BubR1 depends on its association with Bub3 to localize on DNA breaks during mitosis, where it sequesters Cdc20Fizzy and induces the inhibition of the APC/C locally, promoting the faithful segregation of broken chromatids.
Abstract
The presence of DNA double-strand breaks during mitosis is particularly challenging for the cell, as it produces broken chromosomes lacking a centromere. This situation can cause genomic instability resulting from improper segregation of the broken fragments into daughter cells. We recently uncovered a process by which broken chromosomes are faithfully transmitted via the BubR1-dependent tethering of the two broken chromosome ends. However, the mechanisms underlying BubR1 recruitment and function on broken chromosomes were largely unknown. We show that BubR1 requires interaction with Bub3 to localize on the broken chromosome fragments and to mediate their proper segregation. We also find that Cdc20, a cofactor of the E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C), accumulates on DNA breaks in a BubR1 KEN box–dependent manner. A biosensor for APC/C activity demonstrates a BubR1-dependent local inhibition of APC/C around the segregating broken chromosome. We therefore propose that the Bub3–BubR1 complex on broken DNA inhibits the APC/C locally via the sequestration of Cdc20, thus promoting proper transmission of broken chromosomes.
Introduction
Cells are constantly exposed to aggressions that alter their genome during their life cycle. To sustain life, mechanisms have evolved to repair DNA lesions and preserve the genome. The presence of DNA damage, such as double-strand breaks (DSBs), during interphase triggers a conserved DNA damage response (DDR) that promotes DNA repair and delays mitotic entry via activation of the DNA damage checkpoint (Melo and Toczyski, 2002; Jackson and Bartek, 2009). Less is known about how mitotic cells process DSBs. However, given the high frequency of cell division in developing organisms and in adult humans, the occurrence of DSBs in mitosis is likely to be significant. Studies in various model organisms have shown that DSBs in mitosis delay anaphase onset via the activation of the DNA damage and/or spindle assembly checkpoints (SACs) depending on the extent of damage (Melo et al., 2001; Mikhailov et al., 2002; Royou et al., 2005; Choi and Lee, 2008; Kim and Burke, 2008; Dotiwala et al., 2010; Jessulat et al., 2015). Moreover, recent studies in vertebrate mitotic cells found that the presence of DSBs triggers the initiation of the DDR, followed by an inhibition of the downstream pathways that repair the damage, either by nonhomologous end joining (NHEJ) or homology-targeted repair (Mari et al., 2006; Giunta et al., 2010; Gomez-Godinez et al., 2010; Peterson et al., 2011; Orthwein et al., 2014; Silva et al., 2014; Benada et al., 2015). These studies further show that a full DDR is reactivated upon G1 entry (Giunta et al., 2010; Orthwein et al., 2014). Thus, DSBs are detected in mitosis and marked to facilitate their repair after mitotic exit. However, it remains unclear how the cell segregates the two broken chromosome fragments, the centric and acentric fragments resulting from DSB generation. In yeast, chromosome fragments with DSBs remain apposed throughout mitosis, leading to the formation of aneuploid daughter cells (Melo et al., 2001; Kaye et al., 2004). In Drosophila melanogaster, the centric and acentric chromosome fragments generated by the endonuclease I-CreI, which creates DSBs at ribosomal DNA repeats, remain attached via a DNA thread that we refer to as a “tether” (Royou et al., 2010). These tethers facilitate the faithful segregation of sister chromatids, thus preventing aneuploidy. The kinases BubR1, Polo, and Aurora B accumulate on these tethers throughout mitosis. Attenuation of BubR1 or Polo function provoke severe defects in broken chromatid segregation and induce synthetic lethality in combination with I-CreI expression. BubR1 and Polo may facilitate the proper segregation of broken chromatids by maintaining the integrity of the tether, providing an opportunity to repair the damage in the next cell cycle.
BubR1 localizes to unattached kinetochores and plays a conserved role in the SAC that delays anaphase onset until all chromosomes are properly attached to the spindle microtubules (Karess et al., 2013). Anaphase is triggered by the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase that targets Securin and Cyclin B for proteolysis, thereby allowing sister chromatid separation resulting from separase-mediated cleavage of the cohesin complex and mitotic exit via dephosphorylation of Cyclin B/CDK1 substrates. In the presence of unattached kinetochores, BubR1, Bub3, and Mad2 form the mitotic checkpoint complex that sequesters Cdc20, a key subunit of the APC/C, thereby inhibiting its activity globally (Foley and Kapoor, 2013). BubR1 also plays a role in promoting the stabilization of kinetochore–microtubule attachments (Rahmani et al., 2009; Elowe et al., 2010; Karess et al., 2013). We have recently identified a novel localization and function for BubR1 on DSBs in mitosis (Royou et al., 2010). However, the mechanisms that promote BubR1 localization on broken chromatids and its molecular function in facilitating their proper segregation have yet to be elucidated.
Here, we show that BubR1 requires its association with Bub3 to localize on laser-induced DNA breaks and I-CreI–induced tethers in mitosis, and that both proteins are required for the efficient segregation of broken chromatids. In addition, the Bub3–BubR1 complex sequesters Cdc20 on DSBs throughout mitosis via the BubR1 KEN box, thereby inducing a local inhibition of the APC/C at the site of DSBs until late anaphase. A BubR1 KEN box mutant fails to sequester Cdc20 and to locally inhibit the APC/C on DNA breaks. As a result, these mutant cells exhibit severe defects in broken chromatid segregation. Reciprocally, a Cdc20 mutant that perturbs its interaction with the BubR1 KEN box fails to localize on DNA breaks and compromises the faithful segregation of broken chromatids.
Results
The Bub3-binding domain of BubR1 is necessary and sufficient for BubR1 localization on the kinetochore and tether
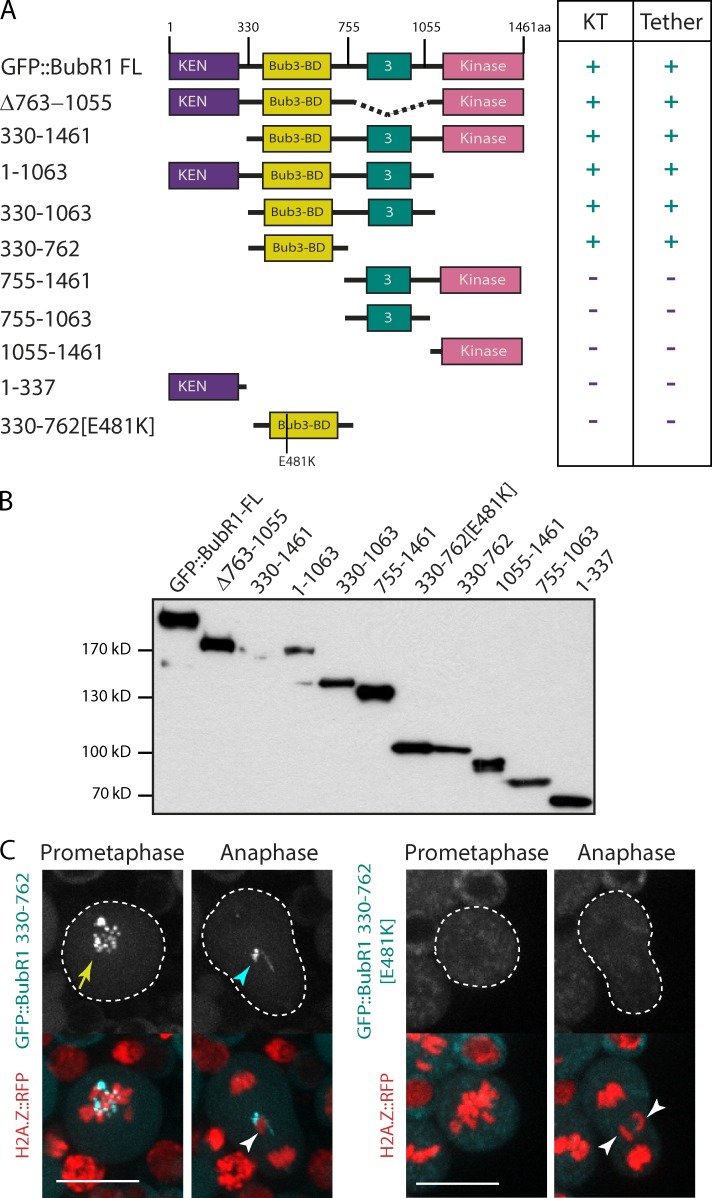
Drosophila BubR1 contains four large domains separated by unstructured regions. The N-terminal domain is well conserved and contains a KEN box (lysine–glutamate–asparagine) and tetratrico peptide repeats required for BubR1 interaction with Cdc20 and its spindle checkpoint activity (Burton and Solomon, 2007; King et al., 2007; Sczaniecka et al., 2008; Rahmani et al., 2009). The second domain, called the Bub3-binding domain (Bub3-BD or GLEBS domain), is conserved and promotes BubR1 recruitment to the kinetochore via its interaction with Bub3 in yeast and vertebrates (Wang et al., 2001; Chen, 2002; Larsen et al., 2007). This domain also contains a kinetochore-associated regulatory domain (KARD) identified in vertebrate BubR1, whose phosphorylation promotes BubR1 association with the PP2A-B56α phosphatase subunit at the kinetochore (Suijkerbuijk et al., 2012; Kruse et al., 2013). Using a secondary structure prediction algorithm (globprot; Linding et al., 2003), we identified a third structured domain with poor conservation (named 3). Finally, the C terminus contains the kinase domain, which is known to play a role in stabilizing kinetochore–microtubule attachments (Rahmani et al., 2009).
To determine which domain of BubR1 is required for its localization on the tether, the DNA thread that links the centric and acentric chromosome fragments, we cloned a series of truncated BubR1 constructs tagged at their N terminus with GFP under the control of a ubiquitous promoter. We cloned 11 BubR1 constructs, shown schematically in Fig. 1 A. The expression of the GFP::BubR1 fusion proteins in adult flies was confirmed by Western blotting (Fig. 1 B). The full-length fusion rescued the lethality of the bubR11 null mutant.
Figure 1.
The Bub3-BD of BubR1 is necessary and sufficient for BubR1 localization on the tether. (A) Scheme of BubR1 full-length (FL) domains identified with a secondary structural prediction algorithm (globprot), and the BubR1 truncated versions along with their localization (+) or not (−) on the kinetochore (KT) or tether. The numbers correspond to the position of the first and last amino acid of the BubR1 truncation construct. The 330–762 [E481K] construct contains a substitution of E481 by K. The BubR1 constructs are fused with GFP on their N terminus. (B) Western blot of the different GFP::BubR1 constructs from transgenic adult flies. (C) Images of live neuroblasts expressing I-CreI and labeled with H2A.Z::RFP and the indicated GFP::BubR1 constructs (also see Video 1 and Video 2). The kinetochore and tether localization of the GFP::BubR1 constructs are indicated with a yellow arrow and a cyan arrowhead. The white arrowheads point to the I-CreI–induced acentric chromatids. The cells are delineated with white dotted lines. Bars, 10 µm.
Neuroblasts from female larvae expressing the truncated GFP::BubR1 constructs and H2A.Z::RFP, a marker of chromosomes, were imaged by time-lapse microscopy after I-CreI induction. I-CreI creates DSBs specifically at one location in the X chromosomes, generating two distinct fragments, a short fragment containing the centromere and a long acentric fragment (Rong et al., 2002; Royou et al., 2010). As expected, full-length GFP::BubR1 signal accumulated on kinetochores at prometaphase and faded during anaphase (Fig. S1). Moreover, as previously observed with a different GFP::BubR1 transgene and with anti-BubR1 antibodies, the fluorescent signal remained on the tether of the lagging I-CreI–induced broken chromatids throughout anaphase (Fig. S1; Royou et al., 2010). The localization of the truncated BubR1 fusions on the kinetochore and tether are summarized in Fig. 1 A. We found that amino acids 330–762, containing the Bub3-BD, were necessary and sufficient for BubR1 localization on both the kinetochore and the tether (Fig. 1, A and C; Fig. S1; and Video 1).
Glutamate 481 in the Bub3-BD is required for BubR1 recruitment on the kinetochore and the tether
Glutamate (E) 406 in the Bub3-BD of murine BubR1 has been identified as a key residue for BubR1 interaction with Bub3 (Harris et al., 2005). E481 in the BubR1 Drosophila sequence is predicted by Clustal alignment to be the equivalent of E406. To determine whether the BubR1 interaction with Bub3 is required for BubR1 localization on the tether, we mutated E481 to lysine (K) in the Bub3-BD construct (330–762 [E481K]) and monitored its dynamics during mitosis (Fig. 1 A). We found that 330–762 [E481K] is expressed in the adult fly (Fig. 1 B), but localized neither on the kinetochore nor on the tether in mitotic cells (Fig. 1 C and Video 2). These data confirmed that E481 is a key conserved residue essential for BubR1 localization on the kinetochore and support the idea that Bub3 mediates BubR1 localization on the kinetochore and tether.
The complex Bub3–BubR1 localizes on I-CreI– and laser-induced DNA breaks
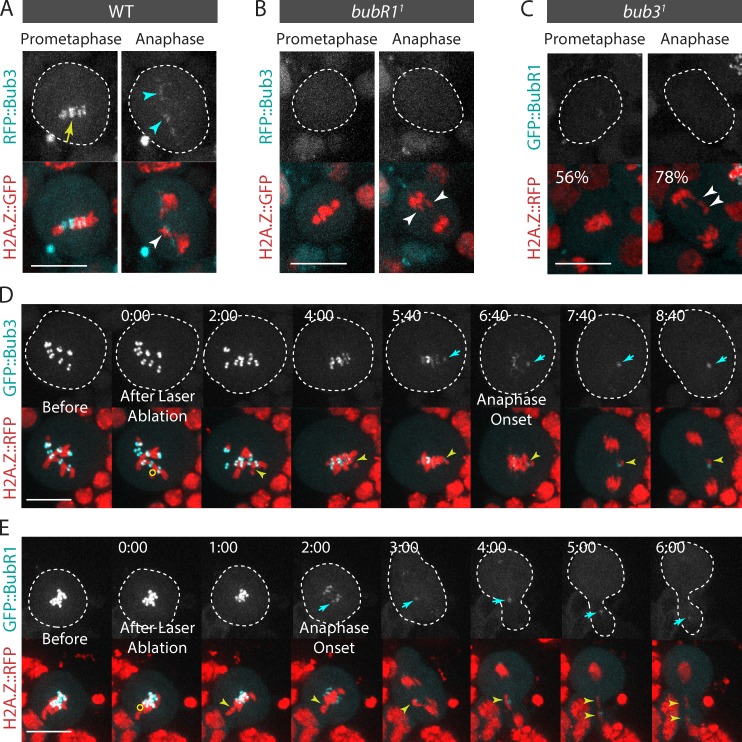
Bub3 interacts with BubR1 at the kinetochore (Taylor et al., 1998; Larsen et al., 2007). To assess the role of Bub3 in mitotic cells with broken chromosomes, we investigated the dynamics of Bub3 in dividing neuroblasts after I-CreI expression. We generated transgenic flies expressing RFP::Bub3 under the control of the Bub3 promoter. The fusion protein rescued the lethality associated with the bub31 mutant allele, consistent with it being functional. We found that RFP::Bub3 localized on the kinetochore during prometaphase and disappeared from the kinetochore progressively during anaphase, in agreement with previous findings using anti-Bub3 antibodies (Fig. 2 A; Basu et al., 1998). However, in I-CreI–expressing cells, RFP::Bub3 signal lingered on the lagging broken chromatids throughout anaphase. Like BubR1, the signal disappeared at telophase (Fig. 2 A and Video 3). Similar results were obtained using a GFP::Bub3 construct (unpublished data).
Figure 2.
Bub3 and BubR1 localize on I-CreI– and laser-induced DNA breaks. (A) Bub3 localizes on the I-CreI–induced tether (also see Video 3). Time-lapse images of WT neuroblasts expressing H2A.Z::GFP and RFP::Bub3 after I-CreI induction. The yellow arrow and cyan arrowheads indicate the localization of RFP::Bub3 on kinetochores and tethers. (B) Bub3 does not localize to the kinetochore or the tether in bubR11 mutants. Time-lapse images of bubR11 mutant neuroblasts expressing H2A.Z::GFP and RFP::Bub3 after I-CreI induction. (C) BubR1 does not localize to the kinetochore or the tether in 56% and 78% of bub31 mutant cells (n = 9). Time-lapse images of bub31 mutant neuroblasts expressing H2A.Z::RFP and GFP::BubR1 after I-CreI induction. The white arrowheads point to the I-CreI–induced acentric chromatids. (D and E) Bub3 (D) and BubR1 (E) localize on laser-induced DNA damage during mitosis. Time-lapse images of neuroblasts expressing H2A.Z::RFP and GFP::Bub3, or GFP::BubR1, before and after laser ablation (also see Video 4). The yellow circles correspond to the zones of laser ablation. The cyan arrows point to the accumulation of Bub3 or BubR1 at the site of the chromosome damage. The yellow arrowheads indicate the laser-induced damage. Time (given in minutes/seconds) 0:00 corresponds to the first acquisition immediately after laser ablation. The cells are delineated with dotted lines. Bars, 10 µm.
To verify that Bub3 and BubR1 recruitment to DSBs is a result of DNA damage, and not specific to I-CreI–induced damage on ribosomal DNA repeats, we examined the dynamics of GFP::Bub3 and GFP::BubR1 after surgical damage of a single chromosome with a 355-nm pulsed laser. Bub3 and BubR1 signal appeared on DNA damage 2–5 min after laser ablation (Fig. 2, D and E; and Video 4).
Next, we tested the dependency relationship between BubR1 and Bub3 for their localization on DNA breaks. First, we analyzed the localization of RFP::Bub3 in bubR11 null mutant neuroblasts after I-CreI expression. RFP::Bub3 signal was not detected on the kinetochore in any of the bubR1 mutant cells monitored, consistent with immunostaining data using anti-Bub3 antibodies (Basu et al., 1998). In addition, we found no accumulation of RFP::Bub3 near the broken chromatids during anaphase, indicating that Bub3 requires BubR1 to accumulate on the tether (Fig. 2 B). Next, we monitored the dynamics of GFP::BubR1 in a bub31 mutant. The bub31 allele carries a point mutation in the coding region, resulting in lethality at the pupal stage (Lopes et al., 2005). In 46% of bub31 mutant cells, the Bub3 mutant protein failed to localize on the kinetochore (Lopes et al., 2005). Indeed, GFP::BubR1 did not localize on the kinetochore in half of bub31 mutant cells (Fig. 2 C). In addition, GFP::BubR1 failed to localize on the tether in the majority of bub31 mutant cells (Fig. 2 C). Collectively, these results indicate that coassociation of BubR1 and Bub3 is required for their robust localization on the kinetochore and tether.
DNA breaks do not induce neokinetochore formation
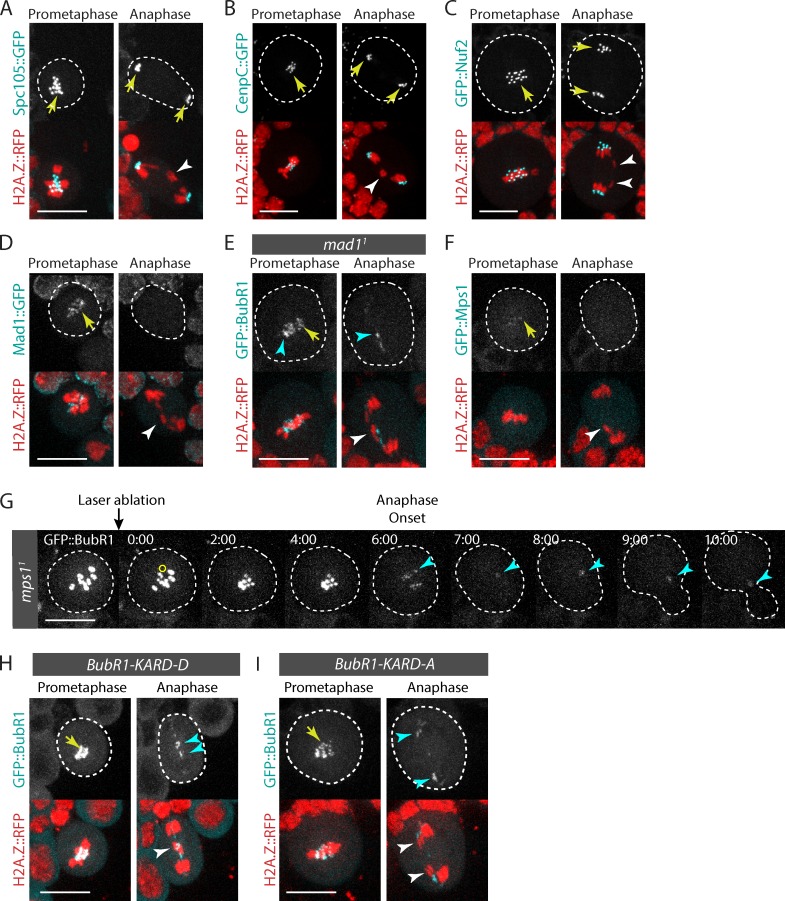
It was recently shown that human KNL1 (also known as Spc105, Spc7, and Blinkin), a component of the core kinetochore KNL1–Mis12–Ndc80 (KMN) network, interacts directly with BubR1 and controls the localization of BubR1 and Bub3 on the kinetochore (Kiyomitsu et al., 2007, 2011; D’Arcy et al., 2010; Bolanos-Garcia et al., 2011). Although Drosophila Spc105 is dispensable for BubR1 recruitment to the kinetochore (Schittenhelm et al., 2009), we tested the idea that it might promote BubR1 and Bub3 association with the tether. Thus, we monitored the dynamics of Spc105::GFP in mitotic cells with broken chromosomes. Spc105::GFP localized to the kinetochore until late telophase (Fig. 3 A; Schittenhelm et al., 2009). However, no signal was detected near the lagging broken chromatids, suggesting that Spc105 is not part of the upstream pathway that recruits BubR1 and Bub3 to the tether (Fig. 3 A).
Figure 3.
DNA breaks do not induce neokinetochore formation. (A–D and F) Time-lapse images of neuroblasts expressing H2A.Z::RFP and Spc105::GFP (A), CenpC::GFP (B), GFP::Nuf2 (C), Mad1::GFP (D), and GFP::Mps1 (F) after I-CreI expression. The white arrowheads point to the I-CreI–induced acentric chromatids. Yellow arrows indicate the localization of the GFP-labeled proteins at the kinetochore. (E) Mad1 is not required for BubR1 localization on broken chromatids. Time-lapse images of a mad11 mutant neuroblast expressing I-CreI and labeled with H2A.Z::RFP and GFP::BubR1. The white arrowhead indicates I-CreI–induced acentric chromatids. The yellow arrow and cyan arrowheads indicate the localization of GFP::BubR1 at the kinetochore and tether. (G) Mps1 is not required for BubR1 localization on broken chromatids. Time-lapse images of mps11 mutant neuroblast labeled with GFP::BubR1 before and after laser ablation. The yellow circle corresponds to the zone of laser ablation. The cyan arrowheads indicate the appearance of GFP::BubR1 at the site of chromosome damage. Time (given in minutes/seconds) 0:00 corresponds to the first acquisition immediately after laser ablation. (H and I) BubR1 localization on DNA breaks does not require Polo-dependent phosphorylation of the BubR1 KARD motif. Time-lapse images of neuroblasts expressing I-CreI labeled with H2A.Z::RFP. The neuroblasts express either the GFP::BubR1-KARD-D mutant, where the putative Polo-dependent phosphorylation sites in the KARD motif are replaced by aspartate (H), or GFP::BubR1-KARD-A, where the same residues are mutated to alanine (I). The white arrowheads indicate the I-CreI–induced acentric chromatids. Yellow arrows and cyan arrowheads point to the localization of GFP::BubR1-KARD-D and GFP::BubR1-KARD-A on the kinetochore and tether. Cells are delineated with white dotted lines. Bars, 10 µm.
Next, we assessed the dynamics of GFP::Nuf2, a component of the KMN network, and CenpC::GFP in mitotic cells with DSBs. We found that they exclusively localized to the kinetochore throughout mitosis (Fig. 3, B and C; Schittenhelm et al., 2007). These results indicate that the tether does not serve as a platform for the assembly of a neokinetochore. Consistent with this idea, Cid, the H3 histone variant that specifically associates with centromeric DNA and is essential for kinetochore assembly and function, does not localize on the tether (Royou et al., 2010).
Next, we tested the localization of other proteins known to be involved in the Bub3–BubR1-dependent SAC response. We first monitored the localization of Mad1::GFP on broken chromosomes. Mad1 has a conserved SAC function on the kinetochore. It recruits Mad2, which exists in two conformations: open (O-Mad2) or closed (C-Mad2). The complex Mad1–C-Mad2 promotes O-Mad2 to C-Mad2 conversion. C-Mad2 then binds Cdc20, BubR1, and Bub3, forming the mitotic checkpoint complex, a diffusible APC/C inhibitory complex (Foley and Kapoor, 2013). We found that Mad1::GFP accumulated on the kinetochore during prometaphase and disappeared from the kinetochore at anaphase onset (Fig. 3 D; Emre et al., 2011). In agreement with our previous findings with GFP::Mad2 (Royou et al., 2010), no Mad1::GFP signal was detected near the lagging broken chromatids during anaphase (Fig. 3 D). Consistently, BubR1 localization on the tether did not depend on Mad1 (Fig. 3 E). These data, and the fact that mad2 mutants are not sensitive to I-CreI expression, collectively confirm our model that the Mad1–Mad2 pathway does not play a role in the proper segregation of broken chromatids (Royou et al., 2010).
Next, we analyzed the dynamics of Mps1 on broken chromosomes. Mps1 functions in the SAC response by controlling the dynamics of Bub3, BubR1, and Cdc20 at the kinetochore (Shepperd et al., 2012; Conde et al., 2013; Espert et al., 2014). We found that the GFP::Mps1 signal was weak, but nevertheless visible on the kinetochore; however, no signal was detected near the lagging broken chromatids during anaphase (Fig. 3 F). In addition, GFP::BubR1 was recruited on laser-induced breaks in the mps11 mutant. Collectively, these results suggest that Mps1 does not mediate BubR1 recruitment to broken chromatids (Fig. 3 G).
We have previously reported that Polo localizes to the tether, where it promotes the proper segregation of broken chromatids (Royou et al., 2010). Recently, studies in mammalian cells show that Polo-like kinase 1 phosphorylates BubR1 on its KARD motif, which promotes BubR1 association with the phosphatase subunit PP2A-B56α at the kinetochore during prometaphase/metaphase (Suijkerbuijk et al., 2012; Kruse et al., 2013; Espert et al., 2014). These observations prompted us to examine the involvement of Polo-mediated phosphorylation of BubR1’s KARD motif in BubR1 recruitment to the tether during mitosis. To test this idea, we replaced the putative Polo-dependent phosphorylation sites, S685, S691, and T695, in the Drosophila BubR1 KARD motif with either alanine (GFP::BubR1-KARD-A) or aspartate (GFP::BubR1-KARD-D) to prevent or mimic the phosphorylation, respectively. The expression of either GFP::BubR1-KARD-D or GFP::BubR1-KARD-A did not rescue the bubR11 null mutant and induced an increase in the mitotic index resulting from an extended prometaphase (unpublished data). This phenotype, similar to the one observed in vertebrates (Kruse et al., 2013), suggests a conserved function for the Drosophila BubR1 KARD motif in promoting kinetochore–microtubule attachments. Both GFP::BubR1-KARD-D and GFP::BubR1-KARD-A constructs remained associated with the tether, suggesting that the polo-mediated phosphorylation of BubR1’s KARD motif does not control BubR1 dynamics on DNA breaks in mitosis (Fig. 3, H and I). These results, however, do not rule out the possibility that Polo directly or indirectly promotes Bub3–BubR1 localization on DSBs.
The DNA damage checkpoint kinases Chk1 and ATM do not mediate BubR1 recruitment to DNA breaks
Drosophila BubR1 localizes on uncapped telomeres, and this recruitment is attenuated in DNA damage checkpoint–compromised cells, including atmtefu and chk1grp mutants (Musarò et al., 2008). Uncapped telomeres resemble DSBs, leading us to posit that a similar pathway could control BubR1 recruitment to both uncapped telomeres and DNA breaks. To test this hypothesis, we monitored the localization of BubR1 on I-CreI–induced breaks in chk1grp and atmtefu mutants. BubR1 signal was visible on the tether in mitosis in both mutants, consistent with previous observations (Fig S2; Royou et al., 2010). These results indicate that attenuation of Chk1 and ATM kinase activity does not prevent BubR1 localization on DSBs.
Fizzy is recruited to the tether in a BubR1 KEN box–dependent manner
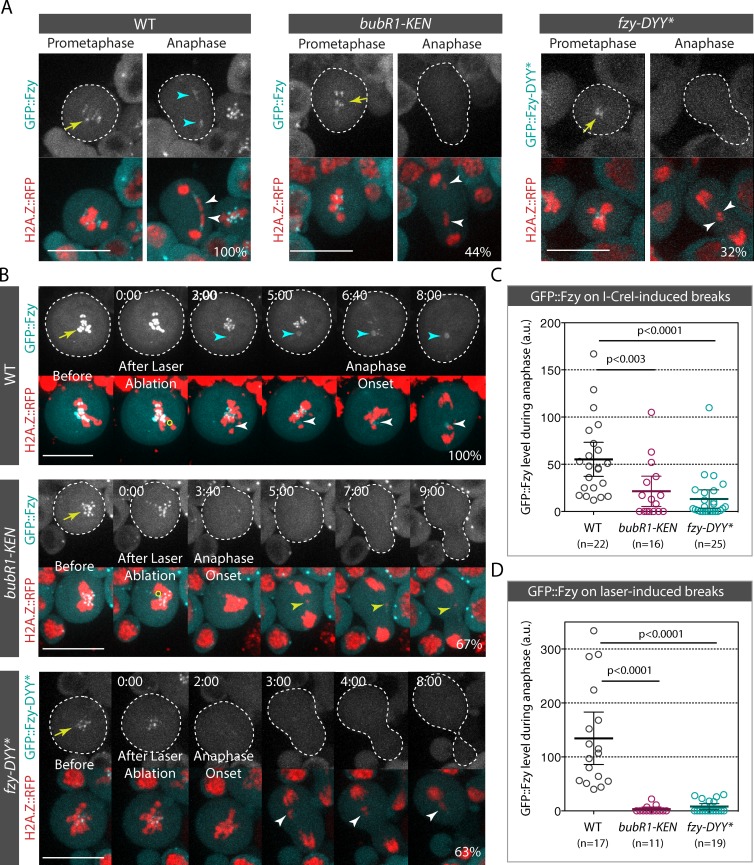
We have previously found that the expression of BubR1 carrying two mutations in its KEN box (bubR1-KEN), which abolishes BubR1 SAC activity (Rahmani et al., 2009), impairs faithful broken chromatid transmission (Royou et al., 2010). Given that the KEN box is required for BubR1 association with Cdc20 (called Fizzy in Drosophila; Rahmani et al., 2009), we reasoned that BubR1 may sequester Fizzy (Fzy) on the tether during mitosis. To test this hypothesis, we first examined the dynamics of GFP::Fzy in live cells carrying broken chromosomes induced by I-CreI expression or laser ablation. GFP::Fzy accumulated on the kinetochore during prometaphase and disappeared progressively from the kinetochore during anaphase (Fig. 4, A and B; Raff et al., 2002; Schittenhelm et al., 2007, 2009). In addition, GFP::Fzy accumulated on I-CreI– and laser-induced breaks from prometaphase to telophase (Fig. 4, A and B; Video 5; and Video 7).
Figure 4.
Fzy is recruited on I-CreI– or laser-induced breaks in a BubR1 KEN box–dependent manner. (A) Time-lapse images of WT (also see Video 5), bubR1-KEN (also see Video 6), and fzy-DYY* neuroblasts expressing I-CreI. WT and bubR1-KEN mutant cells express GFP::Fzy, and fzy-DYY* mutant cells express GFP::Fzy-DYY*. The yellow arrows and cyan arrowheads indicate the localization of GFP::Fzy and GFP::Fzy-DYY* on the kinetochore and tether. The white arrowheads indicate the I-CreI–induced acentric chromatids. 100% of WT cells exhibited GFP::Fzy signal on the tether (n = 22). In contrast, no GFP::Fzy signal on the tether was detected in 44% of bubR1-KEN mutant cells (n = 16). GFP::Fzy-DYY* signal was not visible in 32% of cells (n = 25). (B) Time-lapse images of WT (also see Video 7), bubR1-KEN (also see Video 8), and fzy-DYY* mutant cells before and after laser ablation. The yellow arrows indicate the accumulation of GFP::Fzy and GFP::Fzy-DYY* on kinetochores. The yellow circles correspond to the zones of laser ablation. Time (given in minutes/seconds) 0:00 corresponds to the first acquisition immediately after laser ablation. The white arrowheads indicate laser-induced damage. The cyan arrowheads show the appearance of GFP::Fzy at the site of chromosome damage. For WT cells, the percentage represents the frequency of cells with GFP::Fzy signal on DNA breaks (n = 17). In the bubR1-KEN mutant, the percentage denotes the frequency of cells without GFP::Fzy signal on laser-induced damage (n = 11). Finally, for the fzy-DYY* mutant, the percentage indicates the frequency of cells with no detectable GFP::Fzy-DYY* signal on laser-induced breaks (n = 19). The cells are delineated with dotted lines. Bars, 10 µm. (C) Scatter dot plot of GFP::Fzy and GFP::Fzy-DYY* levels at the site of the I-CreI–induced tether during anaphase in WT, bubR1-KEN, and fzy-DYY* mutants. (D) Scatter dot plot of GFP::Fzy and GFP-Fzy-DYY* levels at the site of the laser-induced breaks during anaphase. (C and D) n = number of cells. The GFP level is normalized to the cytoplasmic GFP signal (see Materials and methods section Image analysis for quantification details). A Mann-Whitney nonparametric test was used to calculate p-values. Error bars represent mean ± 95% confidence interval. a.u., arbitrary units.
We could not detect either Cdh1 (called Fzy-related [Fzr] in Drosophila), the other cofactor of the APC/C, or the APC/C core complex subunits Cdc16 and Cdc27 on DNA breaks during mitosis, suggesting that the pool of Fzy recruited on DNA breaks is not associated with the APC/C core complex (Fig. S3).
To assess whether Fzy recruitment on DNA breaks was dependent on the BubR1 KEN box, we monitored GFP::Fzy dynamics in bubR1-KEN mutants after I-CreI induction or laser ablation. A great proportion of bubR1-KEN mutant cells failed to localize GFP::Fzy on I-CreI–induced tethers and laser-induced breaks (Fig. 4, A–D; Video 6; and Video 8). This result suggests that Fzy recruitment to DNA breaks is mediated via its interaction with the BubR1 KEN box.
To test this idea, we examined the localization of a Fzy mutant that has a reduced affinity for the BubR1 KEN box. The interaction of human Cdc20 with the first KEN box of BubR1 involves residues D184, Y185, and Y186 (Tian et al., 2012). These residues are conserved in Drosophila Fzy and correspond to D210, Y211, and Y212 (Fig. S4 A). We constructed a Fzy mutant in which the DYY residues are mutated to alanine (Fzy-DYY*). The in vitro binding efficiency of Fzy-DYY* to GST-BubR1[1–337], the BubR1 KEN domain including the KEN box, decreased by 50% (Fig. S4 B). This reduced binding efficiency of Fzy-DYY* to the BubR1 KEN domain was associated with a dramatic decrease in GFP::Fzy-DYY* levels on I-CreI– and laser-induced DNA breaks in vivo (Fig. 4, A–D). Collectively, these results are consistent with a model in which Fzy is sequestered on DNA breaks throughout mitosis via an interaction with the BubR1 KEN box.
Impairment of Fzy recruitment to DNA breaks is associated with severe defects in broken chromatid segregation
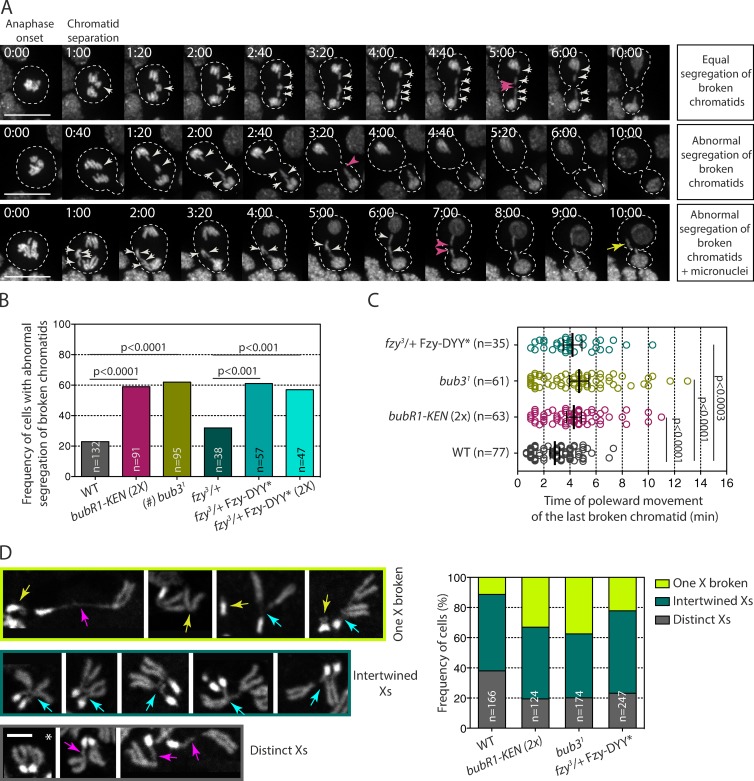
Next, we assessed the physiological consequences on acentric chromatid segregation when Fzy is no longer recruited to DNA breaks. We monitored the frequency of abnormal segregation of acentric fragments, as well as the rate at which the last broken chromatid moved poleward in wild-type (WT), bub31, BubR1-KEN, and fzy-DYY* mutant cells after I-CreI expression. In agreement with our previous observations, the majority of WT cell divisions produced daughter cells with proper genome content as a result of the equal segregation of acentric chromatids (Fig. 5 A, top; Royou et al., 2010). Occasionally, the broken chromatids partitioned abnormally, producing two daughter cells with incorrect genome content (Fig. 5 A, middle and bottom; and Fig. 5 B). The majority of the broken fragments segregated within 4 min after anaphase onset (Fig. 5 C).
Figure 5.
bubR1-KEN, bub31, and fzy-DYY* mutants exhibit severe defects in broken chromatid segregation. (A) Time-lapse images of neuroblasts expressing I-CreI and labeled with H2A.Z::RFP. The top row shows an example of a dividing cell with equal partition of the broken X chromatids (white arrows), which produces euploid daughter cells. The middle and bottom rows show examples of cells with abnormal segregation of broken chromatids, where three X broken chromatids segregate in one daughter cell (white arrows). These divisions will produce two aneusomic daughter cells with, in some cases, visible micronuclei (yellow arrow, bottom). The pink arrowheads indicate the time at which the last acentric fragments move poleward. Time is given in minutes/seconds. Bars, 10 µm. (B) Histogram showing the frequency of neuroblast divisions with abnormal segregation of broken chromatids after I-CreI expression. n = number of cells. A Fisher extract test was used to calculate the p-value. (#) Given that bub31 mutant cells exhibit a high frequency of anaphase with whole lagging chromosomes (Basu et al., 1999; Logarinho et al., 2004; Lopes et al., 2005), cells in which no clear lagging broken chromatids could be followed because of extensive chromosome segregation defects were excluded from our analysis. (C) Scatter dot plot showing the time at which the last acentric chromatid starts moving poleward. The black lines correspond to mean ± 95% confidence interval. Time 0:00 corresponds to anaphase onset. n = number of cells. A Mann-Whitney nonparametric test was used for calculating p-values. (D) DAPI staining of X chromosomes from fixed neuroblasts after I-CreI expression. The bottom row illustrates cells exhibiting two distinct X chromosomes with no apparent tether, indicated by an asterisk, or apparent tether, indicated by pink arrows. The middle row represents intertwined X homologues (cyan arrows), and the top row shows an example of cells with one X broken fragment (yellow arrows). The histogram shows the frequency of cells with either two distinct Xs with or without tethers, intertwined X homologues, or at least one X broken. n = number of cells. Bar, 2 µm. bubR1-KEN(2X) corresponds to bubR11 null cells expressing two doses of the RFP::BubR1-KEN transgene; fzy3/+ corresponds to fzy3 heterozygote mutants cells; and fzy3/+ Fzy-DYY* and fzy3/+ Fzy-DYY* (2X) correspond to the fzy3 heterozygote carrying one or two doses of the GFP::Fzy-DYY* transgene.
Given that Bub3 promotes BubR1 recruitment to DNA breaks and that BubR1 is required for the proper segregation of broken fragments, we expected to observe a high frequency of abnormal acentric segregation in mutant cells that have a reduced level of Bub3. Consistently, the frequency of abnormal segregation of broken chromatids was dramatically increased in bub31 mutants (Fig. 5 B). Moreover, a great proportion of chromatids exhibited slow or no poleward movement in bub31 mutants (Fig. 5 C).
We have previously shown that the expression of one copy of the bubR1-KEN mutant transgene in the bubR11 null background impaired broken chromatid segregation. Although the expression of one copy of the bubR1-KEN transgene was sufficient to rescue the lethality of the bubR11 null allele, it may not have been sufficient to support the segregation of broken chromatids. Alternatively, the severe broken chromatid segregation defects in bubR1-KEN mutants may reflect a specific role for the KEN motif in the mechanism that facilitates the proper poleward movement of the broken fragments. To distinguish between these possibilities, we analyzed the segregation of broken chromatids in bubR1-KEN(2X) mutants in which the transgene is expressed from two copies. We found that the frequency of cells that fail to properly segregate the broken fragments increased significantly in the bubR1-KEN(2X) mutant compared with the WT (60 vs. 23%; Fig. 5 B). This frequency is similar to that observed for one copy of bubR1-KEN (Royou et al., 2010). In addition, a large number of bubR1-KEN(2X) mutant cells exhibited slow or no poleward movement of broken fragments (Fig. 5 C). These results indicate that the KEN box plays a specific role in facilitating the faithful segregation of broken chromatids.
The observation that loss of the BubR1 KEN box impairs Fzy recruitment to DNA breaks prompted us to test whether the high frequency of broken chromatid missegregation in the bubR1-KEN mutant was a direct consequence of a lack of Fzy sequestration on DNA breaks. To do so, we monitored the segregation of broken chromatids in cells expressing the Fzy-DYY* mutant that no longer associates with DNA breaks (Fig. 4). Fzy-DYY* mutants expressed from one or two copies (2X) in a fzy heterozygote mutant exhibited similar frequencies of cells with missegregated acentric fragments as bubR1-KEN mutants. These defects were more severe than the defects observed in fzy heterozygote mutants, reflecting a dominant effect of Fzy-DYY* expression (Fig. 5, B and C). Collectively, these results indicate that the BubR1 KEN box–mediated recruitment of Fzy to DNA breaks plays an important role in the proper transmission of damaged chromosomes.
To gain insight into the possible causes of the elevated acentric missegregation in bub31, bubR1-KEN, and fzy-DYY* mutants, we examined the morphology of the X chromosome after I-CreI expression in fixed prometaphase neuroblasts. As previously reported, three chromosome X configurations were observed in all genotypes, including the WT (Fig. 5 D; Royou et al., 2010). In the first configuration, both X chromosomes were distinct from each other and had normal morphologies, or exhibited loosely condensed chromatin threads that maintained the centric and acentric chromosome fragments together (Fig. 5 D, bottom, pink arrows). This configuration was found in almost 40% of WT cells but was reduced to 20% in bub31, bubR1-KEN, and fzy-DYY* mutants (Fig. 5 D). In the second configuration, at least one X chromosome was broken into two distinct fragments (Fig. 5 D, top, yellow arrows). In agreement with our previous observation, we found that the number of WT cells exhibiting X fragments without a tether was minor (11%; n = 166). In contrast, the frequency of cells with clear broken X fragments was double in the fzy-DYY* mutant and triple in bub31 and bubR1-KEN mutants. These cells are likely to produce aneuploid cells. However, the fact that the frequency of cells displaying a broken chromosome X in prometaphase is lower than the frequency of cells with missegregated acentrics in both the WT and mutants implies that the presence of broken fragments in prometaphase is not the sole cause of acentric missegregation. The last and most prevalent configuration in all genotypes was intertwined X homologues (Fig. 5 D, middle, cyan arrows). Because I-CreI predominantly induces DNA damage during interphase in our assay, this X configuration may be the result of unresolved DNA repair intermediates produced during the preceding interphase. These entanglements may be problematic during the segregation of sister chromatids. The tension applied to the X homologues with entangled tethers during anaphase may result in tether rupture, inducing missegregation of the fragments. Tether breakage may occur more frequently in bub31, bubR1-KEN, and fzy-DYY* mutants than WT cells. All together, these results favor a model in which the structure of the tether is more fragile or unstable when Fzy recruitment to DNA breaks is reduced.
BubR1 induces local APC/C inhibition on the tether during early anaphase
Our results indicate that Fzy is sequestered on breaks/tethers throughout mitosis, largely via its association with the BubR1 KEN motif. Abrogation of this interaction impairs the proper segregation of broken chromatids. This raises the possibility that the BubR1-mediated sequestration of Fzy locally inhibits the APC/C on the tether, and this inhibition is required to maintain the integrity of the tether by preventing the degradation of key APC/C substrates even during anaphase. To test this idea, we monitored the localization of Securin and Cyclin B, two established APC/C substrates. We tagged Securin with GFP on its N or C terminus and analyzed its dynamics in mitotic cells with broken chromosomes. GFP::Securin and Securin::GFP were uniformly cytoplasmic during prometaphase (Fig. S5 A and not depicted). The signal disappeared gradually and globally during anaphase, likely because of its degradation by the proteasome. As reported previously, GFP::Cyclin B localized on the kinetochore, spindle, and centrosome during prometaphase and disappeared progressively during anaphase (Fig. S5 A; Huang and Raff, 1999). Both GFP::Cyclin B and GFP::Securin did not localize specifically on the tether during anaphase. In addition, no difference in the rate of GFP::Cyclin B signal disappearance was observed between cells with or without I-CreI expression (Fig. S5 B). It therefore seems unlikely that Securin and Cyclin B play a direct role in regulating the faithful segregation of broken chromosomes.
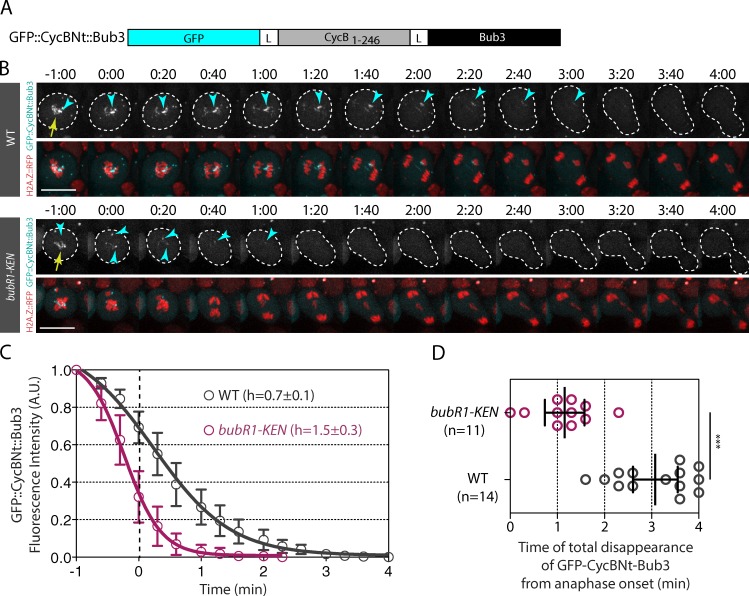
To determine whether BubR1 localization on the tether induces local APC/C inhibition, we created a synthetic APC/C substrate that associated with the tether. We fused the first 246 amino acids of Cyclin B, which contains the DEAD box, the recognition motif for Fzy, and the lysines that are ubiquitinated by the APC/C within full-length Bub3. GFP was fused to the N terminus of the probe to monitor fluorescence disappearance over time (Fig. 6 A). We then measured the signal of the synthetic probe, called GFP::CycBNt::Bub3, on the kinetochore and tether over time in WT and bubR1-KEN mutants after I-CreI expression (Fig. 6, B–D; Fig. S5 D; Video 9; and Video 10). We controlled that the GFP::CycBNt::Bub3 signal disappeared rapidly from the kinetochore in cells without broken chromosomes (WT no heat shock [HS]); Fig. S5, C and D). If tether-associated BubR1 inhibits the APC/C locally via the sequestration of Fzy, we would expect to detect a bubR1-KEN box–dependent delay of GFP::CycBNt::Bub3 degradation on the tether during anaphase. Consistently, in WT cells, GFP::CycBNt::Bub3 disappeared from the tether at a twofold slower rate than in bubR1-KEN mutant cells (h = 0.7 ± 0.1 vs. h = 1.5 ± 0.3; Fig. 6 C). The signal disappeared completely from the tether on average 3 min after anaphase onset in control cells, compared with 1 min in bubR1-KEN mutant cells (Fig. 6 D). These results are consistent with our proposal that tether-associated BubR1 is sufficient to protect APC/C substrates locally from degradation.
Figure 6.
The APC/C synthetic substrate GFP::CycBNt::Bub3 is maintained on the tether during early anaphase in a BubR1 KEN box–dependent manner. (A) Scheme of the APC/C synthetic substrate GFP::CycBNt::Bub3. The N terminus (including the amino acids 1–246) of Cyclin B (CycB1–246) was fused on its N terminus to GFP and on its C terminus to full-length Bub3. The CycBNt sequence was flanked with a 4× glycine–alanine linker (L). (B) Time-lapse images of WT and bubR1-KEN mutant cells after I-CreI induction labeled with H2A.Z::RFP and GFP::CycBNt::Bub3. The kinetochore and tether localization of GFP::CycNt::Bub3 are indicated with yellow arrows and cyan arrowheads, respectively. Bar, 10 µm. (C) Quantitative analysis of the disappearance of the GFP::CycBNt::Bub3 signal on kinetochores and tethers. The graph shows the fluorescence intensity of GFP signal on kinetochores and tethers over time (see Fig. S5 for raw data and Materials and methods section Image analysis for details on the quantification). The fluorescence intensities were normalized to the fluorescence intensity measured at the time point −1 min. The 0 of the x axis corresponds to anaphase onset as defined by the onset of sister chromatid separation. The signal was measured every 20 s. (D) Scatter dot plot showing the time of complete disappearance of the signal of GFP::CycBNt::Bub3 in WT and bubR1-KEN mutant cells expressing I-CreI. The time starts at anaphase onset. The black lines correspond to mean ± 95% confidence interval. A Mann-Whitney nonparametric test was used to calculate p-values (***, P < 0.001). A.U., arbitrary units.
Discussion
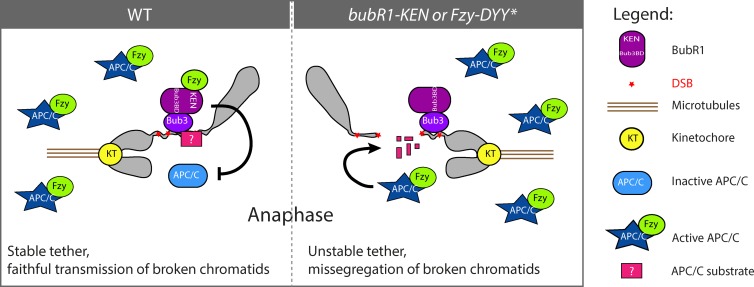
In this work, we showed that BubR1 depends on its association with Bub3 to localize on I-CreI– and laser-induced DNA breaks during mitosis, where it sequesters Fzy and promotes the inhibition of the APC/C locally. Failure to sequester Fzy on DNA breaks by altering Fzy’s interaction with the BubR1 KEN box induces broken chromatid missegregation. We propose that Bub3–BubR1-mediated APC/C inhibition on DNA breaks preserves a key component required to tether the broken fragments, thereby ensuring their proper transmission (Fig. 7).
Figure 7.
Model for Bub3–BubR1 function on the tether/DNA breaks. The BubR1–Bub3 complex sequesters Fzy on broken chromosomes from prometaphase to late anaphase, preventing activation of the APC/C locally and transiently. This inhibition may prevent the APC/C-dependent degradation of a key factor (pink square with question mark) required to maintain the two broken fragments tethered and to facilitate their proper segregation. In bubR1-KEN or fzy-DYY* mutants, the Fzy-mediated activation of the APC/C around the broken fragments allows the premature degradation of the unknown factor, which impairs the tethering of the broken fragments and their correct segregation.
The upstream component that recruits the Bub3–BubR1 complex to DNA breaks has yet to be identified. In yeast and mammals, the interaction of BubR1 and Bub3 with KNL1 plays an important role in their recruitment to the kinetochore (Kiyomitsu et al., 2007, 2011; D’Arcy et al., 2010; Bolanos-Garcia et al., 2011). However, a recent structural study on human BubR1 revealed that mutations in the tetratrico peptide repeats motif that prevent its association with KNL1 do not affect its kinetochore localization (Krenn et al., 2012). Moreover, accumulation of BubR1 to kinetochores in Drosophila spc105 mutant embryos is only mildly reduced (Schittenhelm et al., 2009). These data suggest that an alternative pathway recruits Bub3–BubR1 to the kinetochore. Therefore, it is possible that a similar pathway regulates Bub3–BubR1 localization on DNA breaks. The observation that the dynamics of the Bub3–BubR1 complex on the kinetochore and tether/DSBs are dramatically different does not favor this idea. Indeed, although Bub3 and BubR1 dissociate from kinetochores attached to microtubules in metaphase, they remain abundant on broken chromatids well into anaphase. Similarly, both proteins accumulate rapidly on laser-induced breaks in all stages of mitosis, including anaphase. This suggests that different signaling events control the dynamics of Bub3–BubR1 on the kinetochore and DNA breaks. Alternatively, a similar pathway may recruit and dissociate Bub3–BubR1 at the kinetochore and broken chromosomes, but the signals controlling the dynamics of both proteins on DNA breaks may be more robust and resistant to the global activation of APC/C during anaphase. Although no evidence for BubR1 or Bub3 affinity for naked DNA or uncondensed chromatin has been reported thus far, it is possible that the Bub3–BubR1 complex binds directly to chromatin or DNA at the site of DSBs. The I-CreI– and laser-mediated DNA damage may induce the unwinding of the chromatin at the site of the breaks, thus exposing nucleosomes and DNA to the Bub3–BubR1 complex.
Our work demonstrates that the Bub3–BubR1 complex recruits Fzy to the tether/DSBs. Previous work on SAC activity indicated that Mad2 association with Cdc20 is a prerequisite for efficient Bub3–BubR1 binding to Cdc20 and subsequent inhibition of APC/C (Sudakin et al., 2001; Kulukian et al., 2009). In contrast, we found that the Bub3–BubR1 complex efficiently recruits Fzy to DSBs without Mad2 and induces local APC/C inhibition well into anaphase, a period during which the APC/C is globally active. We propose that Bub3–BubR1–Fzy-mediated local APC/C inhibition around the DSBs protects key components required to tether the two broken fragments, thereby assuring their proper transmission. Our findings, that the failure to recruit Fzy to DSBs led to the premature activation of APC/C around the breaks associated with an elevated frequency of broken chromatid missegregation, are consistent with this model.
The exact nature of the DNA damage that triggers the recruitment of Bub3–BubR1–Fzy complexes is not defined. We also do not know whether I-CreI– and laser-induced damages create different types of DNA structures. However, the findings that DNA damages created during mitosis by either genotoxic agents, endonucleases, x ray, or laser irradiation consistently trigger the accumulation of components involved in DDR signaling support the idea that these insults share common structural characteristics that promote the recruitment of both DDR and Bub3–BubR1–Fzy complexes (Melo et al., 2001; Kaye et al., 2004; Mari et al., 2006; Giunta et al., 2010; Gomez-Godinez et al., 2010; Royou et al., 2010; Peterson et al., 2011; Orthwein et al., 2014; Silva et al., 2014; Benada et al., 2015). It is therefore possible that the APC/C substrates protected by Bub3–BubR1–Fzy may be components of the DDR pathway.
Our findings that tether-associated Bub3–BubR1 inhibits the APC/C well into anaphase support an alternative possibility: that Bub3–BubR1 protects an APC/C substrate that acts to resolve altered DNA structures during anaphase. We found that X homologues are intertwined in the majority of WT and mutant cells after endonuclease expression. These entanglements may produce catenated DNA strands that require resolution during anaphase. Impairing the resolution of these entanglements would result in slow chromatid poleward movements and elevated frequencies of chromatid missegregation resulting from the rupture of one of the strands. Bub3–BubR1-mediated local inhibition of APC/C activity may facilitate the resolution of these entanglements. Consistently, we found that these entanglements had little effect on acentric chromosome segregation in WT cells but were associated with an increased frequency of slow and improper segregation of acentric fragments in bub3, bubR1-KEN, and fzy-DYY* mutants. Studies on catenated DNA structures, called ultra-fine DNA bridges, have greatly improved our understanding of catenated DNA resolution in mitosis and provide clues as to which putative APC/C substrate may be protected by Bub3–BubR1 during anaphase (Liu et al., 2014). The resolution of ultrafine DNA bridges, which originate from unreplicated DNA at fragile loci or catenated centromeric and telomeric DNA, involves the activities of the topoisomerases TopoII and TopoIII, the translocase PICH, the helicase BLM, and the DNA repair component Rif1 (Baumann et al., 2007; Chan et al., 2007, 2009; Barefield and Karlseder, 2012; Biebricher et al., 2013; Hengeveld et al., 2015). Such enzymes may be required to resolve entanglements from damaged chromosomes during anaphase and may be protected from global APC/C activity by Bub3–BubR1 complexes.
Future studies aimed at elucidating the composition of the chromatin structures associated with I-CreI– and laser-induced DNA damage will provide clues to the underlying mechanisms by which Bub3–BubR1 allows the proper segregation of broken chromatids.
Materials and methods
Fly strains
Flies were grown on corn flour and yeast extract medium in standard conditions, at 25°C. The h2a.z>H2A.Z::RFP (previously named H2Av::RFP; Talbert et al., 2012) stock was obtained from the Bloomington Drosophila Stock Center at Indiana University. h2a.z>H2A.Z::GFP was described previously (Clarkson and Saint, 1999). The bub3>EGFP::Bub3 stock was provided by C. Lehner (Institute of Molecular Life Sciences, University of Zurich, Zurich, Switzerland). The hsp70>I-CreI strain has been described previously (Rong et al., 2002). The bubR11 and bub31 mutant alleles were described previously (Basu et al., 1999; Lopes et al., 2005). The bubR1>mRFP::bubR1-KEN mutant has also been previously reported (Rahmani et al., 2009). cenpC>CenpC::EGFP, nuf2>EGFP::Nuf2, spc105>Spc105::EGFP, mad1>Mad1::EGFP, and mps1>EGFP::Mps1 stocks were previously characterized and were provided by C. Lehner and R. Karess (Institut Jacques Monod, Centre National de la Recherche Scientifique, Paris, France; Schittenhelm et al., 2007, 2009; Emre et al., 2011; Althoff et al., 2012). mps11 and mad11 mutants have been characterized previously (Fischer et al., 2004; Emre et al., 2011). The chk1grp-1 allele was described previously (Fogarty et al., 1997). The atmtefu-3 and atmtefu-8 alleles have also been previously described (Silva et al., 2004). ubi>GFP::Fzy, ubi>GFP::Cdc27, ubi>GFP::Cdc16, and ubi>GFP::Fzr were described previously and were provided by J. Raff (Sir William Dunn School of Pathology, University of Oxford, Oxford, England, UK; Raff et al., 2002; Huang and Raff, 2002). GFP::Cyclin B has also been described previously (Huang and Raff, 1999). All the other transgenic stocks used in this work were produced in the laboratory: ubi>GFP::BubR1[], ubi>mRFP::Bub3, ubi>GFP::BubR1-KARD-A, ubi>GFP::BubR1-KARD-D, ubi>GFP::Fzy-DYY*, ubi>GFP::Securin, ubi>securin::GFP, and ubi>GFP::CycBNt::Bub3. The genotypes of the larvae used for each figure are listed in Table S1.
Plasmid cloning
All plasmids were verified by sequencing before being injected into fly embryos to generate transgenic stocks (Bestgene Inc.).
BubR1 truncated plasmid constructs
All constructs were derived from a plasmid containing full-length BubR1 cDNA and cloned using Gateway technology. PCR products using the primers listed in Table S2 were inserted into the pDONR/Zeo vector (Life Technologies) to create entry clones. We generated the construct [Δ763–1,055] by a two-step PCR. The first PCR used four primers (F1, Δ3NTR, Δ3CTF, and R1). Using these products as matrices, a second PCR was performed using F1 and R1 primers. Entry vectors containing the truncated BubR1 sequence were recombined into the pUbi-GFP-GAT vector containing the ubiquitin promoter and an N-terminal GFP tag (a gift from J. Raff). The entry clones containing the BubR1 [1–337] sequence and the destination vector pDEST15 (Invitrogen) were used to generate a GST::BubR1 [1–337] plasmid using standard Gateway recombination.
mRFP::Bub3 cloning
The RFP::Bub3 construct was obtained by multistep cloning. First, 900 nt upstream of the Bub3 initiation codon was amplified from genomic DNA using the Bub3prom-F and Bub3prom-R oligonucleotides, introducing KpnI and XhoI restriction sites, respectively (Table S2). After digestion, the product was ligated into pBluescript KS (pBS KS; Agilent Technologies). The genomic Bub3 sequence with its 3′ untranslated region was amplified with Bub3gen-F and Bub3gen-R primers, introducing SmaI and XbaI sites, and cloned into the pBS KS Bub3 promoter plasmid (Table S2). mRFP was amplified from pCaSpeR4 mRFP::BubR1 (a gift from R. Karess) using mRFP-F and mRFP-R primers, introducing XhoI and SmaI sites and a linker coding for GAGAGAGA after the mRFP sequence. Ligation of this product resulted in the final pBS KS bub3>mRFP::Bub3 vector. This construct was then transferred into the pCaSpeR4 vector using KpnI and XbaI restriction sites.
GFP::CycBNt::Bub3 cloning
The CycBNt sequence was amplified from a pDONR/Zeo-CycBNt vector obtained from GenScript using CycBNt-Gat-F (which contains the AttB1 Gateway sequence) and Bub3-Lnk-CycBNt-R primers (which adds a GAGAGAGA linker to the C terminus of CycBNt; Table S2). The Bub3 genomic sequence containing the GAGAGAGA linker on its N terminus was amplified from the pBS KS g>mRFP-Bub3 vector with Lnk-Bub3-F and Bub3-R primers (with an AttB2 Gateway sequence; Table S2). The complete sequence was amplified from the two PCR products with CycBNt-Gat-F and Bub3-R primers. The construct was recombined with the pUbi-GFP-GAT destination vector to obtain the final plasmid ubi>-GFP::CycBNt::Bub3 using standard Gateway recombination.
GFP::Fzy-DYY* cloning
A pUC57 vector containing the Fzy cDNA sequence in which the D210, Y211, and Y212 are changed to alanine (fzy-DYY*) and flanked with AttB sequences was obtained from GenScript. This construct was then used to create an entry clone subsequently recombined with the pUbi-GFP-GAT destination vector to obtain the final plasmid ubi>GFP::Fzy-DYY*.
BubR1-KARD-A and BubR1-KARD-D cloning
The BubR1-KARD-A construct was generated by a two-step PCR. In the first step, the 5′ and 3′ parts of the BubR1 coding DNA sequence were amplified with the F1/KARD-A(R) and KARD-A(F)/R1 primer pairs, respectively (Table S2). In a second step, the resulting PCR products were used in an overlapping PCR with the flanking F1/R1 primers to generate the whole mutated coding DNA sequence, which was then inserted into the pDONR-Zeo plasmid. The construct was recombined with the pUbi-GFP-GAT destination vector to obtain the final plasmid ubi>GFP::BubR1-KARD-A. The same strategy was used to obtain the ubi>GFP::BubR1-KARD-D plasmid, with the KARD-D(F) and KARD-D(R) primers (Table S2).
GFP::Securin and Securin::GFP cloning
For GFP::Securin fusion Gateway cloning, Drosophila Securin cDNA was amplified from pSecurin vector (LD16810; Drosophila Genomics Resource Center) using Securin-Nt-F and Securin-Nt-R primers for GFP N-terminal tagging and Securin-Ct-F and Securin-Ct-R for GFP C-terminal tagging (Table S2) and then inserted into pDONR/Zeo plasmid. Each plasmid was recombined with either the pUbi-GFP-GAT or pUbi-GAT-GFP destination vector to obtain ubi>GFP::Securin and ubi>Securin::GFP constructs.
Generation of α-GFP antibody
6xHis-tagged GFP was expressed in BL21-CodonPlus cells by IPTG induction at 37°C for 4 h. The fusion protein was subsequently purified by immobilized metal affinity chromatography using Ni-bound Sepharose. Approximately 1 mg of purified protein was used for rabbit immunization. Additional GFP protein was covalently coupled to Affi-gel 10 resin (Bio-Rad) and used for affinity purification of anti-GFP polyclonal antibodies from the sera.
Western blots
Adult flies were ground in 2× sample buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, and 125-mM Tris-Cl, pH 6.8) containing protease inhibitor cocktail (Roche) and 1-mM PMSF (Thermo Fisher Scientific). The extracts were boiled for 5 min, centrifuged, and analyzed by 10% SDS-PAGE. After electrophoresis at 20 mA in running buffer (380-mM glycine, 50-mM Tris base, and 1% wt/vol SDS), proteins were transferred to nitrocellulose membrane (Protran) by Western blotting at 500 mA for 2 h at 4°C in transfer buffer (150-mM glycine, 25-mM Tris base, and 70% ethanol in distilled water). Membrane was blocked in TBS with Tween (TBST; 50-mM Tris-Cl, pH 7.5, 150-mM NaCl, and 0.1% vol/vol Tween 20) containing 5% dried nonfat milk for 1 h. The membrane was incubated overnight at 4°C with mouse anti–α-tubulin (1:750; DM1A; Sigma-Aldrich) and rabbit anti-GFP (1:1,500) primary antibodies diluted in TBST containing 5% dried nonfat milk. After washing in TBST and incubation with appropriate secondary antibodies (peroxidase-conjugated antibodies [DAKO] and fluorescently coupled antibodies [Molecular Probes]) diluted at 1:5,000 in TBST containing 5% dried nonfat milk, the membrane was developed with ECL (Thermo Fisher Scientific) or with Storm equipment with the appropriate lasers (Storm Trio).
GST::BubR1 [1–337] protein purification
Bacteria BL21-AI carrying the expression vector containing the GST::BubR1 [1–337] sequence was grown at 37°C to an optical density of 2.5, and expression was induced overnight at 18°C by the addition of 0.3-mM IPTG and 0.2% l-arabinose. Cells were harvested by centrifugation and frozen in liquid nitrogen. The frozen pellet was ground with dry ice in an electric grinder until a fine powder was obtained. The powder was transferred to a beaker cooled with liquid nitrogen, allowed to warm briefly at room temperature, and resuspended in five volume (wt/vol) lysis buffer (PBS containing 1.0-M NaCl, 0.5% Tween 20, and 1-mM PMSF) at 4°C. After sonication, the lysate was centrifuged at 25,000 rpm for 1 h. 10-mM DTT was then added to the supernatant. This solution was loaded onto a glutathione-beads column previously equilibrated with a five volume column of lysis buffer with 10-mM DTT. The column was washed with PBS containing 0.25-M KCl, 0.1% Tween 20, and 0.5-mM DTT and eluted with a buffer containing 50-mM Tris, pH 8.1, 0.25-mM KCL, and 5-mM reduced glutathione. Peak fractions were pooled and dialyzed in 50-mM Hepes, pH 7.6, 0.25-M KCL, and 30% glycerol. The purified GST-BubR1 [1–337] was stored at −80°C. This protocol is a slightly modified version of the protocol described previously (Carroll et al., 1998).
GFP::Fzy pull-down
24-h collections of embryos expressing either GFP, GFP::Fzy, or GFP::Fzy-DYY* were dechorionated in 2% bleach and stored at −80°C in PBS and 1-mM PMSF. ∼500 mg of embryos were mechanically ground in 200 µl lysis buffer (50-mM Tris-Cl, pH 7.5, 0.1% Tergitol NP-40, 150-mM NaCl, 10-mM EDTA, 3-mM MgCl2, 1-mM PMSF, 1-mM sodium orthovanadate, and containing one tablet of protease inhibitor cocktail [Roche]). Lysates were centrifuged at 17,000 g for 20 min at 4°C, and clarified supernatants were transferred into clean tubes. The total protein concentration of each lysate was evaluated by the Bradford assay. The amount of GFP, GFP::Fzy, and GFP::Fzy-DYY* in each lysate was estimated by Western blotting using mouse anti-GFP antibodies (1:500; Roche). The lysates were subsequently diluted in lysis buffer to obtain an equal concentration of GFP, GFP::Fzy, and GFP::Fzy DYY*. A fraction of each lysate was boiled in one volume of 2× Laemmli sample buffer for 5 min. Normalized extract was added to 50 µg GST::BubR1 [1–337] immobilized on glutathione agarose beads (1 µg/1 µl of beads) in a final volume of 200 µl. The lysates were incubated with the beads for 4 h at 4°C and subsequently centrifuged at 500 g for 2 min at 4°C. The beads were washed four times in lysis buffer without Tergitol NP-40 and finally resuspended in 25 µl of 2× Laemmli before boiling for 5 min. Inputs and beads were analyzed by 12.5% SDS-PAGE. Western blots were probed using mouse anti-GFP antibodies (Roche) and anti-GST antibodies (Jose et al., 2015).
DAPI staining
The third instar larval central nervous system was dissected in PBS. Each brain was treated with 1% sodium citrate for 5 min and then transferred to a 10-µl drop of fixative (45% acetic acid and 2% formaldehyde) on a coverslip and fixed for 5 min. The samples were squashed between a slide and a coverslip and frozen in liquid nitrogen. The slides were rinsed in PBS, dried, and mounted with 10 µl of slow fade with DAPI (Invitrogen).
Live imaging and microscopy
DSBs at the ribosomal DNA locus, located on the X and Y chromosomes, were generated using I-CreI endonuclease under the control of a heat shock 70 promoter. Female third instar larvae were heat shocked for 1 h at 37°C in a water bath to induce I-CreI expression. After at least 1.5 h of recovery at room temperature, the larval central nervous system was dissected in fresh PBS and transferred to a 12-µl drop of PBS on a coverslip. The preparation was slightly squashed between a slide and coverslip by aspiration of the liquid with a piece of paper as described previously (Buffin et al., 2005). The coverslip was sealed with halocarbon oil 700 (Sigma-Aldrich) and observed for 30–40 min under a microscope.
Live imaging was performed at room temperature with either a microscope (Axio-Observer.Z1; Carl Zeiss) and a 100× 1.4 Plan Apochromat oil objective equipped with 491-nm (100 mW; Cobolt Calypso) and 561-nm (100 mW; Cobolt Jive) lasers, a spinning disk (CSUX-A1; Yokogawa), and an electron-multiplying charge-coupled device (emCCD) camera (Evolve; Roper Scientific; all figure panels except Fig. 5 D), or a confocal microscope (SP8; Leica) with a 63× 1.4 Plan Apochromat oil objective equipped with 488-nm and 561-nm lasers and photomultipliers (Hybrid; PicoQuant; Fig. 5 D). The imaging systems were driven by MetaMorph (Molecular Devices) or Leica software.
A 355-nm pulsed laser (passively Q-switched SNV-20F-000) with a 21-kHz repetition rate, 0.8-µJ energy/pulse, 2 kW of peak power, and 400-ps pulse width, powered with an iLas PULSE system (Roper Scientific) and adapted to a microscope (Axio-Observer.Z1; Carl Zeiss), was used at 20% power for one pulse (100 ms) to induce chromosome breaks.
Image analysis
Images in all panels are maximum projections (12 z steps of 0.5-µm depth each). ImageJ (Fiji; National Institutes of Health) was used for image quantification. In Fig. 4 C, the integrated fluorescence intensity of GFP::Fzy on I-CreI–induced DNA breaks was measured on a single Z plane for 12 time points (starting at anaphase onset) and averaged. In Fig. 4 D, the integrated fluorescence intensity of GFP::Fzy on laser-induced DNA breaks was measured on a single Z plane for one time point during anaphase. Only pixels with intensity above the threshold, defined as the maximum pixel intensity in the cytoplasm, were included in the quantification. The GFP::Fzy levels on the tether and laser-induced breaks were normalized to the average intensity in the cytoplasm. In Fig. 6 and Fig. S5, the integrated fluorescence intensity of GFP::CycBNt::Bub3 was measured from a sum projection (5.5 µm) at the kinetochore and tether (signal above the maximum pixel intensity in the cytoplasm) and normalized to the fluorescence intensity at the kinetochore and tether at metaphase (1 min before anaphase onset). Data points were fit to a sigmoid curve (Y = 1/{1 + 10^[(LogEC-X)*Hill slope]}) using Prism software (GraphPad). The Hill slope (h) was used to compare the kinetics of GFP::CycBNt::Bub3 signal disappearance between the WT without I-CreI expression (WT no HS; Fig. S5, C and D), the WT after I-CreI expression (WT), and bubR1-KEN mutant cells after I-CreI expression (bubR1-KEN). In Fig. S5 B, the average fluorescence intensity of GFP::Cyclin B signal in the whole cell was measured over time from a sum projection (5.5 µm). The signal was normalized to the average fluorescence intensity at anaphase onset (time = 0).
Online supplemental material
Fig. S1 demonstrates that the Bub3-BD of BubR1 is required for its localization on both the kinetochore and tether. Fig. S2 shows that BubR1 is recruited on DNA breaks in chk1 and atm mutants. Fig. S3 shows that the APC/C subunits Cdc16 and Cdc27 and the cofactor Fzr do not localize on DNA breaks. Fig. S4 shows the reduced affinity of Fzy-DYY* for BubR1 N-terminal sequence. Fig. S5 shows the dynamics of GFP::Securin and GFP::Cyclin B during anaphase after I-CreI expression. It also shows the dynamics of GFP::CycBNt::Bub3 in cells without I-CreI expression. Video 1 shows the localization of GFP::BubR1 330–762 construct on kinetochore and the I-CreI–induced tether. Video 2 shows the lack of accumulation of GFP::BubR1 330–762 [E481K] on both kinetochore and tether. Video 3 shows the localization of GFP::Bub3 on I-CreI–induced tether. Video 4 shows the localization of GFP::BubR1 on laser-induced DNA breaks. Video 5 shows the localization of GFP::Fzy on the I-CreI–induced tether. Video 6 shows the reduced level of GFP::Fzy on I-CreI–induced DNA breaks in the bubR1-KEN mutant. Video 7 shows the appearance of GFP::Fzy on laser-induced DNA breaks. Video 8 shows the lack of GFP::Fzy accumulation on laser-induced DNA breaks in the bubR1-KEN mutant. Video 9 shows the disappearance of synthetic APC/C substrate GFP::CycBNt::Bub3 signal during anaphase. The signal remains on I-CreI–induced DNA breaks during early anaphase. Video 10 shows the disappearance of synthetic APC/C substrate GFP::CycBNt::Bub3 signal during anaphase in the bubR1-KEN mutant. The signal on DNA breaks disappears rapidly. Table S1 lists the genotypes of the larvae used for each figure. Table S2 lists oligonucleotide sequences. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504059/DC1.
Supplementary Material
Acknowledgments
We thank Zohra Rahmani, Roger Karess, Christian Lehner, Claudio Sunkel, and Jordan Raff for sharing reagents. We thank all laboratory members for technical help and fruitful discussion.
N. Derive was supported by the University of Bordeaux. C. Landmann and E. Montembault were supported by grant ANR-12-PDOC-0020-01/ARC2-ChromSCeD. E. Montembault was also supported by Centre National de la Recherche Scientifique (CNRS). M.-C. Claverie was supported by the University of Bordeaux. N. Founounou was supported by grant ATIP-AVENIR/INCa 2010-291 and Conseil Régional d’Aquitaine (grant 20111301010). D. Goutte-Gattat and P. Pierre-Elies were supported by grant ERC-STG-2012 GA311358-NoAneuploidy. D. McCusker was supported by grant ANR-13-BSV2-0015-01-POLARFLUX and CNRS. A. Royou was supported by grant ERC-STG-2012 GA311358-NoAneuploidy, Conseil Régional d’Aquitaine (grant 20111301010), and CNRS.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- DDR
- DNA damage response
- DSB
- double-strand break
- emCCD
- electron-multiplying charge-coupled device
- HS
- heat shock
- KARD
- kinetochore-associated regulatory domain
- NHEJ
- nonhomologous end joining
- SAC
- spindle assembly checkpoint
- WT
- wild type
References
- Althoff F., Karess R.E., and Lehner C.F.. 2012. Spindle checkpoint-independent inhibition of mitotic chromosome segregation by Drosophila Mps1. Mol. Biol. Cell. 23:2275–2291. 10.1091/mbc.E12-02-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefield C., and Karlseder J.. 2012. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 40:7358–7367. 10.1093/nar/gks407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Logarinho E., Herrmann S., Bousbaa H., Li Z., Chan G.K., Yen T.J., Sunkel C.E., and Goldberg M.L.. 1998. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma. 107:376–385. 10.1007/s004120050321 [DOI] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B.C., Lopes C., Sunkel C.E., and Goldberg M.L.. 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146:13–28. 10.1083/jcb.146.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C., Körner R., Hofmann K., and Nigg E.A.. 2007. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 128:101–114. 10.1016/j.cell.2006.11.041 [DOI] [PubMed] [Google Scholar]
- Benada J., Burdová K., Lidak T., von Morgen P., and Macurek L.. 2015. Polo-like kinase 1 inhibits DNA damage response during mitosis. Cell Cycle. 14:219–231. 10.4161/15384101.2014.977067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebricher A., Hirano S., Enzlin J.H., Wiechens N., Streicher W.W., Huttner D., Wang L.H., Nigg E.A., Owen-Hughes T., Liu Y., et al. 2013. PICH: A DNA translocase specially adapted for processing anaphase bridge DNA. Mol. Cell. 51:691–701. 10.1016/j.molcel.2013.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Lischetti T., Matak-Vinković D., Cota E., Simpson P.J., Chirgadze D.Y., Spring D.R., Robinson C.V., Nilsson J., and Blundell T.L.. 2011. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure. 19:1691–1700. 10.1016/j.str.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., and Karess R.E.. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861. 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Burton J.L., and Solomon M.J.. 2007. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 21:655–667. 10.1101/gad.1511107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Altman R., Schieltz D., Yates J.R., and Kellogg D.. 1998. The septins are required for the mitosis-specific activation of the Gin4 kinase. J. Cell Biol. 143:709–717. 10.1083/jcb.143.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., North P.S., and Hickson I.D.. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26:3397–3409. 10.1038/sj.emboj.7601777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Palmai-Pallag T., Ying S., and Hickson I.D.. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11:753–760. 10.1038/ncb1882 [DOI] [PubMed] [Google Scholar]
- Chen R.H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158:487–496. 10.1083/jcb.200204048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E., and Lee H.. 2008. Chromosome damage in mitosis induces BubR1 activation and prometaphase arrest. FEBS Lett. 582:1700–1706. 10.1016/j.febslet.2008.04.028 [DOI] [PubMed] [Google Scholar]
- Clarkson M., and Saint R.. 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18:457–462. 10.1089/104454999315178 [DOI] [PubMed] [Google Scholar]
- Conde C., Osswald M., Barbosa J., Moutinho-Santos T., Pinheiro D., Guimarães S., Matos I., Maiato H., and Sunkel C.E.. 2013. Drosophila Polo regulates the spindle assembly checkpoint through Mps1-dependent BubR1 phosphorylation. EMBO J. 32:1761–1777. 10.1038/emboj.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcy S., Davies O.R., Blundell T.L., and Bolanos-Garcia V.M.. 2010. Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J. Biol. Chem. 285:14764–14776. 10.1074/jbc.M109.082016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotiwala F., Harrison J.C., Jain S., Sugawara N., and Haber J.E.. 2010. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr. Biol. 20:328–332. 10.1016/j.cub.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Dulla K., Uldschmid A., Li X., Dou Z., and Nigg E.A.. 2010. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J. Cell Sci. 123:84–94. 10.1242/jcs.056507 [DOI] [PubMed] [Google Scholar]
- Emre D., Terracol R., Poncet A., Rahmani Z., and Karess R.E.. 2011. A mitotic role for Mad1 beyond the spindle checkpoint. J. Cell Sci. 124:1664–1671. 10.1242/jcs.081216 [DOI] [PubMed] [Google Scholar]
- Espert A., Uluocak P., Bastos R.N., Mangat D., Graab P., and Gruneberg U.. 2014. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J. Cell Biol. 206:833–842. 10.1083/jcb.201406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M.G., Heeger S., Häcker U., and Lehner C.F.. 2004. The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1. Curr. Biol. 14:2019–2024. 10.1016/j.cub.2004.11.008 [DOI] [PubMed] [Google Scholar]
- Fogarty P., Campbell S.D., Abu-Shumays R., Phalle B.S., Yu K.R., Uy G.L., Goldberg M.L., and Sullivan W.. 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7:418–426. 10.1016/S0960-9822(06)00189-8 [DOI] [PubMed] [Google Scholar]
- Foley E.A., and Kapoor T.M.. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14:25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S., Belotserkovskaya R., and Jackson S.P.. 2010. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190:197–207. 10.1083/jcb.200911156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Godinez V., Wu T., Sherman A.J., Lee C.S., Liaw L.H., Zhongsheng Y., Yokomori K., and Berns M.W.. 2010. Analysis of DNA double-strand break response and chromatin structure in mitosis using laser microirradiation. Nucleic Acids Res. 38:e202 10.1093/nar/gkq836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L., Davenport J., Neale G., and Goorha R.. 2005. The mitotic checkpoint gene BubR1 has two distinct functions in mitosis. Exp. Cell Res. 308:85–100. 10.1016/j.yexcr.2005.03.036 [DOI] [PubMed] [Google Scholar]
- Hengeveld R.C., de Boer H.R., Schoonen P.M., de Vries E.G., Lens S.M., and van Vugt M.A.. 2015. Rif1 is required for resolution of ultrafine DNA bridges in anaphase to ensure genomic stability. Dev. Cell. 34:466–474. 10.1016/j.devcel.2015.06.014 [DOI] [PubMed] [Google Scholar]
- Huang J., and Raff J.W.. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184–2195. 10.1093/emboj/18.8.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.Y., and Raff J.W.. 2002. The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell. Sci. 115:2847–2856. [DOI] [PubMed] [Google Scholar]
- Jackson S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessulat M., Malty R.H., Nguyen-Tran D.H., Deineko V., Aoki H., Vlasblom J., Omidi K., Jin K., Minic Z., Hooshyar M., et al. 2015. Spindle checkpoint factors Bub1 and Bub2 promote DNA double-strand break repair by nonhomologous end joining. Mol. Cell. Biol. 35:2448–2463. 10.1128/MCB.00007-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose M., Tollis S., Nair D., Mitteau R., Velours C., Massoni-Laporte A., Royou A., Sibarita J.B., and McCusker D.. 2015. A quantitative imaging-based screen reveals the exocyst as a network hub connecting endocytosis and exocytosis. Mol. Biol. Cell. 26:2519–2534. 10.1091/mbc.E14-11-1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R.E., Wassmann K., and Rahmani Z.. 2013. New insights into the role of BubR1 in mitosis and beyond. Int. Rev. Cell Mol. Biol. 306:223–273. 10.1016/B978-0-12-407694-5.00006-7 [DOI] [PubMed] [Google Scholar]
- Kaye J.A., Melo J.A., Cheung S.K., Vaze M.B., Haber J.E., and Toczyski D.P.. 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14:2096–2106. 10.1016/j.cub.2004.10.051 [DOI] [PubMed] [Google Scholar]
- Kim E.M., and Burke D.J.. 2008. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 4:e1000015 10.1371/journal.pgen.1000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E.M., van der Sar S.J., and Hardwick K.G.. 2007. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS One. 2:e342 10.1371/journal.pone.0000342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., and Yanagida M.. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676. 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Murakami H., and Yanagida M.. 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31:998–1011. 10.1128/MCB.00815-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., and Musacchio A.. 2012. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 196:451–467. 10.1083/jcb.201110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Zhang G., Larsen M.S., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S.P., and Nilsson J.. 2013. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 126:1086–1092. 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., and Cleveland D.W.. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117. 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N.A., Al-Bassam J., Wei R.R., and Harrison S.C.. 2007. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc. Natl. Acad. Sci. USA. 104:1201–1206. 10.1073/pnas.0610358104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R., Russell R.B., Neduva V., and Gibson T.J.. 2003. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701–3708. 10.1093/nar/gkg519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Nielsen C.F., Yao Q., and Hickson I.D.. 2014. The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet. Dev. 26:1–5. 10.1016/j.gde.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J.M., Lopes C., Amorim I., Antunes-Martins A., and Sunkel C.E.. 2004. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 117:1757–1771. 10.1242/jcs.01033 [DOI] [PubMed] [Google Scholar]
- Lopes C.S., Sampaio P., Williams B., Goldberg M., and Sunkel C.E.. 2005. The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J. Cell Sci. 118:187–198. 10.1242/jcs.01602 [DOI] [PubMed] [Google Scholar]
- Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Brüggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A., Luider T.M., et al. 2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA. 103:18597–18602. 10.1073/pnas.0609061103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J., and Toczyski D.. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237–245. 10.1016/S0955-0674(02)00312-5 [DOI] [PubMed] [Google Scholar]
- Melo J.A., Cohen J., and Toczyski D.P.. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov A., Cole R.W., and Rieder C.L.. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797–1806. 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Musarò M., Ciapponi L., Fasulo B., Gatti M., and Cenci G.. 2008. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat. Genet. 40:362–366. 10.1038/ng.2007.64 [DOI] [PubMed] [Google Scholar]
- Orthwein A., Fradet-Turcotte A., Noordermeer S.M., Canny M.D., Brun C.M., Strecker J., Escribano-Diaz C., and Durocher D.. 2014. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 344:189–193. 10.1126/science.1248024 [DOI] [PubMed] [Google Scholar]
- Peterson S.E., Li Y., Chait B.T., Gottesman M.E., Baer R., and Gautier J.. 2011. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J. Cell Biol. 194:705–720. 10.1083/jcb.201103103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff J.W., Jeffers K., and Huang J.Y.. 2002. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157:1139–1149. 10.1083/jcb.200203035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani Z., Gagou M.E., Lefebvre C., Emre D., and Karess R.E.. 2009. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187:597–605. 10.1083/jcb.200905026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S., Titen S.W., Xie H.B., Golic M.M., Bastiani M., Bandyopadhyay P., Olivera B.M., Brodsky M., Rubin G.M., and Golic K.G.. 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16:1568–1581. 10.1101/gad.986602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Macias H., and Sullivan W.. 2005. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr. Biol. 15:334–339. 10.1016/j.cub.2005.02.026 [DOI] [PubMed] [Google Scholar]
- Royou A., Gagou M.E., Karess R., and Sullivan W.. 2010. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell. 140:235–245. 10.1016/j.cell.2009.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Heeger S., Althoff F., Walter A., Heidmann S., Mechtler K., and Lehner C.F.. 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 116:385–402. 10.1007/s00412-007-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Chaleckis R., and Lehner C.F.. 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28:2374–2386. 10.1038/emboj.2009.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczaniecka M., Feoktistova A., May K.M., Chen J.S., Blyth J., Gould K.L., and Hardwick K.G.. 2008. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C). J. Biol. Chem. 283:23039–23047. 10.1074/jbc.M803594200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B.A., Stambaugh J.R., Yokomori K., Shah J.V., and Berns M.W.. 2014. DNA damage to a single chromosome end delays anaphase onset. J. Biol. Chem. 289:22771–22784. 10.1074/jbc.M113.535955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E., Tiong S., Pedersen M., Homola E., Royou A., Fasulo B., Siriaco G., and Campbell S.D.. 2004. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr. Biol. 14:1341–1347. 10.1016/j.cub.2004.06.056 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., and Yen T.J.. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., Vleugel M., Teixeira A., and Kops G.J.. 2012. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 23:745–755. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Ahmad K., Almouzni G., Ausió J., Berger F., Bhalla P.L., Bonner W.M., Cande W.Z., Chadwick B.P., Chan S.W., et al. 2012. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin. 5:7 10.1186/1756-8935-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., and McKeon F.. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1–11. 10.1083/jcb.142.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Li B., Warrington R., Tomchick D.R., Yu H., and Luo X.. 2012. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc. Natl. Acad. Sci. USA. 109:18419–18424. 10.1073/pnas.1213438109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Babu J.R., Harden J.M., Jablonski S.A., Gazi M.H., Lingle W.L., de Groen P.C., Yen T.J., and van Deursen J.M.. 2001. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J. Biol. Chem. 276:26559–26567. 10.1074/jbc.M101083200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.