from Krt5-CreER;Rosa-tdTm mice treated with Tmx were incubated as described previously (Rock et al., 2009) in 16 U/ml Dispase (BD), epithelial sheets were peeled with forceps, and single cells were obtained by incubation with 0.1% trypsin at 37°C for 20 min. Cells were labeled with 9 µg/ml rabbit anti-NGFR (8875; Abcam) or rabbit IgG for 45 min in 2% FBS and 2% BSA in PBS on ice, washed, and incubated with Alexa Fluor 488–conjugated donkey anti–rabbit in 2% FBS and 2% BSA in PBS for 45 min on ice. Propidium iodide was added to exclude dead cells, and FACS was performed in the Duke Cancer Center shared flow cytometry facility. Tm+ (RFP+) NGFR+ lineage–labeled BCs were collected.
Mouse tracheosphere culture
RFP+ NGFR+ lineage–labeled BCs were resuspended in mouse tracheal epithelial cell (MTEC)/Plus culture (You et al., 2002), mixed 1:1 with growth factor–reduced Matrigel (BD), and seeded at 1,000 cells per well in a 24-well, 0.4-µm pore size polyethylene terephthalate membrane insert (Falcon model 353095; BD). MTEC/Plus was added to the lower chamber and changed every other day. Cultures were maintained at 37°C and 5% CO2. On day 7, MTEC/SF was placed in the lower chamber and changed every other day. The number of spheres per insert was counted at day 14. Samples were set up in duplicate or triplicate from four biological replicates. Spheres were counted, fixed in 4% PFA in PBS, embedded in paraffin, and sectioned.
HBE cell culture
Primary HBE cells were cultured in ALI, and trans-epithelial electrical resistance (TER) measurement was performed as described previously (Gao et al., 2013). In brief, passage (P) 0 cells were seeded into plastic dishes coated with bovine collagen (PureCol model 5005-B; Advanced BioMatrix) in basal epithelial growth medium. At 30% confluence, cultures were infected with lentivirus for 8 h and grown for 48 h before puromycin selection. Confluent cultures were passaged to P1 with puromycin in dishes without collagen.
For ALI culture, 4 × 104 P1 cells, harvested at 70–90% confluence, were seeded onto collagen IV (C7521; Sigma-Aldrich)–coated 0.4-µm pore size polyester membranes in 24-well, 6.5-mm Transwell permeable supports (Corning). ALI culture medium (Fulcher et al., 2005) was changed three times a week. Triplicate inserts were washed, and 500 µl of fresh ALI medium was added before measuring TER every other day using a Millicell ERS2 (EMD Millipore). Given variation between different donor cells in the time at which maximal TER is reached, only data from one donor are shown, but all results were similar.
For bronchosphere culture, a cushion of 100% Matrigel was first added to the apical surface of the same uncoated inserts. 1,200 HBE cells with 24,000 cells of the MRC5 human lung fibroblast cell line in 100 µl of 50% ALI/50% Matrigel per well were seeded on the top of the cushion. ALI medium was added to the lower chamber and changed every other day. Values for CFE and sphere diameter are the mean of experiments using cells from at least three donors.
EdU staining
Cells were incubated in 10-µM EdU for 2 h at 37°C. EdU staining was performed using the Click-iT EdU imaging kit (Invitrogen) according to the manufacturer’s protocol.
CRISPR/Cas9 genome editing
sgRNAs for Grhl2 and its target genes and control sgRNA (that does not correspond to a sequence present in the human genome) were cloned into lentiCRISPR v2 (Addgene plasmid 52961; Sanjana et al., 2014). The sequences of sgRNA are listed in Table S1. Plasmids were transfected into 293T cells with lentiviral packaging vectors Δ8.9 and VSVG using FuGENE 6 Transfection Reagent (Promega). Viral particles were collected and concentrated by centrifugation. Genomic DNA was isolated from transfected and selected HBE cells either 5–7 d after adding puromycin or after 10-d ALI culture with continued puromycin selection. Primers were designed to amplify from genomic DNA the region of mutation for different target genes as listed in Table S2. PCR was performed using Phusion High-Fidelity DNA Polymerase (model M0530; New England Biolabs, Inc.). The Clontech In-Fusion PCR Cloning System (model 639649; Takara Bio Inc.) was used to fuse the ends of the PCR fragment to the homologous ends of a NotI and SalI linearized pCMV-Sport 6 vector. For each mutation, at least five clones were picked for sequencing. Mutant alleles were identified by comparison with the wild-type unmodified sequence.
qRT-PCR
RNA was extracted with the RNeasy Micro kit (QIAGEN), and cDNA was synthesized with SuperScript III (Invitrogen). PCR was performed with SYBR green chemistry in a StepOnePlus Real-Time PCR System (Applied Biosystems). Samples were analyzed in triplicate from two biological replicates, and data were analyzed using the ΔΔCt method.
For detecting GRHL2, SMAGP, and ZNF750 wild-type and mutant transcripts, RNA was extracted from control and mutants cells after ALI culture for 21 d. Paired primers were designed to span the sg region. Primer sequences are listed in Table S2.
Statistical analysis
Student’s t tests were used for all statistical analyses. Data are reported as mean ± SEM.
Online supplemental material
Fig. S1 presents evidence, using tracheas from Tjp1tm1lch mice, that ZO1 is localized between BCs early during regeneration after loss of luminal cells. Fig. S2 is a schematic for the timing and dose of Tmx treatment and FACS strategy used for isolating wild-type and Grhl2 conditionally deleted Ngfr+ BCs. Fig. S3 shows evidence from DNA sequencing of the kind and frequency of mutations that were generated in SMAGP, ZNF750, ANKRD22, PVRL4, VGLL1, RHBDL2, MPZL2, SCRIB, and WDR5B genes using lentiCRISR-sgRNAs. Fig. S4 shows the Grhl2-binding sites in ZNF750 and SMAGP genomic DNA in HBE cells as determined by ChIP-Seq and the CFE and phenotype of spheres generated from mutant HBE cells. Fig. S5 provides evidence that CRISPR-Cas9–induced mutations reduced expression of GRHL2, SMAGP, and ZNF750 RNA and protein in HBE cells. Tables S1, S2, and S3 give the sequences of sgRNA in lentiCRISPR vectors, the primers used in qRT-PCR, and the primers for sequencing genomic DNA, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506014/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201506014.dv.
Supplementary Material
Acknowledgments
We thank the Duke Transgenic Core Facility for generating the Grhl2 floxed mice and Stephen Jane for providing Grhl2-null mutant mice. We also thank Christopher Vockley for suggesting the use of CRISPR/Cas9 technology with HBE cells and for ideas and advice and Michel Bagnat and Hogan laboratory members for critical comments on the manuscript.
This work was supported by National Institutes of Health grants U01HL111018, U01HL110967, and R37HL071303 to B.L.M. Hogan.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ALI
- air–liquid interface
- BC
- basal cell
- CFE
- colony forming efficiency
- ChIP-Seq
- chromatin immunoprecipitation sequencing
- DIC
- differential interference contrast
- dpi
- days postinhalation
- EdU
- 5-ethynl-2′-deoxyuridine
- HBE
- human bronchial epithelial
- hpi
- hours postinhalation
- IHC
- immunohistochemistry
- MTEC
- mouse tracheal epithelial cell
- qRT-PCR
- quantitative RT-PCR
- sg
- short guide
- SMAGP
- small membrane adhesion glycoprotein
- TER
- trans-epithelial electrical resistance
- Tmx
- tamoxifen
References
- Ader M., and Tanaka E.M.. 2014. Modeling human development in 3D culture. Curr. Opin. Cell Biol. 31:23–28. 10.1016/j.ceb.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Aue A., Hinze C., Walentin K., Ruffert J., Yurtdas Y., Werth M., Chen W., Rabien A., Kilic E., Schulzke J.D., et al. 2015. A grainyhead-like 2/ovo-like 2 pathway regulates renal epithelial barrier function and lumen expansion. J. Am. Soc. Nephrol. 10.1681/ASN.2014080759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum R.Y., Zvulunov A., Hallel-Halevy D., Cagnano E., Finer G., Ofir R., Geiger D., Silberstein E., Feferman Y., and Birk O.S.. 2006. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat. Genet. 38:749–751. 10.1038/ng1813 [DOI] [PubMed] [Google Scholar]
- Borthwick D.W., Shahbazian M., Krantz Q.T., Dorin J.R., and Randell S.H.. 2001. Evidence for stem-cell niches in the tracheal epithelium. Am. J. Respir. Cell Mol. Biol. 24:662–670. 10.1165/ajrcmb.24.6.4217 [DOI] [PubMed] [Google Scholar]
- Boxer L.D., Barajas B., Tao S., Zhang J., and Khavari P.A.. 2014. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 28:2013–2026. 10.1101/gad.246579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske P., Przybilla J., Loeffler M., Sachs N., Sato T., Clevers H., and Galle J.. 2012. On the biomechanics of stem cell niche formation in the gut—modelling growing organoids. FEBS J. 279:3475–3487. 10.1111/j.1742-4658.2012.08646.x [DOI] [PubMed] [Google Scholar]
- Castanieto A., Johnston M.J., and Nystul T.G.. 2014. EGFR signaling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. eLife. 3:e04437 10.7554/eLife.04437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers A.D., Lachani K., Shin Y., Sherwood V., Cho K.W., and Papalopulu N.. 2006. Grainyhead-like 3, a transcription factor identified in a microarray screen, promotes the specification of the superficial layer of the embryonic epidermis. Mech. Dev. 123:702–718. 10.1016/j.mod.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Cohen I., Birnbaum R.Y., Leibson K., Taube R., Sivan S., and Birk O.S.. 2012. ZNF750 is expressed in differentiated keratinocytes and regulates epidermal late differentiation genes. PLoS ONE. 7:e42628 10.1371/journal.pone.0042628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.T., Gautrot J.E., Trappmann B., Tan D.W., Donati G., Huck W.T., and Watt F.M.. 2010. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 12:711–718. 10.1038/ncb2074 [DOI] [PubMed] [Google Scholar]
- Danahay H., Pessotti A.D., Coote J., Montgomery B.E., Xia D., Wilson A., Yang H., Wang Z., Bevan L., Thomas C., et al. 2015. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Reports. 10:239–252. 10.1016/j.celrep.2014.12.017 [DOI] [PubMed] [Google Scholar]
- Folgueras A.R., Guo X., Pasolli H.A., Stokes N., Polak L., Zheng D., and Fuchs E.. 2013. Architectural niche organization by LHX2 is linked to hair follicle stem cell function. Cell Stem Cell. 13:314–327. 10.1016/j.stem.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote H.P., Sumigray K.D., and Lechler T.. 2013. FRAP analysis reveals stabilization of adhesion structures in the epidermis compared to cultured keratinocytes. PLoS ONE. 8:e71491 10.1371/journal.pone.0071491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher M.L., Gabriel S., Burns K.A., Yankaskas J.R., and Randell S.H.. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107:183–206. [DOI] [PubMed] [Google Scholar]
- Gao X., Vockley C.M., Pauli F., Newberry K.M., Xue Y., Randell S.H., Reddy T.E., and Hogan B.L.. 2013. Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc. Natl. Acad. Sci. USA. 110:9356–9361. 10.1073/pnas.1307589110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B.L., Barkauskas C.E., Chapman H.A., Epstein J.A., Jain R., Hsia C.C., Niklason L., Calle E., Le A., Randell S.H., et al. 2014. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 15:123–138. 10.1016/j.stem.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holgate S.T. 2011. The sentinel role of the airway epithelium in asthma pathogenesis. Immunol. Rev. 242:205–219. 10.1111/j.1600-065X.2011.01030.x [DOI] [PubMed] [Google Scholar]
- Huebner R.J., Lechler T., and Ewald A.J.. 2014. Developmental stratification of the mammary epithelium occurs through symmetry-breaking vertical divisions of apically positioned luminal cells. Development. 141:1085–1094. 10.1242/dev.103333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Kaji N., Durgan J., and Hall A.. 2008. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 183:625–633. 10.1083/jcb.200807121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight D.A., and Holgate S.T.. 2003. The airway epithelium: structural and functional properties in health and disease. Respirology. 8:432–446. 10.1046/j.1440-1843.2003.00493.x [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., and Knoblich J.A.. 2014. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 345:1247125 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- Li X., Chen Z., and Desplan C.. 2013. Temporal patterning of neural progenitors in Drosophila. Curr. Top. Dev. Biol. 105:69–96. 10.1016/B978-0-12-396968-2.00003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Mahoney J.E., Stupnikov M.R., Paez-Cortez J.R., Szymaniak A.D., Varelas X., Herrick D.B., Schwob J., Zhang H., and Cardoso W.V.. 2015. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development. 142:258–267. 10.1242/dev.116855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A., Law B.M., Tata P.R., Villoria J., Saez B., Mou H., Zhao R., and Rajagopal J.. 2015. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell. 16:184–197. 10.1016/j.stem.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré A., Kim M., Juarez M.T., Brody S., and McGinnis W.. 2012. The functions of grainy head-like proteins in animals and fungi and the evolution of apical extracellular barriers. PLoS ONE. 7:e36254 10.1371/journal.pone.0036254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.K., Bisht B., Darmawan D.O., Chiou R., Ha V.L., Wallace W.D., Chon A.T., Hegab A.E., Grogan T., Elashoff D.A., et al. 2014. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell Stem Cell. 15:199–214. 10.1016/j.stem.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof G., Nanda A., Howden J., Takeichi T., McMillan J.R., Aristodemou S., Ozoemena L., Liu L., South A.P., Pourreyron C., et al. 2014. Mutations in GRHL2 result in an autosomal-recessive ectodermal Dysplasia syndrome. Am. J. Hum. Genet. 95:308–314. 10.1016/j.ajhg.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud D., and Leigh R.. 2011. Epithelial cells and airway diseases. Immunol. Rev. 242:186–204. 10.1111/j.1600-065X.2011.01033.x [DOI] [PubMed] [Google Scholar]
- Pyrgaki C., Liu A., and Niswander L.. 2011. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev. Biol. 353:38–49. 10.1016/j.ydbio.2011.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaee F., and Georas S.N.. 2014. Breaking barriers. New insights into airway epithelial barrier function in health and disease. Am. J. Respir. Cell Mol. Biol. 50:857–869. 10.1165/rcmb.2013-0541RT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y., Parekh V., Wilanowski T., Hislop N.R., Auden A., Ting S.B., Cunningham J.M., and Jane S.M.. 2010. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 345:237–245. 10.1016/j.ydbio.2010.07.017 [DOI] [PubMed] [Google Scholar]
- Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., and Hogan B.L.. 2009. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA. 106:12771–12775. 10.1073/pnas.0906850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., and Hogan B.L.. 2011a Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA. 108:E1475–E1483. 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.R., Gao X., Xue Y., Randell S.H., Kong Y.Y., and Hogan B.L.. 2011b Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 8:639–648. 10.1016/j.stem.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Boxer L.D., Webster D.E., Bussat R.T., Qu K., Zarnegar B.J., Johnston D., Siprashvili Z., and Khavari P.A.. 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell. 22:669–677. 10.1016/j.devcel.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga K., Mostov K.E., Mitaka T., Miyajima A., and Tanimizu N.. 2012. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol. Biol. Cell. 23:2845–2855. 10.1091/mbc.E12-02-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T., Wang Y., Barak L.S., Bai Y., Randell S.H., and Hogan B.L.. 2014. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. USA. 111:E3641–E3649. 10.1073/pnas.1409781111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N., and Mitaka T.. 2013. Role of grainyhead-like 2 in the formation of functional tight junctions. Tissue Barriers. 1:e23495 10.4161/tisb.23495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbé N.G., Rio M.C., and Weidle U.H.. 2004. SMAGP, a new small trans-membrane glycoprotein altered in cancer. Oncogene. 23:3395–3403. 10.1038/sj.onc.1207469 [DOI] [PubMed] [Google Scholar]
- Tata P.R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B.M., Vinarsky V., Cho J.L., Breton S., Sahay A., et al. 2013. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 503:218–223. 10.1038/nature12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S.B., Caddy J., Hislop N., Wilanowski T., Auden A., Zhao L.L., Ellis S., Kaur P., Uchida Y., Holleran W.M., et al. 2005. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 308:411–413. 10.1126/science.1107511 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., and Blanpain C.. 2011. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 479:189–193. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- Walentin K., Hinze C., Werth M., Haase N., Varma S., Morell R., Aue A., Pötschke E., Warburton D., Qiu A., et al. 2015. A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development. 142:1125–1136. 10.1242/dev.113829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., and Samakovlis C.. 2012. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr. Top. Dev. Biol. 98:35–63. 10.1016/B978-0-12-386499-4.00002-1 [DOI] [PubMed] [Google Scholar]
- Watson J.K., Rulands S., Wilkinson A.C., Wuidart A., Ousset M., Van Keymeulen A., Göttgens B., Blanpain C., Simons B.D., and Rawlins E.L.. 2015. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Reports. 12:90–101. 10.1016/j.celrep.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth M., Walentin K., Aue A., Schönheit J., Wuebken A., Pode-Shakked N., Vilianovitch L., Erdmann B., Dekel B., Bader M., et al. 2010. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 137:3835–3845. 10.1242/dev.055483 [DOI] [PubMed] [Google Scholar]
- Wilanowski T., Tuckfield A., Cerruti L., O’Connell S., Saint R., Parekh V., Tao J., Cunningham J.M., and Jane S.M.. 2002. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech. Dev. 114:37–50. 10.1016/S0925-4773(02)00046-1 [DOI] [PubMed] [Google Scholar]
- You Y., Richer E.J., Huang T., and Brody S.L.. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L1315–L1321. 10.1152/ajplung.00169.2002 [DOI] [PubMed] [Google Scholar]
- Zhao R., Fallon T.R., Saladi S.V., Pardo-Saganta A., Villoria J., Mou H., Vinarsky V., Gonzalez-Celeiro M., Nunna N., Hariri L.P., et al. 2014. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev. Cell. 30:151–165. 10.1016/j.devcel.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.