The ability of the conserved ATPase TRIP13PCH-2 to disassemble a Mad2-containing complex is critical to promote the spindle checkpoint response by contributing to the robust localization of Mad2 to unattached kinetochores.
Abstract
The spindle checkpoint acts during cell division to prevent aneuploidy, a hallmark of cancer. During checkpoint activation, Mad1 recruits Mad2 to kinetochores to generate a signal that delays anaphase onset. Yet, whether additional factors contribute to Mad2’s kinetochore localization remains unclear. Here, we report that the conserved AAA+ ATPase TRIP13PCH-2 localizes to unattached kinetochores and is required for spindle checkpoint activation in Caenorhabditis elegans. pch-2 mutants effectively localized Mad1 to unattached kinetochores, but Mad2 recruitment was significantly reduced. Furthermore, we show that the C. elegans orthologue of the Mad2 inhibitor p31(comet)CMT-1 interacts with TRIP13PCH-2 and is required for its localization to unattached kinetochores. These factors also genetically interact, as loss of p31(comet)CMT-1 partially suppressed the requirement for TRIP13PCH-2 in Mad2 localization and spindle checkpoint signaling. These data support a model in which the ability of TRIP13PCH-2 to disassemble a p31(comet)/Mad2 complex, which has been well characterized in the context of checkpoint silencing, is also critical for spindle checkpoint activation.
Introduction
Accurate chromosome segregation is essential to avoid aneuploidy, a hallmark of cancer (Holland and Cleveland, 2012). During mitosis, replicated chromosomes must attach to microtubules emanating from opposite spindle poles (referred to as bi-orientation) so that each daughter cell receives an equivalent complement of chromosomes. To ensure the fidelity of this process, cells use a molecular safety mechanism called the spindle checkpoint. This checkpoint monitors chromosome attachment to the mitotic spindle and delays anaphase until all chromosomes are bi-oriented, allowing time for error correction (London and Biggins, 2014).
Mitotic chromosome segregation is choreographed by kinetochores, macromolecular protein complexes that bridge centromeric DNA with the mitotic spindle and serve as signaling platforms for the spindle checkpoint (Cheeseman and Desai, 2008; Foley and Kapoor, 2013). When sister chromatids fail to bi-orient, spindle checkpoint components including Bub1, Bub3, Mad1, and Mad2 are hierarchically recruited to kinetochores. Kinetochores then catalyze the formation of the soluble mitotic checkpoint complex (MCC) (De Antoni et al., 2005), which in turn inhibits the anaphase-promoting complex, preventing anaphase (Sudakin et al., 2001). Mad1 plays multiple roles in checkpoint activation: It recruits Mad2 to unattached kinetochores (Chen et al., 1996; Ballister et al., 2014; Kuijt et al., 2014) and likely promotes Mad2 activation (Ballister et al., 2014; Heinrich et al., 2014; Kruse et al., 2014), although this second role is less well understood. Kinetochore localization of the Mad1/Mad2 complex, however, appears to be the determining step in checkpoint activation: Artificial tethering of Mad1 to kinetochores is sufficient to both recruit Mad2 and to constitutively activate the checkpoint (Maldonado and Kapoor, 2011; Ballister et al., 2014; Kuijt et al., 2014). Furthermore, the amount of Mad2 localized to kinetochores correlates with checkpoint signal strength (Collin et al., 2013; Heinrich et al., 2013). Mad2 exists in two unique conformational states: a free "open" form (O-Mad2) and a bound "closed" form (C-Mad2) (Luo et al., 2002, 2004; Sironi et al., 2002). Kinetochore bound C-Mad2 acts as a template to activate soluble O-Mad2, converting it to C-Mad2, a significantly more robust anaphase-promoting complex inhibitor (De Antoni et al., 2005). However, whether additional mechanisms regulate Mad2 dimerization at the kinetochore, and therefore the strength of the spindle checkpoint response, remains unknown.
TRIP13 is a highly conserved AAA+ ATPase that contributes to homologue pairing, synapsis, and recombination during meiosis (Wu and Burgess, 2006; Joshi et al., 2009, 2015; Wojtasz et al., 2009; Zanders and Alani, 2009; Roig et al., 2010; Zanders et al., 2011; Chen et al., 2014; Deshong et al., 2014). A large class of AAA+ ATPases is thought to remodel or disassemble protein complexes via ATP hydrolysis (Dougan et al., 2002). Specifically, TRIP13 is thought to remodel proteins containing a HORMA domain, a common structural motif found among checkpoint proteins, including Hop1, Rev7, and Mad2 (Aravind and Koonin, 1998; Börner et al., 2008; Chen et al., 2014; Vader, 2015; Ye et al., 2015). Indeed, budding yeast TRIP13 was shown to disassemble the meiotic axis component Hop1 from a DNA template in vitro (Chen et al., 2014).
Recent studies have established an additional role for TRIP13 in regulating mitosis. These experiments have revealed that TRIP13 collaborates with the spindle checkpoint silencing protein and Mad2 inhibitor, p31(comet), to disassemble the MCC and promote anaphase (Teichner et al., 2011; Tipton et al., 2012; Eytan et al., 2014; Wang et al., 2014). To render MCC disassembly irreversible, TRIP13’s ATPase activity converts C-Mad2 to O-Mad2. However, it can accomplish this only in the presence of p31(comet) (Ye et al., 2015), indicating that although C-Mad2 is the substrate for TRIP13, p31(comet) is a necessary adapter for this reaction. Interestingly, the Caenorhabditis elegans version of TRIP13, PCH-2, shows the same requirement for the presence of both proteins in stimulating its ATPase activity, suggesting a similar role in mitosis (Ye et al., 2015).
Here, we explore the hypothesis that in addition to checkpoint silencing, TRIP13 and p31(comet) contribute to spindle checkpoint activation. Consistent with this idea, both proteins localize to kinetochores in prometaphase (Hagan et al., 2011; Tipton et al., 2012) and TRIP13 colocalizes with Mad2 in the presence of spindle poisons (Tipton et al., 2012). Because p31(comet) can outcompete O-Mad2 for C-Mad2/Mad1 binding in vitro (Vink et al., 2006), one model proposes that p31(comet) may negatively regulate Mad2 dimerization and activation at the kinetochore and this activity must be antagonized at kinetochores during spindle checkpoint activation (Musacchio and Salmon, 2007; Lara-Gonzalez et al., 2012). Given that p31(comet) is bound to C-Mad2 throughout the cell cycle (Date et al., 2014), another possibility is that O-Mad2 may need to be released from p31(comet) to provide a substantial pool of O-Mad2 for a robust spindle checkpoint response (Ye et al., 2015). Given the well-characterized interaction between p31(comet) and TRIP13 during mitotic exit (Tipton et al., 2012; Eytan et al., 2014; Wang et al., 2014) and the biochemical ability of TRIP13 to convert C-Mad2 to O-Mad2, (Ye et al., 2015) we reasoned that TRIP13 might contribute to these regulatory mechanisms during checkpoint activation. We set out to test this possibility in C. elegans.
We report the first genetic analysis of the functions of TRIP13 (PCH-2 in C. elegans) and p31(comet) (CMT-1) during spindle checkpoint activation. Unlike their mammalian counterparts, loss of TRIP13PCH-2 or p31(comet)CMT-1 has no effect on mitotic timing during a normal cell cycle. However, like its human orthologue, TRIP13PCH-2 localizes to unattached kinetochores during spindle checkpoint activation. Furthermore, TRIP13PCH-2 is required for spindle checkpoint activation in two cell types in C. elegans: germline mitotic cells and cells undergoing embryonic divisions. We demonstrate that the function of TRIP13PCH-2 in the checkpoint is to promote Mad2 (MDF-2/MAD-2) localization to kinetochores, as Mad2 levels are markedly reduced at unattached kinetochores in pch-2 mutant embryos. The localization of Mad1, Bub1, and Bub3 (MAD-1/MDF-1, BUB-1, and BUB-3, respectively) are unaffected by mutation of pch-2, indicating that the role for TRIP13PCH-2 in the checkpoint is limited to regulating Mad2. TRIP13PCH-2 modulates Mad2 via p31(comet)CMT-1. TRIP13PCH-2 and p31(comet)CMT-1 physically interact via yeast two-hybrid, and both p31(comet)CMT-1 and Mad2 are required for TRIP13PCH-2 localization to unattached kinetochores. Finally, our data show that TRIP13PCH-2 genetically antagonizes p31(comet)CMT-1 during checkpoint activation: Mutation of cmt-1 partially suppresses the defects in both checkpoint signaling and Mad2 recruitment observed in pch-2 mutants. Collectively, these data suggest a model in which TRIP13PCH-2 regulates spindle checkpoint activation by disassembling a p31(comet)CMT-1/Mad2 complex, promoting Mad2 localization to kinetochores and activation of the checkpoint.
Results
PCH-2 is required for spindle checkpoint activation
First, we tested whether PCH-2 regulates the duration of mitosis in the mitotic region of the C. elegans germline, as has been shown in mammalian cells (Wang et al., 2014). We measured the mitotic index in this region by assaying the number of nuclei positive for phosphorylation of histone H3 serine 10 (phospho-H3S10; Fig. 1 A) in wild type, pch-2 mutants, and mad-1 mutants. A null allele of pch-2, pch-2(tm1458) (Bhalla and Dernburg, 2005), and a hypomorphic allele of mad-1, mdf-1(av19) (Stein et al., 2007), were used for all analyses. The mad-1(av19) allele contains a point mutation in the MAD-2 binding motif that specifically affects MAD-1’s checkpoint function (Stein et al., 2007; Moyle et al., 2014). To ensure that our analysis was limited to mitotic cells, we also stained germlines with an antibody against phosphorylated SUN-1, which delineates meiotic entry (phospho-SUN-1-S8; Fig. 1 A; Penkner et al., 2009; Burger et al., 2013). We did not detect an increase in the mitotic index of pch-2 mutant germlines as compared with wild-type or mad-1 mutants (Fig. 1 B), suggesting that germline mitotic timing is not significantly altered by deletion of pch-2.
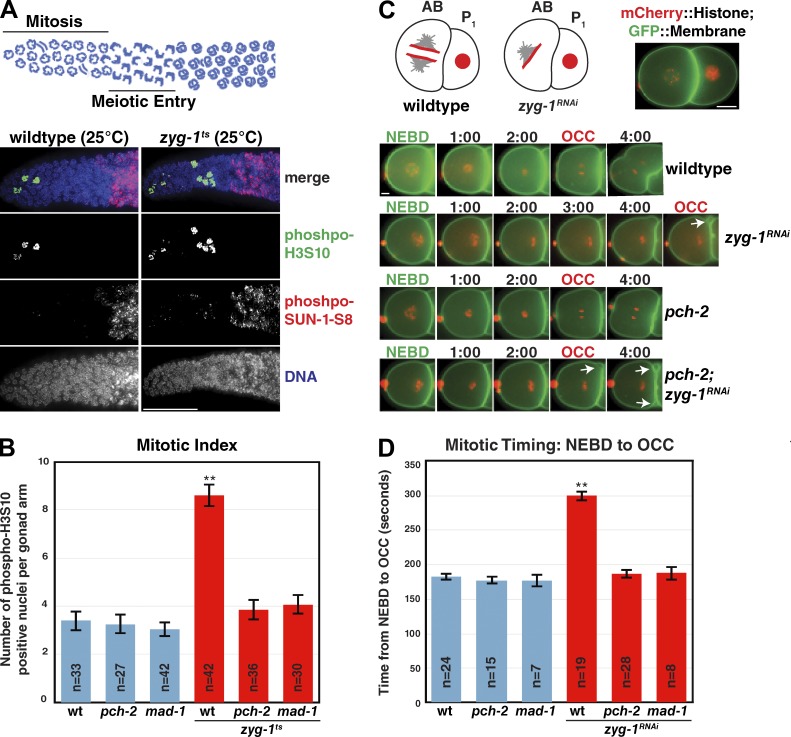
Figure 1.
PCH-2 is required for the spindle checkpoint in C. elegans. (A, top) Schematic of the distal section of the C. elegans germline. (A, bottom) Activation of the spindle checkpoint by shifting zyg-1ts worms to the nonpermissive temperature increases the mitotic index (number of phosph-H3S10–positive nuclei). Representative images of control and zyg-1ts germlines stained with antibodies recognizing phospho-H3S10 and phospho-SUN-1-S8 as well as with DAPI. Bar, 20 µm. (B) Mutation of pch-2 does not affect the mitotic index of germline mitotic nuclei unless zyg-1 is inactivated (zyg-1ts). (C, top) Schematic and image of the two-cell C. elegans embryo. Bar, 10 µm. (C, bottom) Selected frames from wild-type, pch-2, zyg-1RNAi, and pch-2;zyg-1RNAi movies are shown, denoting NEBD and OCC. Arrow indicates persistent membrane blebs between AB and P1. Bar, 5 µm. (D) Mutation of pch-2 has no effect on mitotic timing during an unperturbed mitosis but reduces mitotic timing to wild-type timing when zyg-1 is knocked down by RNAi (zyg-1RNAi). Error bars in all graphs represent SEM. **, P < 0.0001.
Next, we evaluated whether PCH-2 was required for the spindle checkpoint in the mitotic region of the germline. The checkpoint can be activated in this region using the temperature-sensitive allele zyg-1(b1), referred to here as zyg-1ts. ZYG-1 is an essential regulator of centrosome duplication in C. elegans and inactivation of zyg-1ts at the nonpermissive temperature creates monopolar spindles (O’Connell et al., 2001). This defect in spindle formation delays, but doesn’t permanently arrest, mitosis in germline nuclei and is dependent on the spindle checkpoint (Stevens et al., 2013). As a result, the mitotic index of zyg-1ts worms shifted to the nonpermissive temperature for 24 h was significantly increased compared with wild-type worms (Fig. 1, A and B). As expected, this increase was dependent on MAD-1 (Fig. 1 B and Fig. S1 A). Mutation of pch-2 also decreased the mitotic index to wild-type levels in zyg-1ts germlines, mirroring the mad-1 mutant phenotype (Fig. 1 B and Fig. S1 A). Thus, PCH-2 is required for the mitotic delay induced by the spindle checkpoint in the C. elegans germline.
Given that PCH-2 function is well characterized in the germline (Deshong et al., 2014), we were curious whether its checkpoint function is conserved in other cellular contexts. To investigate this possibility, we used the two-cell embryo, as the spindle checkpoint has been well characterized during C. elegans embryogenesis (Encalada et al., 2005; Essex et al., 2009; Moyle et al., 2014). The egg shell renders the embryo largely impenetrable to drug treatment (Carvalho et al., 2011) and spindle checkpoint activation does not appear to occur during normal embryonic divisions (Essex et al., 2009). Thus, we again relied on genetic perturbations to activate the spindle checkpoint. We generated a feeding RNAi vector that inactivated the zyg-1 gene. We fed worms bacteria expressing this vector for 24 h and verified that monopolar spindles were present in the two-cell embryo (Fig. S1 B), consistent with previous analysis (Essex et al., 2009).
To analyze mitotic timing during embryogenesis, we took advantage of an assay developed by Essex et al. (2009): DNA was visualized with an mCherry-tagged version of histone H2B (mCh::H2B) and the plasma membrane with a GFP-tagged plasma membrane marker (GFP::PH). We measured mitotic timing from nuclear envelope breakdown (NEBD), defined as a diffusion of mCh::H2B from the nucleoplasm, to the onset of cortical contractility (OCC), defined by a change in conformation of the plasma membrane from circular to rectangular (Fig. 1 C). In embryos with monopolar spindles induced by zyg-1 RNAi, OCC is defined as the formation of a persistent membrane bleb (or blebs) between the anterior (AB) and posterior (P1) cells (Fig. 1 C, arrows). OCC is concomitant with mitotic exit and is used as a marker for live microscopy (Canman et al., 2000). All further mitotic timing and localization analyses were performed in the AB cell, which enters mitosis before the P1 cell (Bao et al., 2008).
We first tested whether PCH-2 regulates mitotic timing in the two-cell embryo. We found that an unperturbed wild-type mitosis lasted a mean of 183 s (Fig. 1 D and Video 1). Mean mitotic timing in mad-1 mutants (176 s) and pch-2 mutants (177 s; Video 2) was no different than wild type (Fig. 1 D), consistent with our analysis of mitotic index in germline mitotic nuclei (Fig. 1 B). Activation of the spindle checkpoint via zyg-1 RNAi (zyg-1RNAi) caused a statistically significant delay in mean mitotic timing in otherwise wild-type embryos (300 s; P < 0.0001; Video 3 and Fig. 1 D). However, when zyg-1 was knocked down by RNAi in mad-1 or pch-2 mutant embryos, we did not observe a delay in mitotic timing (mean of 188 s and 186 s, respectively; P < 0.0001; Video 4 and Fig. 1 D). Collectively, these data indicate that PCH-2 does not regulate mitotic timing in the mitotic germline or in the developing embryo. Instead, PCH-2 is required for spindle checkpoint activation in two mitotic cell types: the developing embryo and the mitotic germline.
PCH-2 is required for robust accumulation of MAD-2 at unattached kinetochores
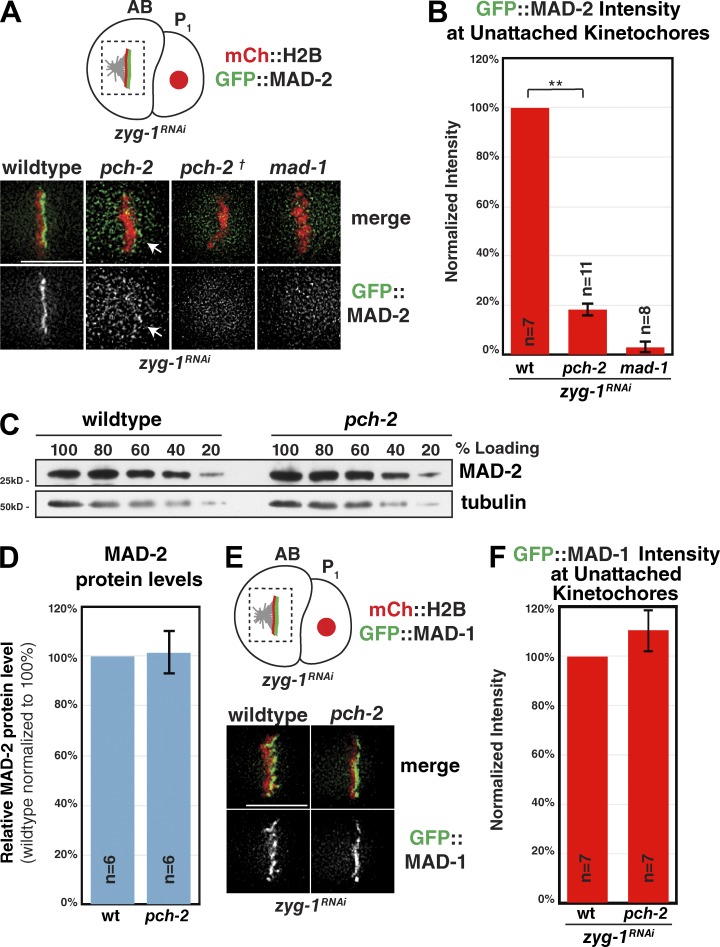
We further investigated the loss of checkpoint function in pch-2 mutants. Given the evidence that PCH-2 regulates HORMA-domain containing proteins (Börner et al., 2008; Chen et al., 2014; Deshong et al., 2014; Ye et al., 2015), we analyzed the localization of the HORMA-domain protein MAD-2. C. elegans chromosomes are holocentric and localize kinetochore proteins and checkpoint components along their entire lengths (Oegema et al., 2001; Essex et al., 2009). MAD-2 only localizes to kinetochores during checkpoint activation and specifically localizes to the unattached side of the pseudo-metaphase plate (Essex et al., 2009; Fig. 2 A). As expected, GFP::MAD-2 showed robust localization to unattached kinetochores when the checkpoint was activated in embryos via zyg-1 RNAi (Fig. 2 A). In pch-2 mutants, we observed little (pch-2) or no (pch-2†) GFP::MAD-2 localized to unattached kinetochores (Fig. 2 A). We quantified this defect and found that kinetochore localized GFP::MAD-2 signal was reduced by a mean of 82% in pch-2 mutants (Fig. 2 B). In contrast, GFP::MAD-2 kinetochore signal was reduced by a mean of 97% in mad-1 mutant embryos (Fig. 2, A and B), indicating that the genetic lesion in mad-1(av19) is sufficient to abolish MAD-2 kinetochore recruitment. Therefore, PCH-2 is required for full recruitment of MAD-2 at unattached kinetochores. However, MAD-2 signal was not completely ablated as in mad-1 mutants.
Figure 2.
PCH-2 is required for robust GFP::MAD-2 localization to unattached kinetochores. (A, top) Schematic showing the localization of GFP::MAD-2 to unattached kinetochores after checkpoint activation. Unattached kinetochores are present on the side of the pseudo-metaphase plate lacking a centrosome. Dashed box indicates area shown in images (A, bottom) GFP::MAD-2 signal after zyg-1 RNAi seen in control worms is nearly absent (pch-2) or completely absent (pch-2†) in pch-2 mutants. Arrows indicate residual GFP::MAD-2 localization. (B) Quantification of kinetochore bound GFP::MAD-2 shows that signal was reduced by a mean of 82% in pch-2 mutants and a mean of 97% in mad-1 mutants. (C) MAD-2 protein levels are unaffected in pch-2 mutants. Whole worm lysates were first normalized for protein concentration and then serial dilutions were analyzed via immunoblot with an anti–MAD-2 antibody and an anti-α-tubulin antibody serving as a loading control. (D) Quantification of MAD-2 protein level in pch-2 mutants across multiple immunoblots indicates that MAD-2 protein level is 102% of wild type. (E) GFP::MAD-1 properly localizes to unattached kinetochores in pch-2 mutants (F) Quantification of GFP::MAD-1 at unattached kinetochores in wild-type and pch-2 mutants. Error bars in all graphs represent SEM. Bars, 5 µm. **, P < 0.0001.
We considered the possibility that PCH-2 may support stability of the MAD-2 protein. To test this, we analyzed MAD-2 protein levels by immunoblotting serial dilutions of whole worm lysates. We reproducibly saw no defect in the level of MAD-2 protein in pch-2 mutants as compared with wild type (Fig. 2 C). We quantified MAD-2 protein levels in pch-2 mutants using multiple immunoblots and found MAD-2 protein levels to be essentially identical to wild type (102% of wild type; Fig. 2 D), indicating that the loss of MAD-2 kinetochore localization in pch-2 mutants is not simply a secondary consequence of decreased protein level.
Loss of MAD-2 at kinetochores in pch-2 mutants could be a result of direct regulation of MAD-2 by PCH-2, or an indirect consequence of failed kinetochore assembly or the failure to recruit other checkpoint components. We explored these latter hypotheses. KNL-1 is a member of the outer kinetochore KMN network, which includes the Knl1, Mis12, and Ndc80 complexes and is responsible for kinetochore–microtubule binding as well as the loading of checkpoint components (Desai et al., 2003; Cheeseman et al., 2006; London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012). We analyzed the localization of KNL-1::GFP in pch-2 mutants and found that KNL-1 loaded properly onto mitotic chromosomes, indicating that pch-2 mutants do not have significant defects in kinetochore assembly (Fig. S2 A). Furthermore, pch-2 mutants are fully viable (Deshong et al., 2014), indicating that gross chromosome segregation defects are unlikely. MAD-1 is the receptor for MAD-2 at kinetochores (Chen et al., 1996, 1998). We analyzed GFP::MAD-1 localization to unattached kinetochores in pch-2 mutants treated with zyg-1 RNAi (Fig. 2 E). Quantification of GFP::MAD-1 signal at unattached kinetochores showed that pch-2 mutants localize similar amounts of GFP::MAD-1 as compared with wild type (110% of wild type; Fig. 2 F). This indicates that the loss of MAD-2 at kinetochores in pch-2 mutants is not due to a failure to localize its receptor. Similarly, localization of the checkpoint components BUB-1::GFP and GFP::BUB-3 were unaffected by deletion of pch-2 (Fig. S2 B). Together, these data demonstrate that PCH-2 is required for robust accumulation of MAD-2 at kinetochores during the spindle checkpoint response and strongly suggest that PCH-2 directly regulates MAD-2 or a MAD-2–containing protein complex during checkpoint activation.
PCH-2 localizes to unattached kinetochores during spindle checkpoint activation
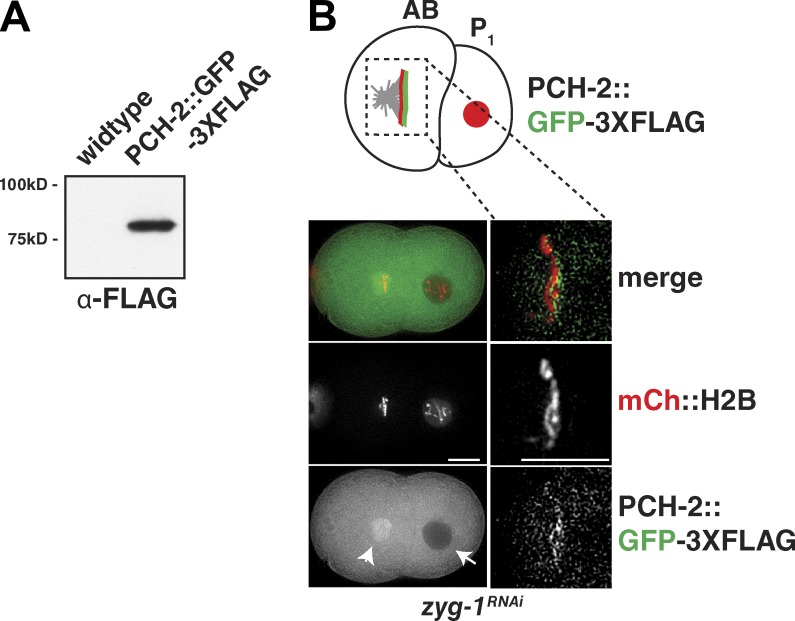
We previously showed that PCH-2 localizes to chromosomes during meiotic prophase (Deshong et al., 2014). However, its expression and localization during mitosis have not been explored in C. elegans. To analyze PCH-2 localization in live embryos, we inserted a C-terminal gfp-3xflag tag at the endogenous pch-2 locus using CRISPR/Cas9 genome editing (Dickinson et al., 2013; Paix et al., 2014). We verified insertion of the tag via immunoblot (Fig. 3 A) and then tested whether embryos expressing PCH-2::GFP-3XFLAG were competent for spindle checkpoint activation using chromosome decondensation (DCON) as a marker of mitotic exit (Fig. S3 A). Mitotic timing in control embryos averaged 281 s and zyg-1 RNAi produced a significant delay (mean, 440 s; P < 0.0001). zyg-1 RNAi also significantly delayed mitosis in embryos expressing PCH-2::GFP-3XFLAG from an mean of 265 s to 420 s (P < 0.0001). Thus, embryos expressing PCH-2::GFP-3XFLAG are competent for spindle checkpoint activation.
Figure 3.
PCH-2::GFP-3XFLAG localizes to unattached kinetochores during spindle checkpoint activation. (A) An immunoblot of PCH-2::GFP-3XFLAG worms shows that the full-length tagged protein is expressed. (B, left) PCH-2:::GFP-3XFLAG is expressed in the mitotic embryo. The arrowhead (left) indicates enrichment of PCH-2::GFP-3XFLAG after NEBD in the AB cell, whereas the single arrow (right) indicates PCH-2::GFP-3XFLAG exclusion from the nucleoplasm before mitotic entry in the P1 cell. Bar, 10 µm. (B, right) Checkpoint activation via RNAi of zyg-1 localizes PCH-2::GFP-3XFLAG to unattached kinetochores. Bar, 5 µm.
Next, we monitored PCH-2::GFP-3XFLAG localization using live microscopy. In embryos exposed to zyg-1 RNAi, PCH-2::GFP-3XFLAG was excluded from nuclei before NEBD (P1 cell; Fig. 3 B, arrow) and then became enriched in the “cloud” surrounding mitotic chromatin after NEBD (AB cell; Fig. 3 B, arrowhead). This localization is identical to that of MAD-2, which is excluded from the nucleoplasm until NEBD and then becomes enriched around chromatin (Essex et al., 2009). We detected a similar localization pattern in wild-type embryos (unpublished data). When chromosomes formed a pseudo-metaphase plate in zyg-1RNAi embryos, PCH-2::GFP-3XFLAG localized to unattached kinetochores (Fig. 3 B, right panels), mirroring the localization of MAD-2 (Fig. 2 A) and MAD-1 (Fig. 2 E). Moreover, this recruitment to unattached kinetochores exhibited similar timing as MAD-1 and MAD-2, occurring a mean of 60 s after NEBD (unpublished data). This is unique from other checkpoint components, such as BUB-1 and BUB-3, which become highly enriched on kinetochores immediately upon NEBD in C. elegans (Essex et al., 2009). Thus, PCH-2 localizes to unattached kinetochores during checkpoint activation, similar to its mammalian counterpart TRIP13, and with kinetics similar to that of MAD-1 and MAD-2.
CMT-1 and MAD-2 are required for PCH-2 localization to unattached kinetochores
Given the failure of pch-2 mutants to localize MAD-2 during checkpoint activation, we were curious whether or not PCH-2 might directly interact with MAD-2. To test this, we performed a directed yeast two-hybrid screen, using PCH-2 as bait, and a library of known kinetochore and checkpoint components as the prey (Moyle et al., 2014). We failed to detect an interaction between PCH-2 and MAD-2 or any other of the other proteins in the library (Table 1).
Table 1. Prey vectors analyzed for interaction with PCH-2 via yeast two-hybrid assay.
| Common names | C. elegans proteins |
|---|---|
| Mis12 complex | MIS-12, KNL3, KBP-1, KBP-2 |
| Ndc80 complex | NDC-80, HIM-10, KBP-3, KBP-4 |
| Knl1 complex | KNL-1, KBP-5 |
| CCAN | HCP-4 |
| CENP-F | HCP-1, HCP-2 |
| RZZ complex | ROD-1, CZW-1, ZWL-1 |
| Spindly | SPDL-1 |
| Checkpoint proteins | BUB-1, BUB-3, MDF-1, MDF-2, SAN-1 |
| Cdc20 | FZY-1 |
| Polo kinase | PLK-1 |
| p31comet | CMT-1 |
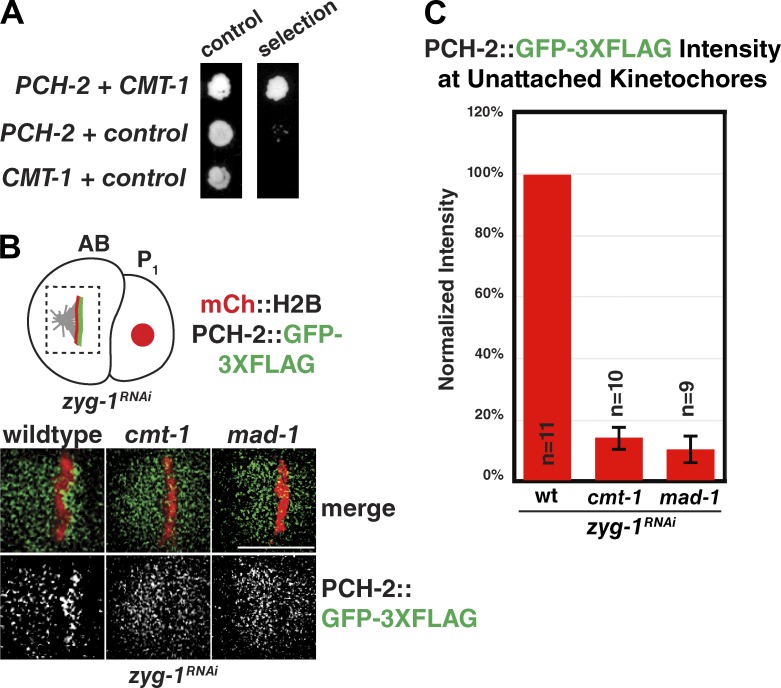
Recently, the mammalian orthologue of PCH-2, TRIP13, was shown to interact with the MAD-2 inhibitor p31(comet) in vitro (Tipton et al., 2012). CMT-1 is the C. elegans orthologue of p31(comet) (Vleugel et al., 2012). Whereas CMT-1 function during C. elegans mitosis has not been well characterized, cmt-1 mutants are viable and fertile (unpublished data), suggesting no major defects during embryogenesis or germline development. CMT-1, like MAD-2, contains a HORMA domain (Yang et al., 2007), suggesting PCH-2 may regulate it directly. Using the yeast two-hybrid assay, we detected a robust interaction between PCH-2 and CMT-1 (Fig. 4 A).
Figure 4.
CMT-1 and MAD-2 are required for PCH-2 localization to unattached kinetochores during checkpoint activation. (A) PCH-2 interacts with CMT-1 by yeast two-hybrid assay. PCH-2 is fused to GAL4 DNA-binding domain (bait protein) and CMT-1 is fused to the GAL4-activation domain (prey protein). Empty prey or bait vectors were used as controls in lanes 2 and 3, respectively. (B) PCH-2::GFP-3XFLAG fails to localize to unattached kinetochores in cmt-1 and mad-1 mutant embryos when zyg-1 is knocked down by RNAi. Bar, 5 µm. (C) Quantification of PCH-2::GFP-3XFLAG at unattached kinetochores indicates that mutation of cmt-1 and mad-2 reduce signal to 14% and 10% of wild type, respectively. Error bars in all graphs represent SEM.
Given this interaction, we reasoned that CMT-1 might be responsible for the kinetochore localization of PCH-2 during checkpoint activation. We analyzed the localization of PCH-2::GFP-3XFLAG in wild-type and cmt-1 mutant embryos treated with zyg-1 RNAi. In contrast to wild-type embryos, which effectively localized PCH-2::GFP-3XFLAG, the enrichment of PCH-2 at unattached kinetochores was lost in cmt-1 mutant embryos (Fig. 4 B). We quantified the PCH-2::GFP-3XFLAG kinetochore signal and found that it was reduced to 14% of wild type in cmt-1 mutants (Fig. 4 C), indicating that CMT-1 is required for the localization of PCH-2 to unattached kinetochores. Given that TRIP13PCH-2, p31(comet)CMT-1, and MAD-2 were shown to form a complex in vitro (Ye et al., 2015), we reasoned that PCH-2 localization to unattached kinetochores might also depend on MAD-2. To test this, we again used the mad-1(av19) allele, which fails to localize MAD-2 to kinetochores (Fig. 2 A). Similar to cmt-1 mutants, PCH-2::GFP-3XFLAG did not become enriched at unattached kinetochores in mad-1(av19) mutants (Fig. 4 B) and quantification indicated that PCH-2 kinetochore signal was reduced to 10% of wild type (Fig. 4 C). Thus, both CMT-1 and MAD-2 are required for PCH-2 localization at unattached kinetochores, consistent with their ability to form a complex in vitro (Ye et al., 2015).
Mutation of cmt-1 suppresses the checkpoint defect of pch-2 mutants
Because the localization of PCH-2 to unattached kinetochores requires CMT-1, we assessed how mutation of cmt-1 affected the spindle checkpoint. We used a null allele of cmt-1, cmt-1(ok2879), for all analyses. We again assessed mitotic timing during embryogenesis. Unlike in mammalian cells, in which p31(comet) is required for efficient mitotic exit (Xia et al., 2004), cmt-1 single mutants exhibited wild-type rates of mitotic timing (Fig. 5 A and Video 5), similar to our analysis of pch-2 mutants. This result indicates that neither PCH-2 nor CMT-1 regulates mitotic timing in a normal cell cycle in C. elegans.
Figure 5.
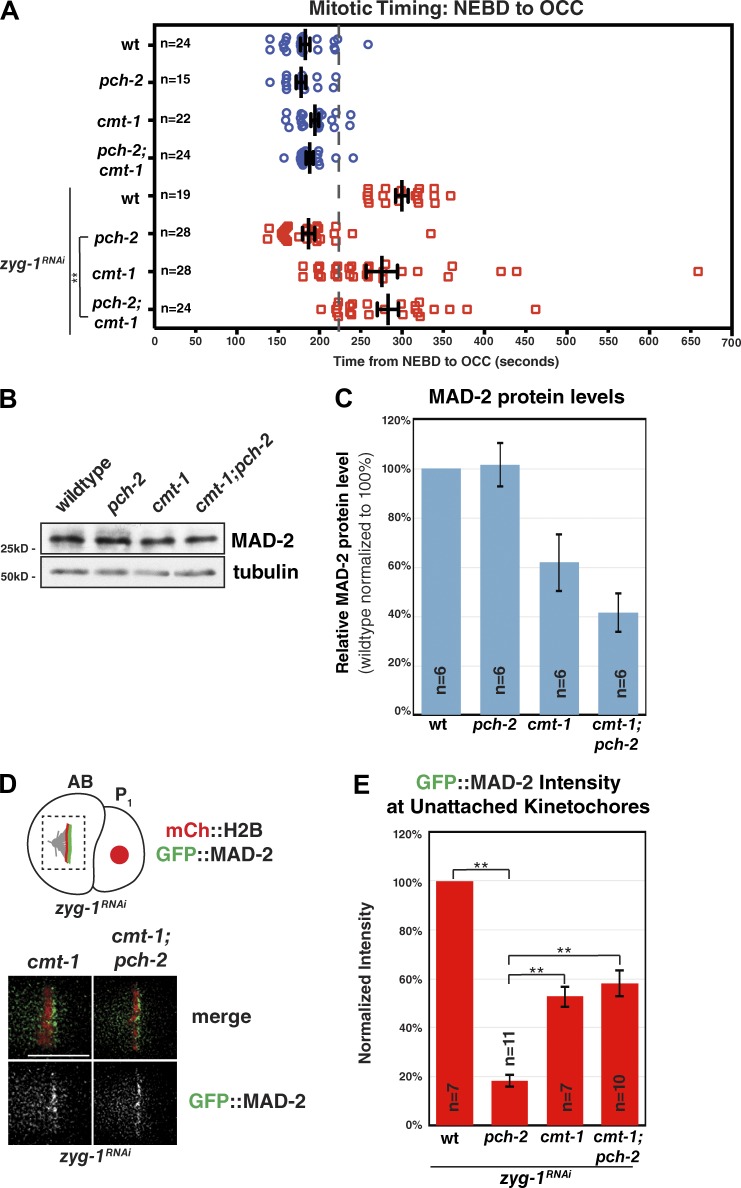
Mutation of cmt-1 suppresses the checkpoint defect of pch-2 mutants. (A) Mutation of cmt-1 restores checkpoint function in pch-2 mutants. A dashed gray line was drawn at 222 s representing the upper limit of wild-type mitotic timing. 95% of wild-type embryos and 0% of zyg-1RNAi embryos displayed mitotic timing at or below this line. Black lines indicate the mean mitotic timing for each genotype; the whiskers indicate SEM. (B). MAD-2 protein levels are reduced in cmt-1 and cmt-1;pch-2 mutants. Whole worm lysates were first normalized for protein concentration and then serial dilutions were analyzed via immunoblot with an anti–MAD-2 antibody and an anti-α-tubulin antibody serving as a loading control. (C) cmt-1 and cmt-1;pch-2 double mutants show significant reductions in MAD-2 protein levels to 61% and 42% of wild type, respectively, after quantification. (D) cmt-1 and cmt-1;pch-2 mutants show GFP::MAD-2 localization to unattached kinetochores. Bar, 5 µm. (E) Quantification of GFP::MAD-2 signal at unattached kinetochores shows that mutation of cmt-1 partially restores GFP::MAD-2 localization in pch-2 mutants. **, P < 0.0001.
Next, we analyzed mitotic timing in cmt-1 mutants in the context of spindle checkpoint activation. Most cmt-1 mutant embryos were competent for checkpoint activation: the mean length of mitosis in cmt-1;zyg-1RNAi embryos was significantly longer than in wild-type embryos (277 s; P < 0.0001; Fig. 5 A and Video 6), similar to zyg-1RNAi embryos (300 s). However, overall, the distribution of mitotic timing in cmt-1;zyg-1RNAi embryos was broader and shifted lower compared with RNAi of zyg-1 alone. Furthermore, a small population (7 of 28 [25%]) of cmt-1;zyg-1RNAi mutant embryos showed wild-type mitotic timing (≤222 s, below the gray dashed line; Video 7), which we never observed in zyg-1RNAi embryos (0 of 19 [0%]). Therefore, mutation of cmt-1 appears to reduce spindle checkpoint robustness. Moreover, because cmt-1 mutants fail to localize PCH-2 to unattached kinetochores (Fig. 4 B), these data also demonstrate that kinetochore localization of PCH-2 is not strictly required for checkpoint activation when CMT-1 is absent.
Because the checkpoint was largely functional in cmt-1 mutants despite the absence of PCH-2::GFP-3XFLAG at unattached kinetochores (Fig. 4 B), we wondered whether checkpoint activation in cmt-1 mutants required PCH-2 at all. We monitored mitotic timing in cmt-1;pch-2 double mutants in the absence and presence of zyg-1 RNAi. Untreated double mutants exhibited wild-type mitotic timing (mean, 188 s; Video 8). Strikingly, most cmt-1;pch-2;zyg-1RNAi mutant embryos were functional for checkpoint activation, exhibiting mean mitotic timing similar to zyg-1RNAi embryos (283 s; Fig. 5 A and Video 9). Again, however, like cmt-1;zyg-1RNAi embryos, the distribution of mitotic timing was broader and shifted lower compared with zyg-1RNAi embryos. Likewise, a small fraction (4 of 24 [16.7%]) of cmt-1;pch-2;zyg-1RNAi embryos went through mitosis in 222 s or less, indicating a subtle checkpoint defect (Video 10). Importantly, however, mean mitotic timing in cmt-1;pch-2;zyg-1RNAi mutants was significantly longer than pch-2;zyg-1RNAi mutants (P < 0.0001). Altogether, our results show that CMT-1 and PCH-2 interact both physically and genetically. More specifically, mutation of cmt-1 partially restores checkpoint function in pch-2 mutants, indicating that CMT-1 function is antagonized by PCH-2 function during spindle checkpoint activation.
We were curious about why checkpoint function was slightly reduced in both cmt-1 and cmt-1;pch-2 mutants (Fig. 5 A). We reasoned that MAD-2 protein levels might be affected by mutation of cmt-1. We qualitatively and quantitatively assessed MAD-2 protein levels in these genetic backgrounds (Fig. 5, B and C; and Fig. S4 A). cmt-1 mutants and cmt-1;pch-2 double mutants showed reductions in MAD-2 protein levels of ∼39% and 58%, respectively (Fig. 5 C). This suggests that CMT-1 may play a secondary role in stabilizing the MAD-2 protein, perhaps through a direct interaction. This reduction in MAD-2 protein level may also explain the defect in checkpoint robustness observed in the cmt-1 genetic background.
Given the reduction in MAD-2 protein levels we detected in cmt-1;pch-2 double mutants, we wondered whether the mitotic delay induced by zyg-1 RNAi that we observed in this background was indeed due to spindle checkpoint activation (Fig. 5 A). To test this, we created cmt-1;pch-2;mad-2 triple mutants for mitotic timing analyses. However, these triple mutants produced no viable embryos for analysis (unpublished data). To circumvent this genetic interaction, we performed sequential feeding RNAi of mad-2 and zyg-1. We verified that this scheme was sufficient for checkpoint activation and that RNAi of mad-2 was sufficient to disable the checkpoint. RNAi of both mad-2 and zyg-1 in wild-type embryos decreased mitotic timing to a mean of 175 s, substantially lower than the mean mitotic timing of embryos in which only zyg-1 was inactivated (Fig. S4 B; P < 0.0001). Similarly, RNAi of both zyg-1 and mad-2 in cmt-1;pch-2 mutants significantly reduced mitotic timing to a mean of 189 s, compared with the mean of 262 s observed in zyg-1RNAi embryos (Fig. S4 B; P = 0.008). These data show that the mitotic delay observed in cmt-1;pch-2;zyg-1RNAi embryos is dependent on MAD-2 and, by extension, the spindle checkpoint. Despite expressing ∼42% of the amount of MAD-2 protein of wild-type embryos, cmt-1;pch-2 double mutants effectively activate the spindle checkpoint in a majority of embryos.
Because mutation of cmt-1 in pch-2 mutant embryos rescued checkpoint function despite the reduced levels of MAD-2, we evaluated whether MAD-2 localization to unattached kinetochores was restored in these mutants as well. GFP::MAD-2 localized to unattached kinetochores in cmt-1;zyg-1RNAi mutants, though less robustly than in zyg-1RNAi embryos (Fig. 5 D). When we quantified the amount of kinetochore bound GFP::MAD-2, cmt-1;zyg-1RNAi mutants exhibited 53% of the level of GFP::MAD-2 at kinetochores observed in zyg-1RNAi embryos (Fig. 5 E). Consistent with our mitotic timing analysis, mutation of cmt-1 partially rescued GFP::MAD-2 localization in pch-2 mutants (Fig. 5 D). Kinetochore bound GFP::MAD-2 levels in cmt-1;pch-2;zyg-1RNAi mutant embryos were 58% of the level of GFP::MAD-2 at kinetochores observed in zyg-1RNAi embryos (Fig. 5 E). Similar to pch-2 mutants, we detected no defects in the localization of GFP::MAD-1 in cmt-1 or cmt-1;pch-2 double mutants (Fig. S5 A). Furthermore, kinetochore assembly, as visualized by KNL-1::GFP loading, was unaffected by mutation of cmt-1 (Fig. S2 C). Finally, BUB-1::GFP and GFP::BUB-3 localized to kinetochores normally in cmt-1 mutants (Fig. S2 B). Thus, although MAD-2 protein levels are most profoundly affected in cmt-1;pch-2 double mutants (Fig. 5 C and Fig. S4 A), they are generally competent for checkpoint activation (Fig. 5 A) and localize functional amounts of GFP::MAD-2 to kinetochores (Fig. 5, D and E). Together, these data strongly argue that the primary role for PCH-2 during checkpoint activation is to antagonize CMT-1 to promote the robust accumulation of MAD-2 at unattached kinetochores.
Our analyses of MAD-2 localization used a GFP::MAD-2 construct driven from a nonnative promoter in a strain that also includes endogenous MAD-2. MAD-2 protein levels are higher in this genetic background, and this increase in MAD-2 levels bypasses the requirement for other checkpoint components, such as MAD-3 (SAN-1 in C. elegans) and BUB-3 (Essex et al., 2009). We were curious about whether overexpression of MAD-2 would also bypass the requirement for PCH-2 in checkpoint activation. To test this, we analyzed mitotic timing in GFP::MAD-2 embryos using DCON as a marker of mitotic exit as the presence of GFP::MAD-2 in this strain prevented us from using GFP::PH. RNAi of zyg-1 induced a statistically significant mitotic delay in GFP::MAD-2 embryos (mean, 460 s; zyg-1RNAi) versus control RNAi (mean, 274 s; P < 0.0001; Fig. S5 A). Mutation of pch-2 in GFP::MAD-2 embryos with zyg-1 RNAi reduced mitotic timing to a mean of 300 s, which was significantly different than GFP::MAD-2;zyg-1RNAi embryos (Fig. S5 A; P < 0.0001). Furthermore, much like endogenous MAD-2, we detected no difference in the level of the GFP::MAD-2 protein in pch-2 mutants (Fig. S5 B). These results indicate that overexpression of MAD-2 in pch-2 mutants is not sufficient to overcome the pch-2 checkpoint defect, lending additional support to our hypothesis that PCH-2 plays a more direct role in localizing MAD-2 to kinetochores.
Finally, given that MAD-2 levels are dramatically reduced in cmt-1 and cmt-1;pch-2 mutants (Figs. 5, B and C; and Fig. S4 A) and the checkpoint appears less robust in these backgrounds (Fig. 5 A), we wondered whether MAD-2 overexpression could restore checkpoint robustness in these strains. Even though endogenous MAD-2 levels were reduced in cmt-1 and cmt-1;pch-2 mutants, GFP::MAD-2 was expressed at similar levels to wild type (Fig. S5 B). However, after zyg-1 RNAi, mitotic timing in both cmt-1 and cmt-1; pch-2, although significantly different than pch-2 mutants (P < 0.0001), was still reduced compared with zyg-1RNAi alone (Fig. S5 A). Furthermore, the distribution of mitotic timing still appeared slightly broader. Collectively, these data indicate that overexpression of MAD-2 in the cmt-1 background may not be sufficient to rescue checkpoint robustness.
Discussion
The spindle checkpoint, once thought to display switch-like “on” or “off” behavior, is now thought to generate a more dynamic response, which can vary in strength (London and Biggins, 2014). In particular, the strength of the checkpoint response correlates with the amount of kinetochore-bound Mad2 (Collin et al., 2013). Here, we have shown that TRIP13PCH-2 regulates Mad2 recruitment to unattached kinetochores, independent of Mad1, by antagonizing p31(comet)CMT-1 during the spindle checkpoint response in C. elegans. Thus, TRIP13PCH-2 represents an ideal candidate as a checkpoint robustness factor through its regulation of the p31(comet)CMT-1/Mad2 complex. Our analysis may appear contradictory to the characterization of these factors as collaborators during checkpoint silencing and mitotic exit. However, these two roles can be reconciled when considering the in vitro biochemical activity of TRIP13PCH-2: disassembly of the p31(comet)/Mad2 complex (Ye et al., 2015). During checkpoint silencing and mitotic exit, this activity contributes to irreversibly inactivating the MCC (Eytan et al., 2014). During checkpoint activation, our data suggest that this same biochemical activity is used in a unique context, promoting Mad2 localization to unattached kinetochores and anaphase delay.
Our data suggest the existence of a regulatory mechanism that follows the initial recruitment of Mad1/Mad2 at unattached kinetochores during checkpoint activation in C. elegans. The pch-2 mutants show reduced kinetochore recruitment of Mad2. However, pch-2 mutants recruit Mad2 at levels higher than in mad-1 mutants, which effectively have no kinetochore-bound Mad2 (Fig. 2 B). According to the template model (De Antoni et al., 2005), kinetochore-associated C-Mad2 is bound to Mad1 and exhibits slower turnover (Shah et al., 2004), providing the template for dimerization and activation of free O-Mad2. This pool of activated C-Mad2 exhibits rapid turnover to generate a potent cytosolic wait anaphase signal (Howell et al., 2004; Shah et al., 2004). Mad2 activation via its dimerization appears to be a conserved mechanism of checkpoint activation (Nezi and Musacchio, 2009). Our quantitative Mad2 analysis suggests that the basal Mad1/Mad2 complex may still be recruited in pch-2 mutants, but that subsequent Mad2 dimerization and turnover may be disrupted. Therefore, we suggest that TRIP13PCH-2 is not simply regulating gross Mad2 kinetochore recruitment but instead the ability of cytosolic O-Mad2 to dimerize with C-Mad2 and convert to C-Mad2 to produce the soluble “wait anaphase” signal.
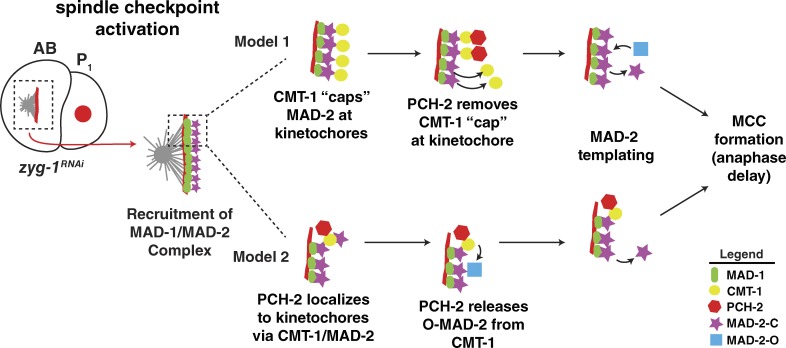
We offer two models to explain how TRIP13PCH-2 may regulate a p31(comet)CMT-1/Mad2 complex to promote spindle checkpoint signaling at unattached kinetochores. Our first model is based on the proposed idea that p31(comet)CMT-1 “caps” the stable Mad1/C-Mad2 complex at unattached kinetochores to limit checkpoint signaling in prometaphase (Musacchio and Salmon, 2007; Lara-Gonzalez et al., 2012). In this model, TRIP13PCH-2 would be responsible for removing the inhibitory p31(comet)CMT-1 “cap” to allow Mad2 dimerization, additional C-Mad2 production, and checkpoint activation (Fig. 6 A, model 1). This model is consistent with our data showing that TRIP13PCH-2 localization to kinetochores during checkpoint activation depends on p31(comet)CMT-1. However, this model does not adequately explain the minor checkpoint defect observed in cmt-1 mutants because removal of the C-Mad2 “cap” would not be predicted to reduce Mad2 stability or checkpoint robustness.
Figure 6.
Models for role of TRIP13PCH-2 in spindle checkpoint activation. Two models for how TRIP13PCH-2 regulates spindle checkpoint activation discussed in the text are depicted. During checkpoint activation, the Mad1/C-Mad2 complex is recruited to unattached kinetochores. In model 1, TRIP13PCH-2 removes an inhibitory p31(comet)CMT-1 “cap” from kinetochore bound Mad1/C-Mad2 to allow free O-Mad2 to dimerize with C-Mad2 at the kinetochore, converting it to C-Mad2, promoting MCC formation and preventing anaphase. In model 2, TRIP13PCH-2 localizes to kinetochores where it interacts with p31(comet)CMT-1/C-Mad2, releasing O-Mad2 from p31(comet)CMT-1. This allows for O-Mad2 to dimerize with Mad1/C-Mad2 at the kinetochore, promoting the generation of additional soluble C-Mad2, MCC production, and an anaphase delay.
Our second model is informed by the recent papers that describe the biochemical roles of p31(comet)CMT-1 and TRIP13PCH-2 in mitotic exit in mammalian cells (Eytan et al., 2014; Ye et al., 2015). Combined, these papers present data that p31(comet)CMT-1 and TRIP13PCH-2 collaborate in a two-step reaction to disassemble C-Mad2 from the MCC via ATP hydrolysis, generating free O-Mad2. In particular, Ye et al. (2015) argue that p31(comet)CMT-1 acts as an adapter, enabling TRIP13PCH-2 to catalyze the conformational switch of C-Mad2 to O-Mad2. Because p31(comet)CMT-1 and Mad2 interact throughout the cell cycle in mammalian cells (Date et al., 2014), it’s possible that this dimer must be disassembled to generate sufficient O-Mad2 to allow for robust checkpoint activation. Thus, our second model proposes that TRIP13PCH-2 specifically disassembles a p31(comet)CMT-1/Mad2 complex to provide free O-Mad2 for the template reaction, thereby amplifying the checkpoint signal (Fig. 6 A, model 2). This model is consistent with the complete lack of checkpoint activity in pch-2 mutants, as CMT-1 may sequester MAD-2 in this mutant background. Furthermore, this model could explain the reduction in Mad2 protein levels in cmt-1 mutants (Fig. 5 B and Fig. S4 A), particularly if the formation of a p31(comet)CMT-1/Mad2 complex contributes to Mad2 stability. This model is also supported by data showing that p31(comet)CMT-1 rapidly cycles on and off of unattached kinetochores in mammalian cells (Hagan et al., 2011), similar to the highly mobile population of Mad2. However, this disassembly reaction may not necessarily need to occur at unattached kinetochores because cmt-1 mutants abolish PCH-2 localization (Fig. 4 C) while maintaining an active spindle checkpoint (Fig. 5 A). In addition, these two models are not mutually exclusive, raising the possibility that TRIP13PCH-2 undertakes both of these tasks to promote checkpoint activation in C. elegans.
The mitotic delay induced by the spindle checkpoint in the C. elegans embryo is relatively short compared with its mammalian counterpart. This may be a consequence of prioritizing the coordination of mitotic divisions over responding to cell cycle defects during embryonic development or due to the large cytoplasmic to nuclear ratio that affects checkpoint signaling in other systems, or both (Minshull et al., 1994; Bao et al., 2008). These factors may also explain why TRIP13PCH-2 and p31(comet)CMT-1 appear to be dispensable for regulating normal mitotic timing in C. elegans. It is formally possible, however, that TRIP13PCH-2 is required for checkpoint silencing in C elegans as in mammalian cells (Eytan et al., 2014). Our ability to detect this function may simply be masked by the lack of a functional checkpoint in the absence of TRIP13PCH-2. Still, our experiments demonstrate that mitotic divisions in C. elegans, both in the context of germline mitosis and embryonic development, are ideal to interrogate the functions of TRIP13PCH-2 and p31(comet)CMT-1 in promoting spindle checkpoint activation. An obvious next question raised by our studies is whether TRIP13 is also required for Mad2 recruitment in mammalian cells. The colocalization of TRIP13 and Mad2 at kinetochores in the presence of spindle poisons suggests this function is likely to be conserved (Tipton et al., 2012).
Despite the lack of a complete mitotic arrest when the checkpoint is activated in C. elegans embryos, Mad2 protein levels still appear to have repercussions on the robustness of the delay induced by zyg-1 RNAi. Overexpression of Mad2 bypasses the requirement for Bub3 and Mad1 in the checkpoint (Essex et al., 2009) and a reduction in Mad2 protein levels, as observed in cmt-1 and cmt-1;pch-2 double mutants (Fig. 5, B and C; and Fig. S4 A), correlates with the inability of a fraction of embryos to activate the checkpoint (Fig. 5 A). However, checkpoint robustness in cmt-1 and cmt-1;pch-2 double mutants appears compromised even when MAD-2 is overexpressed (Fig. S5 A), suggesting that overexpression of GFP::MAD-2 cannot restore functional MAD-2 protein levels or there is an additional layer of complexity in checkpoint regulation, perhaps through modulation of the C-MAD-2/O-MAD-2 equilibrium (Ye et al., 2015).
The precise role of TRIP13PCH-2 in meiosis has been enigmatic and a definitive meiotic substrate for this AAA+–ATPase has been difficult to identify. Biochemical analysis of TRIP13PCH-2 in mitotic exit (Eytan et al., 2014; Ye et al., 2015) combined with our investigation of TRIP13PCH-2 function in checkpoint activation clearly indicates a role for this protein in regulating proteins with HORMA domains. It seems likely that the effects of TRIP13PCH-2 on pairing, synapsis, and recombination also rely on its ability to regulate meiotic HORMA domain–containing proteins, which localize to meiotic chromosomes and are required for pairing, synapsis, and recombination (Zetka et al., 1999; Couteau et al., 2004; Nabeshima et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005; Goodyer et al., 2008). Indeed, recent experiments in C. elegans have revealed a similar requirement for these proteins in regulating the progression of meiotic events (Kim et al., 2014; Silva et al., 2014). Whether these HORMA domain proteins undergo conformational changes similar to Mad2, whether TRIP13PCH-2 regulates these changes, either directly or through an adapter protein, and how the events of meiotic prophase are affected by these changes are open and intriguing questions.
Our data contribute to several recent studies that demonstrate a close relationship between TRIP13PCH-2 and p31(comet)CMT-1, during the activation or the silencing of the spindle checkpoint (Tipton et al., 2012; Eytan et al., 2014; Wang et al., 2014; Ye et al., 2015). This relationship could potentially explain why some organisms don’t rely on TRIP13PCH-2 and/or p31(comet)CMT-1 for spindle checkpoint function. For example, although TRIP13Pch2 is present in budding yeast, its expression is limited to meiosis (San-Segundo and Roeder, 1999). Fission yeast does not have a TRIP13 orthologue (Wu and Burgess, 2006). Both of these model systems also lack p31(comet) (Vleugel et al., 2012). This suggests that mitotic expression of TRIP13PCH-2 may be limited to organisms expressing p31(comet)CMT-1. Given that TRIP13PCH-2 also functions in meiotic prophase (Wu and Burgess, 2006; Joshi et al., 2009, 2015; Wojtasz et al., 2009; Zanders and Alani, 2009; Roig et al., 2010; Zanders et al., 2011; Chen et al., 2014; Deshong et al., 2014), it’s possible that p31(comet)CMT-1 acts as an adaptor protein for TRIP13PCH-2 during chromosome segregation to specifically allow for regulation of Mad2 and its spindle checkpoint function. Future experiments aim to test this hypothesis, to understand whether the ATPase function of TRIP13PCH-2 contributes to checkpoint activation, and to distinguish between our two models for the role of TRIP13PCH-2 in checkpoint activation.
Materials and methods
C. elegans strains and husbandry
The wild-type C. elegans strain background was Bristol N2 (Brenner, 1974). All strains were maintained at 20°C except for those containing the zyg-1(b1) allele (O’Connell et al., 2001), which were maintained at 15°C. See Table S1 for a list of all C. elegans strains used in this study. The strain BHL664, expressing a c-terminal GFP-3XFLAG fusion with PCH-2, was generated by cloning the 3kb genomic region surrounding the pch-2 stop codon with gfp-3xflag in frame into pUC19 using Gibson cloning (Gibson et al., 2009). This plasmid was used as a repair template with the CRISPR/Cas9 system (Dickinson et al., 2013; Paix et al., 2014) with the guide RNA 5′-AATTGCATGAATCTCTTTCTCGAGG-3′ to tag the endogenous protein. The insertion was verified by PCR and live microscopy, and then backcrossed six times to N2 before analysis.
Microscopy and mitotic timing experiments
All immunofluorescence and live microscopy was performed on a DeltaVision Personal DV deconvolution microscope (GE Healthcare) equipped with a 100× NA 1.40 oil-immersion objective (Olympus) resulting in an effective XY pixel spacing of 0.064 or 0.040 µm. Images were captured with a CoolSNAP charge-coupled camera (Roper Scientific). Environmental temperature averaged 21°C during image collection for all experiments. Three-dimensional image stacks were collected at 0.2-µm Z-spacing and processed by constrained, iterative deconvolution. Imaging, image scaling, and analysis were performed using functions in the softWoRx software package (GE Healthcare). Projections were calculated by a maximum intensity algorithm. Composite images were assembled and some false coloring was performed with Adobe Photoshop.
For mitotic timing experiments, Z-sections were acquired with 8 × 2-µm steps using a 100× objective (Olympus) at 20-s intervals. Exposure time was 100 ms for mCherry::H2B and 50 ms for GFP::PH. Mitotic duration was calculated for the AB cell in the presence of monopolar spindles as the interval between NEBD to OCC or the interval between NEBD and DCON. NEBD was defined by the equilibration of mCh::H2B from the nucleus into the cytosol. OCC was defined as the change in conformation of the plasma membrane from circular to rectangular, or with zyg-1 RNAi as the first frame when a persistent membrane bleb formed from the cortex of the embryo. DCON was defined as the loss of punctate mCh::H2B signal within the decondensing chromatin. To minimize bleaching and maximize signal intensity of GFP-tagged SAC and kinetochore components (PCH-2, MAD-2, MAD-1, BUB-1, BUB-3, and KNL-1), imaging was started just after NEBD as visualized by mCh::H2B. Here, 8 × 1-µm steps were captured with 250-ms GFP and 100-ms mCherry exposures at 20-s intervals. All images of GFP::MAD-1, GFP::MAD-2, BUB-1::GFP, GFP::BUB-3, and PCH-2::GFP-3XFLAG are shown at pseudo-metaphase.
Immunofluorescence of gonads was performed as described elsewhere (Bhalla and Dernburg, 2005). For experiments with zyg-1(b1), L4s were picked and incubated at 25°C for 24–26 h before dissection. For live microscopy of two cell embryos, eggs were dissected 18–26 h after L4 into 1× egg buffer (25 mM Hepes, pH 7.4, 118 µM NaCl, 48 mM KCl, 2 mM EDTA, and 0.5 mM EGTA) and mounted on 2% agarose pads for immediate analysis. The following primary antibodies were used for C. elegans immunofluorescence (dilutions in parentheses): guinea pig anti–SUN-1 phosphoserine 8 (1:700; Penkner et al., 2009) and mouse antihistone H3 phosphoserine 10 (1:500; Sigma-Aldrich). Guinea pig anti–SUN-1 phosphoserine 8 was generated against the phosphoepitope of SUN-1Ser8Pi (Penkner et al., 2009). Secondary antibodies were Cy3 anti–guinea pig (Jackson Immunochemicals) and Alexa Fluor 488 anti-mouse (Invitrogen).
Quantification of GFP::MAD-1, GFP::MAD-2, and PCH-2::GFP-3XFLAG
Analysis was performed in Fiji. Quantification of unattached kinetochore signal was performed essentially as described for GFP::MAD-1 quantification in Moyle et al. (2014). Maximum-intensity projections of both mCh::H2B and GFP fusion proteins were made after the pseudometaphase plate was generated. The image was rotated so the metaphase plate was vertical, channels were split, and the maximum GFP pixel was identified using the process function within a box on the unattached side of the metaphase plate. In the same x-plane, the maximum mCh::H2B pixel was found. The width was changed to 12 pixels and the maximum GFP signal intensity was recorded in this 12-pixel window centered at the mCherry maxima. The background GFP signal was calculated by taking the mean GFP intensity of a 4-pixel box in the same x-plane, 8 pixels away from the maximum mCherry on the opposite side of the pseudo-metaphase plate to the maximum GFP (i.e., the attached side). This background GFP was then subtracted from the maximum to measure the kinetochore-bound GFP fusion intensity. This process was repeated at least 7× for each genetic background and the signal was averaged. The mean signal in a wild-type genetic background was set as 100% and relative signals were calculated for other genetic backgrounds as compared with wild type. Significance was assessed using a paired t test.
Feeding RNAi
RNAi was performed by growing relevant worm strains on HT115 bacteria transformed with vectors allowing for IPTG inducible expression of the desired dsRNA. Bacterial strains containing RNAi vectors were cultured overnight at 37°C, centrifuged, and the pellet was resuspended in 1/10 of the original volume. 50 µl of concentrated culture was spotted onto an nematode growth medium (NGM) plate with 1 mM IPTG and 50 µg/μl of kanamycin or carbenicillin and the RNAi spot was allowed to grow overnight at 37°C.
To knock down zyg-1 by feeding RNAi, we used Gateway cloning (Invitrogen) to insert the first 1.5kb of zyg-1 genomic DNA into pDONRT7 (Couteau and Zetka, 2011) using zyg-1_FWD (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCTATGAGCGGTGGGAAGAGTGG-3′) and zyg-1_REV (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGAAGTATAAACAAAAGGATTGTTCGTC-3′). L4 hermaphrodite worms were picked into M9, transferred to RNAi plates, allowed to incubate for 2–3 h, and then transferred to fresh RNAi plates. Live microscopy was performed on embryos 22–26 h after worms were picked to the zyg-1 RNAi plate. HT115 bacteria transformed with pHSG298 (Clontech) was used as a control for zyg-1RNAi.
To knockdown mad-2, feeding RNAi clones from the Ahringer library (Fraser et al., 2000) were used: mad-2RNAi (sjj_Y69A2A_2326.a) and controlRNAi (L4440).
For double RNAi of mad-2 and zyg-1, L4s were picked to mad-2RNAi or control (L4440) plates and allowed to grow for at 20°C. After 4 d, F1 progeny were picked as L4s onto zyg-1RNAI or control (pHSG298) plates, incubated for 22–26 h at 20°C and then dissected for analysis.
Worm lysis and immunoblotting
To make worm lysates, worms of each genotype were grown on 10 NGM plates spread with OP50 bacteria at 20°C. Worms were washed from plates (M9+ 0.1% Triton X-100) and resuspended in 500-µl buffer H (50 mM Hepes, pH 8.0, 2 mM MgCl2, 0.1 mM EDTA, pH 8.0, 0.5 mM EGTA-KOH, pH 8.0, 15% glycerol, 0.1% NP-40, and 500 mM KCl; Akiyoshi et al., 2009) supplemented with protease inhibitors (complete mini tablets without EGTA [Roche], 0.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 5 mM benzamidine, and 10 µg/ml aprotinin). Worms were bead beat (BioSpec) 3 × 30 s with 30-s rest at 4°C and then sonicated 2 × 30 s (Braun). Lysates were spun for 10 min at 14,000 and protein concentration was measured in the supernatant with a Bradford assay (Bio-Rad). 250 µl of 4× sample buffer was added. Equivalent amounts of protein were run for each sample for analysis by immunoblot.
For immunoblotting, samples were run on 12% SDS-PAGE gels; transferred to nitrocellulose using a Trans-Blot SD Semi-Dry system (Bio-Rad); blocked in a PBST + 5% (wt/vol) nonfat milk solution; and then probed with mouse anti-GFP (1:1000; Roche), mouse anti-FLAG M2 (1:2500; Sigma), rabbit anti-MAD-2 (1:10,000; gift from A. Desai, Ludwig Institute for Cancer Research, University of California, San Diego, La Jolla, CA), or mouse anti–α-tubulin (1:3,500; DM1A; Sigma-Aldrich) overnight at 4°C. Blots were washed three times for 10 min in PBS with Tween, probed for 1 h using an HRP-conjugated secondary antibody (rabbit or mouse; GE Healthcare), washed three times for 10 min in PBS with Tween, and then analyzed using a chemiluminescent substrate (Thermo Fisher Scientific).
For quantification of MAD-2 protein levels, the analyze gel function was used in ImageJ. For each genotype, two Western blots from three independent lysate preparations (six total immunoblots) were analyzed and the signal between them was averaged. MAD-2 protein level in a wild-type genetic background was normalized to 100%.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed according to the manufacturer’s protocols (Matchmaker Gold System; Clontech). Control refers to growth on SC -leu/-trp, whereas selection refers to growth on SC -leu/-trp/-his/-ade (high stringency). cDNA for pch-2 was cloned into pGBKT7 (bait vector) and cDNA for cmt-1 was cloned into both pGBKT7 and PGADT7 (prey vector). All other kinetochore and SAC genes in Y2H vectors were a gift from A. Desai.
Online supplemental material
Fig. S1 shows that mutation of mad-1 or pch-2 reduces the mitotic index of zyg-1ts germlines and confirms that feeding RNAi of zyg-1 produces monopolar spindles in the AB cell of dividing embryos. Fig. S2 demonstrates that mutation of pch-2 or cmt-1 has no effect on the localization of KNL-1, BUB-1, BUB-3, or MAD-1 to kinetochores. Fig. S3 presents data that embryos expressing PCH-2::GFP-3XFLAG have a functional spindle checkpoint response. Fig. S4 illustrates that the mitotic delay produced by zyg-1 RNAi in cmt-1;pch-2 mutants is dependent on MAD-2. Fig. S5 provides data that overexpression of GFP::MAD-2 does not rescue the checkpoint defect in pch-2 or cmt-1 mutants. Table S1 lists the genotypes of all the C. elegans strains used in this study. All videos depict mitosis in AB cells of dividing C. elegans embryos. Videos 1 and 2 depict mitosis in wild type and pch-2 mutants, respectively. Videos 3 and 4 depict mitosis in zyg-1RNAi and pch-2;zyg-1RNAi mutants, respectively. Video 5 depicts mitosis in cmt-1 mutants, and Videos 6 and 7 depict mitosis in cmt-1;zyg-1RNAi double mutants. The embryo in Video 6 exhibits a delay in mitosis and the embryo in Video 7 exhibits wild-type mitotic timing. Video 8 depicts mitosis in cmt-1;pch-2 mutants, and Videos 9 and 10 depict mitosis in cmt-1;pch-2;zyg-1RNAi triple mutants. The embryo in Video 9 exhibits a delay in mitosis and the embryo in Video 10 exhibits wild-type mitotic timing. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505114/DC1.
Supplementary Material
Acknowledgments
We would like to thank Arshad Desai, Karen Oegema, and Mark Moyle for valuable C. elegans strains, yeast two hybrid reagents, antibodies, and discussion. We would like to thank Risa Kitagawa for providing the GFP::MAD-1 transgene. We would like to thank Sue Biggins, Kevin Corbett, Bungo Akiyoshi, Doug Kellogg, and the rest of the Bhalla laboratory for their input on the manuscript.
This work was supported by the National Institutes of Health (grants T32GM008646 to C.R. Nelson and R01GM097144 to N. Bhalla). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
The authors declare no competing financial interests.
Author Contributions: C.R. Nelson and N. Bhalla designed the experiments. C.R. Nelson and T. Hwang constructed C. elegans strains. C.R. Nelson performed all experiments, except the yeast two-hybrid assays, which were performed by P.-H. Chen. C.R. Nelson and N. Bhalla analyzed the data. T. Hwang and C.R. Nelson drew the schematics. C.R. Nelson and N. Bhalla wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- AB
- anterior
- C-Mad2
- closed form of Mad2
- CMT
- comet
- DCON
- chromosome decondensation
- MCC
- mitotic checkpoint complex
- NEBD
- nuclear envelope breakdown
- OCC
- onset of cortical contractility
- O-Mad2
- open form of Mad2
- P1
- posterior
References
- Akiyoshi B., Nelson C.R., Ranish J.A., and Biggins S.. 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23:2887–2899. 10.1101/gad.1865909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., and Koonin E.V.. 1998. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23:284–286. 10.1016/S0968-0004(98)01257-2 [DOI] [PubMed] [Google Scholar]
- Ballister E.R., Riegman M., and Lampson M.A.. 2014. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J. Cell Biol. 204:901–908. 10.1083/jcb.201311113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z., Zhao Z., Boyle T.J., Murray J.I., and Waterston R.H.. 2008. Control of cell cycle timing during C. elegans embryogenesis. Dev. Biol. 318:65–72. 10.1016/j.ydbio.2008.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., and Dernburg A.F.. 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 310:1683–1686. 10.1126/science.1117468 [DOI] [PubMed] [Google Scholar]
- Börner G.V., Barot A., and Kleckner N.. 2008. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA. 105:3327–3332. 10.1073/pnas.0711864105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J., Merlet J., Tavernier N., Richaudeau B., Arnold A., Ciosk R., Bowerman B., and Pintard L.. 2013. CRL2(LRR-1) E3-ligase regulates proliferation and progression through meiosis in the Caenorhabditis elegans germline. PLoS Genet. 9:e1003375 10.1371/journal.pgen.1003375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman J.C., Hoffman D.B., and Salmon E.D.. 2000. The role of pre- and post-anaphase microtubules in the cytokinesis phase of the cell cycle. Curr. Biol. 10:611–614. 10.1016/S0960-9822(00)00490-5 [DOI] [PubMed] [Google Scholar]
- Carvalho A., Olson S.K., Gutierrez E., Zhang K., Noble L.B., Zanin E., Desai A., Groisman A., and Oegema K.. 2011. Acute drug treatment in the early C. elegans embryo. PLoS One. 6:e24656 10.1371/journal.pone.0024656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Chen C., Jomaa A., Ortega J., and Alani E.E.. 2014. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc. Natl. Acad. Sci. USA. 111:E44–E53. 10.1073/pnas.1310755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Waters J.C., Salmon E.D., and Murray A.W.. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. 10.1126/science.274.5285.242 [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., and Murray A.W.. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. 10.1083/jcb.143.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin P., Nashchekina O., Walker R., and Pines J.. 2013. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 15:1378–1385. 10.1038/ncb2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., and Zetka M.. 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 19:2744–2756. 10.1101/gad.1348205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., and Zetka M.. 2011. DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Dev. Cell. 20:353–363. 10.1016/j.devcel.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., and Zetka M.. 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14:585–592. 10.1016/j.cub.2004.03.033 [DOI] [PubMed] [Google Scholar]
- Date D.A., Burrows A.C., and Summers M.K.. 2014. Phosphorylation regulates the p31Comet-mitotic arrest-deficient 2 (Mad2) interaction to promote spindle assembly checkpoint (SAC) activity. J. Biol. Chem. 289:11367–11373. 10.1074/jbc.M113.520841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., and Musacchio A.. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshong A.J., Ye A.L., Lamelza P., and Bhalla N.. 2014. A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 10:e1004291 10.1371/journal.pgen.1004291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., and Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 10:1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan D.A., Mogk A., Zeth K., Turgay K., and Bukau B.. 2002. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529:6–10. 10.1016/S0014-5793(02)03179-4 [DOI] [PubMed] [Google Scholar]
- Encalada S.E., Willis J., Lyczak R., and Bowerman B.. 2005. A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell. 16:1056–1070. 10.1091/mbc.E04-08-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A., Dammermann A., Lewellyn L., Oegema K., and Desai A.. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 20:1252–1267. 10.1091/mbc.E08-10-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan E., Wang K., Miniowitz-Shemtov S., Sitry-Shevah D., Kaisari S., Yen T.J., Liu S.T., and Hershko A.. 2014. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet). Proc. Natl. Acad. Sci. USA. 111:12019–12024. 10.1073/pnas.1412901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E.A., and Kapoor T.M.. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14:25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., and Ahringer J.. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330. 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J.D., Boulton S.J., and Zetka M.. 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell. 14:263–274. 10.1016/j.devcel.2007.11.016 [DOI] [PubMed] [Google Scholar]
- Hagan R.S., Manak M.S., Buch H.K., Meier M.G., Meraldi P., Shah J.V., and Sorger P.K.. 2011. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol. Biol. Cell. 22:4236–4246. 10.1091/mbc.E11-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Geissen E.M., Kamenz J., Trautmann S., Widmer C., Drewe P., Knop M., Radde N., Hasenauer J., and Hauf S.. 2013. Determinants of robustness in spindle assembly checkpoint signalling. Nat. Cell Biol. 15:1328–1339. 10.1038/ncb2864 [DOI] [PubMed] [Google Scholar]
- Heinrich S., Sewart K., Windecker H., Langegger M., Schmidt N., Hustedt N., and Hauf S.. 2014. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 15:291–298. 10.1002/embr.201338114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., and Cleveland D.W.. 2012. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 13:501–514. 10.1038/embor.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., and Salmon E.D.. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964. 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Joshi N., Barot A., Jamison C., and Börner G.V.. 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 5:e1000557 10.1371/journal.pgen.1000557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N., Brown M.S., Bishop D.K., and Börner G.V.. 2015. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol. Cell. 57:797–811. 10.1016/j.molcel.2014.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Rosenberg S.C., Kugel C.L., Kostow N., Rog O., Davydov V., Su T.Y., Dernburg A.F., and Corbett K.D.. 2014. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev. Cell. 31:487–502. 10.1016/j.devcel.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Larsen M.S., Sedgwick G.G., Sigurdsson J.O., Streicher W., Olsen J.V., and Nilsson J.. 2014. A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO Rep. 15:282–290. 10.1002/embr.201338101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt T.E., Omerzu M., Saurin A.T., and Kops G.J.. 2014. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma. 123:471–480. 10.1007/s00412-014-0458-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., and Taylor S.S.. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- London N., and Biggins S.. 2014. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15:736–747. 10.1038/nrm3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906. 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., and Yu H.. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 9:59–71. 10.1016/S1097-2765(01)00435-X [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Xia G., Wassmann K., Matsumoto T., Rizo J., and Yu H.. 2004. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 11:338–345. 10.1038/nsmb748 [DOI] [PubMed] [Google Scholar]
- Maldonado M., and Kapoor T.M.. 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 13:475–482. 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., and Villeneuve A.M.. 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19:2727–2743. 10.1101/gad.1338505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J., Sun H., Tonks N.K., and Murray A.W.. 1994. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 79:475–486. 10.1016/0092-8674(94)90256-9 [DOI] [PubMed] [Google Scholar]
- Moyle M.W., Kim T., Hattersley N., Espeut J., Cheerambathur D.K., Oegema K., and Desai A.. 2014. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 204:647–657. 10.1083/jcb.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., and Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A.M., and Hillers K.J.. 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 168:1275–1292. 10.1534/genetics.104.030700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezi L., and Musacchio A.. 2009. Sister chromatid tension and the spindle assembly checkpoint. Curr. Opin. Cell Biol. 21:785–795. 10.1016/j.ceb.2009.09.007 [DOI] [PubMed] [Google Scholar]
- O’Connell K.F., Caron C., Kopish K.R., Hurd D.D., Kemphues K.J., Li Y., and White J.G.. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. 10.1016/S0092-8674(01)00338-5 [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H.E., Lee C.Y., Calidas D., Lu T., Smith J., Schmidt H., Krause M.W., and Seydoux G.. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 198:1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., and Jantsch V.. 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 139:920–933. 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Roig I., Dowdle J.A., Toth A., de Rooij D.G., Jasin M., and Keeney S.. 2010. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 6:6 10.1371/journal.pgen.1001062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo P.A., and Roeder G.S.. 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 97:313–324. 10.1016/S0092-8674(00)80741-2 [DOI] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., and Cleveland D.W.. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952. [DOI] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N., Ferrandiz N., Barroso C., Tognetti S., Lightfoot J., Telecan O., Encheva V., Faull P., Hanni S., Furger A., et al. 2014. The fidelity of synaptonemal complex assembly is regulated by a signaling mechanism that controls early meiotic progression. Dev. Cell. 31:503–511. 10.1016/j.devcel.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.T., and Musacchio A.. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506. 10.1093/emboj/21.10.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K.K., Davis E.S., Hays T., and Golden A.. 2007. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 175:107–123. 10.1534/genetics.106.059105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D., Oegema K., and Desai A.. 2013. Meiotic double-strand breaks uncover and protect against mitotic errors in the C. elegans germline. Curr. Biol. 23:2400–2406. 10.1016/j.cub.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., and Yen T.J.. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichner A., Eytan E., Sitry-Shevah D., Miniowitz-Shemtov S., Dumin E., Gromis J., and Hershko A.. 2011. p31comet Promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc. Natl. Acad. Sci. USA. 108:3187–3192. 10.1073/pnas.1100023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton A.R., Wang K., Oladimeji P., Sufi S., Gu Z., and Liu S.T.. 2012. Identification of novel mitosis regulators through data mining with human centromere/kinetochore proteins as group queries. BMC Cell Biol. 13:15 10.1186/1471-2121-13-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G. 2015. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma. 124:333–339. 10.1007/s00412-015-0516-y [DOI] [PubMed] [Google Scholar]
- Vink M., Simonetta M., Transidico P., Ferrari K., Mapelli M., De Antoni A., Massimiliano L., Ciliberto A., Faretta M., Salmon E.D., and Musacchio A.. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766. 10.1016/j.cub.2006.03.057 [DOI] [PubMed] [Google Scholar]
- Vleugel M., Hoogendoorn E., Snel B., and Kops G.J.. 2012. Evolution and function of the mitotic checkpoint. Dev. Cell. 23:239–250. 10.1016/j.devcel.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Wang K., Sturt-Gillespie B., Hittle J.C., Macdonald D., Chan G.K., Yen T.J., and Liu S.T.. 2014. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J. Biol. Chem. 289:23928–23937. 10.1074/jbc.M114.585315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C.R., Cooke H.J., Jasin M., Keeney S., et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5:e1000702 10.1371/journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., and Burgess S.M.. 2006. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr. Biol. 16:2473–2479. 10.1016/j.cub.2006.10.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G., Luo X., Habu T., Rizo J., Matsumoto T., and Yu H.. 2004. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23:3133–3143. 10.1038/sj.emboj.7600322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yang M., Li B., Tomchick D.R., Machius M., Rizo J., Yu H., and Luo X.. 2007. p31comet blocks Mad2 activation through structural mimicry. Cell. 131:744–755. 10.1016/j.cell.2007.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Rosenberg S.C., Moeller A., Speir J.A., Su T.Y., and Corbett K.D.. 2015. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife. 4:4 10.7554/eLife.07367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S., and Alani E.. 2009. The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 5:e1000571 10.1371/journal.pgen.1000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S., Sonntag Brown M., Chen C., and Alani E.. 2011. Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics. 188:511–521. 10.1534/genetics.111.129031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M.C., Kawasaki I., Strome S., and Müller F.. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13:2258–2270. 10.1101/gad.13.17.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.