TRIM3 regulates synaptic γ-actin levels. TRIM3-deficient mice consequently have higher hippocampal spine densities, increased long-term potentiation, and enhanced contextual fear memory consolidation, indicating that temporal control of ACTG1 levels by TRIM3 is required to constrain hippocampal plasticity within physiological boundaries.
Abstract
Synaptic plasticity requires remodeling of the actin cytoskeleton. Although two actin isoforms, β- and γ-actin, are expressed in dendritic spines, the specific contribution of γ-actin in the expression of synaptic plasticity is unknown. We show that synaptic γ-actin levels are regulated by the E3 ubiquitin ligase TRIM3. TRIM3 protein and Actg1 transcript are colocalized in messenger ribonucleoprotein granules responsible for the dendritic targeting of messenger RNAs. TRIM3 polyubiquitylates γ-actin, most likely cotranslationally at synaptic sites. Trim3−/− mice consequently have increased levels of γ-actin at hippocampal synapses, resulting in higher spine densities, increased long-term potentiation, and enhanced short-term contextual fear memory consolidation. Interestingly, hippocampal deletion of Actg1 caused an increase in long-term fear memory. Collectively, our findings suggest that temporal control of γ-actin levels by TRIM3 is required to regulate the timing of hippocampal plasticity. We propose a model in which TRIM3 regulates synaptic γ-actin turnover and actin filament stability and thus forms a transient inhibitory constraint on the expression of hippocampal synaptic plasticity.
Introduction
Plasticity of synapses depends to a large extent on rapid changes in shape and composition of the actin cytoskeleton (Cingolani and Goda, 2008). Both filamentous actin (F-actin) and globular actin (G-actin) are highly enriched at dendritic spines (Matus, 2000; Okamoto et al., 2004; Yuste and Bonhoeffer, 2004). Dynamic actin remodeling contributes to the structural changes observed during synaptic plasticity (Matsuzaki et al., 2004; Okamoto et al., 2007) and plays a key role in membrane insertion, clustering, and internalization of postsynaptic receptors (Allison et al., 1998; Charrier et al., 2006), all of which are essential to the expression of functional plasticity, i.e., long-term potentiation (LTP) or long-term depression (Malinow and Malenka, 2002; Choquet and Triller, 2003; Shepherd and Huganir, 2007). Calcium entry into dendritic spines triggers intracellular signaling mechanisms that favor F-actin over G-actin, including direct calcium-dependent changes in actin polymerization and depolarization rates (Bergeron et al., 2010), local translation of actin (Tiruchinapalli et al., 2003), posttranslational modification of actin (Karakozova et al., 2006), and recruitment of regulatory actin-binding proteins (Schubert et al., 2006).
Two isoforms of actin are constitutively expressed in neurons: β-actin and γ-actin (Rubenstein, 1990). Both isoforms are almost identical, differing only at four amino acid positions in the N terminus. Synaptic actin regulatory mechanisms are often ascribed to β-actin, but recent studies indicate that γ-actin can contribute synaptic plasticity in a very specific manner. First, it has been suggested that high γ-/β-actin ratios stabilize actin filaments, whereas filaments with low γ-/β-actin ratios are more dynamic (Belyantseva et al., 2009; Bergeron et al., 2010). Moreover, γ-actin is translated at a much slower rate than β-actin, allowing a hidden lysine residue in the N-terminal part of the protein to become uniquely polyubiquitylated and targeted for proteasomal degradation (Zhang et al., 2010). The regulatory mechanisms of γ-actin ubiquitylation, as well as its role in synaptic plasticity, are still unknown.
Ubiquitin-mediated protein degradation plays an important role in the regulation of synaptic plasticity, learning, and memory (Ehlers, 2003; Tai and Schuman, 2008; Mabb and Ehlers, 2010). It involves, in addition to an E1 ubiquitin-activating enzyme and an E2 ubiquitin-conjugating enzyme, an E3 ubiquitin-protein ligase that transfers activated ubiquitin to one or more specific substrate proteins. Polyubiquitylated protein substrates are then recognized and degraded by the proteasome. Several synaptic E3 ubiquitin ligases have been identified that regulate the levels of synaptic proteins, including scaffold proteins (Colledge et al., 2003), neurotransmitter receptors (Schwarz et al., 2010), and signaling molecules (Mabb et al., 2014). However, no E3 ligases have been identified that directly target neuronal actins for proteasomal degradation.
TRIM (tripartite motif) proteins are a large family of structurally related proteins sharing an N-terminal RBCC (RING finger/B-box/coiled coil) region with E3 ubiquitin ligase activity (Reymond et al., 2001). TRIM2, TRIM3, and TRIM32 belong to a subfamily of TRIM proteins sharing a C-terminal ABP280 domain followed by six NHL repeat domains (Short and Cox, 2006). Whereas TRIM2 polyubiquitylates neurofilament light chain in cerebellar Purkinje cells (Balastik et al., 2008), and TRIM32 polyubiquitylates muscle actin (Kudryashova et al., 2005) and c-Myc in neuronal progenitors (Schwamborn et al., 2009), the substrate and function of TRIM3 are still unclear. Various functions have been proposed. First, TRIM3 may be part of the CART (cytoskeleton-associated recycling or transport) complex that regulates rapid recycling of membrane receptors (Yan et al., 2005). Second, TRIM3 may control GABA-A receptor trafficking and inhibitory synaptic transmission in cortical neurons (Cheung et al., 2010). Finally, TRIM3 was shown to reduce the levels of postsynaptic scaffold proteins GKAP and SHANK1 and alter spine morphology in hippocampal neurons in vitro (Hung et al., 2010); however, activity-dependent removal of GKAP from synapses in vivo does not appear to depend on TRIM3 (Shin et al., 2012). Importantly, none of these studies investigated the direct effects of TRIM3 deficiency on synaptic protein levels, synaptic plasticity, or learning and memory in vivo.
Here, we show that Trim3−/− mice have enhanced contextual fear memory acquisition and increased LTP and spine densities in the hippocampus. We demonstrate that neither GKAP nor SHANK1 but instead γ-actin protein levels are increased in hippocampal neurons of Trim3−/− mice. Furthermore, γ-actin is polyubiquitylated by TRIM3 in response to plasticity-inducing stimulation. Our findings thus identify the E3 ubiquitin ligase TRIM3 as a regulator of the synaptic actin cytoskeleton and propose that the temporal control of synaptic γ-actin levels by TRIM3 is a critical setscrew in neuronal plasticity, learning, and memory.
Results
TRIM3 is a dendritically localized ubiquitin ligase in the hippocampus
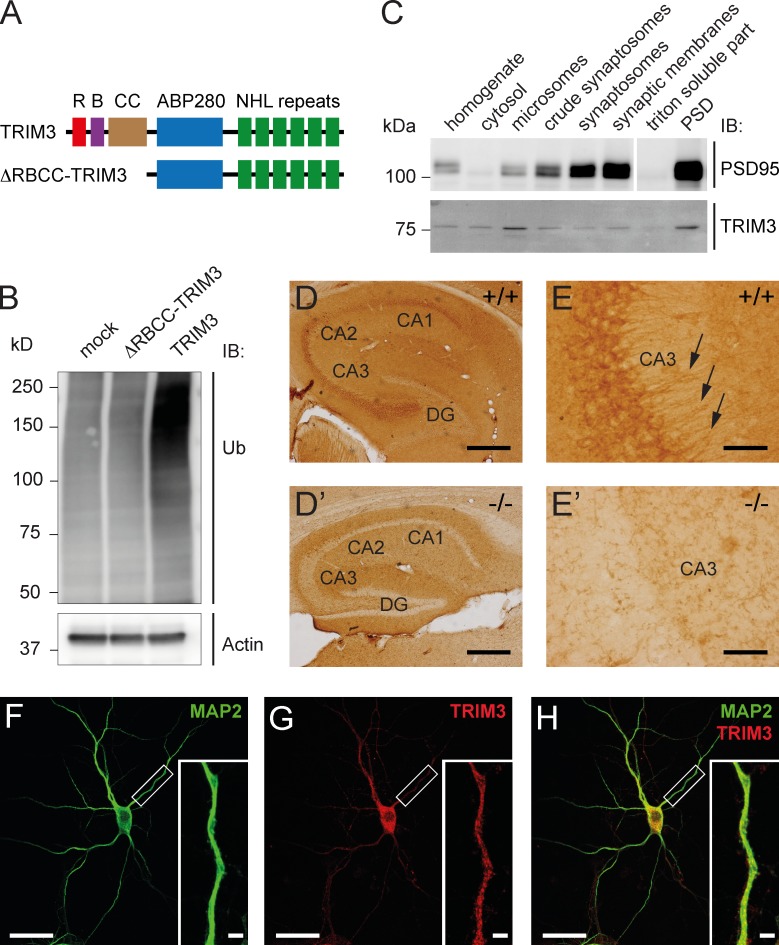
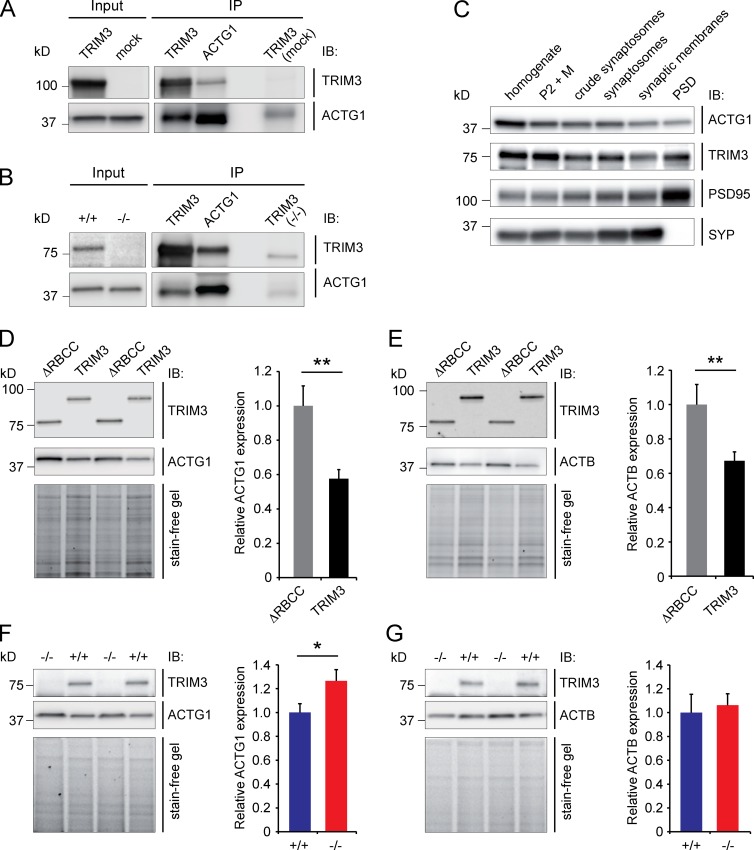
Full-length TRIM3 and mutant TRIM3 lacking the RING/B-box/coiled-coil domain (ΔRBCC-TRIM3; Fig. 1 A) were expressed in HEK293 cells. Immunoblots of the cell lysates showed a strong increase in high-molecular-weight ubiquitin staining specifically in TRIM3-expressing cells (Fig. 1 B), indicating that TRIM3, but not ΔRBCC-TRIM3, has ubiquitin ligase activity in vitro. Biochemical enrichment analysis of TRIM3 in different fractions of hippocampal tissue revealed relatively high TRIM3 protein levels in microsomes and in the postsynaptic density (Fig. 1 C). Immunohistochemical staining of hippocampal sections subsequently showed high expression of TRIM3 throughout the CA1-3 regions of the hippocampus (Fig. 1 D). CA pyramidal cells showed a particular strong expression of TRIM3 also at the dendrite level (Fig. 1 E; for negative control staining of Trim3−/− sections, see Fig. 1, D′ and E′). Staining cultured hippocampal neurons revealed a punctate labeling for TRIM3 in MAP2-positive dendrites (Fig. 1, F–H), consistent with microsomal and/or synaptic localization. Together, these findings establish that TRIM3 is a ubiquitin ligase that is localized to dendritic compartments, including postsynaptic sites, as previously shown by Hung et al. (2010).
Figure 1.
TRIM3 is a dendritically localized ubiquitin ligase in the hippocampus. (A) Schematic diagram of full-length TRIM3 and ΔRBCC-TRIM3 constructs. R, RING; B, B-box; CC, coiled-coil; ABP280, actin-binding protein 280 repeat; NHL repeat, NCL-1/HT2A/LIN-41 repeat. (B) TRIM3, but not ΔRBCC-TRIM3, induces polyubiquitylation in HEK293 cells. HEK293 cell lysates were subjected to immunoblot (IB) analysis using an anti-ubiquitin (Ub) antibody. Increased polyubiquitylation is indicated by a strong increase in high-molecular-weight ubiquitin staining. Actin staining is shown as loading control. (C) Biochemical fractionation of hippocampal tissue followed by immunoblot analysis shows that TRIM3 protein is enriched in the postsynaptic density fraction and in microsomes. Synaptic enrichment is shown by PSD-95 staining of the same protein samples. (D and E) Immunohistochemistry shows that TRIM3 protein is expressed throughout the CA regions of the hippocampus, but less in the dentate gyrus (DG; D). TRIM3 protein was primarily detected in CA pyramidal cell bodies, but also in dendrites (arrows in E). Staining of hippocampal sections from Trim3−/− animals confirmed the specificity of the TRIM3 labeling (D′ and E′). Bars: (D) 400 µm; (E) 50 µm. (F–H) Immunocytochemistry revealed a punctuate staining of TRIM3 in dendrites of cultured primary hippocampal neurons. Neurons were labeled with antibodies against MAP2 (green; F) and TRIM3 (red; G). TRIM3-positive puncta are observed inside MAP2-positive dendrites (H). Bars: 25 µm; (insets) 2.5 µm.
Trim3−/− mice show enhanced hippocampal memory consolidation
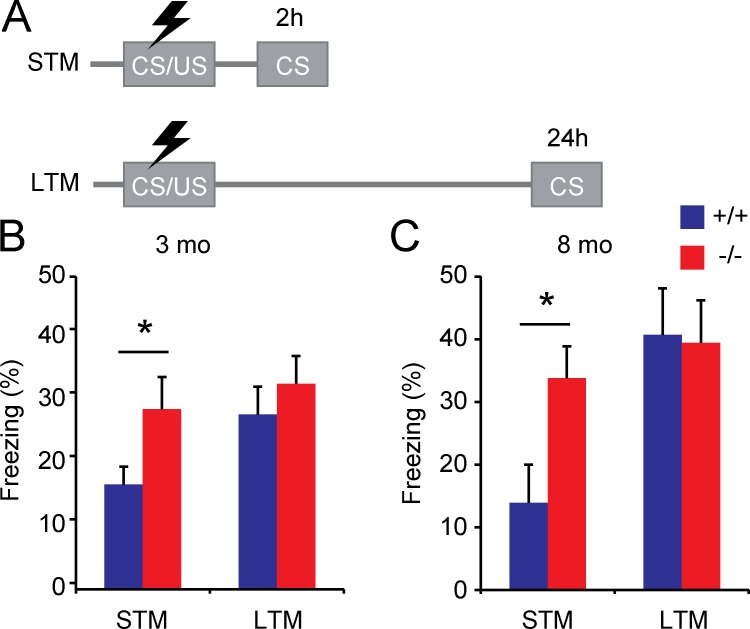
Behavioral analysis of 3-mo-old Trim3−/− mice (Labonté et al., 2013) did not reveal important alterations in anxiety and locomotor behavior using open field, dark-light box, and elevated plus maze tests (Table 1). However, in a hippocampus-dependent contextual fear memory task, Trim3−/− mice showed enhanced memory acquisition. Freezing was measured independently at 2 h (short-term memory) and 24 h (long-term memory) after the shock using different cohorts of animals (Fig. 2 A). Importantly, none of the animals showed baseline freezing before the shock. At 24 h, Trim3−/− mice and wild-type littermates showed a similar increase in freezing behavior (wild type: 27.6% ± 4.4% freezing; Trim3−/−: 32.4% ± 4.3% freezing; Fig. 2 B), indicating that long-term memory consolidation had occurred in both genotypes. However, at 2 h after the shock, Trim3−/− mice showed significantly higher freezing levels compared with wild-type controls (wild type: 16.6% ± 2.8% freezing; Trim3−/−: 28.4% ± 5.1% freezing; P < 0.05; Fig. 2 B). Notably, the level of freezing in Trim3−/− mice was already at the level observed for both genotypes at 24 h after the shock.
Table 1. Behavioral analysis of Trim3−/− mice at 3 mo of age.
| Test/test measure | Unit | Wild type (n = 12) | Knockout (n = 12) | P-value |
|---|---|---|---|---|
| Open field/Ethovision | ||||
| Number of entries into the center area | 84.83 ± 4.07 | 91.67 ± 6.36 | 0.375 | |
| Time spent in the center area | s | 314.5 ± 18.0 | 308.0 ± 18.4 | 0.803 |
| Total distance traveled | cm | 5,602 ± 264 | 5,819 ± 345 | 0.622 |
| Open field/SEE analysis/velocity | ||||
| Lingering progression threshold speed | cm/s | 12.05 ± 0.61 | 12.89 ± 0.83 | 0.418 |
| Median of move segment maximum speed | cm/s | 24.03 ± 0.69 | 25.01 ± 1.00 | 0.425 |
| Quantile 95 of move segment maximum speed | cm/s | 35.80 ± 0.89 | 35.54 ± 1.03 | 0.850 |
| Lingering mean speed | cm/s | 1.65 ± 0.10 | 1.66 ± 0.10 | 0.988 |
| Median of lingering segment maximum speed | cm/s | 3.17 ± 0.40 | 2.28 ± 0.60 | 0.231 |
| Open field/SEE analysis/acceleration | ||||
| Latency to maximum half speed | s | 8.65 ± 3.84 | 5.88 ± 1.62 | 0.513 |
| Median segment acceleration to maximum speed | cm/s2 | 15.77 ± 0.69 | 15.44 ± 0.57 | 0.715 |
| Open field/SEE analysis/other | ||||
| Number of progression segments | segments | 143.3 ± 10.7 | 142.0 ± 10.2 | 0.934 |
| Median length of progression segments | cm | 24.73 ± 0.96 | 28.40 ± 2.08 | 0.123 |
| Median duration of progression segments | s | 1.55 ± 0.05 | 1.63 ± 0.07 | 0.369 |
| Quantile 5 of duration of progression segments | s | 0.55 ± 0.03 | 0.57 ± 0.04 | 0.755 |
| Quantile 95 of duration of progression segments | s | 3.38 ± 0.10 | 3.62 ± 0.14 | 0.172 |
| Number of stops per distance | segments/cm | 0.03 ± 0.00 | 0.03 ± 0.00 | 0.386 |
| Time proportion of lingering episodes | 0.59 ± 0.03 | 0.58 ± 0.03 | 0.651 | |
| Median radius of turn | cm | 77.69 ± 3.74 | 81.05 ± 3.41 | 0.513 |
| Median turn rate | degrees/s | 14.06 ± 0.58 | 13.83 ± 0.48 | 0.761 |
| Light-dark box | ||||
| Number of entries into the light compartment | 29.50 ± 3.30 | 38.00 ± 3.67 | 0.099 | |
| Time spent in the light compartment | s | 301.1 ± 17.2 | 305.2 ± 16.7 | 0.863 |
| Elevated plus maze | ||||
| Number of entries into the closed arms | 22.00 ± 1.93 | 23.50 ± 1.81 | 0.576 | |
| Number of entries into the open arms | 28.42 ± 2.94 | 32.75 ± 2.66 | 0.287 | |
| Number of entries into the middle | 25.92 ± 2.53 | 29.58 ± 2.18 | 0.284 | |
| Time spent in the closed arms | s | 419.8 ± 15.0 | 395.7 ± 7.9 | 0.171 |
| Time spent in the open arms | s | 159.7 ± 12.6 | 175.2 ± 8.3 | 0.315 |
| Time spent in the middle | s | 84.64 ± 7.37 | 110.07 ± 7.87 | 0.028 |
| Total distance moved | cm | 1,552 ± 86 | 1,608 ± 76 | 0.632 |
Entries show mean ± SEM values unless otherwise indicated.
Figure 2.
Short-term contextual fear memory is enhanced in Trim3−/− mice. (A) Animals received a foot shock (unconditioned stimulus; US) in a novel context (conditioned stimulus; CS). Memory retrieval was tested either 2 h (short-term memory; STM) or 24 h (long-term memory; LTM) after conditioning in two independent cohorts of mice. (B) Freezing behavior was significantly increased in 3-mo-old Trim3−/− mice compared with wild-type littermates when reexposed to the conditioned context 2h after receiving the shock (STM), but not at 24 h (LTM), indicating enhanced consolidation of fear memory (means ± SEM, two-tailed t test, *, P < 0.05, n = 8/10 per genotype). (C) A similar increase in freezing behavior at 2 h, but not at 24 h, after the shock was observed in 8-mo-old Trim3−/− mice (means ± SEM, two-tailed t test, *, P < 0.05, n = 5/7 per genotype).
Because Trim2−/− mice show a progressive age-dependent neurological phenotype (Balastik et al., 2008), behavioral analyses of Trim3−/− mice were also performed in an independent cohort of 8-mo-old animals. Anxiety and locomotor behavior were unaffected in Trim3−/− mice at this age (Table S1), and enhanced consolidation of contextual fear memory was still observed (Fig. 2 C). Freezing levels were slightly higher than in 3-mo-old mice at 24 h after the shock (wild type: 40.7% ± 7.4% freezing; Trim3−/−: 39.4% ± 6.7% freezing) but did not differ between genotypes. At 2 h after the shock, however, significantly higher freezing levels were observed again in Trim3−/− mice (wild type: 13.9% ± 6.1% freezing; Trim3−/−: 33.8% ± 5.1% freezing). Collectively, these findings show that Trim3−/− mice show enhanced hippocampal memory consolidation and that this is a robust and specific behavioral phenotype and not the result of age-dependent neurological alterations.
TRIM3 is present in messenger RNP (mRNP) particles but is not essential for mRNP particle trafficking
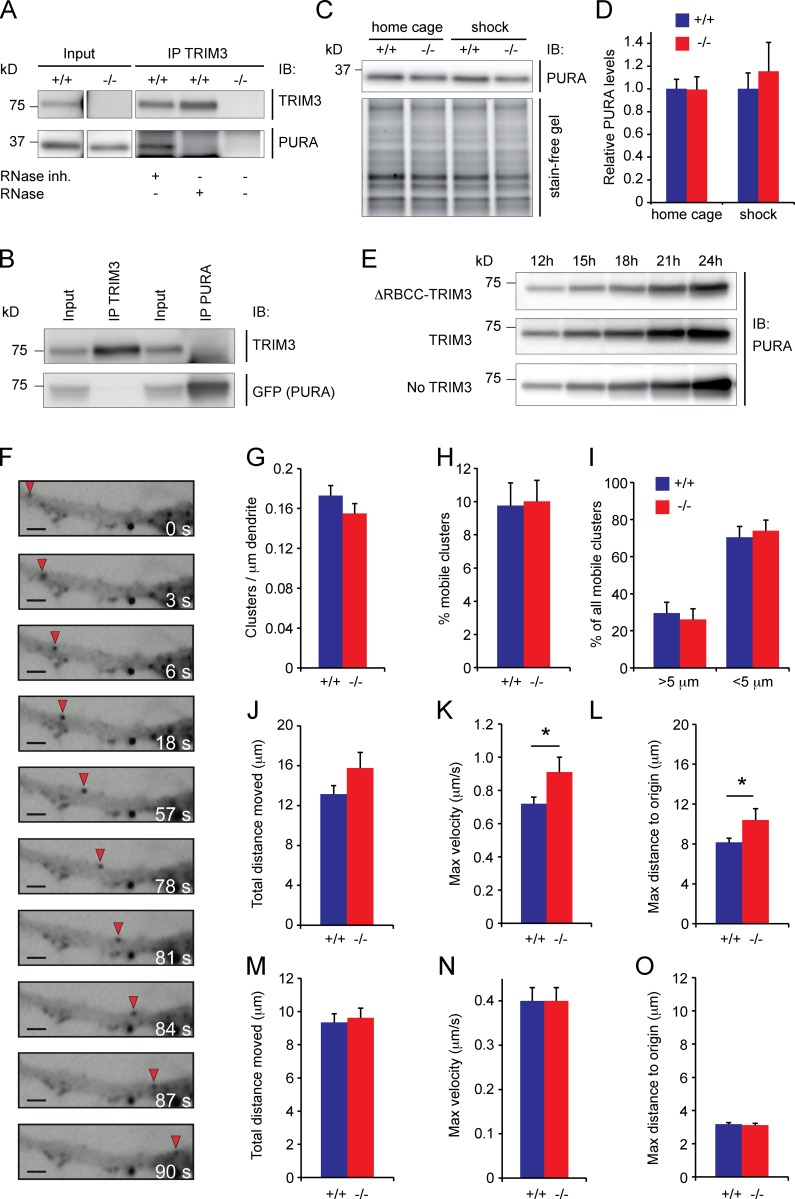
Previous studies suggested that TRIM3 functions in dendritic trafficking, which would be consistent with its dendritic and microsomal localization (Fig. 1). Specifically, TRIM3 interacts with the tubulin-based motor KIF21B (Labonté et al., 2013) and the actin-based motor myosin V (El-Husseini and Vincent, 1999; Yan et al., 2005). Myosin V regulates dendritic trafficking of mRNP particles that transport mRNAs to synapses (Ohashi et al., 2002; Yoshimura et al., 2006), and both TRIM2 and TRIM3 are among the most abundant protein constituents of RNase-sensitive mRNP particles in neurons (Kanai et al., 2004). Immunoprecipitation of TRIM3 from hippocampal synapse-enriched fractions revealed that TRIM3 and PUR-α (PURA), one of the most abundant proteins in mRNP particles (Ohashi et al., 2002; Kanai et al., 2004; Elvira et al., 2006), interact in an RNase-sensitive manner (Fig. 3 A), confirming that they coexist in mRNP particles and that their interaction depends on the presence of RNA. When coexpressing TRIM3 and PURA in HEK293 cells, PURA did not coimmunoprecipitate TRIM3 and vice versa (Fig. 3 B), indicating that they do not interact directly. We then investigated whether PURA levels are regulated by TRIM3 and found no difference in PURA levels in Trim3−/− mice compared with wild-type controls in hippocampal synapse-enriched fractions, neither under basal conditions nor 2 h after fear conditioning when enhanced memory consolidation is observed (wild-type home cage: 1 ± 0.08; Trim3−/− home cage: 0.99 ± 0.11; wild-type shock: 1 ± 0.14; Trim3−/− shock: 1.15 ± 0.25; Fig. 3, C and D). When TRIM3 and PURA were coexpressed in HEK293, we observed a stable expression of PURA over time, independent of whether full-length TRIM3 or ΔRBCC-TRIM3 was expressed. Collectively, these data demonstrate that PURA levels are not regulated by TRIM3.
Figure 3.
TRIM3 is present in mRNP particles but is not essential for mRNP particle trafficking. (A) TRIM3 interacts with PURA in an RNA-dependent manner. TRIM3 was immunoprecipitated from hippocampal synapse-enriched fractions and samples were immunoblotted (IB) and stained for TRIM3 and PURA. PURA was detected in RNase inhibitor–treated samples, but not in RNase-treated samples. Samples prepared from Trim3−/− mice served as negative control for the immunoprecipitation. (B) TRIM3 and PURA do not interact when expressed in HEK293 cells. HEK293 cells were cotransfected with full-length TRIM3 and GFP-PURA. TRIM3 and PURA were immunoprecipitated from lysates, immunoblotted and stained for TRIM3 and GFP (PURA). PURA did not coimmunoprecipitate with TRIM3, and TRIM3 did not coimmunoprecipiate with PURA. (C and D) PURA levels are not altered in Trim3−/− mice. Hippocampal synapse-enriched fractions were prepared from wild-type and Trim3−/− mice under control conditions (home cage) and 2 h after contextual fear conditioning (shock). Samples were immunoblotted and stained for PURA. Normalized PURA levels did not differ between conditions (means ± SEM, n = 4 per genotype). (E) TRIM3 does not alter PURA levels in HEK293 cells. HEK293 cells were cotransfected with PURA and TRIM3, PURA and ΔRBCC-TRIM3, or PURA alone. No differences in PURA levels were observed at any time point after transfection. (F) TRIM3 is not essential for PURA trafficking. Example time-lapse images of a mobile GFP-PURA cluster (red arrow) over a timespan of 90 s are shown. Bars, 2 µm. (G and H) Trim3−/− neurons and wild-type control neurons expressed equal amounts of PURA clusters (G) and had equal fractions of mobile clusters (H). (I) Trim3−/− neurons and wild-type control neurons expressed equal amounts of short-distance and long-distance clusters. (J and K) Long-distance PURA clusters show slightly increased kinetics in Trim3−/− neurons. The total distance moved (J) did not change, but the maximum velocity (K) and the maximum distance reached from origin (L) were significantly increased in Trim3−/− neurons (means ± SEM, two-tailed t test, *, P < 0.05, n = 27/40 clusters per genotype). (M–O) Short-distance PURA clusters show unaltered kinetics in Trim3−/− neurons. The total distance moved (M), the maximum velocity (N), and the maximum distance reached from origin (O) did not differ significantly in Trim3−/− neurons compared with wild-type neurons (means ± SEM, n = 76/80 clusters per genotype).
We next expressed GFP-tagged PURA in cultured hippocampal neurons derived from Trim3−/− mice or wild-type controls and quantified the mobility of GFP-positive PURA-containing mRNP particles (Fig. 3 F). The total number of particles observed was not different between genotypes (wild type: 0.173 ± 0.01 clusters/µm, Trim3−/−: 0.155 ± 0.01 clusters/µm; Fig. 3 G), nor was the percentage of mobile clusters (wild type: 9.76% ± 1.37%, Trim3−/−: 10.01% ± 1.27%; Fig. 3 H). Clusters were then separated on the basis of their mobility into those that moved >5 µm over the 5-min imaging period (long-distance clusters) and those that moved <5 µm (short-distance clusters). No difference was observed in the percentage of long- and short-distance moving clusters between genotypes (long-distance, wild type: 29.56% ± 5.83%, Trim3−/−: 26.09% ± 5.8%; short-distance, wild type: 70.44% ± 5.83%, Trim3−/−: 73.91% ± 5.8%; Fig. 3 I). When we analyzed the kinetics of long-distance moving clusters separately, we observed no difference in the total distance moved (wild type: 13.15 ± 0.86 µm, Trim3−/−: 15.77 ± 1.57 µm; Fig. 3 J) and a small but significant increase in the maximum velocity of clusters in Trim3−/− neurons (wild type: 0.72 ± 0.04 µm/s, Trim3−/−: 0.91 ± 0.09 µm/s; P < 0.05; Fig. 3 K) and the maximum distance traveled (wild type: 8.15 ± 0.43 µm, Trim3−/−: 10.39 ± 1.16 µm; P < 0.05; Fig. 3 L). In contrast, for short-distance moving clusters none of these parameters was significantly affected in Trim3−/− neurons (total distance moved, wild type: 9.34 ± 0.53 µm, Trim3−/−: 9.62 ± 0.58 µm; maximum velocity, wild type: 0.4 ± 0.03 µm/s, Trim3−/−: 0.4 ± 0.03 µm/s; maximum distance to origin, wild type: 3.17 ± 0.11 µm, Trim3−/−: 3.12 ± 0.11 µm; Fig. 3, M–O). Thus, although TRIM3 is present in mRNP particles and may affect some kinetic parameters of a subpopulation of particles, it is not essential for mRNP trafficking.
ACTG1 is a potential synaptic substrate of TRIM3
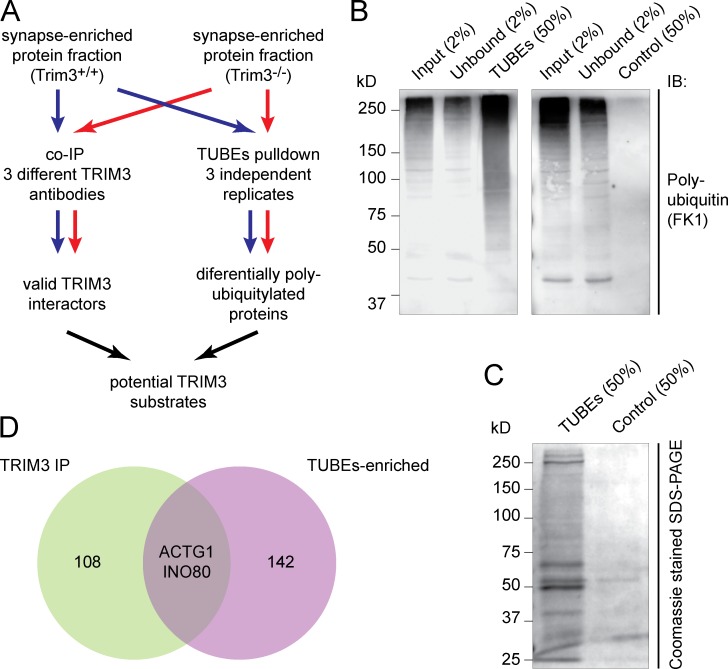
Previous studies suggested that the postsynaptic scaffold proteins GKAP and SHANK1 are TRIM3 substrates in vivo (Hung et al., 2010). We quantified GKAP and SHANK1 levels in hippocampal synapse-enriched fractions, both under basal conditions and 2 h after fear conditioning, and observed no increase in Trim3−/− mice as would be expected if they undergo TRIM3-dependent proteasomal degradation (Fig. S1). We then reasoned that a bona fide substrate should (a) physically interact with TRIM3, at least transiently, and (b) be less polyubiquitylated in Trim3−/− mice. We designed a substrate identification strategy (Fig. 4 A) in which we immunoprecipitated TRIM3 from hippocampal synapse-enriched fractions using three different antibodies and using Trim3−/− hippocampal fractions as negative control. In parallel, we performed affinity purification of polyubiquitylated proteins from Trim3−/− and wild-type hippocampal synapse-enriched fractions using tandem ubiquitin-binding entities (TUBEs) in triplicate. The efficacy of the TUBEs approach was confirmed by showing that TUBEs pull-down, but not a pull-down with empty agarose beads, resulted in a strong enrichment of polyubiquitylated proteins detected by immunoblotting (Fig. 4 B). Moreover, the Coomassie-stained gel revealed that the TUBEs-enriched protein fraction contained many individual proteins that were not detected in the control pull-down samples (Fig. 4 C). Using mass spectrometry, 110 candidate TRIM3-interacting proteins were identified that immunoprecipitated specifically from wild-type samples and not from Trim3−/− samples, and 144 candidate TRIM3 substrate proteins were identified that were TUBEs-enriched in wild-type samples compared with Trim3−/− samples (Table S2). Only two proteins were identified in both experiments, of which γ-actin (ACTG1) ranked as the second most abundant candidate interactor and the first most abundant candidate substrate (Fig. 4 D).
Figure 4.
Identification of potential TRIM3 substrate proteins. (A) Strategy for identifying TRIM3 substrates. Hippocampal synapse-enriched protein fractions were used as input for both immunoprecipitation (left) and polyubiquitin affinity pull-down (right). Samples from Trim3−/− mice served as controls. All samples were analyzed by quantitative mass spectrometry and potential TRIM3 substrates were identified by ranking and comparing results from all six samples. (B) Validation of the TUBEs pull-down approach. Input, unbound, TUBEs pull-down (50%) and control pull-down (agarose beads only) fractions were resolved on SDS-PAGE and immunoblotted for polyubiquitin. A strong enrichment for polyubiquitylated proteins (50–250 kD) was observed in the TUBEs pull-down fraction (left), but not in the control pull-down fraction (right). (C) TUBEs pull-down samples were highly enriched in proteins compared with control pull-down samples. (D) Venn diagram indicating the proteins that were identified as potential TRIM3 substrates. Coimmunoprecipitation identified 110 TRIM3-interacting proteins, whereas TUBEs pull-down identified 144 proteins that were more polyubiquitylated in wild-type mice compered to Trim3−/− mice (Table S2). Only two proteins were identified in both experiments: γ-actin (ACTG1) and DNA helicase INO80.
TRIM3 interacts with ACTG1 and regulates ACTG1 levels
Expression of TRIM3 in HEK293 cells and immunoprecipitation of either TRIM3 or endogenously expressed ACTG1 resulted in coimmunoprecipitation of ACTG1 and TRIM3, respectively, whereas mock-transfected cells did not show coimmunoprecipitation (Fig. 5 A). Similarly, immunoprecipitating TRIM3 from wild-type hippocampal synapse-enriched fractions, but not from Trim3−/− fractions, resulted in coimmunoprecipitation of ACTG1 and vice versa (Fig. 5 B). Importantly, the two antibodies that we used against ACTG1 are highly specific (see Fig. 9). Cellular fractionation of hippocampal tissue followed by immunoblotting for TRIM3, ACTG1, and markers for pre- and postsynaptic structures showed that TRIM3 and ACTG1 colocalize in all fractions, including postsynaptic structures (Fig. 5 C). Thus, TRIM3 and ACTG1 physically interact, both in HEK293 cells and in hippocampal synaptic protein fractions, and both proteins are localized at synaptic sites. We then asked whether TRIM3 also regulates ACTG1 protein levels. HEK293 cells that expressed TRIM3 showed a significant downregulation of ACTG1 protein levels compared with ΔRBCC-TRIM3-expressing cells (ΔRBCC-TRIM3: 1 ± 0.11, TRIM3: 0.57 ± 0.05; P < 0.01; Fig. 5 D). We also observed a significant reduction in β-actin (ACTB) levels (ΔRBCC-TRIM3: 1 ± 0.08, TRIM3: 0.67 ± 0.05; P < 0.01; Fig. 5 E), indicating that TRIM3-dependent protein degradation is not specific for ACTG1 when TRIM3 is overexpressed in HEK293 cells. However, in hippocampal neurons, a specific increase was observed in ACTG1 levels in Trim3−/− neurons compared with wild-type control neurons (wild type: 1 ± 0.07, Trim3−/−: 1.26 ± 0.10; P < 0.05; Fig. 5 F), whereas ACTB levels were not affected (wild type: 1 ± 0.16, Trim3−/−: 1.06 ± 0.10; Fig. 5 G). These data show that ACTG1 protein levels are specifically downregulated in the presence of TRIM3.
Figure 5.
TRIM3 interacts with ACTG1 and regulates ACTG1 levels. (A) HEK293 cells were transfected with TRIM3 or mock transfected. TRIM3 and ACTG1 were immunoprecipitated (IP) from the lysates, immunoblotted and stained for TRIM3 and ACTG1. TRIM3 can be observed in the ACTG1 IP and ACTG1 in the TRIM3 IP. No or significantly less TRIM3 or ACTG1 protein was detected in IPs on the mock-transfected cell lysate. (B) TRIM3 and ACTG1 were immunoprecipitated from hippocampal synapse-enriched fractions, immunoblotted, and stained for TRIM3 and ACTG1. TRIM3 can be observed the ACTG1 IP and ACTG1 in the TRIM3 IP. No or significantly less TRIM3 or ACTG1 protein was detected in the TRIM3 IP on the Trim3−/− tissue sample. (C) ACTG1 and TRIM3 are colocalized in hippocampal synapses. Biochemical fractionation of hippocampal tissue followed by immunoblot analysis showed that ACTG1 and TRIM3 are both detected in synaptic fractions. Enrichment of synapses is indicated by PSD-95 and SYP staining. (D and E) TRIM3 regulates actin levels in HEK293 cells. HEK293 cells were transfected with TRIM3 or ΔRBCC-TRIM3. Lysates were immunoblotted and stained for TRIM3 and ACTG1 (D) or TRIM3 and ACTB (E). Normalized protein levels of both ACTG1 and ACTB were significantly reduced in TRIM3-expressing cells compared with cells expressing ΔRBCC-TRIM3 (means ± SEM, two-tailed t test, **, P < 0.01, n = 8 cultures per condition. (F and G) TRIM3 regulates ACTG1 levels, but not ACTB levels, in hippocampal neurons. Cultured hippocampal neurons were lysed, immunoblotted, and stained for TRIM3 and ACTG1 (F) or TRIM3 and ACTB (G). Normalized protein levels of ACTG1, but not ACTB, were significantly higher in neurons from Trim3−/− mice compared with wild-type control neurons (means ± SEM, two-tailed t test, *, P < 0.05, n = 8 cultures per genotype).
Figure 9.
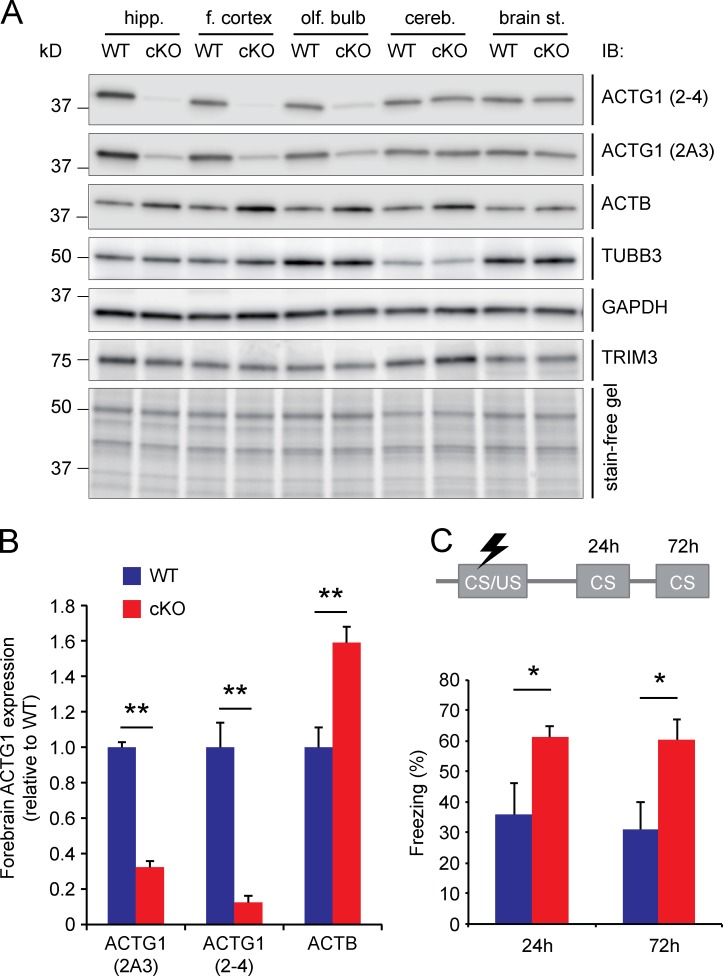
Fear memory consolidation is increased in Actg1 cKO mice. (A) Actg1 was conditionally deleted in forebrain regions by crossing Actg1lox/lox mice. Immunoblotting of total protein lysates confirmed a reduction of ACTG1 protein levels and a compensatory upregulation of ACTB levels in hippocampus, frontal cortex, and olfactory bulb of Actg1 cKO mice. No changes were observed in the levels of TUBB3, GAPDH, or TRIM3, and no reduction in ACTG1 protein levels was observed in brainstem or cerebellum. (B) ACTG1 protein levels in forebrain areas (hippocampus, frontal cortex, and olfactory bulb) of Actg1 cKO mice were reduced 70–90% relative to wild-type control tissue, whereas ACTB levels were 60% increased (means ± SEM, two-tailed t test, **, P < 0.01). (C) Hippocampal memory performance was tested at 3 mo of age in a contextual fear memory task. Freezing behavior was significantly increased in Actg1 cKO mice compared with wild-type littermates when reexposed to the conditioned context 24 h or 72 h after receiving the shock (means ± SEM, two-tailed t test, *, P < 0.05, n = 5/6 per genotype).
TRIM3 polyubiquitylates ACTG1
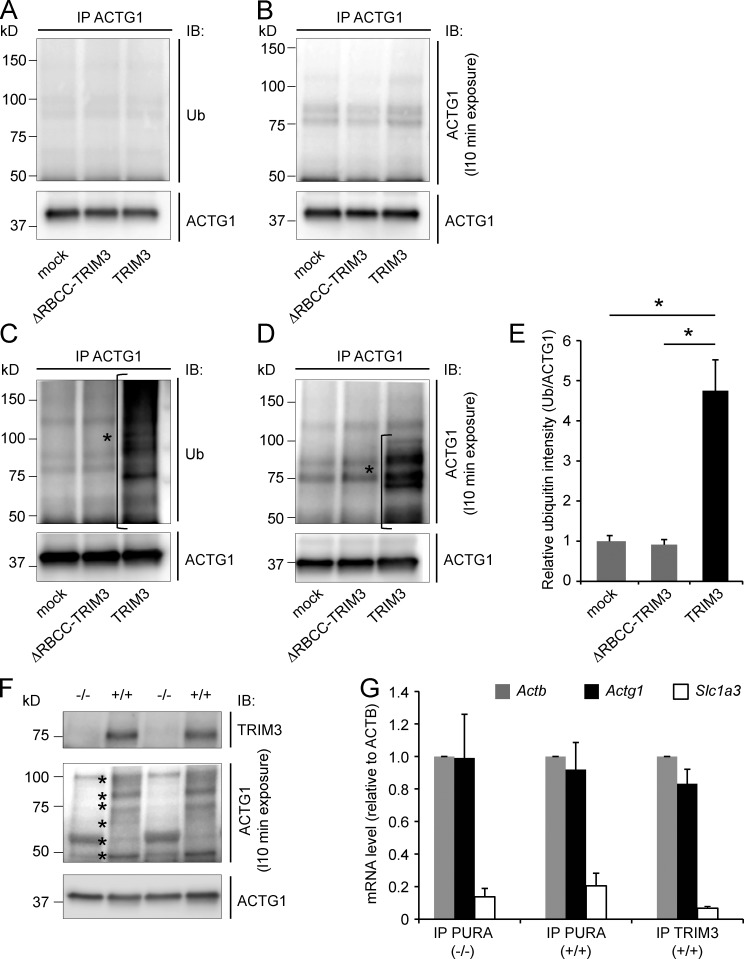
Full-length TRIM3 or ΔRBCC-TRIM3 was expressed in HEK293 cells. 2 d later, proteasomal degradation was blocked by adding MG132 for 4 h, cell lysates were harvested, and ACTG1 was immunoprecipitated. The immunoprecipitates were subsequently immunoblotted for ubiquitin and ACTG1. Unmodified ACTG1 was detected at the expected molecular weight of 42 kD, and we did not detect any increase in high-molecular-weight ubiquitin (Fig. 6 A) or high-molecular-weight ACTG1 species (Fig. 6 B) as a result of TRIM3 expression. However, when blocking proteasomal degradation and at the same time stimulating cells with the phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA; also known as phorbol 12-myristate 13-acetate, or PMA), a PKC activator that induces protein synthesis and synaptic plasticity (Herbert et al., 2000; Carriere et al., 2011; Kim et al., 2013), a strong increase was observed in high molecular weight ubiquitin staining after immunoprecipitation of ACTG1 (Fig. 6 C). ACTG1 staining confirmed the presence of 50–100-kD modified ACTG1 species (Fig. 6 D) specific for TRIM3-expressing cells, whereas it was absent in ΔRBCC-TRIM3– or mock-transfected cells. The increase in ACTG1-specific ubiquitin staining in TRIM3-expressing cells was reproducible and statistically significant (mock: 1 ± 0.14, ΔRBCC-TRIM3: 0.90 ± 0.14, TRIM3: 4.76 ± 0.76; P < 0.05; Fig. 6 E). In cultured hippocampal wild type, but not Trim3−/−, neurons treated with TPA and MG132 for 4 h, a similar increase was observed in 50–100 kD modified ACTG1 species (Fig. 6 F). The discrete banding pattern of these high molecular weight ACTG1 species is consistent with ubiquitin modifications consisting of one or more ubiquitin moieties.
Figure 6.
TRIM3 polyubiquitylates ACTG1. (A–D) TPA stimulation induces ACTG1 polyubiquitylation by TRIM3 in HEK293 cells. HEK293 cells were transfected with TRIM3 or ΔRBCC-TRIM3. After incubation for 4 h in the presence of MG132, cells were lysed and ACTG1 was immunoprecipitated from the lysates, resolved on SDS-PAGE, and immunoblotted. Blots were first stained for polyubiquitin (A) and then stripped and restained for ACTG1 (B). Under these basal conditions, only unmodified ACTG1 was detected. However, when cells were incubated in the presence of MG132 and TPA, a significant increase in high-molecular-weight polyubiquitylated forms of ACTG1 (indicated with *) was observed specifically in TRIM3-transfected cells (C–E; means ± SEM, two-tailed t test, *, P < 0.05, n = 3 cultures per condition). (F) TPA induces ACTG1 polyubiquitylation by TRIM3 in hippocampal neurons. Cultured hippocampal neurons from wild-type and Trim3−/− mice were treated with MG132 and TPA for 4 h. Lysates were immunoblotted and stained for ACTG1. TPA induced the appearance of multiple high molecular weight (50–100 kD) bands (indicated with *) consistent with polyubiquitylation and specifically in neurons from wild-type mice and not from Trim3−/− mice. (G) Actg1 mRNA is present in TRIM3/PURA containing mRNP granules. mRNP granules were immunoprecipitated from hippocampal lysates with antibodies against TRIM3 or PURA. mRNA was isolated from immunoprecipitates, reverse transcribed into cDNA, and used for real-time quantitative PCR. Actg1 and Actb mRNA was detected at equal levels in all precipitates, whereas the negative control mRNA Slc1a3 was ∼10-fold lower detected (means ± SEM, n = 3 immunoprecipitations per condition).
It was previously reported that polyubiquitylation of ACTG1 occurs cotranslationally at Lys-18, which becomes inaccessible for modification once the protein is fully synthesized (Zhang et al., 2010). In that respect, it is interesting to reiterate that TRIM3 is found in mRNP particles (Fig. 3), as these might codeliver TRIM3 and Actg1 mRNA to synaptic sites, allowing TRIM3 to polyubiquitylate ACTG1 during local translation. In contrast to Actb mRNA, Actg1 mRNA was not previously reported to be present at synapses. To test whether Actg1 mRNA is available for local translation, mRNP particles were immunoprecipitated from hippocampal synapse-enriched fractions using antibodies against TRIM3 or PURA and the levels of different mRNAs quantified using quantitative real-time PCR. We found that Actg1 and Actb mRNAs were present in all immunoprecipitates at similar levels, whereas a negative control transcript (Slc1a3) was detected at ∼10 times lower levels (Fig. 6 G). The same results were obtained when mRNP particles were immunoprecipitated from Trim3−/− protein fractions using the PURA antibody, suggesting that TRIM3 is not required for the incorporation of Actg1 mRNA into mRNP particles. Thus, these data show that TRIM3 can polyubiquitylate ACTG1 and that it probably does so cotranslationally at synaptic sites.
Trim3−/− mice show increased hippocampal LTP and spine densities
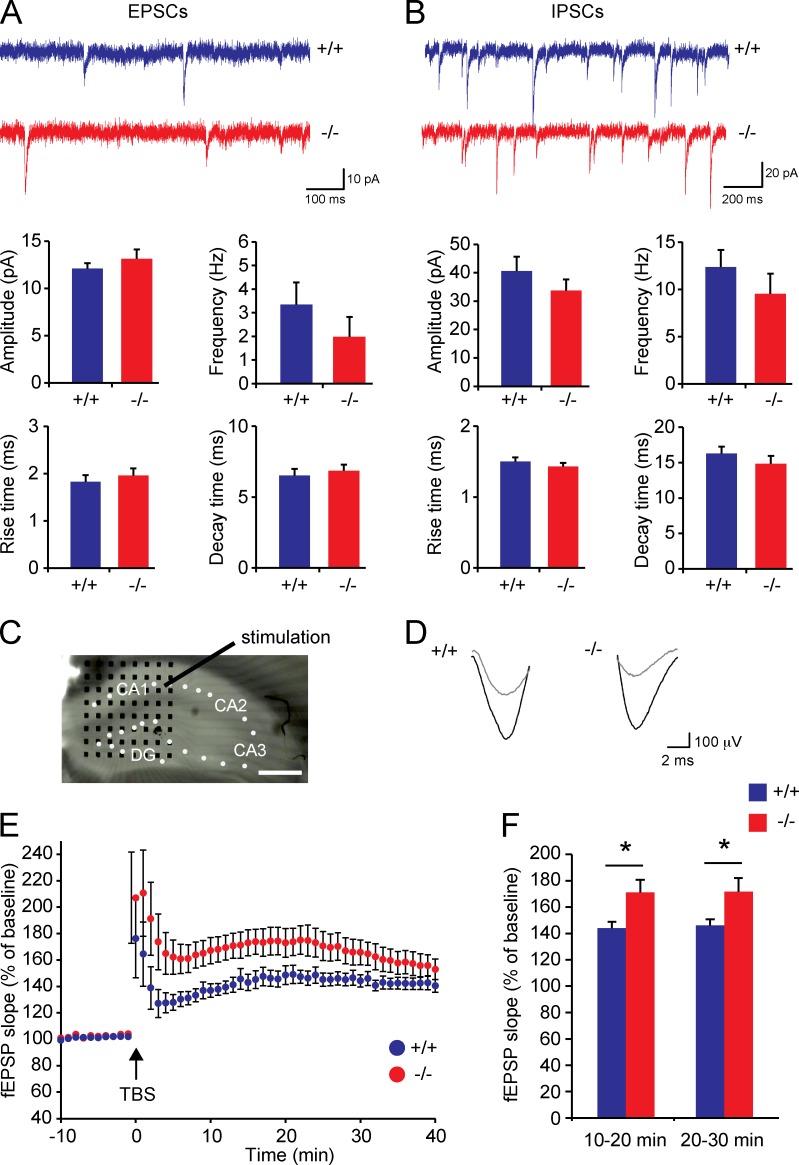
Spontaneous synaptic activity measurements in hippocampal slice preparations did not reveal significant differences between Trim3−/− mice and wild-type controls in either excitatory postsynaptic current (EPSC; Fig. 7 A) or inhibitory postsynaptic current (IPSC; Fig. 7 B) amplitude (EPSC, wild type: 12.11 ± 0.55 pA, Trim3−/−: 13.13 ± 1.01 pA; IPSC, wild type: 40.6 ± 5.1 pA, Trim3−/−: 33.71 ± 3.99 pA), frequency (EPSC, wild type: 3.34 ± 0.95 Hz, Trim3−/−: 1.98 ± 0.83 Hz; IPSC, wild type: 12.37 ± 1.81 Hz, Trim3−/−: 9.52 ± 2.14 Hz), rise time (EPSC, wild type: 1.82 ± 0.14 ms, Trim3−/−: 1.96 ± 0.15 ms; IPSC, wild type: 1.50 ± 0.06 ms, Trim3−/−: 1.42 ± 0.05 ms) or decay time (EPSC, wild type: 6.52 ± 0.47 ms, Trim3−/−: 6.85 ± 0.43 ms; IPSC, wild type: 16.26 ± 0.97 ms, Trim3−/−: 14.84 ± 1.12 ms). However, hippocampal LTP was significantly enhanced in slices from Trim3−/− mice. LTP was induced in acute hippocampal slices by theta burst stimulation of the Schaffer collateral pathway using a single stimulation electrode and recorded in CA1 using an 8 × 8 multielectrode array (Fig. 7 C). Potentiation was readily detected in slices from both genotypes (Fig. 7 D), with Trim3−/− slices showing on average a stronger potentiation (Fig. 7 E). This increase in LTP was significant at 10–20 min after induction (wild type: 144% ± 4.8%, Trim3−/−: 171.2% ± 9.6%; P < 0.05) as well as 20–30 min after induction (wild type: 146.1% ± 4.6%, Trim3−/−: 171.7% ± 10.4%; P < 0.05; Fig. 7 F).
Figure 7.
Hippocampal LTP is increased in Trim3−/− mice. (A and B) Spontaneous excitatory (A) and inhibitory (B) synaptic transmission are unaffected in Trim3−/− mice. Excitatory synaptic currents (mEPSC) and inhibitory synaptic currents (mIPSCs) were recorded from hippocampal slices. Frequency, amplitude, rise time, and decay time of both excitatory and inhibitory currents were unaffected in Trim3−/− mice compared with wild-type controls (means ± SEM, n = 6–8 for EPSCs, n = 10 for IPSCs). (C) LTP was induced in acute hippocampal slices using a single stimulation electrode in the Schaffer collateral pathway, and field excitatory postsynaptic potentials (fEPSPs) were recorded using an 8 × 8 multielectrode array (black dots) in the CA1 area. Bar, 400 µm. (D) Example field EPSP traces from a Trim3−/− mice and a wild-type control before (gray) and after (black) tetanus stimulation indicate stronger potentiation in Trim3−/− mice. (E) Averaged fEPSP data show enhanced LTP in Trim3−/− animals, in particular in the first 30 min after theta burst stimulation (TBS). (F) Quantification of the mean amount of potentiation at 10–20 min and 20–30 min after TBS show a significant increase in Trim3−/− mice (means ± SEM, two-tailed t test, *, P < 0.05, n = 15/16 slices obtained from 9/10 animals per genotype).
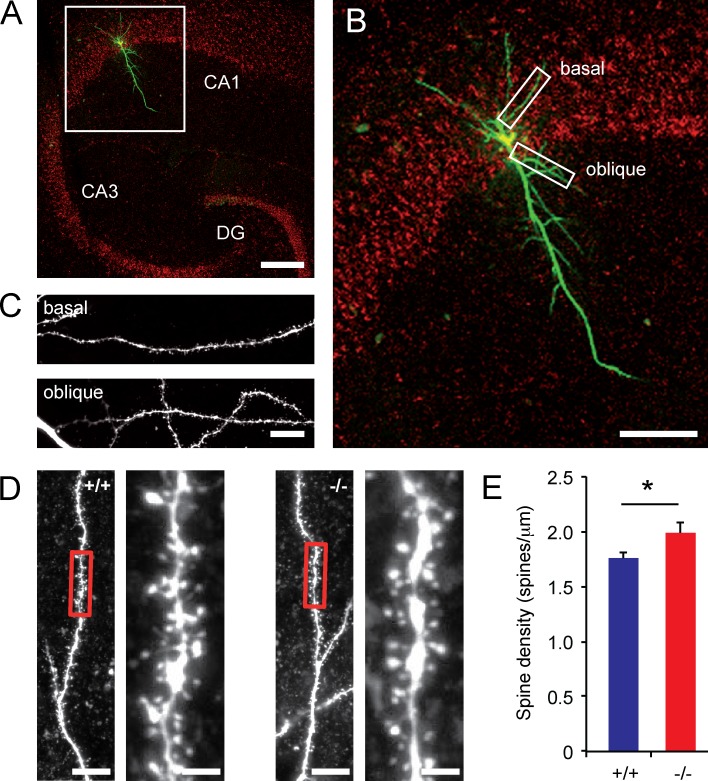
Histochemical analysis revealed no differences between Trim3−/− mice and wild-type controls at the anatomical level (Fig. S2). We next filled hippocampal CA1 neurons in slices with biocytin and collected high-resolution confocal images of the first basal and oblique dendrite of each neuron (Fig. 8, A–C). Quantification of spine densities revealed that Trim3−/− dendrites on average have a higher spine density (wild type: 1.76 ± 0.05 spines/µm, Trim3−/−: 1.99 ± 0.10 spines/µm; P < 0.05; Fig. 8, D and E). Together, these data show that increased synaptic ACTG1 levels in Trim3−/− mice correlate with increased LTP and spine densities in the hippocampus.
Figure 8.
Hippocampal spine densities are increased in Trim3−/− mice. (A) Example image of a hippocampal CA1 pyramidal cell filled with biocytin and imaged using confocal microscopy. Bar, 200 µm. (B and C) The first basal and the first oblique dendrite of each cell were selected for spine density measurements. Bars, 100 µm (B) and 10 µm (C). (D) Representative images of dendrite segments from Trim3−/− mice and wild-type controls. Bars, 10 µm (overview) and 2 µm (enlargement). (E) Mean spine density was significantly higher in Trim3−/− mice compared with wild-type controls (means ± SEM, two-tailed t test, *, P < 0.05, n = 6/11 cells per genotype).
ACTG1 regulates memory formation
We generated Actg1 conditional knockout (cKO) mice by crossing Actg1lox/lox mice (Perrin et al., 2010) with mice that express Cre-recombinase under control of the Camk2a promoter, which is activated in hippocampal neurons from the first postnatal week on (Mantamadiotis et al., 2002). Actg1 cKO mice showed significantly reduced levels of ACTG1 in the hippocampus (Fig. 9 A and B). We also observed a compensatory increase in ACTB expression, which is in accordance with previously published literature (Belyantseva et al., 2009). No changes were observed in the expression of TUBB3, GAPDH, or TRIM3 (Fig. 9 A). We next tested these mice in a contextual fear conditioning paradigm. We reasoned that if increased ACTG1 levels caused by TRIM3 deletion would result in faster consolidation of contextual fear memory (Fig. 2), deletion of ACTG1 might delay or extend the contextual fear memory response. Freezing was therefore measured at 24 and 72 h after the shock. At both time points, Actg1 cKO mice showed significantly higher levels of freezing compared with wild-type controls (24 h, wild type: 35.7% ± 10.3% freezing, Actg1 cKO: 61.2% ± 3.8% freezing; 72 h, wild type: 31.1% ± 9.0% freezing, Actg1 cKO: 60.6% ± 6.7% freezing; P < 0.05; Fig. 9 C).
Discussion
In this study, we identified TRIM3 as a ubiquitin ligase that targets ACTG1 for proteasomal degradation. Trim3−/− mice have increased levels of ACTG1 at hippocampal synaptic sites, higher spine densities in the hippocampus, increased hippocampal LTP, and enhanced short-term contextual fear memory. Interestingly, mice with a hippocampal deletion of Actg1 showed increased long-term fear memory. Our data thus support the hypothesis that temporal control of ACTG1 levels by TRIM3 is required to regulate the timing of hippocampal plasticity and constrain plasticity within physiological boundaries.
Studies in heterologous cells demonstrated that ACTG1 is polyubiquitylated cotranslationally (Zhang et al., 2010). Noncoding sequences in the Actg1 mRNA cause ACTG1 to be synthesized at a much slower rate than ACTB, allowing a hidden lysine residue in the N terminus to be exposed long enough to be ubiquitylated. For TRIM3 to be responsible for cotranslational ACTG1 ubiquitylation, TRIM3 protein and Actg1 mRNA should be in close proximity. Moreover, because polyubiquitylated ACTG1 is rapidly degraded (Zhang et al., 2010), translation should occur close to the site of action for ACTG1 to be functionally incorporated into F-actin. In this respect, it is interesting to note that Kanai et al. (2004) found TRIM3 among a core set of 42 proteins that constitute a detergent-resistant, RNase-sensitive neuronal transport granule that associates with KIF5. These so-called mRNP granules transport mRNAs toward synapses (Buchan, 2014). We confirmed the presence of TRIM3 in mRNP granules by showing that TRIM3 coimmunoprecipitates with PURA, the major component of mRNP granules (Elvira et al., 2006), in an RNase-sensitive manner. We also established that Actg1 mRNA is present in mRNP granules that are immunoprecipitated with antibodies against either TRIM3 or PURA. Actg1 mRNA lacks the zip code sequence that is known to target Actb mRNA to dendrites (Eom et al., 2003; Poon et al., 2006). However, a recent deep sequencing study showed that both Actg1 and Actb transcript are identified with high confidence as dendritic/axonal transcripts in the hippocampal neuropil (Cajigas et al., 2012). Our findings confirm that Actg1 mRNA levels in TRIM3- or PURA-positive immunoprecipitates are comparable with those of Actb. Thus, TRIM3 and Actg1 mRNA colocalize in mRNP granules that target mRNAs to dendritic spines for local translation, suggesting that TRIM3-mediated cotranslational ubiquitylation of ACTG1 occurs at synaptic sites.
Consistent with our finding that TRIM3 protein and Actg1 mRNA colocalize in mRNP granules trafficking to synaptic sites, proteomics screens identified ACTG1 as a high-confidence TRIM3 substrate in hippocampal synapse-enriched fractions. Moreover, TRIM3-dependent polyubiquitylated forms of ACTG1 are detected in hippocampal neurons after stimulation with TPA, a PKC activator that induces protein synthesis and synaptic plasticity (Herbert et al., 2000; Carriere et al., 2011; Kim et al., 2013), and ACTG1 levels are significantly increased in Trim3−/− neurons. Together, these findings demonstrate that TRIM3 regulates synaptic ACTG1 levels in an activity-dependent manner, most likely via ubiquitin-dependent proteasomal degradation. It is unlikely that ACTG1 is locally synthesized only to be immediately broken down. We propose that polyubiquitylated ACTG1 can be incorporated transiently into F-actin. Possibly, TRIM3 plays an active role in this, as TRIM3 interacts with several actin-binding proteins, including myosin V (El-Husseini and Vincent, 1999) and α-actinin-4 (El-Husseini et al., 2000), and is involved in actin-based transport (Yan et al., 2005). TRIM3 may thus facilitate incorporation of ACTG1 into F-actin while at the same time controlling ACTG1 degradation. Although actin filament turnover in dendritic spines is too fast to be determined by local translation only (Star et al., 2002), a transient increase in ACTG1 levels caused by local translation and degradation might create a window of opportunity for ACTG1 to contribute to activity-induced synaptic plasticity. Interestingly, enhanced LTP in Trim3−/− mice is confined to the early phase of LTP (i.e., the first 30 min after induction), which was previously demonstrated to also depend on protein synthesis (Fonseca et al., 2006). LTP maintenance is normal, suggesting that ACTG1 turnover may not be abolished completely in the absence of TRIM3 or that compensatory mechanisms that depend on gene expression and global protein synthesis (Kelleher et al., 2004) normalize plasticity at later time points.
What might be the specific contribution of ACTG1 to actin dynamics in dendritic spines in relation to synaptic plasticity? Our findings show that increased synaptic ACTG1 protein levels correlate with an increase in spine density, LTP, and short-term contextual fear memory. Moreover, we demonstrate that postnatal deletion in hippocampal neurons of Actg1 itself causes an increase in long-term fear memory. Although these findings do not provide definite proof that the plasticity and memory phenotypes in Trim3−/− mice are caused by increased ACTG1 levels only, they do indicate that ACTG1 contributes to hippocampal plasticity and that TRIM3 is involved as an upstream regulator, controlling the timing of the plasticity response. In contrast to ACTB (Cheever et al., 2012), CNS-specific deletion of ACTG1 does not result in detectable morphological or histological abnormalities (Cheever and Ervasti, 2013), indicating that ACTG1 has no major role in gross brain development. Several studies have hinted toward specific and unique roles for ACTG1 in adulthood. For instance, whereas ACTG1 and ACTB are each dispensable for the normal development of auditory hair cells in the cochlea (Perrin et al., 2010), ACTG1-deficient mice cannot properly maintain hair cell stereocilia actin cores and suffer from progressive age-dependent hearing loss (Belyantseva et al., 2009). The authors observed that ACTG1 accumulates at sites of stereocilia core disruptions and conclude that ACTG1 is required for the reinforcement of long-term stability of actin filaments and the remodeling and repair of stereocilia cores after mechanical stress-induced damage. One possible explanation for these findings is that ACTG1-containing filaments have different biochemical and biophysical properties than filaments that lack ACTG1. Recent studies seem to confirm that ACTG1 may indeed contribute to more stable actin filaments (Bergeron et al., 2010). Although polymerization rates of ACTB and ACTG1 were indistinguishable in the Mg2+-bound state, which is considered to be the physiologically relevant state in most cells, turnover rates of ACTB in its Ca2+-bound state were considerably faster than those of ACTG1. These findings indicate that under high-Ca2+ conditions, e.g., in activated spines, ACTG1 is more stable than ACTB. Moreover, because ACTB and ACTG1 readily copolymerized, it probably is the ACTB/ACTG1 ratio in actin filaments that determines their stability in the presence of Ca2+ (Bergeron et al., 2010). By regulating local ACTG1 levels, TRIM3 may thus play a crucial role in controlling actin stability during synaptic plasticity. Changes in actin stability would directly affect spine morphogenesis (Sekino et al., 2007) and synaptic AMPA receptor recycling (Zhou et al., 2001; Osterweil et al., 2005) and anchoring (Allison et al., 1998; Kuriu et al., 2006), thus explaining the spine density, LTP, and memory phenotypes observed in Trim3−/− mice.
In conclusion, we identified a novel mechanism by which TRIM3 and ACTG1 together control synaptic plasticity. Our data are consistent with a model in which TRIM3 provides an inhibitory constraint on the expression of synaptic plasticity by regulating the availability of ACTG1 to transiently alter actin dynamics in dendritic spines.
Materials and methods
Animals
The generation of Trim3−/− mice was described previously (Labonté et al., 2013). Actg1lox/lox mice (Perrin et al., 2010; gift of J. Ervasti, University of Minnesota, Minneapolis, MN) were crossed with Camk2a-Cre mice (Mantamadiotis et al., 2002) to generate Actg1 cKO mice. All mice were maintained on a C57BL/6J background. In all experiments, wild-type littermates were used as controls. Mice were individually housed on sawdust in standard Makrolon type II cages (26.5 cm long, 20.5 cm wide, and 14.5 cm high) enriched with cardboard nesting material under 12 h/12 h dark/light cycle with access to water and food ad libitum. All animal experiments were approved by the animal ethics committee of the VU University Amsterdam.
Expression constructs
Constructs expressing myc-tagged or EGFP-tagged full-length TRIM3 and ΔRBCC-TRIM3 (amino acids 283–744) were generated in pCDNA3.1-Myc (Invitrogen) and pEGFP-N1 (Clontech), respectively. The EGFP-PURA construct was a gift of J.L. Napoli (University of California, Berkeley, Berkeley, CA; Chen et al., 2008).
Antibodies
The following antibodies were used for immunohistochemistry/immunofluorescence: chicken anti-MAP2 (AB5543, Abcam; 1:5,000) and mouse anti-TRIM3/BERP (clone 27, BD Biosciences; 1:1,000). The following antibodies were used for immunoblotting: mouse anti-ACTB (clone AC-15, Sigma-Aldrich; 1:10,000), mouse anti-ACTG1 (clone 2–4, Santa Cruz Biotechnology; 1:200), mouse anti-ACTG1 (clone 2A3, Abcam; 1:10,000), rabbit anti-GAPDH (AB9485, Abcam; 1:500), mouse anti-GFP (clone N86/8; Neuromab; 1:2,000), mouse anti-GKAP (clone N238/31, Neuromab; 1:1,000), mouse anti–PSD-95 (clone K28/43, Neuromab; 1:10), rabbit anti-PURA (AB79936, Abcam; 1:2,000), mouse anti-SHANK1 (clone N22/21, Neuromab; 1:1,000), rabbit anti-SYP (H-93, Santa Cruz Biotechnology; 1:2,000), mouse anti-TRIM3/BERP (clone 27, BD Biosciences; 1:250), rabbit anti-TRIM3 (custom-made epitope CLRPGDLPPSPDDVK; GenScript; 1:1,000), rabbit anti-TRIM3 (custom-made epitope QAPEGAHDPEDPHPC, GenScript; 1:500), mouse anti-TUBB3 (clone SDL.3D10, Sigma-Aldrich; 1:2,000), mouse antiubiquitin (clone FK2, Enzo Life Sciences; 1:100), and mouse antipolyubiquitin (clone FK1, Enzo Life Sciences; 1:1,000). The following antibodies were used for immunoprecipitation: mouse anti-ACTG1 (clone 2–4, Santa Cruz Biotechnology; 10 µg), mouse anti-ACTG1 (clone 2A3, Abcam; 25 µg), mouse anti-PURA (clone 2B7, Millipore; 10 µg), goat anti-TRIM3/RNF22 (AB4215, Abcam; 10 µg), rabbit anti-TRIM3 (epitope CLRPGDLPPSPDDVK, GenScript; 10 µg), and rabbit anti-TRIM3 (epitope QAPEGAHDPEDPHPC, GenScript; 10 µg).
Immunohistochemistry
Mice were intracardially perfused with ice-cold PFA (4% in PBS, pH 7.4). Brains were removed, postfixed, embedded in gelatin (Mallinckrodt Baker), and cryostat-sectioned at 40 µm. Sections were stained with anti-TRIM3/BERP (BD Biosciences), and staining was visualized using VECTASTAIN ABC standard kit (Vector Laboratories). Nissl staining was used for gross morphological characterization of brain slices.
Golgi-cox staining
Golgi-cox staining was performed using the FD Rapid GolgiStain kit (FD NeuroTechnologies). After staining, brains were microtome-sectioned at 100 µm. Neurons were photographed at 100× magnification, and Z stacks were generated using Image Pro Plus 6.2 software (Media Cybernetics).
Primary neuron cultures
Hippocampi were dissected from E18 wild-type mice, collected in HBSS (Sigma-Aldrich) buffered with 7 mM Hepes and incubated for 30 min in HBBS containing 0.25% trypsin (Invitrogen) at 37°C. After washing, neurons were triturated with fire-polished Pasteur pipettes, counted, and plated in Neurobasal medium supplemented with 2% B-27, 1.8% Hepes, 1% glutamax, 1% penicillin/streptomycin (all from Life Technologies), and 0.2% 14.3 mM β-mercapto-ethanol. Cultures were plated on glass coverslips coated in poly-d-lysine (Sigma-Aldrich) and treated with 5% heat-inactivated horse serum (Life Technologies). Neurons were plated at a seeding density of 250 cells/mm2. Half of the medium was replaced with fresh medium every week. After 14 d in culture, neurons were either lysed in SDS loading buffer directly or first incubated with 20 µM MG132 (Tocris Bioscience) and 500 nM TPA (Sigma-Aldrich) for 4 h and then lysed.
Immunofluorescence
Primary hippocampal neurons were fixed after 14 d in culture using 4% PFA and 10% saccharose in PBS, pH 7.4. Neurons were stained with anti-TRIM3/BERP (BD Biosciences) and anti-MAP2 (Abcam). Antigens were visualized using Alexa Fluor 488 and 568 (Molecular Probes; 1:400). Confocal microscopy was performed using an LSM 510 (Carl Zeiss).
Time-lapse imaging
Hippocampal primary neurons were cultured in glass-bottom dishes and transfected with GFP-PURA at day 8–10 in culture using calcium phosphate as previously described (Labonté et al., 2013). After 15–24 h of GFP-PURA expression, cells were imaged in prewarmed Hepes buffer using an inverse fluorescence microscope (Axiovert 200M; Carl Zeiss) in an incubation chamber (Harnischmacher-Labortechnik) at 37°C. MetaVue software (v6.2r6; Molecular Devices) was used to record cells for 5 min at 3–5-s intervals. Analyses were done with the Metamorph software (v6.3r7; Molecular Devices). Significance was determined using a two-tailed t test.
Human embryonic kidney cell cultures
Human embryonic kidney 293 (HEK293) cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (all from Life Technologies) at 37°C and 5% CO2. Cells were transfected at 60–80% confluence using polyethylenimine. At 48 h after transfection, cells were washed with ice-cold PBS and lysed in SDS loading buffer for subsequent immunoblot analysis or in extraction buffer (1% Triton X-100, 150 mM NaCl, and 25 mM Hepes, pH 7.4) for subsequent immunoprecipitation. For ubiquitylation assays, cells were first incubated with 20 µM MG132 (Tocris Bioscience) and 500 nM TPA (Sigma-Aldrich) for 4 h and then lysed.
Cellular fractionation
Hippocampi were collected and cellular fractionation was performed as previously described (Klemmer et al., 2009). In brief, hippocampi were homogenized in a glass Potter homogenizer containing 5 ml ice-cold homogenization buffer (320 mM sucrose in 5 mM Hepes, pH 7.4) at 900 rpm. Cell debris and nuclei were removed by centrifugation at 1,000 g for 10 min. The supernatant (S1) was centrifuged at 16,000 g for 30 min to obtain crude synaptosomes (P2 fractions) or at 100,000 g for 2 h to obtain crude synaptosomes plus microsomes (P2+M fractions). For synaptosome and synaptic membrane isolations, S1 supernatant was loaded on a 0.85–1.2 M sucrose gradient. After ultracentrifugation at 100,000 g at 4°C for 2 h, the synaptosome fraction was recovered at the 0.85/1.2 M sucrose interface. Synaptosomes were then diluted with an equal volume of 5 mM Hepes, pH 7.4, adjusted to 15 ml with homogenization buffer, and centrifuged at 25,000 g for 30 min. The pellet was resuspended in Hepes buffer and osmotically shocked while stirring slowly on ice for 15 min. The resulting synaptic membrane fraction was recovered using 0.85–1.2 M sucrose gradient ultracentrifugation at 100,000 g at 4°C for 2 h.
Immunoprecipitation
For hippocampal tissue, 10 mg P2+M was extracted in an equal volume of 2% Triton X-100, 150 mM NaCl, and 25 mM Hepes, pH 7.4. For HEK293 cells, the extraction buffer contained 1% Triton X-100. Protein extracts were supplemented with complete protease inhibitor cocktail (Roche Diagnostics) on a rotator at for 1 h at 4°C. Insoluble material was pelleted at 20,000 g for 20 min at 4°C. For hippocampal extracts, the pellet was reextracted in 1% Triton X-100, 150 mM NaCl, and 25 mM Hepes, pH 7.4, supplemented with protease inhibitors for 1 h. Insoluble material was again pelleted at 20,000 g for 20 min. Supernatants from the first and second extraction were then pooled and centrifuged at 20,000 g for 20 min, and the final supernatant served as input for immunoprecipitation. For demonstrating polyubiquitylation of ACTG1, samples were boiled for 10 min in 2% SDS, 150 mM NaCl, 2 mM DTT, and 25 mM Hepes, pH 7.4, before ACTG1 immunoprecipitation to ensure that polyubiquitin signals are actually derived from ACTG1 itself. For HEK293 cells, a single round of extraction was used. In some experiments, input samples were split in half, and one half was treated with RNase inhibitors (40 U/ml; Thermo Fisher Scientific) and the other half with RNase A (∼0.2 µg/µl; Sigma-Aldrich). After 24 h, 10–25 µg antibody was added to each sample and incubated overnight on a rotator at 4°C. Next day, 30 µl of protein A/G beads (Santa Cruz Biotechnology) was washed three times in washing buffer (0.1% Triton X-100, 150 mM NaCl, and 25 mM Hepes, pH 7.4) and added to each immunoprecipitation. Beads were incubated for 120 min on a rotator at 4°C, pelleted at 2,500 g, and washed four times in ice-cold washing buffer. Proteins were eluted-off the beads by adding SDS loading buffer and boiling for 5 min. Samples were then separated on SDS-PAGE and immunoblotted.
TUBEs affinity purification
TUBEs immobilized to agarose beads were purchased from LifeSensors. For each affinity purification experiment, 20 mg hippocampal P2+M protein fraction was added to 100 µl TUBEs slurry. Purification of bound proteins was performed according to the manufacturer’s protocol with minor modifications. Not NP-40 but a combination of 0.5% Na-deoxycholate and 1% n-dodecyl-β-d-maltoside in 25 mM Hepes and 150 mM NaCl, pH 7.2, was used as solubilizing detergent. Deubiquitylation and proteasomal degradation were minimized by adding 10 mM N-ethylmaleimide, 20 µM MG132 (Tocris Bioscience), and protease inhibitors (Roche) to all extraction buffers. Tris was replaced by Hepes as the buffering agent. Purified proteins were washed three times in extraction buffer, once in high salt buffer (500 mM NaCl), and once in no salt buffer. Proteins were then eluted in 0.15% trifluoroacetic acid (TFA) and lyophilized for further analysis.
Immunoblotting
Protein samples were loaded on a 4–20% gradient SDS-PAGE gel (Criterion TGX Stain-Free Gel; Bio-Rad Laboratories). Total protein amounts were imaged after electrophoresis using Criterion Stain Free Imager (Bio-Rad Laboratories). Proteins were then transferred to polyvinylidene difluoride membranes (Immun-Blot, Bio-Rad Laboratories), which were subsequently blocked by immersion for 1 h in TBS containing 0.025% Tween and 5% nonfat dry milk. For ubiquitin staining, membranes were blocked with 0.5% BSA instead of nonfat dry milk. Blots were then incubated with primary antibody for 1 h at RT, followed by a 1-h incubation with a secondary antibody conjugated to HRP. Proteins were visualized using the SuperSignal West Femto Chemiluminescent Substrate kit (Thermo Fisher Scientific) and scanned using an Odyssey Fc imager (Li-Cor). Protein quantities were calculated by dividing background-corrected signal intensity of each protein band (ImageStudio Software v2.0.38; Li-Cor) by the background-corrected signal intensity of the corresponding lane on the stain-free gel (Image Lab v3.0; Bio-Rad). Significance was tested using a two-tailed t test.
Trypsin digestion
Immunoprecipitated protein samples were separated on SDS-PAGE and in-gel trypsin-digested as previously described (Chen et al., 2011). TUBEs-purified proteins were trypsin-digested in solution. Purified proteins were resolubilized in 50 mM NH4HCO3/10% acetonitrile, brought to 2 mM DTT and heated to 60°C for 20 min. Proteins were then alkylated with 8 mM iodoacetamide for 1 h. Alkylation was stopped by bringing samples to 8 mM DTT. Samples were then brought to a final concentration of 25 mM NH4HCO3, 8% acetonitrile, and 2 mM DTT and digested with trypsin (mass spectrometry grade; Promega) overnight at RT. Digestion was stopped by adding 1% TFA. Peptides were purified over a C18 column and lyophilized.
Nano-LC MS/MS
Peptides were redissolved in 20 µl 0.1% TFA and subjected to nano-liquid chromatography (nano-LC) tandem mass spectrometry (MS/MS). Peptides were delivered with a FAMOS autosampler at 30 µl/min to a Pepmap C18 trap column (5 mm × 300 µm i.d.; Dionex) and separated on an Alltima analytical capillary C18 column (150 mm × 100 µm i.d.) at 400 nl/min using the LCPacking Ultimate system. The peptides were separated using a linearly increasing concentration of acetonitrile from 5–35% in 80 min, from 35–45% in 7 min, from 45–90% in 2 min, and then held at 90% for an additional 11 min. The eluent was mixed with matrix (7 mg α-cyano-hydroxycinnaminic acid in 1 ml 70% acetonitrile, 0.1% trifluoroacetic acid, 10 mM ammonium monobasic phosphate) delivered at a flow rate of 1.5 µl/min and deposited offline to the Applied Biosystems metal target every 15 s for a total of 384 spots, using an automatic robot (Probot; Dionex). A 5800 proteomics analyzer (Applied Biosystems) was used for peptide analysis. CID was performed at 2 kV (the collision gas was nitrogen). MS/MS spectra were collected from 2,500 laser shots. The peptides with signal to noise ratio above 50 at the MS mode were selected for the MS/MS experiment; a maximum of 25 MS/MS were allowed per spot. The precursor mass window was 200 relative resolution (FWHM).
Protein identification, quantification, and selection strategy
MS/MS spectra were annotated against a concatenated target-decoy database of Uniprot mouse reference proteome sequences (version 02/2012) by ProteinPilot software (v3.0; Applied Biosystems) using the Paragon algorithm (v3.0.0.0; Shilov et al., 2007). Database searches were performed with trypsin-specificity and alkylation of cysteine residues by iodoacetamide. For each spectrum, the best-scoring peptide sequence was selected as spectrum annotation. Protein inference was performed using IsoformResolver software (Meyer-Arendt et al., 2011) aiming for consistent protein assignment of peptides across experiments. False discovery rates (FDRs) for peptide and protein identification were established using Mayu (Reiter et al., 2009). Peptide-spectrum matches were truncated at 5% FDR and protein identifications were truncated at 1% FDR. Proteins were quantified according to their normalized spectral intensity value (Griffin et al., 2010). Spectral intensity values contributed by peptides shared by different proteins were apportioned according to the spectral intensity values obtained from unique peptides of the respective proteins. Apportioned spectral intensity values were then transformed as follows: all values were divided by the lowest value in the sample, one was added to each value to allow log transformation, and all resulting values were log2 transformed. Candidate TRIM3 interactors were defined as all proteins that were immunoprecipitated with at least two out of three antibodies from wild-type tissue and not detected in immunoprecipitates from Trim3−/− tissue by the same antibodies. For candidate TRIM3 substrates, we selected all proteins that were identified in at least two out three TUBEs affinity purification replicates from wild-type tissue, and had a mean enrichment of at least twofold (log2) compared with samples obtained from Trim3−/− tissue. Candidate TRIM3 interactors and substrates were then ranked based on mean protein intensities in wild-type samples.
Quantitative real-time PCR
Immunoprecipitations were performed using antibodies against TRIM3 or PURA in the presence of RNase inhibitors (40 U/ml; Thermo Fisher Scientific). RNA was extracted from immunoprecipitated complexes using TRIzol reagent (Life Technologies), DNase treated, and reverse transcribed to cDNA using random hexanucleotide primers. Quantitative PCR was performed using an ABI PRISM 7700 Sequence Detection System (Life Technologies) with SYBR green as the reporter dye. The following primers were used: Actg1 (forward: 5′-ACCAACAGCAGACTTCCAGGAT-3′, reverse: 5′-AGACTGGCAAGAAGGAGTGGTAA-3′), Actb (forward: 5′-AAGATCAAGATCATTGCTCCTCCTG-3′, reverse 5′-AGCTCAGTAACAGTCCGCCT-3′) and Slc1a3 (forward: 5′-GTGGACTGGTTTCTGGACCG-3′, reverse: 5′-TCATGTCGGGACAAGTGCTC-3′). Relative mRNA expression levels were calculated using the ΔΔCτ method (Livak and Schmittgen, 2001).
Open field test
Mice were introduced into a corner of the white square open field (50 × 50 cm, walls 35 cm high) illuminated with a single white fluorescent light bulb from above (1,30l×), and exploration was tracked for 10 min (12.5 frames/s; EthoVision 3.0, Noldus Information Technology). EthoVision was used to calculate the total distance traveled as well as the time spent and the number of entries into the center square area (20 × 20 cm). SEE (Software for the Exploration of Exploration; Kafkafi et al., 2003) was used to smooth path shape and calculate specific measures of locomotor behavior. SEE uses the distribution of speed peaks to parse the locomotor data slow local movements (so-called lingering episodes) and progression segments.
Dark-light box
Mice were introduced into the dark compartment (<10 lux; 25 × 25 × 30 cm, length × width × height) of a dark-light box and the lid was closed. After 1 min, the bright light was switched on, and the door to the brightly lit compartment (320 lux; same dimensions) was opened. Exploratory behavior of eight mice was video tracked simultaneously for 10 min (12.5 frames/s; EthoVision 3.0; Noldus Information Technology). The number of transitions into and the total time spent in the light compartment were calculated using the hidden zone feature in EthoVision with the border of crossing into the light compartment defined 2 cm away from the door in the light compartment.
Elevated plus maze
Mice were introduced into the closed arm of an elevated plus maze (arms 30 × 6 cm, walls 35 cm high, elevated 50 cm above the ground), all facing the same closed end of the arm. The maze was illuminated with a single white fluorescent light bulb from above (130 lux), and exploratory behavior was video tracked for 5 min (12.5 frames/s, EthoVision 3.0; Noldus Information Technology). The border between center and arm entries was defined at 2 cm into each arm, producing the number of entries and total duration in the open arms, closed arms and the center platform.
Fear conditioning
Fear conditioning experiments were performed as described previously (Stiedl et al., 2000; Misane et al., 2005) using a computerized fear conditioning system (TSE, 303410). In brief, mice were placed in an acrylic cage mounted into a constant illuminated (450 lux) fear conditioning box. After a 180-s exploration period, mice received a 2-s foot shock (0.7 mA) delivered through a metal floor grid. Mice were returned to their home cages 30 s after termination of the shock. Activity, exploration and freezing (lack of movement aside from respiration and heartbeat) were measured automatically via infrared lasers before, during, and after the shock. Contextual fear memory was measured as the time spent freezing after reexposure of the animals to the conditioned context at various time points after receiving the shock.
Spontaneous synaptic transmission
Mice were decapitated and brains rapidly removed and dissected in ice-cold carbogenated (95% O2/5% CO2) slicing solution (containing in mM: 110 choline chloride, 26 NaHCO3, 10 d-glucose, 11.6 Na-ascorbate, 7 MgCl2, 3.1 Na-pyruvate, 2.5 KCl, 1.25 NaH2PO4, and 0.5 CaCl2). Horizontal hippocampal slices (300 µm) were prepared as previously described (Dawitz et al., 2011). Slices were stored for at least 1 h in a submerged-style holding chamber containing carbogenated artificial cerebrospinal fluid (aCSF; in mM: 125 NaCl, 26 NaHCO3, 10 d-glucose, 3 KCl, 2.5 MgCl2, 1.6 CaCl2, 1.25 NaH2PO4). Slices were then transferred to a recording chamber and perfused continuously with carbogenated aCSF (as in the holding chamber, but with 1.5 mM MgCl2). CA1 pyramidal neurons were identified on the basis of cell soma location and basic firing properties recorded using the whole-cell patch-clamp technique. For recording excitatory synaptic currents, patch pipettes (borosilicate glass, 3–5 MΩ resistance) were filled with intracellular solution (containing in mM: 148 K-gluconate, 1 KCl, 10 Hepes, 4 Mg-ATP, 4 K2-phosphocreatine, 0.4 GTP, and 0.2% biocytin, adjusted with KOH to pH 7.3). For inhibitory currents, intracellular solution contained (in mM) 70 K-gluconate, 70 KCl, 10 Hepes, 4 Mg-ATP, 4 K2phosphocreatine, 0.4 GTP, and 0.2% biocytin, adjusted with KOH to pH 7.3. Neurons were allowed to stabilize for at least 5 min after a whole-cell configuration was achieved. For all measurements, 8-min gap-free recordings were made at −70 mV holding potential in voltage-clamp mode. Spontaneous EPSCs were measured in the presence of the GABAa receptor blocker gabazine (10 µM), and spontaneous IPSCs were measured in the presence of glutamate receptor blockers DAPV (50 µM) and CNQX (10 µM). EPSCs and IPSCs were detected using Minianalysis software (Synaptosoft). Amplitude, frequency, and rise and decay times of excitatory and inhibitory currents were measured and statistical significance was determined using a two-tailed t test.
Spine analysis
Hippocampal slices were prepared from 8–10-wk-old mice, and CA1 neurons were recorded as described in the previous section. Cells were kept in whole-cell configuration for 10–15 min to fill with biocytin, after which a Giga-seal was obtained when retracting the pipette. After retraction of the pipette, slices were immediately fixed in 4% PFA in PBS for 1–2 d at 4°C. Slices were then washed four times for >10 min in PBS and incubated with 0.5% Triton X-100 in PBS, after which they were incubated for 2 d with 1:500 streptavidin–Alexa Fluor 488 conjugate (Life Technologies) and 0.1% Triton X-100 in PBS. To-Pro-3 (Life Technologies) was added at 1:10,000 for the last 10 min of incubation to counterstain for nuclei. Slices were then washed four times for >15 min in PBS and mounted in Mowiol on glass slides. Labeled cells were imaged using a Nikon Ti-E A1R (Nikon) confocal microscope using a 10× 0.45 NA air objective to check the position of the cell in CA1. Spines were imaged using a 100× 1.49 NA oil immersion objective at a resolution of 80 (x) × 80 (y) × 125 (z) nm/px. Approximately 80 µm of the first oblique dendrite and 80 µm of the first basal dendrite (either clockwise or counter clockwise from the apical dendrite) were imaged. For each labeled neuron, one oblique and one apical dendrite were analyzed using NeuronStudio software (v0.9.92; Rodriguez et al., 2008). Automatic dendrite tracing and spine recognition were used, and spine densities were calculated. Statistical significance was determined using a two-tailed t test.
LTP
A planar multielectrode recording setup (MED64 system; Alpha Med Sciences) was used to record field EPSP (fEPSP) and elicit LTP as previously described (Shimono et al., 2002). Animals were decapitated and brains were rapidly removed and placed in ice-cold slice buffer containing (in mM) 124 NaCl, 3.3 KCl, 1.2 KH2PO4, 7 MgSO4, 0.5 CaCl2, 20 NaHCO3 and 10 glucose), which was constantly gassed with 95% O2/5% CO2. Coronal hippocampal slices were prepared using a vibrating microtome at 400 µm and then placed in a chamber containing aCSF (in mM: 124 NaCl, 3.3 KCl, 1.2 KH2PO4, 1.3 MgSO4, 2.5 CaCl2, 20 NaHCO3, and 10 glucose; constantly gassed with 95% O2/5% CO2). Slices were allowed to recover for 1 h and then placed on 8 × 8 multielectrode arrays containing P5155 probes (Alpha Med Sciences; interelectrode distance, 150 µm) precoated with polyethylenimine (Sigma-Aldrich). After addition of 500 µl aCSF, the array was placed in a moist chamber that was constantly gassed with 95% O2/5% CO2 for at least 1 h before recording. Correct placement of the electrodes at the CA3–CA1 region was done manually, monitored by a microscope (SZ61; Olympus). During recording, slices were constantly perfused with oxygenated aCSF containing 10 µM glycine at a flow rate of 2 ml/min at RT. fEPSPs were recorded from multiple electrodes in the stratum radiatum of CA1. An external concentric bipolar electrode (CBCBG75; FH-Company) in the Schaffer collateral pathway was used as the stimulating electrode using a homemade model 440b isolated Bipolar Current Stimulator. Based on the stimulus–response curve, a stimulation intensity was used that evoked fEPSPs with a magnitude of 50% of the maximum response (usually ∼100 µA). After allowing a stable baseline of 10 min, LTP was evoked by a 2× 100 Hz stimulus of 1 s each with a 15-s interval and fEPSP responses were recorded for 1 h after the tetanus. LTP was expressed as the change in the slope of the fEPSP relative to baseline and averaged for multiple electrodes (usually five) located in the stratum radiatum. Statistical significance was determined using a two-tailed t test on the fEPSP change from 10–20 min or 20–30 after LTP induction.
Online supplemental material
Fig. S1 shows that GKAP and SHANK1 levels are not affected in Trim3−/− mice. Fig. S2 shows that hippocampal morphology and cytoarchitecture are not affected in Trim3−/− mice. Table S1 shows behavioral analysis of Trim3−/− mice at 8 mo of age. Table S2 shows a list of proteins identified as potential TRIM3 substrates. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506048/DC1.
Supplementary Material
Acknowledgments
We thank Jaap Timmerman and Elize Haasdijk for technical assistance. Actg1 mice were a gift from Dr. James Ervasti. The EGFP-PURA construct was a gift from Dr. Joseph Napoli.
J. Schreiber was supported through the EU Marie Curie Training Network CerebNet, and J. Dawitz was supported through the EU Marie Curie Training Network BrainTrain. A.B. Smit and K.W. Li received funding from the EU-FP7 framework Health program (SynSys; grant 242167).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- aCSF
- artificial cerebrospinal fluid
- cKO
- conditional knockout
- EPSC
- excitatory postsynaptic current
- FDR
- false discovery rate
- IPSC
- inhibitory postsynaptic current
- LC
- liquid chromatography
- LTP
- long-term potentiation
- mRNP
- messenger RNP
- MS/MS
- tandem mass spectrometry
- TFA
- trifluoroacetic acid
- TPA
- 12-O-tetradecanoylphorbol 13-acetate
- TUBE
- tandem ubiquitin-binding entity
References
- Allison D.W., Gelfand V.I., Spector I., and Craig A.M.. 1998. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J. Neurosci. 18:2423–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balastik M., Ferraguti F., Pires-da Silva A., Lee T.H., Alvarez-Bolado G., Lu K.P., and Gruss P.. 2008. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc. Natl. Acad. Sci. USA. 105:12016–12021. 10.1073/pnas.0802261105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva I.A., Perrin B.J., Sonnemann K.J., Zhu M., Stepanyan R., McGee J., Frolenkov G.I., Walsh E.J., Friderici K.H., Friedman T.B., and Ervasti J.M.. 2009. Gamma-actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. USA. 106:9703–9708. 10.1073/pnas.0900221106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron S.E., Zhu M., Thiem S.M., Friderici K.H., and Rubenstein P.A.. 2010. Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J. Biol. Chem. 285:16087–16095. 10.1074/jbc.M110.110130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11:1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas I.J., Tushev G., Will T.J., tom Dieck S., Fuerst N., and Schuman E.M.. 2012. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 74:453–466. 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere A., Romeo Y., Acosta-Jaquez H.A., Moreau J., Bonneil E., Thibault P., Fingar D.C., and Roux P.P.. 2011. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem. 286:567–577. 10.1074/jbc.M110.159046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C., Ehrensperger M.V., Dahan M., Lévi S., and Triller A.. 2006. Cytoskeleton regulation of glycine receptor number at synapses and diffusion in the plasma membrane. J. Neurosci. 26:8502–8511. 10.1523/JNEUROSCI.1758-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever T.R., and Ervasti J.M.. 2013. Actin isoforms in neuronal development and function. Int. Rev. Cell Mol. Biol. 301:157–213. 10.1016/B978-0-12-407704-1.00004-X [DOI] [PubMed] [Google Scholar]
- Cheever T.R., Li B., and Ervasti J.M.. 2012. Restricted morphological and behavioral abnormalities following ablation of β-actin in the brain. PLoS One. 7:e32970 10.1371/journal.pone.0032970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Onisko B., and Napoli J.L.. 2008. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J. Biol. Chem. 283:20841–20847. 10.1074/jbc.M802314200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., van der Schors R., and Smit A.B.. 2011. A 1D-PAGE/LC-ESI linear ion trap orbitrap MS approach for the analysis of synapse proteomes and synaptic protein complexes. In Neuroproteomics. Li K.W., editor. Humana Press, New York: 159–167. 10.1007/978-1-61779-111-6_12 [DOI] [Google Scholar]
- Cheung C.C., Yang C., Berger T., Zaugg K., Reilly P., Elia A.J., Wakeham A., You-Ten A., Chang N., Li L., et al. 2010. Identification of BERP (brain-expressed RING finger protein) as a p53 target gene that modulates seizure susceptibility through interacting with GABA(A) receptors. Proc. Natl. Acad. Sci. USA. 107:11883–11888. 10.1073/pnas.1006529107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D., and Triller A.. 2003. The role of receptor diffusion in the organization of the postsynaptic membrane. Nat. Rev. Neurosci. 4:251–265. 10.1038/nrn1077 [DOI] [PubMed] [Google Scholar]
- Cingolani L.A., and Goda Y.. 2008. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 9:344–356. 10.1038/nrn2373 [DOI] [PubMed] [Google Scholar]
- Colledge M., Snyder E.M., Crozier R.A., Soderling J.A., Jin Y., Langeberg L.K., Lu H., Bear M.F., and Scott J.D.. 2003. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 40:595–607. 10.1016/S0896-6273(03)00687-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawitz J., Kroon T., Hjorth J.J., and Meredith R.M.. 2011. Functional calcium imaging in developing cortical networks. J Vis Exp. 3550:3550 10.3791/3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M.D. 2003. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 6:231–242. 10.1038/nn1013 [DOI] [PubMed] [Google Scholar]
- El-Husseini A.E., and Vincent S.R.. 1999. Cloning and characterization of a novel RING finger protein that interacts with class V myosins. J. Biol. Chem. 274:19771–19777. 10.1074/jbc.274.28.19771 [DOI] [PubMed] [Google Scholar]
- El-Husseini A.E., Kwasnicka D., Yamada T., Hirohashi S., and Vincent S.R.. 2000. BERP, a novel ring finger protein, binds to alpha-actinin-4. Biochem. Biophys. Res. Commun. 267:906–911. 10.1006/bbrc.1999.2045 [DOI] [PubMed] [Google Scholar]
- Elvira G., Wasiak S., Blandford V., Tong X.K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A.W., Boismenu D., et al. 2006. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 5:635–651. 10.1074/mcp.M500255-MCP200 [DOI] [PubMed] [Google Scholar]
- Eom T., Antar L.N., Singer R.H., and Bassell G.J.. 2003. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 23:10433–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R., Nägerl U.V., and Bonhoeffer T.. 2006. Neuronal activity determines the protein synthesis dependence of long-term potentiation. Nat. Neurosci. 9:478–480. 10.1038/nn1667 [DOI] [PubMed] [Google Scholar]
- Griffin N.M., Yu J., Long F., Oh P., Shore S., Li Y., Koziol J.A., and Schnitzer J.E.. 2010. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28:83–89. 10.1038/nbt.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert T.P., Kilhams G.R., Batty I.H., and Proud C.G.. 2000. Distinct signalling pathways mediate insulin and phorbol ester-stimulated eukaryotic initiation factor 4F assembly and protein synthesis in HEK 293 cells. J. Biol. Chem. 275:11249–11256. 10.1074/jbc.275.15.11249 [DOI] [PubMed] [Google Scholar]
- Hung A.Y., Sung C.C., Brito I.L., and Sheng M.. 2010. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One. 5:e9842 10.1371/journal.pone.0009842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafkafi N., Lipkind D., Benjamini Y., Mayo C.L., Elmer G.I., and Golani I.. 2003. SEE locomotor behavior test discriminates C57BL/6J and DBA/2J mouse inbred strains across laboratories and protocol conditions. Behav. Neurosci. 117:464–477. 10.1037/0735-7044.117.3.464 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., and Hirokawa N.. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 43:513–525. 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Karakozova M., Kozak M., Wong C.C., Bailey A.O., Yates J.R. III, Mogilner A., Zebroski H., and Kashina A.. 2006. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 313:192–196. 10.1126/science.1129344 [DOI] [PubMed] [Google Scholar]
- Kelleher R.J. III, Govindarajan A., and Tonegawa S.. 2004. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 44:59–73. 10.1016/j.neuron.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Kim E.C., Lee M.J., Shin S.Y., Seol G.H., Han S.H., Yee J., Kim C., and Min S.S.. 2013. Phorbol 12-myristate 13-acetate enhances long-term potentiation in the hippocampus through activation of protein kinase Cδ and ε. Korean J. Physiol. Pharmacol. 17:51–56. 10.4196/kjpp.2013.17.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemmer P., Smit A.B., and Li K.W.. 2009. Proteomics analysis of immuno-precipitated synaptic protein complexes. J. Proteomics. 72:82–90. 10.1016/j.jprot.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Kudryashova E., Kudryashov D., Kramerova I., and Spencer M.J.. 2005. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 354:413–424. 10.1016/j.jmb.2005.09.068 [DOI] [PubMed] [Google Scholar]
- Kuriu T., Inoue A., Bito H., Sobue K., and Okabe S.. 2006. Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J. Neurosci. 26:7693–7706. 10.1523/JNEUROSCI.0522-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté D., Thies E., Pechmann Y., Groffen A.J., Verhage M., Smit A.B., van Kesteren R.E., and Kneussel M.. 2013. TRIM3 regulates the motility of the kinesin motor protein KIF21B. PLoS One. 8:e75603 10.1371/journal.pone.0075603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mabb A.M., and Ehlers M.D.. 2010. Ubiquitination in postsynaptic function and plasticity. Annu. Rev. Cell Dev. Biol. 26:179–210. 10.1146/annurev-cellbio-100109-104129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb A.M., Je H.S., Wall M.J., Robinson C.G., Larsen R.S., Qiang Y., Corrêa S.A., and Ehlers M.D.. 2014. Triad3A regulates synaptic strength by ubiquitination of Arc. Neuron. 82:1299–1316. 10.1016/j.neuron.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R., and Malenka R.C.. 2002. AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci. 25:103–126. 10.1146/annurev.neuro.25.112701.142758 [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T., Lemberger T., Bleckmann S.C., Kern H., Kretz O., Martin Villalba A., Tronche F., Kellendonk C., Gau D., Kapfhammer J., et al. 2002. Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 31:47–54. 10.1038/ng882 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M., Honkura N., Ellis-Davies G.C., and Kasai H.. 2004. Structural basis of long-term potentiation in single dendritic spines. Nature. 429:761–766. 10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. 2000. Actin-based plasticity in dendritic spines. Science. 290:754–758. 10.1126/science.290.5492.754 [DOI] [PubMed] [Google Scholar]
- Meyer-Arendt K., Old W.M., Houel S., Renganathan K., Eichelberger B., Resing K.A., and Ahn N.G.. 2011. IsoformResolver: A peptide-centric algorithm for protein inference. J. Proteome Res. 10:3060–3075. 10.1021/pr200039p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misane I., Tovote P., Meyer M., Spiess J., Ogren S.O., and Stiedl O.. 2005. Time-dependent involvement of the dorsal hippocampus in trace fear conditioning in mice. Hippocampus. 15:418–426. 10.1002/hipo.20067 [DOI] [PubMed] [Google Scholar]
- Ohashi S., Koike K., Omori A., Ichinose S., Ohara S., Kobayashi S., Sato T.A., and Anzai K.. 2002. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 277:37804–37810. 10.1074/jbc.M203608200 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Nagai T., Miyawaki A., and Hayashi Y.. 2004. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7:1104–1112. 10.1038/nn1311 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Narayanan R., Lee S.H., Murata K., and Hayashi Y.. 2007. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc. Natl. Acad. Sci. USA. 104:6418–6423. 10.1073/pnas.0701656104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil E., Wells D.G., and Mooseker M.S.. 2005. A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J. Cell Biol. 168:329–338. 10.1083/jcb.200410091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin B.J., Sonnemann K.J., and Ervasti J.M.. 2010. β-actin and γ-actin are each dispensable for auditory hair cell development but required for Stereocilia maintenance. PLoS Genet. 6:e1001158 10.1371/journal.pgen.1001158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon M.M., Choi S.H., Jamieson C.A., Geschwind D.H., and Martin K.C.. 2006. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J. Neurosci. 26:13390–13399. 10.1523/JNEUROSCI.3432-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter L., Claassen M., Schrimpf S.P., Jovanovic M., Schmidt A., Buhmann J.M., Hengartner M.O., and Aebersold R.. 2009. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics. 8:2405–2417. 10.1074/mcp.M900317-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151. 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Ehlenberger D.B., Dickstein D.L., Hof P.R., and Wearne S.L.. 2008. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 3:e1997 10.1371/journal.pone.0001997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein P.A. 1990. The functional importance of multiple actin isoforms. BioEssays. 12:309–315. 10.1002/bies.950120702 [DOI] [PubMed] [Google Scholar]
- Schubert V., Da Silva J.S., and Dotti C.G.. 2006. Localized recruitment and activation of RhoA underlies dendritic spine morphology in a glutamate receptor-dependent manner. J. Cell Biol. 172:453–467. 10.1083/jcb.200506136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn J.C., Berezikov E., and Knoblich J.A.. 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 136:913–925. 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz L.A., Hall B.J., and Patrick G.N.. 2010. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J. Neurosci. 30:16718–16729. 10.1523/JNEUROSCI.3686-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekino Y., Kojima N., and Shirao T.. 2007. Role of actin cytoskeleton in dendritic spine morphogenesis. Neurochem. Int. 51:92–104. 10.1016/j.neuint.2007.04.029 [DOI] [PubMed] [Google Scholar]
- Shepherd J.D., and Huganir R.L.. 2007. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23:613–643. 10.1146/annurev.cellbio.23.090506.123516 [DOI] [PubMed] [Google Scholar]
- Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P., Hunter C.L., Nuwaysir L.M., and Schaeffer D.A.. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics. 6:1638–1655. 10.1074/mcp.T600050-MCP200 [DOI] [PubMed] [Google Scholar]
- Shimono K., Baudry M., Ho L., Taketani M., and Lynch G.. 2002. Long-term recording of LTP in cultured hippocampal slices. Neural Plast. 9:249–254. 10.1155/NP.2002.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.M., Zhang N., Hansen J., Gerges N.Z., Pak D.T., Sheng M., and Lee S.H.. 2012. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat. Neurosci. 15:1655–1666. 10.1038/nn.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K.M., and Cox T.C.. 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 281:8970–8980. 10.1074/jbc.M512755200 [DOI] [PubMed] [Google Scholar]
- Star E.N., Kwiatkowski D.J., and Murthy V.N.. 2002. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat. Neurosci. 5:239–246. 10.1038/nn811 [DOI] [PubMed] [Google Scholar]
- Stiedl O., Misane I., Spiess J., and Ogren S.O.. 2000. Involvement of the 5-HT1A receptors in classical fear conditioning in C57BL/6J mice. J. Neurosci. 20:8515–8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H.C., and Schuman E.M.. 2008. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9:826–838. 10.1038/nrn2499 [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli D.M., Oleynikov Y., Kelic S., Shenoy S.M., Hartley A., Stanton P.K., Singer R.H., and Bassell G.J.. 2003. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 23:3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Sun W., Kujala P., Lotfi Y., Vida T.A., and Bean A.J.. 2005. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol. Biol. Cell. 16:2470–2482. 10.1091/mbc.E04-11-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Fujii R., Watanabe Y., Okabe S., Fukui K., and Takumi T.. 2006. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 16:2345–2351. 10.1016/j.cub.2006.10.024 [DOI] [PubMed] [Google Scholar]
- Yuste R., and Bonhoeffer T.. 2004. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 5:24–34. 10.1038/nrn1300 [DOI] [PubMed] [Google Scholar]
- Zhang F., Saha S., Shabalina S.A., and Kashina A.. 2010. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 329:1534–1537. 10.1126/science.1191701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Xiao M., and Nicoll R.A.. 2001. Contribution of cytoskeleton to the internalization of AMPA receptors. Proc. Natl. Acad. Sci. USA. 98:1261–1266. 10.1073/pnas.98.3.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.