VAcUL-1 and Dsc, two different E3 ubiquitin ligase complexes coexist on the vacuole membrane to regulate different subsets of membrane proteins via the vReD pathway in response to different environmental cues.
Abstract
Cells must regulate the abundance and activity of numerous nutrient transporters in different organelle membranes to achieve nutrient homeostasis. As the recycling center and major storage organelle, lysosomes are essential for maintaining nutrient homeostasis. However, very little is known about mechanisms that govern the regulation of its membrane proteins. In this study, we demonstrated that changes of Zn2+ levels trigger the downregulation of vacuolar Zn2+ transporters. Low Zn2+ levels cause the degradation of the influx transporter Cot1, whereas high Zn2+ levels trigger the degradation of the efflux channel Zrt3. The degradation process depends on the vacuole membrane recycling and degradation pathway. Unexpectedly, we identified a RING domain–containing E3 ligase Tul1 and its interacting proteins in the Dsc complex that are important for the ubiquitination of Cot1 and partial ubiquitination of Zrt3. Our study demonstrated that the Dsc complex can function at the vacuole to regulate the composition and lifetime of vacuolar membrane proteins.
Introduction
Maintaining nutrient homeostasis is essential for all living organisms. For example, defects in metal ion homeostasis are associated with diseases. Iron deficiency causes anemia in humans, whereas excessive iron intake leads to iron-overload diseases in parenchymal tissues (De Domenico et al., 2008). In addition, Zn2+ deficiency leads to many defects in humans, such as growth retardation, cognitive disorders, and infertility (Jeong and Eide, 2013). Conversely, too much Zn2+ is toxic to humans as well and can result in death (Grissinger, 2011). At the cellular level, nutrient homeostasis is achieved by regulating numerous transporters in different organelle membranes. Many of these transporters are highly conserved from yeast to human. For example, two Zn2+ transporter families exist in eukaryotes, including fungi, plants, and mammals, to regulate the level of Zn2+ in the cytoplasm. The ZIP family (14 members in humans) is responsible for transporting Zn2+ into the cytoplasm, either from the plasma membrane or from other internal organelles (Lichten and Cousins, 2009). On the other hand, the ZnT family (10 members in humans) is responsible for removing Zn2+ from the cytoplasm, either into the extracellular space or into organelles like the lysosome (Zhao and Eide, 1996a,b; Lichten and Cousins, 2009). Mutations in these transporters lead to many diseases or even lethality in humans and other mammals. Mutations in human ZIP4 disrupt Zn2+ absorption at the intestine and cause the disease acrodermatitis enteropathica (Fukada et al., 2011; Chimienti, 2013). Homozygous deletion of mouse ZnT1 leads to early embryonic lethality (Lichten and Cousins, 2009).
In yeast, the vacuole (counterpart of the mammalian lysosome) is essential for maintaining nutrient homeostasis because it is the recycling center and the major storage organelle for nutrients, such as amino acids, fatty acids, and metal ions (Blaby-Haas and Merchant, 2014). Not surprisingly, numerous nutrient transporters have been identified on the vacuole surface. For example, three Zn2+ transporters exist on the vacuole membrane. Zrc1 and Cot1 (members of the ZnT family) are responsible for the influx of Zn2+ into the vacuole for storage, and Zrt3 (a member of the ZIP family) mediates the efflux of Zn2+ from the vacuole (Fig. 1 A; Kamizono et al., 1989; Conklin et al., 1992; MacDiarmid et al., 2000). These transporters not only participate in the nutrient homeostasis by transporting nutrients across the lysosomal membrane but also function as nutrient sensors to regulate the mTORC1 activity, which controls a myriad of cellular processes, including protein synthesis, autophagy, and cellular growth (Zoncu et al., 2011; Efeyan et al., 2015; Rebsamen et al., 2015; Wang et al., 2015). Despite their importance in maintaining nutrient homeostasis and regulating signaling pathways, little is known about the regulation of the quantity and quality of these lysosomal membrane transporters.
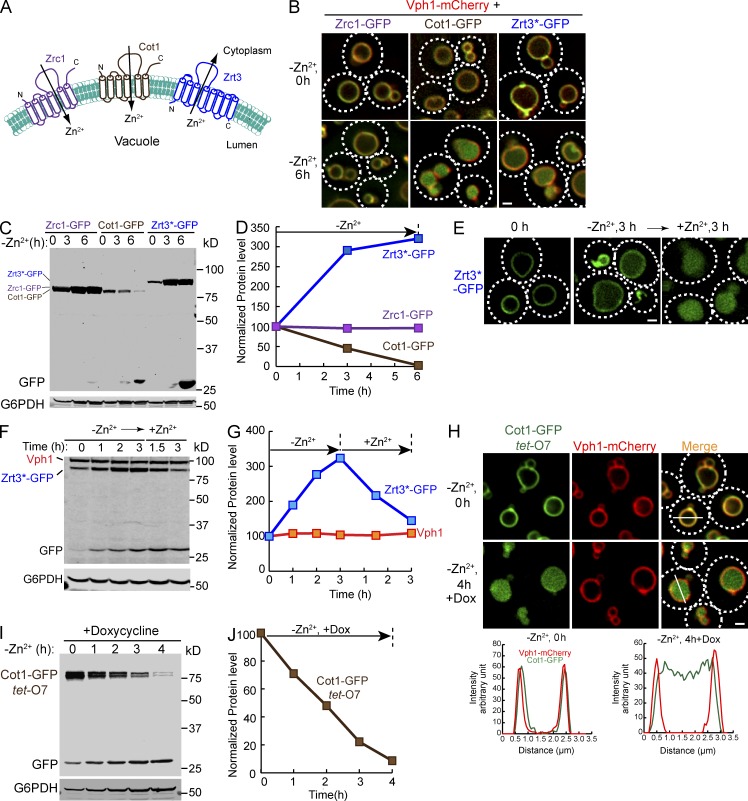
Figure 1.
Vacuolar Zn2+ transporters can be downregulated in response to changes in Zn2+ concentration. (A) A schematic diagram illustrating the topology of vacuolar Zn2+ transporters, including Zrc1, Cot1, and Zrt3. (B) Localization of chromosomally tagged Zrc1-GFP, Cot1-GFP, Zrt3*-GFP and Vph1-mCherry, before and 6 h after Zn2+ withdrawal in YNB media. Dashed lines highlight the cell surface. (C) Western blot analysis of chromosomally tagged Zrc1-GFP, Cot1-GFP, and Zrt3*-GFP over the course of 6 h of Zn2+ withdrawal in YNB media. The samples were blotted with an anti-GFP antibody. G6PDH was used as a loading control. Same volume of cells was loaded in each lane, with 1 OD600 cells loaded at 0 h. (D) Quantification of 1C. All protein levels were normalized using G6PDH level. (E) Localization of Zrt3*-GFP under different Zn2+ treatments. Mid-log cells grown in YPD (0 h) were transferred into YNB without Zn2+ and incubated for 3 h (−Zn2+ 3 h). Then, 2 mM ZnCl2 was added and cells were incubated for another 3 h (+Zn2+ 3 h). (F) Western blot analysis of Zrt3*-GFP and Vph1 levels when yeast cells were shifted from YNB without Zn2+ to YNB with excessive Zn2+ (2 mM ZnCl2). G6PDH was used as a loading control. 1 OD600 cells were loaded in each lane. (G) Quantification of F. All protein levels were normalized using G6PDH level. (H) Localization of tet-O7 Cot1-GFP and Vph1-mCherry before and after Zn2+ withdrawal in YNB media. The incubation was performed in the presence of 2 µg/ml doxycycline (Dox). Right: Line-scan analysis of the image. (I) Degradation kinetics for tet-O7 Cot1-GFP after addition of 2 µg/ml Dox and Zn2+ withdrawal in YNB media. G6PDH was used as a loading control. Same volume of cells were loaded, with 1 OD600 cells loaded at 0 h. (J) Quantification of I. All protein levels were normalized using G6PDH level. Bar, 1 µm.
We recently identified a novel vacuole membrane recycling and degradation (vReD) pathway to downregulate vacuole membrane transporters (Li et al., 2015). As a founding member of the pathway, Ypq1, a cationic amino acid transporter on the vacuole membrane, is degraded in the vacuole lumen upon lysine withdrawal. The degradation process requires protein ubiquitination by the vacuole-anchored ubiquitin ligase complex 1 (VAcUL-1; composed of the ubiquitin ligase Rsp5 and its membrane adaptor Ssh4), sorting into a new class of carriers that bud off the vacuole, transporting to an intermediate compartment, and internalization of the ubiquitinated Ypq1 into intraluminal vesicles by the endosomal sorting complexes required for transport (ESCRT) machinery. However, whether the vReD pathway is a general mechanism to down-regulate vacuole membrane proteins remains to be addressed.
In this study, we establish that the vReD pathway is a general pathway for the turnover of vacuolar transporters. In addition to Ypq1, it mediates the turnover of vacuolar Zn2+ influx and efflux transporters. High concentration of Zn2+ triggers the degradation of the efflux channel Zrt3, whereas low Zn2+ triggers the degradation of the influx transporter Cot1. Both degradation processes follow the vReD pathway, as indicated by the internalization of the GFP-tagged proteins into the lumen of the vacuole and their dependence on the ESCRT machinery. Further analysis revealed a second E3 ubiquitin ligase complex, the defective for SREBP cleavage (Dsc) complex, that can localize to the vacuole membrane to selectively regulate a subset of vacuolar membrane proteins, including Cot1 and Zrt3.
Results
Changes in Zn2+ levels trigger the downregulation of vacuolar membrane Zn2+ transporters
We have shown that lysine levels regulate Ypq1, the vacuolar lysine influx transporter (Sekito et al., 2014; Li et al., 2015). We reasoned that other vacuolar transporters may also be regulated by their substrates/ligands. To test this hypothesis, we attempted to chromosomally tag 33 vacuolar transporters with GFP (Table S2; Huh et al., 2003; Wiederhold et al., 2009; Riffle and Davis, 2010). Out of 33 proteins, 12 proteins exhibited no GFP signal, 9 proteins accumulated in the ER, and 1 protein was delivered to the vacuole lumen (Table S2). 11 proteins, including Cot1, Fet5, Fth1, Ncr1, Vcx1, Ycf1, Ypl162c, Ypq2, Zrc1, and Zrt3, were efficiently sorted to the vacuole membrane (Table S2). Of the 11 vacuole membrane proteins, 8 proteins (Cot1, Fet5, Fth1, Ncr1, Vcx1, Ycf1, Zrc1, and Zrt3) have been suggested to be either nutrient or ion transporters and not characterized by our previous study (Li et al., 2015; Table S2). Therefore, we tested whether manipulating substrate concentrations triggers their downregulation. Among these transporters, Cot1 and Zrt3 were strongly responsive to substrate concentration changes. We thus focused on the vacuolar Zn2+ transporters Zrt3 and Cot1 for subsequent studies.
Zrt3 was predicted to have eight transmembrane helices, with both the N and C termini facing the vacuole lumen (Fig. 1 A; Li et al., 2015). To shift the GFP tag to the cytoplasmic side of the membrane and avoid exposure of the GFP tag to proteases in the vacuole lumen that can release it from the membrane and ultimately degrade the GFP, we deleted the last 23 amino acids of the protein, including the eighth transmembrane helix (Zrt3*). This mutant protein was sorted properly to the vacuole membrane and it also responded to changes in Zn2+ concentration (Fig. 1, B–E). Thus, we used this mutant Zrt3*-GFP for our subsequent work. As shown in Fig. 1 (B and C), withdrawing Zn2+ from the growth medium triggered the internalization and degradation of Cot1-GFP, the influx Zn2+ transporter. The efflux Zn2+ channel, Zrt3*, was upregulated by this treatment, with a threefold increase after 3 h of Zn2+ starvation, as previously observed (Fig. 1, C and E; MacDiarmid et al., 2000). Intriguingly, we observed conversion of a portion of the Zrt3*-GFP fusion protein to a 26-kD fragment (free GFP) and an accumulation of a luminal GFP signal in the vacuole after 6 h of Zn2+ starvation (Fig. 1, B and C), suggesting that high expression levels may result in a biosynthetic sorting of Zrt3*-GFP to the vacuole lumen or the Zrt3*-GFP may have a relatively short lifespan in the vacuole membrane. Addition of 2 mM Zn2+ back to the growth medium triggered the sorting of Zrt3*-GFP to the vacuole lumen for degradation (Fig. 1, F and G). In contrast to Cot1-GFP and Zrt3*-GFP, the level of Zrc1-GFP was unchanged after withdrawing Zn2+ (Fig. 1, C and D) and the protein stayed on the vacuole membrane (Fig. 1 B). Furthermore, Zn2+ withdrawal did not change the localization of Vph1-mCherry (part of the v-ATPase complex; Fig. 1 B), indicating the response to Zn2+ is a highly selective process.
The expression of Zrt3*-GFP was repressed by the addition of excessive Zn2+ (MacDiarmid et al., 2000). Therefore, the accumulation of the luminal GFP signal was likely due to the degradation of preexisting vacuolar Zrt3*-GFP and not the modest pool of newly synthesized Zrt3*-GFP. To confirm that Cot1-GFP at the vacuole membrane is sorted into the lumen, we expressed the protein under the control of a Tet-off system (tet-O7 Cot1-GFP; Garí et al., 1997). After 20 min of preincubation with 2 µg/ml doxycycline, we withdrew Zn2+ from the growth medium and continued the treatment in the presence of doxycycline for 4 h. As shown in Fig. 1, H–J, withdrawing Zn2+ triggered the internalization and degradation of most preexisting Cot1-GFP, whereas Vph1-mCherry did not respond to the Zn2+ withdrawal. To ensure that the degradation happens within the vacuole, we performed degradation assays for both Cot1-GFP and Zrt3*-GFP in a pep4Δ strain that is defective for vacuolar proteases activities. As shown in Fig. S1, although the internalization of both proteins was normal, the degradation process was reduced drastically by the deletion of PEP4, indicating that degradation of these two proteins depends on the vacuolar proteases.
Together, these data showed that changes of the Zn2+ concentration trigger the downregulation of vacuolar Zn2+ transporters. The Zn2+ influx transporter Cot1 is internalized and degraded inside the vacuole under Zn2+ starvation, whereas the efflux channel Zrt3 is targeted for degradation when excessive Zn2+ is present. Although the activity of these transporters may also be regulated by the Zn2+ levels (possibly via protein phosphorylation/dephosphorylation), we restricted our analysis to the mechanism of protein sorting and turnover in this study.
Degradation of Cot1 and Zrt3 depends on the vReD pathway, but not the autophagy pathway
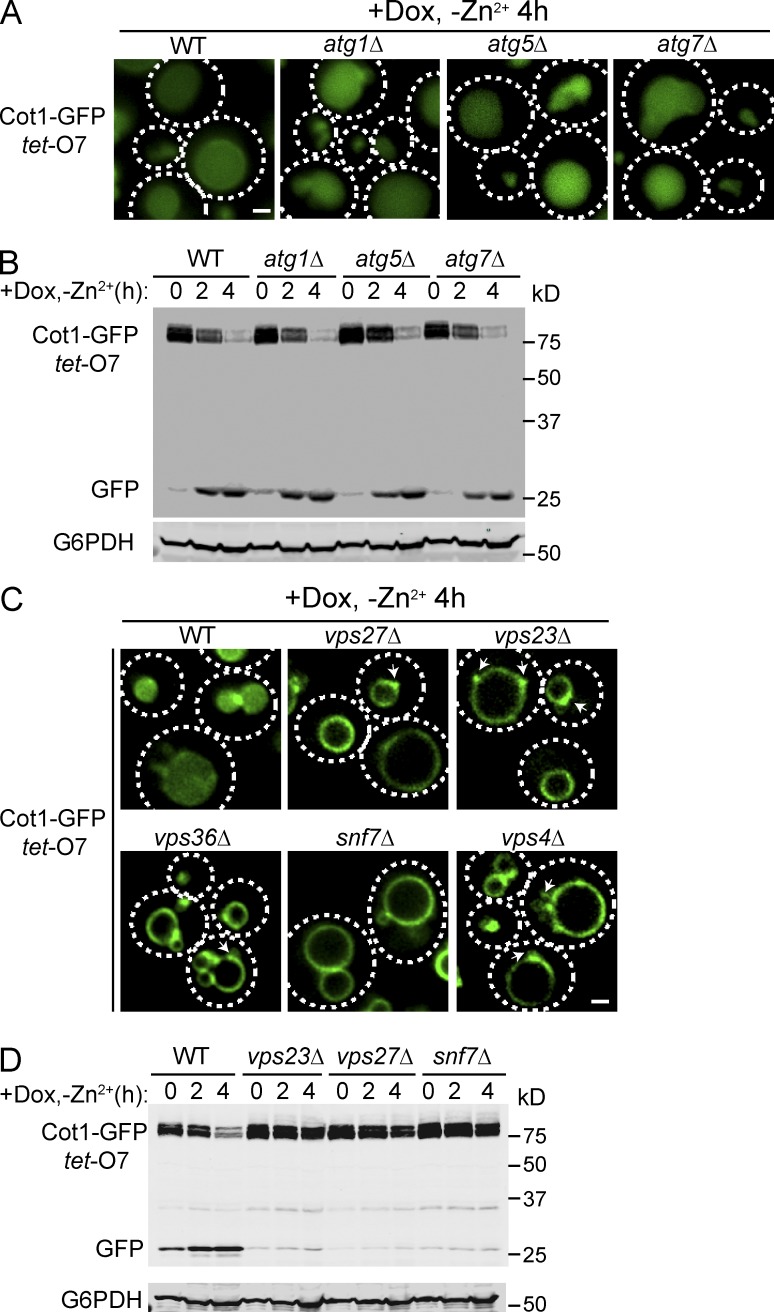
Recently, we identified a vReD pathway for the selective down-regulation of vacuolar membrane transporters (Li et al., 2015). This pathway is initiated by ubiquitination via a vacuole membrane anchored VAcUL-1 complex, composed of the E3 ligase Rsp5 and its membrane adaptor, Ssh4. After protein ubiquitination, the internalization of the cargo depends on the ESCRT machinery, rather than autophagy. The fact that both Cot1-GFP and Zrt3*-GFP were internalized into the vacuole lumen for degradation suggested that the vReD pathway was the mechanism for their degradation. To test this hypothesis, we investigated whether autophagy or the ESCRT machinery was important for the degradation of Cot1-GFP and Zrt3*-GFP. Deletion of the autophagy genes ATG1, ATG5, and ATG7 had no effect on the internalization of either Cot1-GFP (Fig. 2 A) or Zrt3*-GFP (Fig. S2 A), nor did it affect the degradation kinetics of either protein (Figs. 2 B and S2 B). In contrast, deleting genes encoding key components of the ESCRT complexes, including VPS27 (ESCRT-0), VPS23 (ESCRT-I), VPS36 (ESCRT-II), and SNF7 (ESCRT-III), stabilized both Cot1-GFP and Zrt3*-GFP on the vacuole membrane, with a portion of the protein associated with small punctae, presumably the aberrant endosomal compartments that accumulate in ESCRT mutants (the class E compartments; Figs. 2 C and S2 C, arrows). Western blot analysis revealed that no degradation of the fusion proteins was detected in the ESCRT deletion mutant strains (Figs. 2 D and S2 D). Collectively, our data support a model in which the vReD pathway is responsible for the sorting and degradation of both Cot1-GFP and Zrt3*-GFP, suggesting that this pathway may define a general mechanism for the downregulation of many vacuolar membrane proteins.
Figure 2.
ESCRTs, but not the autophagy machinery, are required for the degradation of Cot1-GFP. (A) Localization of tet-O7 Cot1-GFP in WT and autophagy mutants 4 h after Zn2+ withdrawal from YNB media and addition of 2 µg/ml Dox. (B) Degradation kinetics for tet-O7 Cot1-GFP in WT and autophagy mutants. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. (C) Localization of tet-O7 Cot1-GFP 4 h after Zn2+ withdrawal in WT and ESCRT mutants. Arrows highlight the aberrant endosomes. (D) Degradation kinetics for tet-O7 Cot1-GFP in WT and ESCRT mutants. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. Bar, 1 µm.
Identification of the E3 ligases responsible for Cot1 and Zrt3 ubiquitination
Because the ESCRT machinery functions to selectively sort ubiquitinated cargoes, we tested whether Cot1-GFP and Zrt3*-GFP are ubiquitinated during their degradation process. Indeed, as shown in Fig. S3, withdrawing Zn2+ from the growth medium triggered the polyubiquitination of Cot1-GFP, whereas addition of 2 mM Zn2+ triggered the polyubiquitination of Zrt3*-GFP.
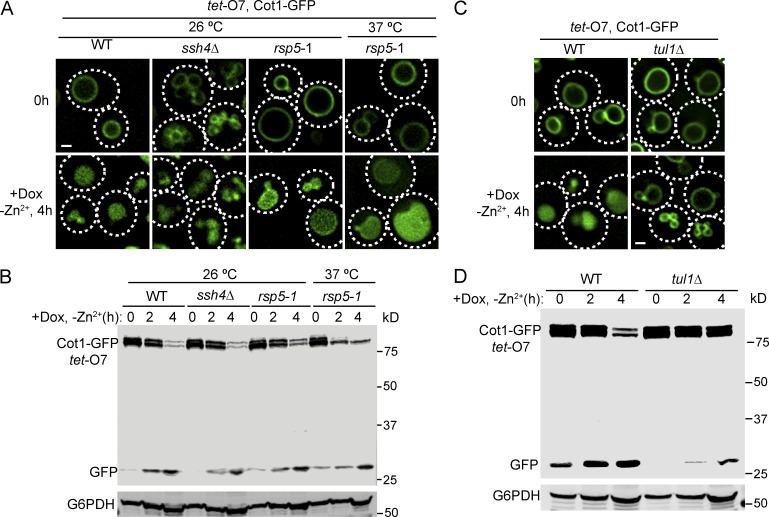
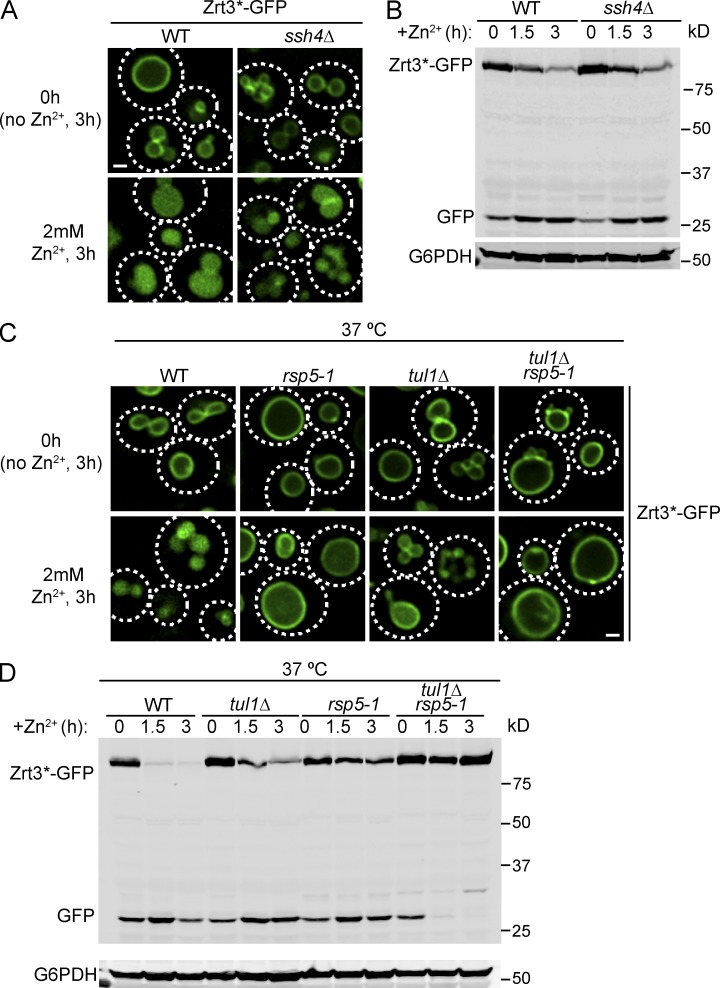
The VAcUL-1 complex is localized on the vacuole membrane, where it can ubiquitinate Ypq1 after lysine withdrawal and thereby trigger its sorting and degradation (Li et al., 2015). We hypothesized that Zrt3 and Cot1 may also be ubiquitinated by the same complex. To test this, we deleted SSH4 and tested whether the degradation of Cot1-GFP or Zrt3*-GFP was impaired. To our surprise, the degradation of both Cot1-GFP and Zrt3*-GFP was unaffected in the ssh4Δ mutant strain (Fig. 3, A and B; and Fig. 4, A and B). SSH4 deletion caused a mild fragmentation of the vacuole, which was exacerbated by the Zn2+ withdrawal (Fig. 3, A and B). Nevertheless, Western blot analysis showed that the degradation kinetics of both Cot1-GFP and Zrt3*-GFP was not affected in the ssh4Δ strain (Figs. 3 B and 4 B). Further analysis using a temperature-sensitive mutant of the ubiquitin ligase Rsp5 (rsp5-1) showed that Rsp5 was not required for Cot1-GFP degradation (Fig. 3, A and B), and the degradation of Zrt3*-GFP was significantly delayed, but not blocked, in the rsp5-1 strain at 37°C (Fig. 4, C and D), suggesting the existence of a second ubiquitin ligase system on the vacuole membrane.
Figure 3.
Tul1, but not the VAcUL-1 complex, is required for Cot1-GFP degradation. (A) ssh4Δ and rsp5-1 mutants did not block the internalization of Cot1-GFP into the vacuole lumen after Zn2+ withdrawal from the YNB media. (B) Degradation kinetics for tet-O7 Cot1-GFP in WT, ssh4Δ and rsp5-1 mutants. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. (C) The internalization of Cot1-GFP into the vacuole lumen was blocked in tul1Δ strain after Zn2+ withdrawal from the YNB media. (D) Degradation kinetics for tet-O7 Cot1-GFP in WT and ssh4Δ strains. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. Bar, 1 µm.
Figure 4.
Both Tul1 and Rsp5 contribute to the degradation of Zrt3*-GFP. (A) The internalization of Zrt3*-GFP into the vacuole lumen was not blocked in ssh4Δ strain after the addition of 2mM ZnCl2 to the YNB media. Cells were pretreated with YNB without Zn2+ for 3 h. (B) Degradation kinetics for Zrt3*-GFP in WT and ssh4Δ strains. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. (C) Localization of Zrt3*-GFP in WT, tul1Δ, rsp5-1, and tul1Δ rsp5-1 mutant strains before and after addition of 2mM ZnCl2 to YNB media. Cells were pretreated with YNB without Zn2+ for 3 h at 26°C. Then, the culture was shifted to 37°C for 30 min before the addition of ZnCl2 and the incubation was continued for another 3 h at 37°C. (D) Degradation kinetics for Zrt3*-GFP in WT, tul1Δ, rsp5-1, and tul1Δ rsp5-1 mutant strains. The cells were pretreated with YNB media without Zn2+ for 3 h at 26°C. Then, the culture was shifted to 37°C for 30 min before the addition of ZnCl2 to 2 mM and the incubation was continued for another 3 h at 37°C. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. Bar, 1 µm.
To identify the unknown E3 ligase in the Cot1 and Zrt3 sorting pathway, we screened all yeast nonessential E3 ligases using the yeast knockout collection (Giaever et al., 2002; Table S3). The screening was conducted by transforming a Tet-off COT1-GFP plasmid into each knockout mutant and performing the Zn2+ withdrawal experiment in the presence of 2 µg/ml doxycycline. Of the 38 nonessential E3 ligases we tested, only the TUL1 deletion strain showed a strong defect in Cot1-GFP degradation. In this mutant, the degradation of Cot1-GFP was much reduced and most of the protein was stabilized on the vacuole membrane after 4 h of Zn2+ starvation (Fig. 3, C and D). Unfortunately, because TUL1 and DOA4 (a prerequisite to stabilized ubiquitinated Cot1) double deletion is synthetic lethal (Tong et al., 2014), we were unable to directly test if TUL1 deletion eliminates Cot1 polyubiquitination. The Zrt3*-GFP degradation triggered by the addition of 2 mM Zn2+ was also significantly delayed in the TUL1 deletion mutant strain (Fig. 4, C and D). Importantly, the rsp5-1 tul1Δ double mutation blocked Zrt3*-GFP degradation and stabilized the protein on the vacuole membrane (Fig. 4, C and D), indicating that both Tul1 and Rsp5 can ubiquitinate Zrt3*.
Tul1 was initially identified as a RING domain–containing E3 ligase localized to the Golgi membrane in budding yeast (Reggiori and Pelham, 2002). It contains a predicted signal sequence at the N terminus, followed by a large luminal domain that contains eight predicted glycosylation sites, seven transmembrane helices, and a C-terminal RING domain that is facing the cytoplasm. The RING domain directly interacts with the E2 ubiquitin conjugase Ubc4, which is required for its E3 ligase activity (Reggiori and Pelham, 2002). It has been shown to ubiquitinate two artificial substrates, Pep12D and a nonpalmitylation mutant form of Tlg1, and therefore has been proposed to be a part of the quality-control system in the Golgi (Reggiori and Pelham, 2002; Valdez-Taubas and Pelham, 2005). However, no native substrates have been reported in budding yeast.
Phylogenetic analyses using the position-specific iterated BLAST (PSI-BLAST) algorithm revealed that homologues of Tul1 widely exist in eukaryotes, including fungi, plants, and many unicellular eukaryotes such as oomycetes, amoebozoa, apicomplexans, diatoms, and dinoflagellates. The Arabidopsis thaliana homologue, FLY1, was reported to be important for the regulation of the methylesterification degree of pectin in the primary cell wall, although the direct substrates for this E3 ligase remain to be identified (Voiniciuc et al., 2013). Strikingly, further PSI-BLAST analysis using just the seven-transmembrane region and the RING domain revealed that Tul1 is related to Hrd1, an important RING domain–containing E3 ligase in the ER-associated degradation (ERAD) pathway (Altschul et al., 1997; Stewart et al., 2011).
The Dsc complex is important for Cot1-GFP degradation
The homologue of Tul1 in fission yeast, Dsc1, was reported to be essential for the proteolytic activation of the fungal SREBP transcription factor in the Golgi (Stewart et al., 2011). Through a combination of genetic and biochemical analysis, Dsc1 was shown to be part of a stable protein complex containing Dsc1 and 4 other proteins (Dsc2–5; Stewart et al., 2011). The integrity of the Dsc complex is important for SREBP cleavage, as deletion of any Dsc component abolished SREBP processing. In addition, the AAA ATPase Cdc48 has been shown to be critical for the processing of SREBP (Stewart et al., 2012). Dsc5 contains a Ubx domain that directly interacts with Cdc48. Intriguingly, the Dsc complex is similar to the Hrd1 E3 ligase complex that functions in the ERAD pathway (Stewart et al., 2011, 2012).
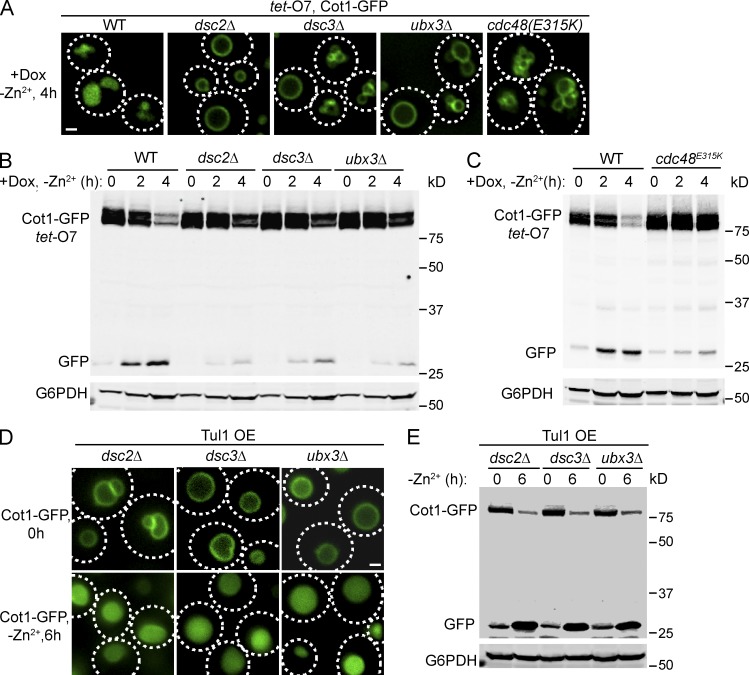
Homologues of Dsc2, Dsc3, and Dsc5, but not Dsc4, exist in budding yeast (Tong et al., 2014). We deleted DSC2, DSC3, or UBX3 (the DSC5 homologue) and tested if these mutants affect Cot1-GFP degradation. As shown in Fig. 5 (A and B), deletion of any Dsc component stabilized Cot1-GFP on the vacuole membrane, underscoring the importance of maintaining the integrity of the Dsc complex for Cot1-GFP degradation. A Cdc48 mutant, cdc48E315K, blocks SREBP processing in fission yeast (Stewart et al., 2012). We tested if this mutation also affects Cot1 degradation in budding yeast. Indeed, Cot1-GFP was stabilized on the vacuole membrane after 4 h of Zn2+ withdrawal in the cdc48E315K mutant (Fig. 5, A and C). Intriguingly, Western blot analysis indicated that the Cot1-GFP level was significantly elevated in the cdc48E315K strain relative to the wild-type (WT) strain at 0 h, suggesting that Cdc48 may play a role in the sorting and degradation of Cot1-GFP during its biosynthetic trafficking to the vacuole membrane (Fig. 5 C).
Figure 5.
The Dsc complex and Cdc48 are important for Cot1-GFP degradation. (A) Localization of tet-O7 Cot1-GFP after Zn2+ withdrawal in WT, deletion mutants for the Dsc complex and the cdc48E315K allele. (B) Degradation kinetics for tet-O7 Cot1-GFP in WT, dsc2Δ, dsc3Δ, and ubx3Δ mutant strains. G6PDH was used as a loading control. Same volume of cells was loaded, with 1 OD600 cells loaded at 0 h. (C) Degradation kinetics for tet-O7 Cot1-GFP in WT and cdc48E315K mutant strains. G6PDH was used as a loading control. Same volume of cells was loaded; with 1 OD600 cells loaded at 0 h. (D) Localization of chromosomally tagged Cot1-GFP before and after Zn2+ withdrawal in dsc deletion mutants overexpressing Tul1. (E) Degradation kinetics of chromosomally tagged Cot1-GFP in dsc deletion mutants overexpressing Tul1. G6PDH was used as a loading control. 1 OD600 of cells were loaded in each lane. Bar, 1 µm.
Because Tul1 is an E3 ligase, we tested if overexpression of Tul1 can bypass the requirement of other Dsc complex components. As shown in Fig. 5, D and E, overexpression of Tul1 did rescue the DSC deletion mutants. The Cot1-GFP fusion protein expressed under its endogenous promoter was largely degraded after 6 h of Zn2+ withdrawal. This suggests that the other Dsc complex subunits may regulate the localization, activity, or stability of Tul1. Interestingly, a similar observation was also made for the Hrd1 E3 ligase complex in the ERAD pathway. Overexpression of Hrd1 bypasses the requirement of Hrd3, Usa1, and Der1 (Carvalho et al., 2010). Together, these findings demonstrate that the Dsc complex is important for the down-regulation of Cot1-GFP.
Overexpression of Tul1 causes the down-regulation of a subset of vacuole membrane proteins
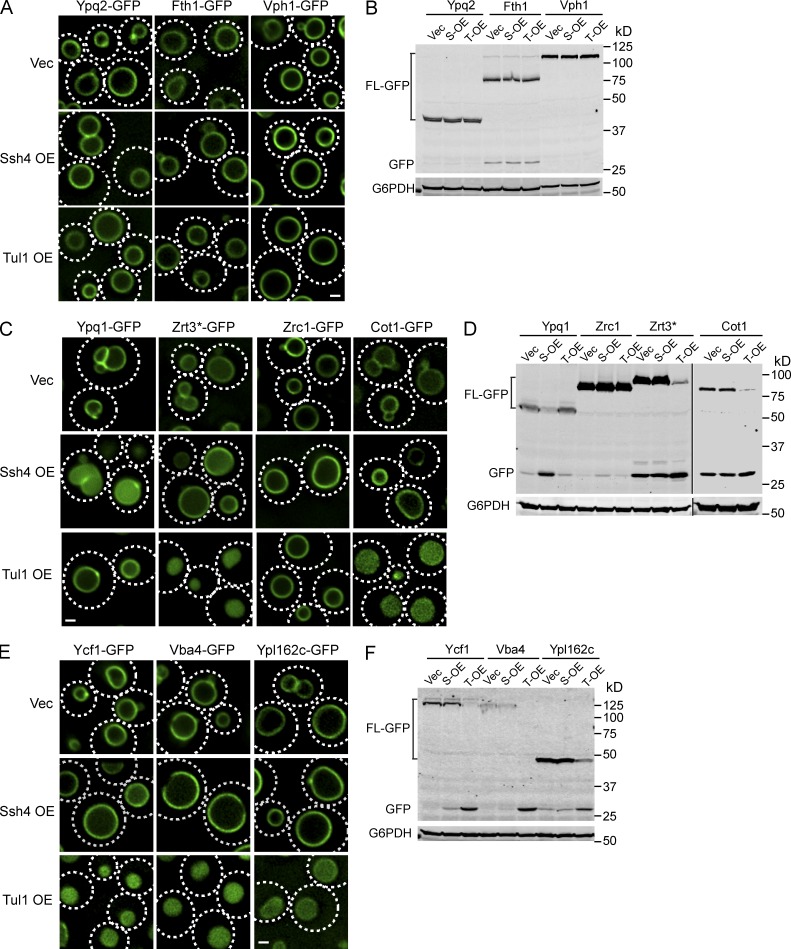
Previously, we have shown that overexpression of Ssh4 caused a partial degradation of Ypq1 (Li et al., 2015). Encouraged by this observation and the result that overexpression of Tul1 bypasses the requirement of other Dsc components, we hypothesized that overexpression of Tul1 or Ssh4 may result in the down-regulation of other vacuolar membrane proteins (possible natural substrates of these ubiquitin ligases).
We tested 10 different vacuolar membrane proteins that have been successfully tagged with a C-terminal GFP tag (Fig. 6). Among the tested vacuolar membrane proteins, Ypq2, Fth1, Zrc1, and Vph1 did not respond to overexpression of either Ssh4 or Tul1 (Fig. 6, A–D). Furthermore, only Ypq1 is internalized and degraded by Ssh4 overexpression (Fig. 6, C and D), confirming the downregulation of vacuole membrane proteins is a selective process. Overexpression of Tul1, however, caused various levels of degradation of five vacuolar membrane proteins (Cot1, Zrt3, Ycf1, Ypl162c, and Vba4; Fig. 6, C–F), suggesting they may be natural substrates of Tul1. Importantly, Cot1 and Zrt3 are the substrates of Tul1 that respond to changes in the Zn2+ concentration, suggesting Ycf1, Ypl162c, and Vba4 might be regulated by Tul1 as well, although conditions that trigger the ubiquitination of these substrates remain to be identified.
Figure 6.
Overexpression of Tul1 causes a constitutive degradation of a subset of vacuolar membrane proteins. (A) Localization of chromosomally tagged Ypq2-GFP, Fth1-GFP, and Vph1-GFP in cells overexpressing Ssh4, Tul1, or the empty vector. (B) Western blot analysis of protein levels for chromosomally tagged Ypq2-GFP, Fth1-GFP, and Vph1-GFP in cells overexpressing Ssh4 (S-OE), Tu1(T-OE), or the empty vector (Vec). G6PDH was used as a loading control. 1 OD600 of cells were loaded for Ypq2 and Fth1 samples, 0.4 OD600 of cells were loaded for Vph1 samples owing to the high expression level of Vph1-GFP. (C) Localization of chromosomally tagged Ypq1-GFP, Zrt3*-GFP, Zrc1-GFP, and Cot1-GFP in cells overexpressing Ssh4, Tul1, or the empty vector. (D) Western blot analysis of protein levels for chromosomally tagged Ypq1-GFP, Zrt3*-GFP, Zrc1-GFP, and Cot1-GFP in cells overexpressing Ssh4, Tul1, or only the empty vector. G6PDH was used as a loading control. 1 OD600 of cells were loaded for each lane. (E) Localization of chromosomally tagged Ycf1-GFP, Vba4-GFP, and Ypl162c-GFP in cells overexpressing Ssh4, Tu1, or the empty vector. (F) Western blot analysis of protein levels for chromosomally tagged Ycf1-GFP, Vba4-GFP, and Ypl162c-GFP in cells overexpressing Ssh4, Tul1, or the empty vector. G6PDH was used as a loading control. 1 OD600 cells were loaded for each lane. Bar, 1 µm.
Localization of the Dsc complex
As stated in the previous section, Tul1 was reported to be a Golgi-localized E3 ligase in budding yeast (Reggiori and Pelham, 2002). Using indirect immunostaining and a C-terminally GFP-tagged allele, Tul1 was shown to colocalize with two different Golgi markers, Tlg1 and Sed5, but not with the aberrant endosomes in a vps4Δ strain, strongly arguing that Tul1 localizes to the Golgi, but not to the endosome or vacuole. We wondered how a Golgi-localized E3 ubiquitin ligase could ubiquitinate vacuole membrane proteins like Cot1. To address this, we analyzed the localization of Tul1 and the Dsc complex in greater detail.
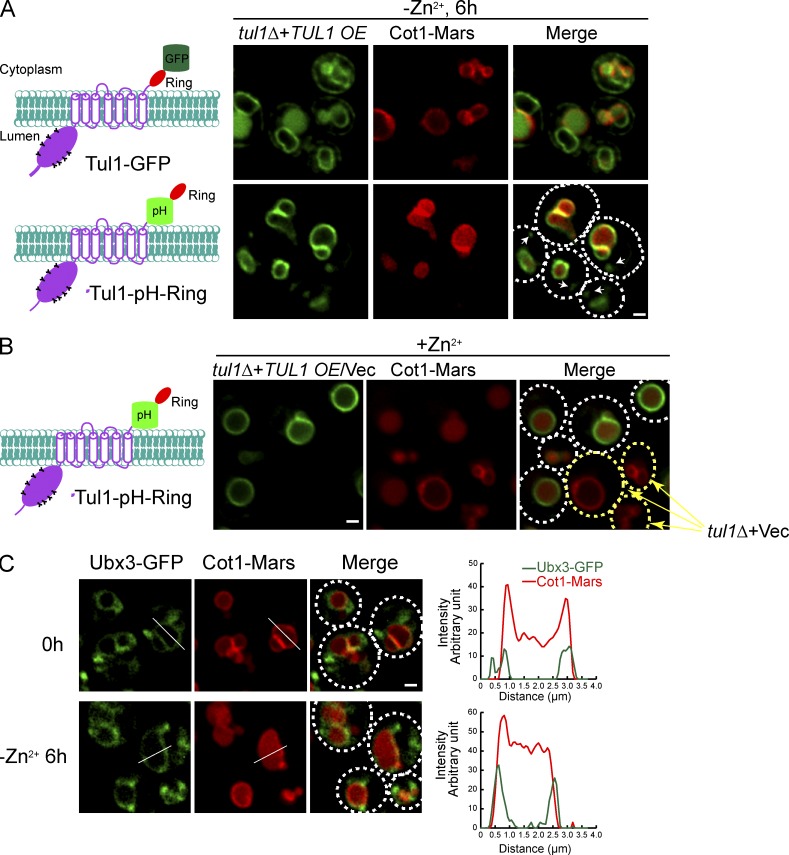
Chromosomally tagging Tul1 with GFP at the C terminus failed to produce any fluorescence signal, nor was this allele able to support the downregulation of Cot1 after Zn2+ withdrawal (unpublished data). An independent large-scale GFP localization study also did not detect any fluorescence for Tul1-GFP (Huh et al., 2003). Recently, Tong et al. also reported the difficulty of generating a functional tagged form of Tul1 (Tong et al., 2014). We reasoned that Tul1-GFP expressed under the endogenous promoter might be too weak to be detected. Therefore, we overexpressed Tul1-GFP under the tet-O7 promoter. Under this condition, we were able to observe Tul1-GFP; however, most of the Tul1-GFP was localized to the ER, with a significant amount of the GFP signal localized to the vacuole lumen (Fig. 7 A). We did not observe localization to the Golgi. Importantly, even after overexpression, Tul1-GFP failed to rescue the tul1Δ mutant. As shown in Fig. 7 A, Cot1-Mars was still on the vacuole membrane in the tul1Δ strain overexpressing Tul1-GFP, even after 6 h of Zn2+ withdrawal. We conclude that Tul1-GFP is not functional and its trafficking is impaired by tagging the protein with a C-terminal GFP.
Figure 7.
A significant fraction of the Dsc complex localizes to the vacuole membrane. (A) Left: Schematics illustrating the position where GFP (or Phluorin) is inserted. Tul1 contains an N-terminal luminal domain with multiple glycosylation sites, seven transmembrane helices, and a C-terminal RING domain. Right: Localization of Tul1-GFP, Tul1-pH-Ring, and Cot1-Mars 6 h after Zn2+ withdrawal from the YNB media. Both Tul1-GFP and Tul1-pH-Ring were overexpressed under the tet-O7 promoter, and Cot1-Mars was chromosomally tagged. White arrows highlight the punctae outside the vacuole. (B) Cot1-Mars was constitutively internalized into the vacuole lumen in cells overexpressing Tul1-pH-Ring, whereas Cot1-Mars stayed on the vacuole membrane in cells transformed with an empty vector (yellow arrows). Tul1-pH-Ring was overexpressed under the tet-O7 promoter, and Cot1-Mars was chromosomally tagged. (C) Localization of chromosomally tagged Ubx3-GFP and Cot1-Mars before and 6 h after removing Zn2+ from the YNB media. White lines highlight the position for line-scanning analysis, which was shown on the right. Bar, 1 µm.
To generate a functional tagged form of the Tul1 protein, we inserted a pH-sensitive GFP variant (pHluorin) in front of the RING domain, between residues Gly689 and Gly690, and expressed the chimera under the control of tet-O7 promoter (Tul1-pH-Ring). This insertion did not interfere with the normal function of Tul1, as demonstrated by its ability to complement the tul1Δ mutant to internalize Cot1-Mars (Fig. 7, A and B). In fact, overexpression of Tul1-pH-Ring in tul1Δ cells triggered a constitutive degradation of Cot1-Mars, bypassing the requirement of Zn2+ withdrawal (Fig. 7 B, white circles). In contrast, Cot1-Mars was stabilized on the vacuole membrane in tul1Δ mutant cells transformed with the empty vector (Fig. 7 B, yellow circles). Importantly, most of the Tul1-pH-Ring signal was localized to the vacuole membrane. After enhancing the image intensity, we also detected a few punctae outside the vacuole in 50% of the cells (n = 50; Fig. 7 A; and Fig. S4, A and B, arrows). To determine the identity of these punctae, we compared them to FM4-64–labeled endosomes (stained for 5 min) and Sec7-Mars labeled Golgi compartments. As shown in Fig. S4, A and B, a fraction of punctae colocalized with either FM4-64–labeled endosomes or Sec7-labeled Golgi, indicating that Tul1-pH-Ring is broadly distributed on the Golgi, endosomes, and the vacuole, with the majority localizing to the vacuole membrane.
To confirm the Dsc complex, expressed at the endogenous level, maintains a similar distribution, we attempted to tag other Dsc components with GFP at their endogenous chromosomal sites. Only Ubx3-GFP exhibited a detectable fluorescence signal. Ubx3-GFP still supports the internalization of Cot1-Marts after Zn2+ withdrawal, albeit at a slower rate (Fig. 7 C). As shown in Fig. 7 C, Ubx3-GFP localized to the vacuole surface, with a few punctae (one to five per cell) in the cytoplasm. Consistent with the colocalization study for Tul1-pH-Ring, further analysis showed that a fraction of these punctae colocalized with either FM4-64–labeled endosomes or Sec7-labeled Golgi (Fig. S4, C and D, arrows).
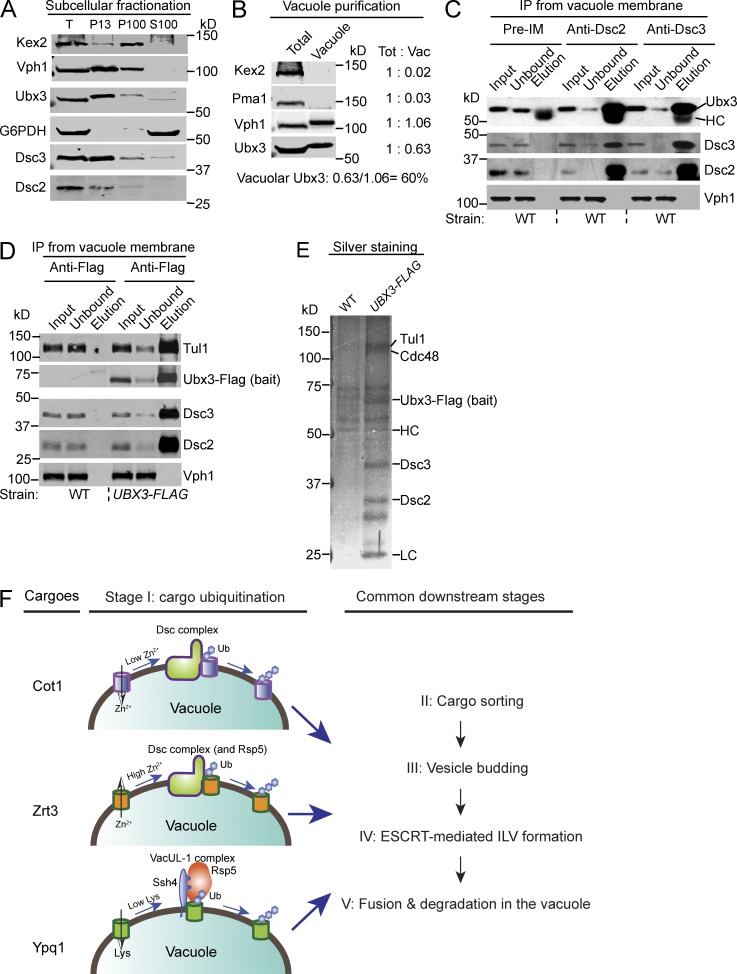
As an alternative approach to determine the distribution of the Dsc complex, we performed subcellular fractionation assays. Mid-log WT yeast cells were spheroplasted, lysed, and separated by differential centrifugation. As shown in Fig. 8 A, components of the Dsc complex, including Dsc2, Dsc3, and Ubx3, were enriched in the P13 fraction (membrane pelleted at 13,000 g). Their distribution patterns were similar to Vph1, a VO subunit of the vacuolar ATPase, suggesting a significant fraction of the Dsc complex is localized to the vacuole membrane. In contrast, Kex2, a transmembrane serine protease that cycles between the Golgi and endosomes, is enriched in the P100 fraction (membrane pelleted at 100,000 g). The cytosolic protein, G6PDH, exists only in the S100 fraction (supernatant after spinning at 100,000 g). Collectively, we conclude that a significant fraction of the Dsc complex is localized to the vacuole membrane.
Figure 8.
Biochemical evidence for the vacuolar localization of the Dsc complex and a model for the regulation of vacuolar membrane proteins. (A) Subcellular fractionation of WT yeast cells. The whole-cell lysate (T) was separated into P13, P100, and S100 fractions by differential centrifugation and probed with the indicated antibodies. (B) Purified vacuole membrane fraction was compared with the whole-cell lysate (total) by Western blot analysis. Samples that contain an equal amount of Vph1 were loaded in each lane. Approximately 60% of Ubx3 was estimated to localize to the vacuole membrane. (C) Immunoprecipitation experiments from the WT vacuole membrane fraction using preimmune, Dsc2, and Dsc3 antibodies. The immunoprecipitation reaction was probed with the indicated antibodies. (D) Immunoprecipitation experiment from the UBX3-FLAG vacuole membrane fraction using the M2 Flag antibody. The immunoprecipitation reaction was probed with indicated antibodies. (E) Silver staining analysis of the eluates from Fig. 7 D. (F) Two distinct E3 ligase systems converge on the vacuole membrane to regulate different vacuolar membrane transporters via the vReD pathway.
To estimate the amount of the Dsc complex that is vacuole localized, we purified vacuoles and measured protein levels with Western blotting (Fig. 8 B). The vacuole membrane fraction lacked detectable Kex2 (Golgi protein) and Pma1 (PM protein), indicating it was highly enriched (Fig. 8 B). Assuming nearly all Vph1 is localized to the vacuole membrane, we estimated that ∼60% of the Dsc complex is localized to the vacuole membrane, as represented by the recovery of Ubx3 (Fig. 8 B).
To ensure that the intact Dsc complex exists on the vacuole membrane, we performed reciprocal immunoprecipitation experiments from the purified vacuole membrane fraction using antibodies against the Dsc complex. As shown in Fig. 8 (C and D), immunoprecipitation of one component of the Dsc complex, including Dsc2, Dsc3, and chromosomally tagged Ubx3-Flag, led to the coprecipitation of the whole Dsc complex. In contrast, Vph1, the most abundant vacuole membrane protein, did not coprecipitate with any tested component of the Dsc complex. Finally, we used silver staining to directly visualize the eluates from the Ubx3-Flag immunoprecipitation reaction. Ubx3-Flag is functional, as it supports the degradation of Cot1-GFP, similar to cells expressing untagged Ubx3 (Fig. S5). As shown in Fig. 8 E, all known components of the Dsc complex, including Tul1, Cdc48, Dsc2, and Dsc3, were coprecipitated with Ubx3-Flag stoichiometrically, demonstrating that components of the Dsc complex form a stable complex on the vacuole membrane.
Taken all together, we conclude that the Dsc complex is broadly distributed on the Golgi, endosomes, and the vacuole, with a significant fraction (∼60%) localized to the vacuole membrane to regulate the composition of vacuole membrane proteins.
Discussion
In this study, we uncovered a second E3 ligase complex, the Dsc complex, in the vacuole membrane to regulate the protein level of vacuolar membrane transporters via the vReD pathway (Fig. 8 F). The low-Zn2+ condition triggers the degradation of the influx transporter Cot1, whereas the high-Zn2+ condition triggers the degradation of the efflux channel Zrt3 (Fig. 8 F). Further analysis indicated that the Dsc complex is broadly distributed in the endomembrane trafficking pathway, including Golgi, endosomes, and the vacuole membrane. Thus, the Dsc complex and the vReD pathway play an important role in the regulation of nutrient homeostasis by regulating the abundance of the vacuolar membrane transporters.
Homologues of Tul1 exist in many eukaryotes, including fungi, plants, and unicellular eukaryotes. In addition, at least two transmembrane RING domain containing E3 ligases, RNF152 and RNF167, have been reported to localize to human lysosomes (Zhang et al., 2010; Lussier et al., 2012; Deng et al., 2015). Furthermore, homologues of Dsc2 and Ubx3 are also widely distributed in eukaryotes, including fungi, plants, and metazoans. Together, these data suggest that the regulation of lysosomal membrane transporters via a vRed-like pathway may be conserved from yeast to humans.
It is not uncommon for two ubiquitination systems to coexist on the same organelle membrane. The Doa10 and Hrd1 E3 ligase complexes are important players of the ERAD pathway (Hampton et al., 1996; Bordallo et al., 1998; Carvalho et al., 2006; Denic et al., 2006). The Doa10 complex recognizes and ubiquitinates ER proteins with misfolded cytoplasmic domains (ERAD-C substrates), whereas the Hrd1 complex ubiquitinates ER proteins with misfolded luminal or transmembrane domains (ERAD-L and ERAD-M substrates; Vembar and Brodsky, 2008; Christianson and Ye, 2014). Ubiquitinated substrates from both complexes converge on the downstream events, including recognition by the AAA ATPase, Cdc48, and retrotranslocation from the ER before being degraded by the proteasome in the cytoplasm (Ye et al., 2001; Wolf and Stolz, 2012).
Here, we demonstrated that the VAcUL-1 complex and the Dsc complex regulate different subsets of the vacuole membrane proteins in response to changes in nutrients and metal ion availability. The VAcUL-1 complex is responsible for the downregulation of Ypq1 in response to lysine starvation. On the other hand, the Dsc complex is responsible for Cot1 degradation after Zn2+ starvation. Interestingly, both Rsp5 and Tul1 contribute to the down-regulation of Zrt3, indicating there is some overlap or crosstalk between the two E3 ligase systems. It is unclear, at this point, how these two complexes recognize their substrates. Rsp5 is a cytosolic protein that is recruited to the vacuole membrane via a PY-motif containing adaptor, which in the VAcUL-1 complex is Ssh4. Ssh4 is a type I transmembrane protein, with the bulk of the protein facing the cytoplasm. The cytosolic domain of Ssh4 contains a SPRY domain that has been implicated in the protein-protein interaction by serving as an interaction scaffold (Perfetto et al., 2013). Considering the bulk of the VAcUL-1 complex is in the cytoplasm, it is possible that this complex recognizes signals/modifications on the cytoplasmic domain of its substrates. On the other hand, components of the Dsc complex are all integral membrane proteins (Stewart et al., 2011, 2012). Strikingly, components of the Dsc complex are all distant homologues to the Hrd1 complex in the ERAD-M pathway. Tul1 (similar to Hrd1) is a 7-TM RING domain–containing E3 ligase. Dsc2 (similar to Der1) contains a rhomboid pseudoprotease domain in addition to a C-terminal ubiquitin-associated domain. Dsc3 (similar to Usa1) contains two predicted transmembrane helices in addition to a N-terminal ubiquitin-like domain (Lloyd et al., 2013). Ubx3 is a homologue to Ubx2 that recruits Cdc48. This striking similarity suggests that the Dsc complex might also recognize signals in the transmembrane region. In line with this hypothesized mechanism, introducing a charged residue into the transmembrane helix of Pep12 (pep12D mutant) leads to the ubiquitination and degradation of the protein by the Dsc complex (Reggiori and Pelham, 2002; Tong et al., 2014). Future studies will be needed to address the specific sequence or structural information that is recognized by the VAcUL-1 and Dsc complexes.
After protein ubiquitination, vReD cargoes are sorted into carrier vesicles, which appear to fuse with an intermediate compartment where the cargo is sorted and internalized into intraluminal vesicles by the ESCRT machinery and eventually delivered into the vacuole lumen for degradation (Li et al., 2015). This is in direct contrast to the ERAD substrates, which are retrotranslocated into the cytoplasm and degraded by the proteasome. Thus far, no evidence has been found that the proteasome plays a direct role in vacuole membrane protein degradation. The addition of MG132 does not affect the degradation kinetics or vacuole lumen sorting of Cot1-GFP (unpublished data). Yet, Cdc48 and its adaptor proteins (Ubx3 for vReD and Ubx2 for ERAD) are critical for both pathways. In the ERAD pathway, Cdc48 has been shown to bind ubiquitinated cargoes and provide a driving force for their retrotranslocation (Ye et al., 2001, 2005). This process also requires two Cdc48 cofactors, Ufd1 and Npl4, to interact with the cargo (Ye et al., 2003). The vReD pathway, however, does not extract the vacuolar membrane protein cargo. In addition, Ufd1 and Npl4 are not required for the vReD pathway (unpublished data). Cdc48 might facilitate cargo ubiquitination by partially extracting vacuole membrane proteins to make them more accessible for the Tul1 ubiquitin ligase. Identifying the natural substrates for the Dsc complex should allow us to test this hypothesis.
Although Tul1 was discovered to be a transmembrane E3 ligase over a decade ago (Reggiori and Pelham, 2002), its cellular function has remained a mystery. No natural substrates had been identified in budding yeast. Its homologue in fission yeast, Dsc1, is critical for the proteolytic processing of the SREBP transcription factor (Stewart et al., 2011). However, no homologue of SREBP exists in the budding yeast. Using a proteomic approach, Tong et al. obtained 11 candidate substrates for Tul1, but further analysis is required to confirm if they are indeed the Tul1 substrates (Tong et al., 2014). No vacuole membrane proteins were identified by this study, which is surprising considering our data indicate the Dsc complex exists on the vacuole membrane. One possible explanation is that Tul1 functions during stress response, such as changes in the Zn2+ levels. It will be interesting to test whether other stress conditions can trigger a Tul1 dependent down-regulation of vacuole membrane proteins.
Vacuolar membrane proteins are under the constant “threat” of digestion by the luminal proteases. It seems likely that these membrane proteins will eventually be damaged, which demands a quality-control mechanism to remove the damaged membrane proteins to maintain the integrity and function of the vacuole. In addition, as the “degradation and recycling center” of the cell, the vacuole serves as an acceptor for numerous transport intermediates via endocytosis, autophagy, and biosynthetic sorting pathways. As these transport intermediates fuse with the vacuole, they deliver large quantities of exogenous membrane lipids and membrane proteins to the vacuole membrane, which will result in an increase in the size of the vacuole and they may alter the stability/identity of the vacuole membrane. The vReD pathway may protect the vacuole by rapidly recycling the exogenous lipids and proteins off the vacuole surface. Clearly, there is still much to understand about the lysosome, an essential and very dynamic organelle.
Materials and methods
Yeast strains, plasmids, media, and growth conditions
All yeast strains and plasmids used in this study are listed in Table S1. Both Difco YPD broth (reference no. 242810) and Difco Yeast Nitrogen Base w/o Amino Acids (reference no. 291920) were purchased from Thermo Fisher Scientific and prepared according to the manufacturer’s protocol. The Zn2+ minus YNB media was prepared by mixing individual components of the YNB media except ZnSO4. The detailed formula for YNB without amino acids is described in the vendor’s manual. For Cot1-GFP degradation assay, yeast cells were grown in YPD media or YNB media to mid-log phase (OD600: 0.4∼0.8) before being collected at 2,500 g for 5 min. After two washes with water, the yeast cells were immediately resuspended in YNB media without Zn2+ and incubated for an appropriate amount of time (typically, 2–6 h) before being collected for further analysis. For Zrt3*-GFP degradation assay, mid-log cells were first starved for Zn2+ for 3 h to induce the expression of Zrt3*-GFP. Then, 2 mM ZnCl2 was added to trigger the degradation of Zrt3*-GFP. Most experiments were performed at 26°C. For the temperature-sensitive mutants expressing Cot1-GFP, after yeast cells reached the mid-log phase at 26°C, the culture was shifted to 37°C for 20 min before being exchanged into YNB media without Zn2+ (preincubated at 37°C) to induce the degradation. In the case of Zrt3*-GFP degradation, after zinc starvation at 26°C for 3 h, the cells were shifted to 37°C for 20 min before the addition of ZnCl2. All later experiments were performed at 37°C.
Western blot and antibodies
Total cell lysates were prepared from seven OD600 cultures by incubating on ice for 1 h in 10% trichloroacetic acid. After two acetone wash-sonication cycles, samples were bead-beated for 5 min in 70 µl 2× urea buffer (150 mM Tris, pH 6.8, 6 M urea, and 6% SDS) and incubated 5 min at 37°C. After addition of 70 µl 2× sample buffer (150 mM Tris, pH 6.8, 2% SDS, 100 mM DTT, and bromophenol blue), samples were bead-beated for 5 min and heated for 5 min at 37°C. Samples were run on 10% polyacrylamide gels and transferred to nitrocellulose membranes. Antibodies used for blotting were G6PDH (A9521; Sigma-Aldrich), GFP (B2; Santa Cruz Biotechnology, Inc.), and Vph1 (10D7, Invitrogen). Antibodies against Dsc2, Dsc3, Ubx3, and Tul1 were provided by P. Espenshade (Johns Hopkins University, Baltimore, MD).
Subcellular fractionation
A total of 20 OD600 of mid-log yeast cells were centrifuged and resuspended in spheroplasting media (20 mM Tris-HCl, pH 7.5, 1× YNB, 1 M sorbitol, 1× amino acids, and 2% glucose) containing zymolyase to digest the cell wall. The spheroplasts were washed once and resuspended in 1 ml lysis buffer (50 mM Tris, pH 7.4, 10 mM EDTA, 200 mM sorbitol, and protease inhibitor cocktail) before being ruptured on ice with 20× strokes in a Dounce homogenizer. For subcellular fractionation assays, lysate was centrifuged for 5 min at 300 g at 4°C to remove cell debris. The supernatant was then collected into a fresh eppendorf tube and a 10 min 13,000 g spin was done at 4°C to separate the P13 from the S13 fraction. The S13 fraction was further separated into pellet (P100) and supernatant (S100) fractions by spinning at 100,000 g for 30 min at 4°C. Then, P13, P100, and S100 fractions were precipitated in 10% trichloroacetic acid. Samples were boiled in sample buffer and analyzed by Western blot.
Vacuole isolation and immunoprecipitation of the Dsc complex
The vacuole isolation procedures were modified from a published protocol (Cabrera and Ungermann, 2008). Essentially, 1 liter of yeast culture (OD600 ∼1) was harvested by spinning at 4,000 g for 10 min, resuspended in 50 ml softening buffer (100 mM Tris-HCl, pH 8.8, and 10 mM DTT) and incubated at 30°C for 15 min to “soften” the cell wall. The cells were washed once with 20 ml spheroplasting media, resuspended with 25 ml spheroplasting media containing 100 µl of 10 mg/ml Zymolyase, and incubated at RT for 30 min with gentle rocking. The spheroplasts were pelleted by spinning at 4,000 g for 10 min at 4°C and gently resuspended with 3.5 ml of 15% Ficoll solution in PS buffer (10 mM Pipes-KOH, pH 6.8, and 200 mM sorbitol, with protease inhibitors). Then, 200 µl of 0.4 mg/ml DEAE-Dextran solution in PS buffer was added to the spheroplasts and the cells were lysed by incubation on ice for 5 min, 30°C for 2 min, and back to ice. The lysate was transferred to the bottom of a SW41 ultracentrifuge tube and a discontinuous Ficoll gradient was set up by sequentially laying 2.5 ml of 8%, 2.5 ml of 4%, and 2.5 ml of 0% Ficoll solutions on the top. After a spin of 110,000 g for 90 min at 4°C, intact vacuoles were collected at the interface between 0% and 4% Ficoll.
Purified vacuole was resuspended in 2 ml of 50 mM Hepes-KOH, pH 6.8, and ruptured on ice by 10 strokes in a Dounce homogenizer. The membrane fraction was collected by a 10-min 13,000-g spin at 4°C before being resuspended in 100 µl immunoprecipitation buffer (50 mM Hepes-KOH, pH 6.8, 150 mM KOAc, MgOAc, 1 mM CaCl2, and 15% glycerol) supplemented with protease inhibitors. Then, resuspended membrane was dissolved in 5 ml immunoprecipitation buffer containing 1% Triton X-100 at 4°C for 20 min, with gentle rocking. Insoluble material was removed by spinning at 100,000 g for 20 min. The resulting lysate was incubated with either 30 µl M2 anti-FLAG resin (A2220; Sigma-Aldrich) or antibodies against Dsc2 and Dsc3 at 4°C for 2 h, with gentle rocking. The resin was then washed six times with 0.1% Triton X-100 in immunoprecipitation buffer, and bound proteins were eluted either using 200 µl 3×FLAG peptide (100 µg/ml, dissolved in immunoprecipitation buffer containing 0.1% Triton X-100; F4799; Sigma-Aldrich), or 100 µl sample buffer. The resulting eluates were analyzed by either Western blotting or silver staining.
Immunoprecipitation and detection of cargo ubiquitination
To stabilize ubiquitinated cargoes, the deubiquitinating enzyme Doa4 was deleted. Transient overexpression of Myc-Ub, which was under the control of the GAL1 promoter, was induced by addition of 100 nM β-estradiol (Howell et al., 2012) 1 h before the cells were treated with excess Zn2+ (2 mM, for Zrt3*-GFP) or no Zn2+ (for Cot1-GFP) conditions to trigger cargo ubiquitination. After 2 h of zinc treatment in the presence of 100 nM β-estradiol, ∼50 OD600 cells were collected for the immunoprecipitation experiment.
The immunoprecipitation assay was adapted from Breslow et al. (2010), with some modifications. Essentially, yeast cells were resuspended in 500 µl immunoprecipitation buffer with 0.1% digitonin, supplemented with protease inhibitors and 20 mM NEM (n-ethylmalemide). Whole-cell lysates were prepared by bead beating at 4°C for 10 min, followed by addition of 500 µl of 1.9% digitonin in immunoprecipitation buffer. Membranes were solubilized by nutating lysates at 4°C for 50 min. The insoluble material was removed by spinning at 100,000 g for 20 min. The resulting lysate was incubated with 25 µl GFP-TRAP resin (Chromo Tek) at 4°C for 2 h. The GFP-TRAP resin was then washed four times with 0.1% digitonin in immunoprecipitation buffer, and bound proteins were eluted by incubating resin with sample buffer at 37°C for 5 min. The eluates were then analyzed by SDS-PAGE and probed with Myc antibody.
Microscopy and image processing
Microscopy was performed with a DeltaVision RT system (Applied Precision), equipped with a microscope (IX71; Olympus), a Photometrics CoolSNAP HQ Camera, a 100× objective (1.40 NA), and a DeltaVision RT Standard Filter Set (FITC for GFP/pHluorin and RD-TR-Cy3 for mCherry). Image acquisition, deconvolution, and maximum projection analysis were performed with the program Softworx. Yeast cells were briefly washed with water and immediately imaged in water at RT. The image cropping and adjustment of image intensity were performed using the ImageJ software (National Institutes of Health).
Online supplemental material
Fig. S1 shows the degradation of both Cot1-GFP and Zrt3*-GFP depends on vacuolar proteases. Fig. S2 shows the ESCRT machinery, but not autophagy, is required for the degradation of Zrt3*-GFP. Fig. S3 shows both Cot1-GFP and Zrt3*-GFP are polyubiquitinated during their degradation. Fig. S4 shows a fraction of the Dsc complex localizes to Golgi and endosomes. Fig. S5 shows Ubx3-Flag is functional. Table S1 lists all strains and plasmids used in this study. Table S2 lists all tested vacuolar membrane transporters. Table S3 lists all tested E3 ubiquitin ligases. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505062/DC1.
Supplementary Material
Acknowledgments
We thank members of the Emr laboratory, including Mike Henne, Steven Tang, Lu Zhu, Matt Baile, Jeff Jorgensen, and Ya-Shan Chuang, for helpful discussion and comments about the manuscript. We are also grateful to Chris Fromme for helpful discussion and critical reading of the manuscript. We are indebted to Peter Espenshade for generously sharing antibodies against components of the Dsc complex. We are also thankful to Erich Schwartz for advice regarding the bioinformatics analysis.
This work was supported by a Cornell University Research Grant (S.D. Emr).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Dsc
- defective for SREBP cleavage
- ERAD
- ER-associated degradation
- ESCRT
- endosomal sorting complexes required for transport
- SREBP
- sterol regulatory element–binding protein
- VAcUL-1
- vacuole-anchored ubiquitin ligase complex 1
- vReD
- vacuole membrane recycling and degradation
- WT
- wild type
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., and Lipman D.J.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas C.E., and Merchant S.S.. 2014. Lysosome-related organelles as mediators of metal homeostasis. J. Biol. Chem. 289:28129–28136. 10.1074/jbc.R114.592618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J., Plemper R.K., Finger A., and Wolf D.H.. 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 9:209–222. 10.1091/mbc.9.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., and Weissman J.S.. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463:1048–1053. 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., and Ungermann C.. 2008. Purification and in vitro analysis of yeast vacuoles. Methods Enzymol. 451:177–196. 10.1016/S0076-6879(08)03213-8 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., and Rapoport T.A.. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373. 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Stanley A.M., and Rapoport T.A.. 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 143:579–591. 10.1016/j.cell.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimienti F. 2013. Zinc, pancreatic islet cell function and diabetes: new insights into an old story. Nutr. Res. Rev. 26:1–11. 10.1017/S0954422412000212 [DOI] [PubMed] [Google Scholar]
- Christianson J.C., and Ye Y.. 2014. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat. Struct. Mol. Biol. 21:325–335. 10.1038/nsmb.2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin D.S., McMaster J.A., Culbertson M.R., and Kung C.. 1992. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:3678–3688. 10.1128/MCB.12.9.3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Domenico I., McVey Ward D., and Kaplan J.. 2008. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat. Rev. Mol. Cell Biol. 9:72–81. 10.1038/nrm2295 [DOI] [PubMed] [Google Scholar]
- Deng L., Jiang C., Chen L., Jin J., Wei J., Zhao L., Chen M., Pan W., Xu Y., Chu H., et al. 2015. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell. 58:804–818. 10.1016/j.molcel.2015.03.033 [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E.M., and Weissman J.S.. 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 126:349–359. 10.1016/j.cell.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Efeyan A., Comb W.C., and Sabatini D.M.. 2015. Nutrient-sensing mechanisms and pathways. Nature. 517:302–310. 10.1038/nature14190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada T., Yamasaki S., Nishida K., Murakami M., and Hirano T.. 2011. Zinc homeostasis and signaling in health and diseases: Zinc signaling. J. Biol. Inorg. Chem. 16:1123–1134. 10.1007/s00775-011-0797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E., Piedrafita L., Aldea M., and Herrero E.. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 13:837–848. [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Grissinger M. 2011. A fatal zinc overdose in a neonate: confusion of micrograms with milligrams. P&T. 36:393–409. [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y., Gardner R.G., and Rine J.. 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 7:2029–2044. 10.1091/mbc.7.12.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A.S., Jin M., Wu C.F., Zyla T.R., Elston T.C., and Lew D.J.. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 149:322–333. 10.1016/j.cell.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jeong J., and Eide D.J.. 2013. The SLC39 family of zinc transporters. Mol. Aspects Med. 34:612–619. 10.1016/j.mam.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono A., Nishizawa M., Teranishi Y., Murata K., and Kimura A.. 1989. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 219:161–167. 10.1007/BF00261172 [DOI] [PubMed] [Google Scholar]
- Li M., Rong Y., Chuang Y.S., Peng D., and Emr S.D.. 2015. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol. Cell. 57:467–478. 10.1016/j.molcel.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Lichten L.A., and Cousins R.J.. 2009. Mammalian zinc transporters: nutritional and physiologic regulation. Annu. Rev. Nutr. 29:153–176. 10.1146/annurev-nutr-033009-083312 [DOI] [PubMed] [Google Scholar]
- Lloyd S.J., Raychaudhuri S., and Espenshade P.J.. 2013. Subunit architecture of the Golgi Dsc E3 ligase required for sterol regulatory element-binding protein (SREBP) cleavage in fission yeast. J. Biol. Chem. 288:21043–21054. 10.1074/jbc.M113.468215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M.P., Herring B.E., Nasu-Nishimura Y., Neutzner A., Karbowski M., Youle R.J., Nicoll R.A., and Roche K.W.. 2012. Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc. Natl. Acad. Sci. USA. 109:19426–19431. 10.1073/pnas.1217477109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid C.W., Gaither L.A., and Eide D.. 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19:2845–2855. 10.1093/emboj/19.12.2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfetto L., Gherardini P.F., Davey N.E., Diella F., Helmer-Citterich M., and Cesareni G.. 2013. Exploring the diversity of SPRY/B30.2-mediated interactions. Trends Biochem. Sci. 38:38–46. 10.1016/j.tibs.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Rebsamen M., Pochini L., Stasyk T., de Araújo M.E., Galluccio M., Kandasamy R.K., Snijder B., Fauster A., Rudashevskaya E.L., Bruckner M., et al. 2015. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 519:477–481. 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., and Pelham H.R.. 2002. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4:117–123. 10.1038/ncb743 [DOI] [PubMed] [Google Scholar]
- Riffle M., and Davis T.N.. 2010. The Yeast Resource Center Public Image Repository: A large database of fluorescence microscopy images. BMC Bioinformatics. 11:263 10.1186/1471-2105-11-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T., Nakamura K., Manabe K., Tone J., Sato Y., Murao N., Kawano-Kawada M., and Kakinuma Y.. 2014. Loss of ATP-dependent lysine uptake in the vacuolar membrane vesicles of Saccharomyces cerevisiae ypq1∆ mutant. Biosci. Biotechnol. Biochem. 78:1199–1202. 10.1080/09168451.2014.918489 [DOI] [PubMed] [Google Scholar]
- Stewart E.V., Nwosu C.C., Tong Z., Roguev A., Cummins T.D., Kim D.U., Hayles J., Park H.O., Hoe K.L., Powell D.W., et al. 2011. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Mol. Cell. 42:160–171. 10.1016/j.molcel.2011.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E.V., Lloyd S.J., Burg J.S., Nwosu C.C., Lintner R.E., Daza R., Russ C., Ponchner K., Nusbaum C., and Espenshade P.J.. 2012. Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J. Biol. Chem. 287:672–681. 10.1074/jbc.M111.317370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., Kim M.S., Pandey A., and Espenshade P.J.. 2014. Identification of candidate substrates for the Golgi Tul1 E3 ligase using quantitative diGly proteomics in yeast. Mol. Cell. Proteomics. 13:2871–2882. 10.1074/mcp.M114.040774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J., and Pelham H.. 2005. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 24:2524–2532. 10.1038/sj.emboj.7600724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S.S., and Brodsky J.L.. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957. 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C., Dean G.H., Griffiths J.S., Kirchsteiger K., Hwang Y.T., Gillett A., Dow G., Western T.L., Estelle M., and Haughn G.W.. 2013. Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell. 25:944–959. 10.1105/tpc.112.107888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tsun Z.Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., Yuan E.D., Jones T.D., Chantranupong L., Comb W., et al. 2015. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 347:188–194. 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold E., Gandhi T., Permentier H.P., Breitling R., Poolman B., and Slotboom D.J.. 2009. The yeast vacuolar membrane proteome. Mol. Cell. Proteomics. 8:380–392. 10.1074/mcp.M800372-MCP200 [DOI] [PubMed] [Google Scholar]
- Wolf D.H., and Stolz A.. 2012. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta. 1823:117–124. 10.1016/j.bbamcr.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., and Rapoport T.A.. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656. 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., and Rapoport T.A.. 2003. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162:71–84. 10.1083/jcb.200302169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., and Rapoport T.A.. 2005. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 102:14132–14138. 10.1073/pnas.0505006102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wu W., Wu Y., Zheng J., Suo T., Tang H., and Tang J.. 2010. RNF152, a novel lysosome localized E3 ligase with pro-apoptotic activities. Protein Cell. 1:656–663. 10.1007/s13238-010-0083-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., and Eide D.. 1996a The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA. 93:2454–2458. 10.1073/pnas.93.6.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., and Eide D.. 1996b The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271:23203–23210. 10.1074/jbc.271.38.23203 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.